Abstract
Achieving sufficient enrichment of ligands from DNA-encoded libraries for detection can be difficult, particularly for low affinity ligands within highly complex libraries. To address this challenge, we present an approach for crosslinking DNA-linked ligands to target proteins using electrophilic or photoreactive groups. The approach involves the teathering of a ssDNA oligonucleotide to a DNA-encoded molecule to enable attachment of a reactive group post-synthetically via DNA hybridization. Crosslinking traps ligand-protein complexes while in solution and allows for stringent washing conditions to be applied in the subsequent purification. Five reactive groups (tosyl, NHS ester, sulfonyl fluoride, phenyl azide, and diazirine) were tested for crosslinking efficiency and specificity with three DNA-linked ligands to their target proteins. In a model selection, crosslinking resulted in improved enrichment of both high and a low affinity ligands in comparison to a selection with a solid-phase immobilized protein.
Introduction
DNA-encoded combinatorial libraries have become useful sources of drug lead and molecular probe compounds.1,2 A critical step in these campaigns is the in vitro selection, where binders are distinguished from non-binders. The signal to noise of this binding assay can make or break the ligand discovery success from these highly complex libraries.
Enrichment of ligands from DNA-encoded libraries generally involves immobilization of a purified target protein onto a physical matrix (e.g. biotinylated protein on streptavidin magnetic beads, His-tagged protein on Ni-NTA resin, or chemical modification of resin/beads with protein), incubation of the target with the library, washing of the support, and finally elution of bound ligands.3 While solid-phase selections have been successful in a number of applications, in certain (and typically unreported) cases, such selections fail to yield enrichments significant enough to indicate potential ligands using next-generation DNA sequencing. This approach has several limitations: background binding to the support matrix, potential for multivalent binding, limited control over protein concentration, and loss of native properties of the target protein upon immobilization. In addition, the required washing steps make solid-phase selections particularly challenging for enrichment of low affinity ligands.3 In order to address these limitations, a number of approaches have been developed including isolation in kinetic capillary electrophoresis,4 interaction dependent PCR/primer extension,5,6 exonuclease protection through DNA-programmed affinity crosslinking,7 and co-compartmentalization with DNA-linked protein targets in emulsion droplets.8
Herein, we explore the potential for crosslinking of DNA-linked ligands to their binding proteins to facilitate the enrichment of ligands from DNA-encoded libraries. Covalent crosslinking of transiently interacting biomolecules is a common approach to enable the identification of binder pairs by co-purification. Protein-protein complexes are often trapped using formaldehyde9 or synthetic bifunctional crosslinkers10 prior to immunoprecipitation. Crosslinking of protein-nucleic acid complexes is generally performed with formaldehyde11–12 or UV light13,14 and is a key component of RNA-binding protein interactome capture,15 CLIP/CLIP-seq,16,17 and XChIP-seq.18 In addition, photocrosslinking has been employed to improve discovery of ssDNA apatamers using photoSELEX.19 Given the utility of these techniques, particularly in characterizing protein-DNA interactions, we hypothesized that crosslinking of small molecule-DNA conjugates to target binding proteins would facilitate the enrichment of ligands from DNA-encoded libraries.
In this paper, we test five reactive groups (both photoreactive and electrophilic) for crosslinking efficiency with three model ligand-receptor systems. In a model library enrichment with DNA-encoded ligands of varying affinity to a protein target, we demonstrate the ability to co-purify crosslinked conjugates after protein denaturation and stringent washing conditions.
Results and discussion
Our approach for affinity crosslinking involves appending a small molecule ligand (or DNA-encoded library member) from the 5′ end of a double-stranded DNA construct and the display of a reactive group from the 3′ end of the opposite DNA strand (Scheme 1). This approach offers a number of conveniences for assembly of a DNA-encoded small molecule with a DNA-linked reactive group. Reactive groups with moderate stability can be synthesized on oligonucleotides directly before use and assembled quickly by DNA hybridization. Also, the modularity of the approach allows various reactive groups to be readily tested with a single ligand construct. This dual display of functional moieties on a DNA scaffold was inspired by prior work with bivalent protein binding agents20 and with dual pharmacophore DNA-encoded libraries.21 This approach has been recently used by Li and coworkers7,22,23 with DNA-programmed photoaffinity crosslinking probes and by Rosen et al.24 for affinity crosslinking of His-tagged proteins with activated ester-modified oligonucleotides.
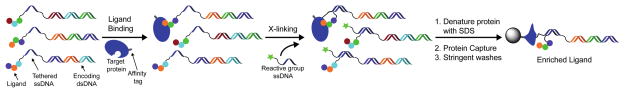
Scheme 1.
Enrichment of DNA-linked ligands by crosslinking to a target protein. All library members contain the same 20-mer ssDNA sequence tethered to the unique dsDNA (ligand-encoding) construct. This allows a reactive group-ssDNA to be added by DNA hybridization after library synthesis and binding to a protein target. Following crosslinking, proteins can be denatured without impairing DNA hybridization and immobilized onto a solid support via an affinity tag (e.g. biotin). Stringent washing conditions can then be applied to remove non-ligands and maximize the enrichment of ligands.
As outlined in Scheme 1, the implementation of this crosslinking approach involves appending small molecule library members via a ssDNA tether on a dsDNA-encoding construct. After equilibration with a protein target, a reactive group containing ssDNA complementary to the tethered ssDNA is added to allow for crosslinking of the DNA-linked small molecule to the protein target. After crosslinking, the protein can be denatured, captured via an affinity tag (e.g. biotin), and washed stringently prior to detection by qPCR or DNA sequencing.
