Abstract
Follistatin-like protein 1 (FSTL-1) possesses several newly identified roles in mammalian biology, including IL-17 driven inflammation, though the mechanism underlying FSTL-1 influence on IL-17 mediated cytokine production is unknown. Using parallel in vitro bone marrow stromal cell models of FSTL-1 suppression we employed unbiased microarray analysis to identify FSTL-1 regulated genes and pathways that could influence IL-17 dependent production of IL-6 and G-CSF. We discovered that FSTL-1 modulates Il17rc gene expression. Specifically, FSTL-1 was necessary for Il17rc gene transcription, IL-17RC surface protein expression and IL-17-dependent cytokine production. This work identifies a mechanism by which FSTL-1 influences IL-17 driven inflammatory signaling in vitro and reveals a novel function for FSTL-1, as a modulator of gene expression. Thus, enhanced understanding of the interplay between FSTL-1 and IL-17 mediated inflammation may provide insight into potential therapeutic targets of IL-17 mediated diseases and warrants ongoing study of in vivo models and clinical scenarios of FSTL-1-influenced diseases.
Introduction
Follistatin-like protein 1 (FSTL-1) is a 306 amino acid glycoprotein belonging to the SPARC/BM-40/osteonectin family was originally cloned from the osteoblast cell line MC3T3-E1 as a TGFβ inducible molecule1. Murine and human FSTL-1 share 91% a.a. sequence homology, with five conserved domains: the eponymous follistatin-like domain, a Kazal serine protease domain, two EF-hand calcium binding domains and a von Willebrand domain, though the functionality of these domains remain unclear. FSTL-1 has been identified in both secreted and intracellular compartments, suggesting differential cell trafficking regulation. Highly expressed in mammalian cells of mesenchymal lineage, several studies have implicated FSTL-1 with functions in a myriad of settings including embryonic development, cardiac and lung dysfunction and repair, as well as tumorigenesis and metastasis and inflammation2–9. In vivo modeling has been limited as FSTL-1 germline knockout mice display a perinatal lethal phenotype associated with multiple lung, skeletal and urogenital defects. However, murine models have employed neutralizing antibodies, siRNA, adenoviral-gene transfer, partial gene disruption (FSTL-1 hypomorphs) and FSTL-1 transgenic models to begin characterizing in vivo functions of FSTL-12, 3, 5, 6, 8, 10, 11.
The role of FSTL-1 in inflammation and immunity remains unclear. We had shown that FSTL-1 expression positively correlated with degree of inflammation and joint disease severity in the collagen induced arthritis and Lyme arthritis models10, 12–14. Further, FSTL-1 facilitates cytokine production in response to a number of inflammatory signals. However the specific mechanism(s) involved and cell populations of significance are actively being explored. For example, we have previously shown that in macrophages/monocytes, FSTL-1 mediates NLRP3-inflammasome activation15. We have also observed that IL-17A in cooperation with TNFα acts on bone marrow stromal cells in an FSTL-1-dependent manner to induce IL-6 and MCP-1 production13, suggesting that novel mechanisms may be involved in FSTL-1 mediated inflammatory pathways.
Interleukin-17 (IL-17), the eponymous cytokine produced by Th17 cells, signals via the IL-17 receptor complex. IL-17 mediated immune responses are central to both host defense against fungal and bacterial infection as well as autoimmune diseases, including ankylosing spondylitis, psoariasis and psoariatic arthritis16–24. IL-17 is not a potent mediator of inflammatory signaling in its own right, but signals potently with a variety of other stimuli including TNFα, FGFR and IFN-γ25. Bone marrow stromal cells (BMSC) represent a unique source of multipotent cells important for bone and joint homeostasis. They are also critically important for immune regulation and the host inflammatory response, in part through the IL-17 signaling axis26–28. Though FSTL-1 participates in several biologic and inflammatory processes the interaction between FSTL-1 and IL-17 signaling in bone marrow stromal cells remains unknown. We sought to explore the interaction of FSTL-1 and IL-17 mediated inflammatory signaling in vitro in order to identify pathways that could be potential targets for therapeutic intervention and further investigation.