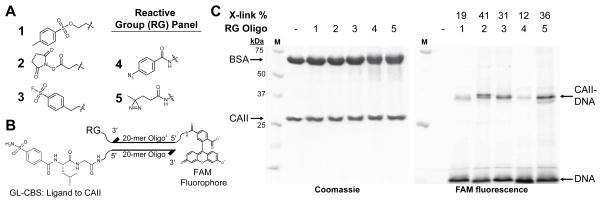
A wide variety of reactive groups have been employed in affinity-based crosslinking approaches.25 To assess suitability for application with DNA-linked small molecules, several reactive groups were tested using modified oligonucleotides. Any of three electrophiles (tosyl 1, N-hydroxysuccinimide (NHS) ester 2, sulfonyl fluoride 3) or two photoreactive groups (diazirine 4 and phenyl azide 5) (Fig. 1A) were synthesized on a 3′-modified, 20-mer oligonucleotide ssDNA. These reactive group-containing oligos also contained a 5′-fluorescein amide (FAM) modification to facilitate detection of crosslinking. Ligands were synthesized separately on a 5′-modified, complementary ssDNA (Fig. 1B). The ligand receptor pairs employed included glycylleucine carboxybenzylsulfonamide (GL-CBS) to bovine carbonic anhydrase II (CAII),26 staurosporine (STS) to protein kinase A (PKA),27 and a peptide ligand to the chromodomain of chromobox protein 8 (CBX8).28
Figure 1.
Ligand-directed crosslinking of DNA to CAII. (A) Structures of reactive groups (RG) (tosyl 1, NHS ester 2, sulfonyl fluoride 3, phenyl azide 4, diazirine 5). (B) Structure of 5′-GL-CBS-ssDNA hybridized to 3′-reactive group-5′-FAM-ssDNA′. (C) SDS-PAGE analysis of reactive group crosslinking efficiencies with GL-CBS ligand to CAII (10 μM CAII, 10 μM BSA, 1.0 μM ligand-ssDNA, 0.75 μM RG-ssDNA′). Crosslinking yields were determined from ratio of the FAM fluorescence of crosslinked CAII (CAII-DNA) to the total crosslinked and non-crosslinked 5′-FAM-ssDNA (DNA).
Crosslinking efficiencies of the reactive group oligos to target proteins were assessed when hybridized to ligand-containing oligos. Ligand-ssDNAs were first incubated with protein to allow for equilibration prior to addition of the reactive group ssDNA. After incubation (and irradiation at 354 nm for 4 and 5), crude reaction mixtures were analyzed by SDS-PAGE. The crosslinking efficiency of the protein to DNA for each reactive group was determined by comparing the FAM fluorescence intensity of the free oligonucleotide to the crosslinked, gel-shifted oligo-protein complex. In the ligand directed crosslinking of CAII (Fig. 1C), efficiencies observed were as high as 36% for 5, and reactive groups 2, 3, and 5 performed similarly. In reactions with the CBX peptide ligand-ssDNA and CBX8 protein (ESI Fig. S5), both 2 and 3 gave significant crosslinking, 45% and 71%, respectively. With STS-ssDNA and PKA (ESI Fig. S6), 2 was the only reactive group to give substantial crosslinking. Overall for these three ligand-receptor pairs, 2 and 3 reacted much more efficiently than 1. Of the photoreactive groups, 5 consistently gave greater crosslinking yields than 4.
The crosslinking efficiency comparisons suggest that the NHS ester is generally the highest yielding reactive groups for crosslinking and perhaps the best choice when approaching a protein target de novo. The electrophilic groups were explored primarily due to the highly selective and efficient affinity crosslinking previously demonstrated with tosyl groups by Hamachi and coworkers29,30 in both live cells and animals. Crosslinking with the acyl imidazole group (also developed by Hamachi and coworkers)31 was explored briefly, however, the group was found to be reactive with the fluorescein tag in the preparation of a modified oligo. Because crosslinking with the electrophiles involves a properly placed, suitable nucleophile on the protein, a concern was that yields would be very case dependent. In exo-mechanism affinity labeling, tosyl groups have shown to react with His, Glu, and Tyr side chains,29 NHS esters with Lys,32 and sulfonyl fluorides with Ser, Tyr, and Lys.33 In contrast, the highly reactive nitrene and carbene species generated by photoactivation of the aryl azide and diazirine, respectively, can insert in a number of bonds (C-H or N-H) on the protein.32
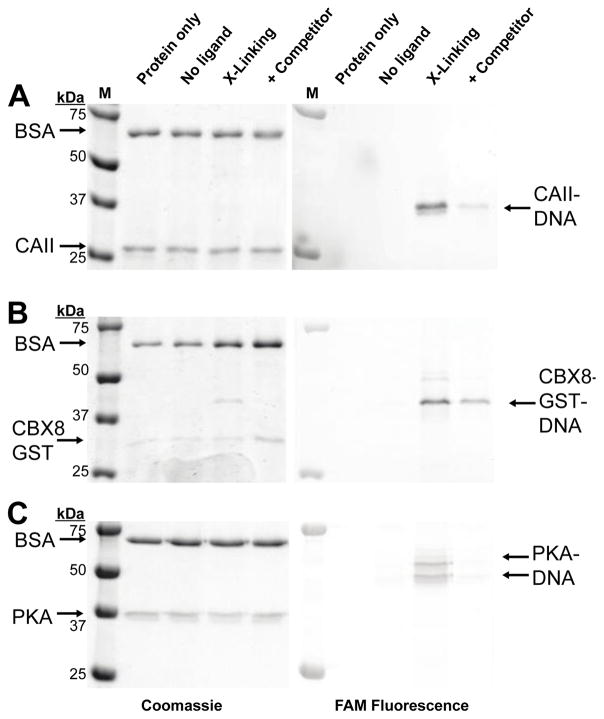
In addition to labeling efficiency, the ligand dependency of crosslinking is critical. Non-specific, ligand-independent crosslinking of the target protein would produce background signal in a binding assay or library selection. To assess ligand dependency of crosslinking, the three proteins were incubated with the reactive group oligos paired with a non-ligand oligo, ligand oligo, or the ligand oligo in the presence of a competitive ligand, Fig. 2A–C. Little to no crosslinking to the target protein was observed with the non-ligand oligo, and suppression in crosslinking was observed in all three systems with the addition of a competitive ligand. To assess ligand dependency of crosslinking, bovine serum albumin (BSA) was included in the crosslinking reactions and little, if any, crosslinking to BSA was observed.
Figure 2.
Ligand dependency of DNA-protein crosslinking. All DNA constructs were excluded from the protein only lane. No-ligand lanes used 5′-OH-ssDNA in place of ligand-ssDNA. Crosslinking of (A) CAII with GL-CBS-ssDNA using 3, (B) CBX8 with CBX peptide-ssDNA using 3, (C) PKA with STS-ssDNA using 2. Competitor ligands used were 10 μM methazolamide for CAII, 5 mM SK(me3)LAF peptide for CBX8, 10 μM STS for PKA.
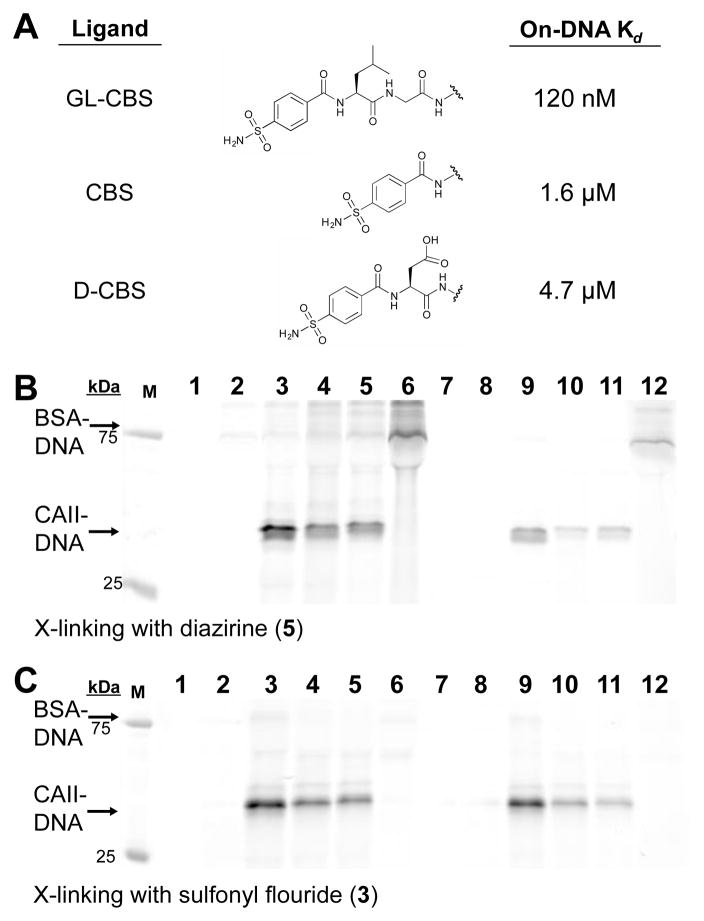
To determine if crosslinking levels were dependent on ligand affinity (particularly for the long-lived electrophile 3), we prepared three ligands on DNA with varying affinities for CAII (Fig. 3A) and applied them in reactions using either 5 or 3 (Fig. 3B and 3C). The on-DNA affinities were consistent with published reports of the free molecules.26,34,35 The differences in crosslinking observed at both 10 μM and 1 μM CAII for each ligand were consistent with the target protein concentrations and ligand Kd’s with both reactive groups. In these tests, significant levels of BSA crosslinking were observed when reactive group 5 was paired with the GL-CBS ligand oligo in reactions that lacked the target protein (Fig. 3B, Lanes 6 and 12). This crosslinking likely reflects the propensity of BSA to bind to hydrophobic small molecules and potentially to sulfonamides specifically36,37. Thus, this labeling may still be considered ligand-dependent. Bio-layer inferometry analysis, however, with the GL-CBS ligand and BSA at 10 μM did not indicate binding (Figure S4).
Figure 3.
Affinity-based crosslinking of DNA to CAII with ligands of varied affinity. (A) Three on-DNA ligands with Kd’s to CAII as determined by bio-layer interferometry (ESI Tables 1–3). (B) FAM fluorescence of SDS-PAGE analysis from crosslinking with 5. Lanes 1, 7: CAII and BSA only; lanes 2, 8: Non-ligand ssDNA (5′-OH-ssDNA); lanes 3, 9: 1 μM GL-CBS-ssDNA; lanes 4, 10: 1 μM CBS-ssDNA, lanes 5, 11: 1 μM D-CBS-ssDNA; lanes 6, 12: no CAII. Lanes 1–6: 10 μM CAII and 10 μM BSA, lanes 7–12: 1 μM CAII and 1 μM BSA. (C) FAM fluorescence of SDS-PAGE analysis from crosslinking with 3. Lane assignments are the same as (B).