We first validated that FSTL-1 mediates IL-17A/TNFα induced cytokine production using parallel strategies of FSTL-1 suppression in two in vitro bone marrow stromal cell systems. We then employed microarray analysis of these systems to identify candidate FSTL-1 influenced gene targets. Unexpectedly, we found that the IL-17 receptor C (IL-17RC), an essential subunit of the heterodimeric IL-17 receptor complex, was upregulated by FSTL-1. FSTL-1 KO cells were refractory to IL-17 signaling. Ectopic Il17rc expression via plasmid transfection rescued IL-17-stimulated cytokine production in the FSTL-1 KO cells. Further, plasmid Fstl1 transfection also rescued Il17rc gene expression and concomitant IL-17A stimulated cytokine production. Mechanistically, FSTL-1 dependent Il17rc transcript abundance was due to de_novo_gene transcription and not mRNA stability, identifying for the first time a role for FSTL-1 in transcriptional regulation. Together, these studies confirm that FSTL-1 regulation of IL-17RC is a novel mechanism by which FSTL-1 modulates IL-17 stimulated cytokine production and a potential target for therapeutic intervention in IL-17 mediated diseases.
Results
Follistatin-like protein 1 suppression by shRNA reduces IL-17/TNFα mediated cytokine production
To examine the role of FSTL-1 in bone marrow stromal cells, we targeted Fstl1 mRNA expression using shRNA in the ST2 bone marrow stromal cell line following IL-17A/TNFα stimulation, as previously described13. Using this approach, we observed reduced Fstl1 mRNA abundance with or without IL-17A stimulation. Additionally we found that IL-17A stimulation increased Fstl1 mRNA suggesting that FSTL-1 was, at least in part, inducible by the IL-17 receptor signaling axis (Fig. 1A). Knockdown with shRNA caused a significant reduction in secreted FSTL-1 protein both at baseline and following IL-17A stimulation (Fig 1B). To assess the effect of Fstl1 mRNA suppression on IL-17A driven cytokine production, we evaluated the IL-17A dependent cytokines Csf3/G-CSF and Il6/IL-6. Again, we observed that IL-17A stimulated Csf3/G-CSF was reduced in ST2 cells transfected with shRNA targeting the Fstl1 mRNA compared to control shRNA (Fig. 1C and 1D). Additionally, IL-17A stimulated Il6/IL-6 production was substantially reduced in the FSTL-1 knockdown ST2 cells compared to control shRNA (Fig. 1E and 1F).
Figure 1. Follistatin-like protein 1 suppression by shRNA reduces IL-17/TNFα mediated cytokine production.
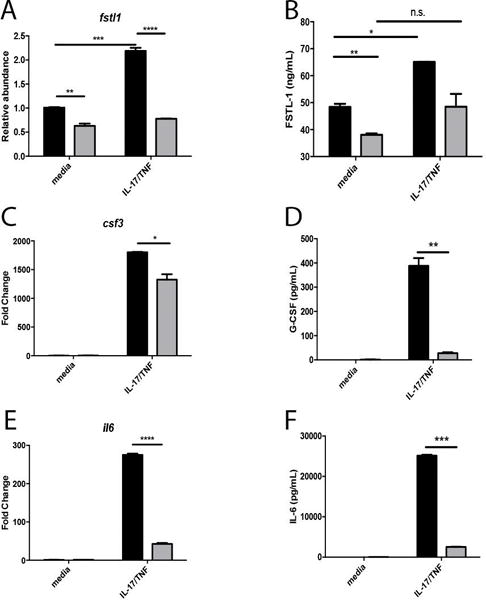
In the ST2 cell line (A) Fstl1 transcript abundance and (B) FSTL-1 protein production increased following IL-17/TNFα stimulation, but was reduced in cells transfected with shRNA targeting Fstl1 (gray bars) compared to control shRNA (black bars) IL-17/TNFα dependent cytokine production is reduced by Fstl1 inhibition as determined by (C, E) transcript induction and protein secretion (D, F). ** p<0.01, ***p<0.001, ****p<0.0001
Follistatin-like protein 1 suppression by gene deletion reduces IL-17/TNFα mediated cytokine production
To further validate the effect of FSTL-1 in IL-17A/TNFα driven cytokine production, we employed a parallel approach to attenuate FSTL-1 production in primary bone marrow stromal cells. As previously described29, BMSCs from FSTL-1 knockout (KO) embryonic mice had markedly reduced Fstl1 transcription and protein secretion (Fig. 2A and 2B) with and without stimulation in wild-type BMSCs. The IL-17A/TNFα stimulated production of Csf3/G-CSF was significantly reduced in FSTL-1 KO BMSCs (Fig. 2C and 2D), as was Il6/IL-6 (Fig. 2E and 2F). Collectively, these data reveal that IL-17 receptor signaling is attenuated in the context of Fstl1 suppression.