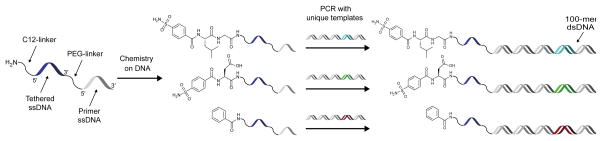
Encouraged by the results of crosslinking efficiency and ligand-dependency, we sought to apply this crosslinking strategy to enrich ligands on encoding DNA sequences in a model selection. Appending a reactive group on the 3′ end opposite a 5′ end DNA-linked small molecule presented a challenge when considering the construct architecture and workflow of many DNA-encoded library approaches. Libraries generated by DNA-programmed combinatorial chemistry (DPCC) are constructed on ssDNA but are subsequently duplexed in a primer extension reaction to minimize any effects of DNA secondary structure in selections.38 Libraries prepared using DNA ligation also yield dsDNA encoding sequences.39 To address this issue, a starting oligonucleotide could be used that contains a polyethylene glycol (PEG) spacer between two oligo segments to result in a dsDNA encoding sequence tethered to a ssDNA sequence via the spacer. This ssDNA would contain the small molecule at its 5′ end and would be available for hybridization to a complementary, reactive group oligo. The spacer would prevent read-through of the appended ssDNA by DNA polymerase in a primer extension, and the 5′ and 3′-end modifications of the tethered ssDNA would prevent any interference in encoding by DNA ligation.
To prepare constructs for test selections, two CAII ligands and a non-ligand control (Scheme 2) were synthesized on a 5′-amine-modified, 40-mer oligonucleotide, which contained a PEG spacer between two 20-mer sequences. Using these oligos as primers in a PCR with unique 100-mer templates gave 100-mer dsDNA products tethered to the ligand-containing 20-mer ssDNA. DNA templates assigned to each ligand contained a unique, internal 20-mer sequence for use as a specific priming site to enable qPCR analysis of ligand enrichments. A mock library was then prepared by mixing ligand constructs GL-CBS and D-CBS at 0.1 nM each with an excess of control, non-ligand construct (Bz) (10 nM).
Scheme 2.
Preparation of ligands on DNA for crosslinking selections. Ligands were synthesized separately on a linkered ssDNA 40-mer and then used as primers in PCR to attach each ligand to a unique 100-mer DNA template.
Using this mixture, test selections were conducted using the crosslinking approach with the sulfonyl fluoride oligo or using a traditional solid-phase selection with CAII immobilized on magnetic beads. For both selections, an approximate protein concentration of 1 μM was used to pose a challenge for the enrichment of the low affinity ligand, D-CBS (Kd = 4.5 μM).
For the crosslinking selection, the DNA-conjugate mixture was equilibrated with a biotinylated CAII in solution. Then a 3′-modified ssDNA with 3 complementary to the tethered ssDNA was added in slight excess of all DNA constructs (150 nM) for crosslinking to the target protein. After overnight incubation, the proteins were denatured with SDS (while maintaining DNA hybridization),40 and the target protein was captured with streptavidin magnetic beads. Taking advantage of the crosslinking, extensive and stringent washes of the beads was performed. Using qPCR to quantify the mixtures before and after selection, the crosslinking approach yielded 17,000-fold enrichment of the high affinity ligand, GL-CBS, and 1700-fold enrichment of the low affinity ligand, D-CBS, relative to the non-ligand, Bz, construct (Table 1). The trend in enrichment was consistent with the differences in crosslinking yields observed at 1 μM CAII in Fig. 3C. The enrichment of both ligands was dependent on the crosslinking. A replica experiment lacking the reactive group (3′-OH ssDNA) failed to produce significant enrichments or ligand-DNA recovery (Table 1).
Table 1.
Enrichment and recovery of small molecule-DNA conjugates from sulfonyl fluoride (3)-based crosslinking or solid-phase affinity selections for ligands binding to B. CAII. Enrichment is given as the fold change in concentration of each ligand-DNA construct relative to the non-ligand (Bz) DNA construct. Recovery is given as the amount of DNA recovered of the initial DNA mixture.
| SDS + Stringent Washes | Solid-Phase Affinity Pulldown | |||
|---|---|---|---|---|
|
| ||||
| X-linking with 3 1 μM CAII Enrichment (Recovery) |
Without X-linking 1 μM CAII Enrichment (Recovery) |
1 μM CAII Enrichment (Recovery) |
17 μM CAII Enrichment (Recovery) |
|
|
| ||||
| GL-CBS (Kd 120 nM) | 17,000 (11 %) | 22 (0.021 %) | 720 (52 %) | 610 (89 %) |
| D-CBS (Kd 4.5 μM) | 1,700 (1.1 %) | 28 (0.026 %) | 2.2 (0.16 %) | 25 (3.6 %) |
| Bz (non-ligand) | (0.00062 %) | (0.00094 %) | (0.073 %) | (0.15 %) |
In comparison, the traditional solid-phase affinity selection enriched the high affinity ligand 720-fold, but did not enrich the low affinity ligand significantly. Failure to enrich the D-CBS ligand was anticipated in this selection given the dissociation constant is ~5-fold above the protein concentration used. With this selection containing an initial binding step and five bead washes (6 total partitioning cycles), the best case recovery of this ligand could be estimated at 0.002% (0.17^6),41 which is well below the observed background recovery of non-ligands. In contrast, the crosslinking selection involves just a single partitioning step, which is slightly less efficient due to crosslinking yields.