Figure 2. Follistatin-like protein 1 suppression by gene deletion reduces IL-17/TNFα mediated cytokine production.

In primary bone marrow stromal cells (A) Fstl1 transcript abundance and (B) FSTL-1 protein production increased following IL-17A and TNFα stimulation, but was reduced in cells from FSTL-1 KO mice (gray bars) compared to wild-type littermate control mice (black bars) IL-17/TNFα dependent cytokine production is reduced by Fstl1 inhibition as determined by (C, E) transcript induction and protein secreton (D, F) *p<0.05. ** p<0.01, ***p<0.001, ****p<0.0001
Microarray and gene expression analysis of the role of FSTL-1 in IL-17 signaling
To evaluate potential pathways and gene targets underlying these FSTL-1 mediated events, we used an unbiased approach to evaluate gene expression by Affymetrix microarray. We assessed gene expression in ST2 cells and primary BMSCs, comparing control (wild-type) cells in which FSTL-1 was attenuated (ST2 shFSTL-1 or FSTL-1 KO, respectively) under baseline conditions and following stimulation with IL-17A/TNFα. We reasoned that bona fide candidate targets of FSTL-1 mediated IL-17A cytokine production would be differentially regulated in both the ST2 cell system and primary BMSCs under both conditions. To determine FSTL-1 regulated target genes independent of IL-17A stimulation we identified transcripts upregulated (Fig. 3A) or downregulated (Fig. 3B) under unstimulated and IL-17A/TNFα-stimulated conditions. Validating this workflow construct, Fstl1 was downregulated in ST2 cells under unstimulated (4.99 fold, p=0.018) and IL-17A stimulated (3.1 fold, p=0.0095) conditions. Fstl1 was also downregulated in unstimulated (17.99 fold, p=0.033) and stimulated (23.29 fold, p=0.021) BMSCs, as expected. Indeed, 46 genes were upregulated (Fig. 3A) and 23 genes were downregulated (Fig 3B) in this dataset. Notably, we observed that Il17rc expression was downregulated in the ST2 unstimulated (1.1 fold, p=0.034) and IL-17A stimulated (1.34 fold, p=0.0086) system as well as in the primary BMSC unstimulated (2.34 fold, p=0.0057) and stimulated (2.39 fold, p=0.0336) system. These data suggest that Il17rc expression may be a significant mediator of FSTL-1-dependent IL-17A cytokine regulation.
Figure 3. Microarray and pathway analysis of the role of FSTL-1 in IL-17 signaling.
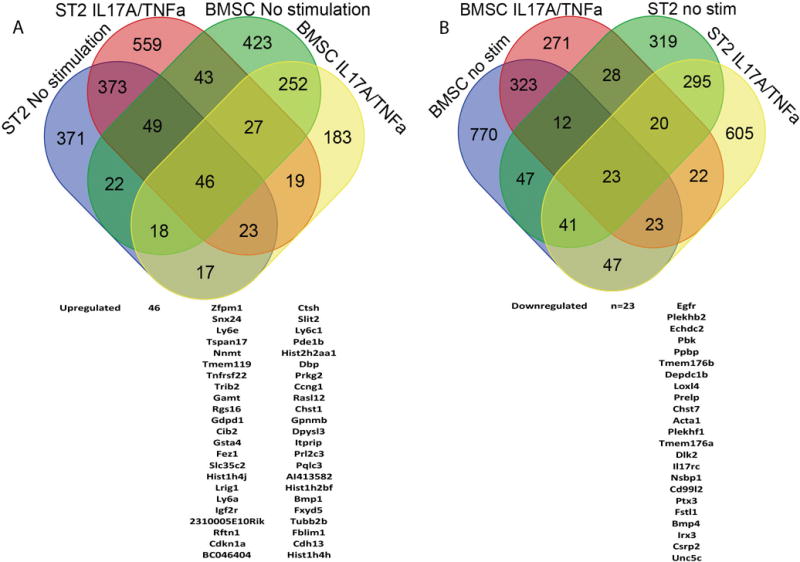
cDNA from ST2 shControl and shFSTL-1 cells, as well as primary WT and FSTL-1 KO BMSCs, with and without IL-17A/TNFa stimulation underwent Microarray analysis. Differential expression (≥1.1 fold-change) for FSTL-1 influenced targets were identified for each condition by t-test with Bonferroni correction (p<0.05) of non-transformed data. (A) Forty-six symmetrically upregulated and (B) twenty-three downregulated genes in both cell systems with and without IL-17A/TNFa stimulation are displayed schematically in a Venn diagram corresponding to Table 1.
FSTL-1 attenuation is associated with reduced IL-17RC mRNA and protein expression
To verify the microarray finding that Il17rc is decreased in an FSTL-1 dependent manner, we assessed its expression by qRT-PCR for this transcript in both ST2 cells (Fig. 4A) and primary BMSCs (Fig. 4B). These results indeed confirmed that Il17rc transcript was downregulated in the absence of FSTL-1. We then examined whether the decrease in transcript abundance was associated with reduced surface expression of IL-17RC protein by FACS. A significantly higher percentage of WT cells expressed the IL-17RC protein compared to FSTL-1 KO cells (Fig. 4C). Additionally, WT cells had a significantly greater mean fluorescence intensity (MFI) compared to FSTL-1 KO BMSCs (Figs. 4D, E). Together, these data demonstrate that attenuation of FSTL-1 results in reduced Il17rc transcription and IL-17RC protein surface expression.
Figure 4. Follistatin-like protein 1 attenuation is associated with reduced IL-17RC mRNA and protein expression.

Il17rc transcript abundance is reduced in (A) ST2 cells with shRNA FSTL-1 suppression as well as (B) primary BMSCs from FSTL-1 KO mice when compared to controls. Surface expression of IL-17RC is reduced as measured by FACS determined by (C) %IL17RC+ and (D) mean fluorescence intensity as noted by (E) representative histograms of isotype (dark grey), FSTL-1 KO (light grey) and WT BMSCs (dashed line). *p<0.05, **p<0.01.
Ectopic Il17rc expression restores IL-17-dependent signaling in FSTL-1 KO cells
Having confirmed that IL-17RC was decreased in an FSTL-1 dependent manner, we next sought to determine whether complementation with IL-17RC could rescue this effect. Accordingly, FSTL-1 KO BMSCs were transfected with a plasmid expressing murine Il17rc. This approach yielded increased Il17rc (Fig. 5A), and transfection of Il17rc partially rescued Il6 (Fig. 5B) and Csf3 (Fig. 5C) production following IL-17A/TNFα stimulation in FSTL-1 BMSCs at or above the level of WT control-transfected cells. These data demonstrate that IL-17RC complementation in the FSTL-1 KO BMSCs is capable of partial rescue of IL-17A stimulated cytokine suppression.
Figure 5. Ectopic Il17rc expression rescues cytokine production in FSTL-1 KO cells.
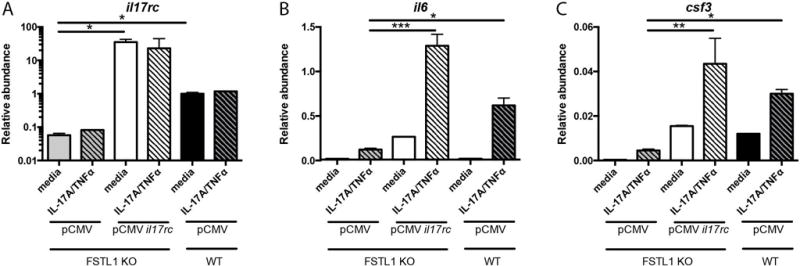
FSTL-1 KO BMSC transfection with pCMV-il17rc rescues Il17rc transcript abundance (A). pCMV-Il17rc complementation in FSTL-1 KO BMSCs rescues IL-17A stimulated transcript levels of (B) Il6 and (C) Csf3. * p<0.05, ** p<0.01, ** p<0.001
Ectopic Fstl1 expression rescues Il17rc expression and cytokine production in FSTL-1 KO cells
To investigate the relationship between FSTL-1 and IL-17RC regulation, we transfected FSTL-1 KO BMSCs with Fstl1 treated with or without IL-17A/TNFα stimulation (Fig. 6A). Indeed, Fstl1 transfection significantly increased Il17rc transcript abundance (Fig. 6B), confirming a causal relationship between Fstl1 expression and Il17rc modulation. Moreover, Fstl1 complementation conferred increased production of Csf3 and Il6 (Figs 6C and 6D) upon stimulation, confirming that FSTL-1 regulated IL-17RC expression is sufficient to rescue IL-17 stimulated cytokine production.