An additional solid-phase test selection was performed at much higher (estimated 17 μM) protein concentration (Table 1). In this case, enrichment of both ligands was detected, and the relative recovery of the GL-CBS and D-CBS ligands observed was consistent with their Kd’s, the number of partitioning cycles, and the estimated protein concentration. While the recovery of ligand-DNA was as good or greater than as observed with crosslinking, the non-ligand recovery was also greater due to the less stringent washing conditions, which lessened the overall enrichment.
In the crosslinking-based test selection, a biotin affinity tag introduced by NHS-coupling on CAII was used to purify DNAs crosslinked to the protein target. The stability of the biotin-streptavidin interaction to relatively high levels of SDS makes this system particularly suitable for this application. While acylation of proteins with biotin is a commonly employed approach for immobilization of selection targets, it is not suitable for many proteins. A milder alternative would be the BirA tag, which allows for enzymatic biotinylation through a short peptide tag.42 Additionally, other affinity tags can be used under denaturing conditions, such as His6-Ni-NTA. Also, immunopurifications using various epitope tags can be performed on denatured proteins after appropriate dilution of denaturants.43
Both the absolute recovery and relative enrichment of ligands are key considerations in the development of selection strategies. While the traditional, solid-phase selection (Table 1) did provide approximately 5-fold greater recovery of the high affinity GL-CBS ligand compared to the crosslinking selection, the overall enrichment was 20-fold lower. The lower recovery in the crosslinking case is likely a result of the crosslinking and protein capture efficiencies. The crosslinking approach clearly benefited from stringent washes, which reduced the background recovery of the non-ligand, Bz, construct 100-fold over the standard selection. Due to the typically high complexity of DNA-encoded libraries, selection methods must produce a high level of enrichment of ligands over non-ligands. The enrichment required is case dependent and is a function of the library complexity size and the number of DNA sequence reads obtainable. As each member in a DNA-encoded library may only be present at thousands of molecules each, high enrichment should not be achieved at the expense of ligand recovery. Large losses of DNA-linked ligands would lead to under sampling of the population. Since the concentration of each library member in a selection is insignificantly low, the free ligand to protein-bound ligand ratio is equal to the ratio of the Kd to the protein concentration. As this ratio becomes much greater than 1, significant enrichment becomes difficult to achieve with solid-phase selections without incurring dramatic losses of ligands.41 Since DNA-encoded libraries are an expensive resource, minimizing the amount used is desirable.
The model selection results demonstrate the potential for crosslinking to improve selections of DNA-encoded libraries. This approach could be particularly useful in cases where the dissociation constants (Kd’s) of ligands are significantly greater than the target protein concentration. This may arise because of difficulties in obtaining a concentrated target protein. Many proteins are prone to aggregation at high concentration. Targeting of unpurified, dilute proteins directly in cell lysates, where the context may be critical for protein function, would be desirable. Additionally, crosslinking may allow discovery of very low affinity (Kd > 10 μM) fragment ligands from DNA-encoded libraries.
This approach to crosslinking could also show utility in various DNA-based assay platforms for detecting and characterizing binding to proteins. Validation and qualitative ranking of protein binding for ligands on DNA could be conducted by simple gel-based assays (analogous to gel-shift assays with DNA-binding proteins)44, which require only pmol or less quantities for detection. This crosslinking approach could be applied in recently developed methods for highly multiplexed protein interaction detection by parallel DNA sequencing, such as single-molecular-interaction sequencing (SMI-seq)45 or parallel analysis of translated ORFs (PLATO).46
Conclusions
In summary, we developed an approach for applying crosslinking to the selection of ligands from DNA-encoded libraries. Employing a tethered ssDNA construct allows for a reactive group to be synthesized separately and appended to DNA-encoded ligands after equilibration with protein targets. The sufficient crosslinking yields and the ability to perform stringent washes after protein denaturation resulted in improved enrichments of DNA-linked ligands in a model selection. The technique is amenable to DNA-encoded libraries produced from a number of platforms and shows particular promise for enrichment of low affinity ligands and for protein targets obtainable only at low concentrations.
Experimental
Unless otherwise noted, reagents were used as provided from commercial sources. Water used in all experiments and analyses was purified by a Millipore Milli-Q RO water purification system. Oligonucleotides were purchased from IDT and used as provided (sequences and specific modifications are given in ESI). Bovine carbonic anhydrase II was purchased from Sigma-Aldrich (C3934). DNA conjugates were purified on a Varian Pro Star HPLC system and analytical analysis on an Agilent 1100 series HPLC system, both using Agilent Microsorb-MV 300-5 C18 250 x 4.6 mm reverse phase columns. Analytical separations were performed using 100 mM triethylammonium acetate (TEAA), pH 5.5 in water (A) or 100 mM triethylammonium acetate in 90 % MeCN (B) with a linear gradient from 5 – 60 % B over 22 minutes. Purifications were performed using the previous or 0.75 % (v/v) 1,1,1,3,3,3-hexafluoropropan-2-ol, and 10 μM EDTA to pH 7.0 with triethylamine in water (A) or 90 % MeOH (B) with a linear gradient from 5 – 50 % B over 22 minutes. ESI-MS analysis was completed using an ABSciex 4500 QTrap from fractions collected from HFIP-based HPLC purifications. MALDI analysis was completed on an Applied Biosystems Voyager DE PRO instrument operated by the Purdue University Campus-Wide Mass Spectrometry Center after TEAA-based purification, ethanol precipitation, and ZipTip cleanup. On-DNA ligand dissociation constants were determined using a ForteBio Octet Red384. NMR analysis were completed on a Bruker ARX300 instrument as part of the Purdue Interdepartmental NMR Facility. All gel images were recorded by a GE Healthcare Typhoon Trio+ with gel band quantifications determined using ImageJ software.
General procedure for acylation of amine-modified ssDNA
Acylation of amine-modified ssDNA was completed using a general procedure, modified from Halpin et al.47 A solution of 1 nmol of amine-modified ssDNA in 1 mL of DEAE bind buffer (10 mM HOAc, 0.005 % Triton X-100) was immobilized onto 200 μL of 50% DEAE Sepharose slurry, pre-washed with DEAE Bind Buffer on a DNA solid-phase cartridge using a vacuum manifold. The immobilized DNA-containing cartridge was washed with 3 mL MeOH on the vacuum manifold and then the cartridge was placed between two 1 mL syringes. The carboxylic acid coupling reaction mixture of 40 % DMF in MeOH with 50 mM carboxylic acid, 50 mM EDC-HCl, and 5 mM HOAt was pulled up by one syringe and passed back and forth through the column several times and then incubated for 30 minutes at RT, after which the reaction mixture was eluted on the vacuum manifold. A fresh reaction mixture was prepared and added to the cartridge and incubated for 30 minutes at RT. After elution of the second reaction mixture, the cartridge was washed with 3 mL DMF, 3 mL MeOH, and 1 mL DEAE bind buffer. The modified oligo was then eluted with 1 mL DEAE elution buffer (1.5 M NaCl, 100 mM TEAA, pH 5.5, 0.005 % Triton X-100) and purified by HPLC.
General procedure for alkynyl reactive group coupling to ssDNA′-3′-N3
Using a modified procedure from Hong et al.48 The general conditions are as follows: 1.0 μM ssDNA′-3′-N3 was added to 50 mM sodium phosphate, pH 7.4, 150 mM NaCl, 5 mM aminoguanidinium hydrochloride, 5 % (v/v) 5:1 50mM THPHA:50mM CuSO4 (premixed), 1 mM alkyne, and 5 mM sodium ascorbate with a final concentration of 5 % (v/v) DMSO. The mixture was incubated for 20 minutes at RT and then concentrated and excess organics removed through 1-butanol extractions. The resulting aqueous mixture was used directly for crosslinking experiments.
General procedure for electrophilic crosslinking
The target protein (1.0 μM) and BSA (1.0 μM) in 0.1 M sodium phosphate, pH 8, 0.25 M NaCl, 0.02% (v/v) Tween-20, was combined with the ligand-ssDNA conjugate (1.0 μM) and incubates for 30 minutes prior to the addition of the reactive group-ssDNA (0.75 μM). Electrophilic crosslinking was allowed to proceed 16 h at RT, quenched by the addition of 6X SDS-loading buffer, and directly analyzed by SDS-PAGE. Gels were imaged immediately for FAM fluorescence and then Coomassie stained.
General procedure for photocrosslinking
Photocrosslinking was performed as described above, except the system was allowed to incubate with both the ligand-ssDNA and reactive group-ssDNA’ for 30 minutes at RT. Irradiation was completed by exposure to a 4W 356 nm UV light source at 4 °C for 1 hour, quenched by the addition 6X SDS-loading buffer, and directly analyzed by SDS-PAGE. Gels were imaged immediately for FAM fluorescence and then Coomassie stained.
General procedure for enrichment of ligands via crosslinking (SDS + stringent washes)
A premix of ligand-dsDNA and non-ligand-dsDNA (0.11 nM and 10 nM, respectively) was added to 1.0 μM biotinylated B. CAII and 10 ;M BSA in 0.1 M sodium phosphate, pH 8.0, 0.25 M NaCl, 0.02 % (v/v) Tween-20, 1.0 mg/mL tRNA and incubated at RT for 30 min. Meanwhile, the reactive group ssDNA was prepared as described above (General procedure for alkynyl reactive group coupling to ssDNA′-3′-N3). The ssDNA′-3′-reactive group was added in 15x excess of ligand and non-ligand DNA to the protein/DNA mixture and incubated 16 h at RT. SDS was added to a final concentration of 5.0 % (w/v) and the mixture was incubated for 30 min at RT, diluted with the above buffer to a final SDS concentration of 1.0 % (w/v) and incubated with pre-washed Nanolink Streptavidin Magnetic Beads (1.5x based on capacity) for 2 h. The magnetic beads were then separated and supernatant removed. The magnetic beads were then washed with the above buffer + 0.1 % (w/v) SDS five times. Following the final wash, the magnetic beads were eluted by suspending in 10 μL water and heated at 95 °C for 5 minutes. qPCR analysis of the premix and magnetic bead elution was completed by comparison of CT standard curves of identical dsDNA constructs.
General procedure for enrichment of ligands via solid-phase affinity pulldowns
Biotinylated B. CAII (1.2x based on magnetic bead capacity) was immobilized onto pre-washed Nanolink Streptavidin Magnetic Beads by incubating in 0.1 M sodium phosphate, pH 8.0, 0.25 M NaCl, 0.02 % (v/v) Tween-20, 1.0 mg/mL tRNA, 10 μM BSA for 2 h at RT. The magnetic beads were then separated and supernatant removed. The CAII-bound magnetic beads were then washed with the above buffer three times and a premix of ligand-dsDNA and non-ligand-dsDNA (0.1 nM and 10 nM, respectively) in the above buffer (in the appropriate volume to give the desired effective protein concentration) was added and incubated at RT for 1.5 h. The magnetic beads were then separated and DNA supernatant removed. The CAII-bound magnetic beads were then washed with the above buffer five times, maintaining the same effective protein concentration in each wash. After the final wash, the magnetic beads were suspended in 10 μL water and heated at 95 °C for 5 min. qPCR analysis of the premix and magnetic bead elution was completed by comparison of CT standard curves of identical dsDNA constructs.
Supplementary Material
Acknowledgments
This work was supported by Purdue University, the Ralph W. and Grace M. Showalter Research Trust, and the Purdue University Center for Cancer Research’s Jim and Diann Robbers Cancer Research Grant. The Purdue University shared Mass Spectrometry Shared Resource and Purdue University Interdepartmental NMR Facility were supported by P30 CA023168 and funding for use of the ForteBio Octet RED384 was provided by S10OD20087 through the Bindley Bioscience Center Biophysical Analysis Lab. Purified CBX8-GST was graciously provided by Prof. Emily Dykhuizen. We thank Jia Ma for assistance with BLI experimental design. We also thank Dr. Rachael Jetson and Dr. Dongwook Kim for helpful suggestions and Ivanna Zhilinskaya for assistance with staurosporine derivatization.
Footnotes
The authors declare no competing interests.
Electronic Supplementary Information (ESI) available: Data for Kd determination of ligands on-DNA (Fig S1–4, Tables S1–3), Figures for CBX8 (Fig. S5) and PKA crosslinking (Fig, S6), oligonucleotide sequences and structural modifications, experimental procedures for synthesis and characterization of DNA conjugates and small molecules. See DOI: 10.1039/x0xx00000x
References
- 1.Mullard A. Nature. 2016;530:367–369. doi: 10.1038/530367a. [DOI] [PubMed] [Google Scholar]
- 2.Salamon H, Klika Škopić M, Jung K, Bugain O, Brunschweiger A. ACS Chem Biol. 2016;11:296–307. doi: 10.1021/acschembio.5b00981. [DOI] [PubMed] [Google Scholar]
- 3.Hale SP. In: A Handbook for DNA-Encoded Chemistry. Goodnow RA, editor. John Wiley & Sons, Inc; 2014. pp. 281–317. [Google Scholar]
- 4.Drabovich AP, Berezovski MV, Musheev MU, Krylov SN. 2009;81:490–494. doi: 10.1021/ac8023813. [DOI] [PubMed] [Google Scholar]
- 5.McGregor LM, Gorin DJ, Dumelin CE, Liu DR. J Am Chem Soc. 2010;132:15522–15524. doi: 10.1021/ja107677q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mcgregor LM, Jain T, Liu DR. J Am Chem Soc. 2014;136:3264–3270. doi: 10.1021/ja412934t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao P, Chen Z, Li Y, Sun D, Gao Y, Huang Y, Li X. Angew Chemie - Int Ed. 2014;53:10056–10059. doi: 10.1002/anie.201404830. [DOI] [PubMed] [Google Scholar]
- 8.Blakskjaer P, Heitner T, Hansen NJV. Curr Opin Chem Biol. 2015;26:62–71. doi: 10.1016/j.cbpa.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Nadeau OW, Carlson GM. Cold Spring Harb Protoc. 2007;2007 pdb.prot4634. [Google Scholar]
- 10.Nadeau OW, Carlson GM. Cold Spring Harb Protoc. 2007. p. 2007. pdb.prot4587. [Google Scholar]
- 11.Zanton SJ, Pugh BF. Proc Natl Acad Sci United States Am. 2004;101:16843–16848. doi: 10.1073/pnas.0404988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon MJ, Larsen PL, Varshavsky A. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 13.Biggin MD. Methods Enzymol. 1999;304:496–515. doi: 10.1016/s0076-6879(99)04029-x. [DOI] [PubMed] [Google Scholar]
- 14.Chodosh LA. Curr Protoc Mol Biol. 2001;Chapter 12(Unit 12.5) doi: 10.1002/0471142727.mb1206s36. [DOI] [PubMed] [Google Scholar]
- 15.Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. Nat Protoc. 2013;8:491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 16.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ule J, Jensen K, Mele A, Darnell RB. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Tian B, Yang J, Brasier AR. Methods Mol Biol. 2012;809:105–120. doi: 10.1007/978-1-61779-376-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golden MC, Collins BD, Willis MC, Koch TH. J Biotechnol. 2000;81:167–178. doi: 10.1016/s0168-1656(00)00290-x. [DOI] [PubMed] [Google Scholar]
- 20.Sprinz KI, Tagore DM, Hamilton AD. Bioorganic Med Chem Lett. 2005;15:3908–3911. doi: 10.1016/j.bmcl.2005.05.094. [DOI] [PubMed] [Google Scholar]
- 21.Melkko S, Scheuermann J, Dumelin CE, Neri D. Nat Biotechnol. 2004;22:568–74. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Liu Y, Liu Y, Chen L, Wu S, Liu Y, Li X. Angew Chemie - Int Ed. 2013;52:9544–9549. doi: 10.1002/anie.201302161. [DOI] [PubMed] [Google Scholar]
- 23.Li G, Liu Y, Yu X, Li X. Bioconjug Chem. 2014;25:1172–1180. doi: 10.1021/bc500195w. [DOI] [PubMed] [Google Scholar]
- 24.Rosen CB, Kodal ALB, Nielsen JS, Schaffert DH, Scavenius C, Okholm AH, Voigt NV, Enghild JJ, Kjems J, Tørring T, Gothelf KV. Nat Chem. 2014;6:804–809. doi: 10.1038/nchem.2003. [DOI] [PubMed] [Google Scholar]
- 25.Takaoka Y, Ojida A, Hamachi I. Angew Chemie - Int Ed. 2013;52:4088–4106. doi: 10.1002/anie.201207089. [DOI] [PubMed] [Google Scholar]
- 26.Mincione F, Starnotti M, Menabuoni L, Scozzafava A, Casini A, Supuran CT. Bioorganic Med Chem Lett. 2001;11:1787–1791. doi: 10.1016/s0960-894x(01)00303-1. [DOI] [PubMed] [Google Scholar]
- 27.Shi H, Cheng X, Sze SK, Yao SQ. Chem Commun. 2011;47:11306. doi: 10.1039/c1cc14824a. [DOI] [PubMed] [Google Scholar]
- 28.Simhadri C, Daze KD, Douglas SF, Quon TTH, Dev A, Gignac MC, Peng F, Heller M, Boulanger MJ, Wulff JE, Hof F. J Med Chem. 2014;57:2874–2883. doi: 10.1021/jm401487x. [DOI] [PubMed] [Google Scholar]
- 29.Tamura T, Tsukiji S, Hamachi I. J Am Chem Soc. 2012;134:2216–2226. doi: 10.1021/ja209641t. [DOI] [PubMed] [Google Scholar]
- 30.Tsukiji S, Miyagawa M, Takaoka Y, Tamura T, Hamachi I. Nat Chem Biol. 2009;5:341–343. doi: 10.1038/nchembio.157. [DOI] [PubMed] [Google Scholar]
- 31.Fujishima SH, Yasui R, Miki T, Ojida A, Hamachi I. J Am Chem Soc. 2012;134:3961–3964. doi: 10.1021/ja2108855. [DOI] [PubMed] [Google Scholar]
- 32.Hermanson GT. 2013:229–258. [Google Scholar]
- 33.Yasueda Y, Tamura T, Fujisawa A, Kuwata K, Tsukiji S, Kiyonaka S, Hamachi I. J Am Chem Soc. 2016 doi: 10.1021/jacs.6b02254. jacs.6b02254. [DOI] [PubMed] [Google Scholar]
- 34.Sigal GB, Whitesides GM. Bioorganic Med Chem Lett. 1996;6:559–564. [Google Scholar]
- 35.West GM, Tang L, Fitzgerald MC. Anal Chem. 2008;80:4175–4185. doi: 10.1021/ac702610a. [DOI] [PubMed] [Google Scholar]
- 36.Jardetzky O, Wade-Jardetzky NG. Mol Pharmacol. 1965;230:214–230. [PubMed] [Google Scholar]
- 37.Hsu P-L, Ma JKH, Luzzi LA. :63. [Google Scholar]
- 38.Halpin DR, Harbury PB. PLoS Biol. 2004;2:1015–1021. doi: 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella Pa, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, Ding Y, Franklin GJ, Franzen KD, Gefter ML, Hale SP, Hansen NJV, Israel DI, Jiang J, Kavarana MJ, Kelley MS, Kollmann CS, Li F, Lind K, Mataruse S, Medeiros PF, Messer Ja, Myers P, O’Keefe H, COliff M, Rise CE, Satz AL, Skinner SR, Svendsen JL, Tang L, van Vloten K, Wagner RW, Yao G, Zhao B, Morgan Ba. Nat Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 40.Rose K, Mason JO, Lathe R. Biotechniques. 2002;33:54–58. doi: 10.2144/02331st01. [DOI] [PubMed] [Google Scholar]
- 41.Satz AL. ACS Chem Biol. 2015;10:2237–2245. doi: 10.1021/acschembio.5b00378. [DOI] [PubMed] [Google Scholar]
- 42.Fairhead M, Howarth M. In: Site-Specific Protein Labeling: Methods and Protocols. Gautier A, Hinner JM, editors. Springer; New York, New York, NY: 2015. pp. 171–184. [Google Scholar]
- 43.Tansey WP. CSH Protoc. 2007:2007. doi: 10.1101/pdb.prot4641. pdb.prot4619. [DOI] [PubMed] [Google Scholar]
- 44.Buratowski S, Chodosh La. Curr Protoc Mol Biol. 2001;Chapter 12(Unit 12.2) doi: 10.1002/0471142727.mb1202s36. [DOI] [PubMed] [Google Scholar]
- 45.Gu L, Li C, Aach J, Hill DE, Vidal M, Church GM. Nature. 2014;515:554–557. doi: 10.1038/nature13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Larman HB, Gao G, Somwar R, Zhang Z, Laserson U, Ciccia A, Pavlova N, Church G, Zhang W, Kesari S, Elledge SJ. Nat Biotechnol. 2013;31:331–4. doi: 10.1038/nbt.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halpin DR, Lee JA, Wrenn SJ, Harbury PB. PLoS Biol. 2004:2. doi: 10.1371/journal.pbio.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong V, Presolski SI, Ma C, Finn MG. Angew Chemie - Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.