Figure 6. Ectopic Fstl1 expression rescues Il17rc expression and cytokine production in FSTL-1 KO cells.
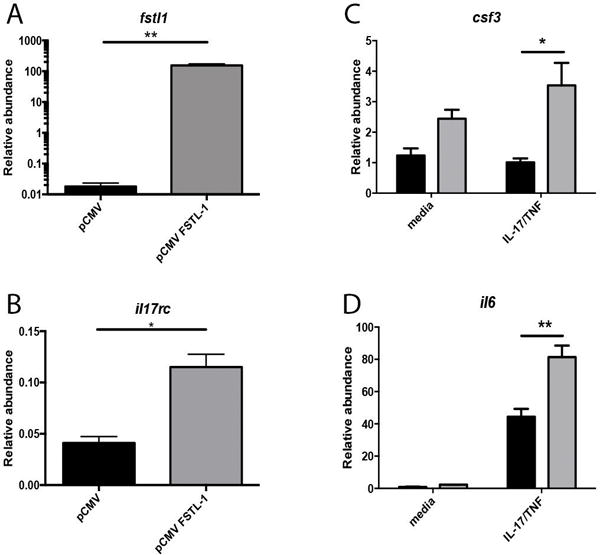
FSTL-1 KO BMSC transfection with pCMV-fstl1 rescues (A) Fstl1 and (B) Il17rc transcript abundance. pCMV-Fstl1 complementation in FSTL-1 KO BMSCs rescues IL-17A/TNF stimulated transcript levels of (C) Il6 and (D) Csf3. *p<0.05, ** p<0.01
Fstl1 influences Il17rc transcript abundance via transcriptional regulation
To better understand the mechanism by which FSTL-1 influences mRNA transcript abundance and subsequent protein surface expression, we stimulated WT and FSTL-1 KO BMSCs under conditions that inhibit new RNA synthesis (ActinomycinD), and separately using a nascent RNA labeling approach. To assess whether Fstl1 regulates mRNA stability of Il17rc, we incubated WT and FSTL-1 KO cells with 5ug/ml ActinomycinD, a potent inhibitor of gene transcription, followed by RNA isolation at various timepoints (Figs. 7A, B). Before ActinomycinD treatment and all subsequent timepoints, there was a reduction of Fstl1 and Il17rc mRNA in FSTL-1 KO BMSCs. However, the rate of decay of Il17rc transcript was similar for both WT and FSTL-1 KO BMSCs (Fig. 7C), suggesting that the half-life of Il17rc transcript abundance was not FSTL-1 dependent.
Figure 7. FSTL-1 influences Il17rc transcript abundance via transcriptional regulation.
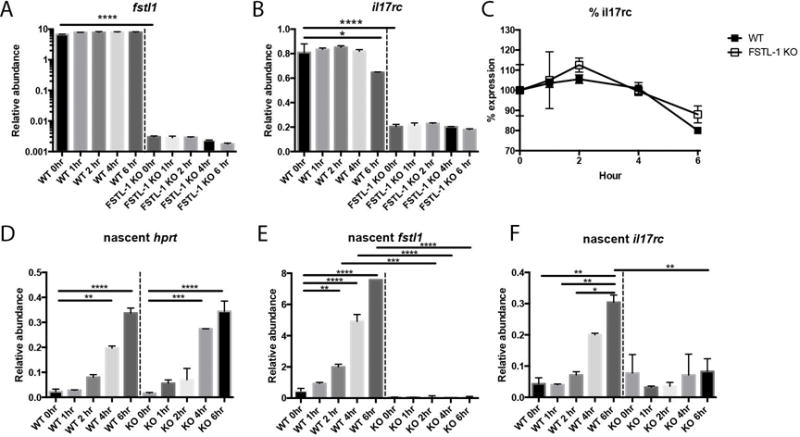
Following ActinomycinD treatment, FSTL-1 KO cells had reduced (A) Fstl1 and (8) Il17rc transcript abundance before ActinomycinD treatment and at 1, 2, 4 and 6 hours post-treatment compared with WT BMSCs. Linear regression of Il17rc transcript abundance (C) at various timepoints following ActinomycinD treatment showed similar slopes for WT and FSTL-1 KO BMSCs (−3.254±1.200 and −2.457±1.429, p=0.6748). Newly synthesized mRNA (5-EU labeled) was similar for (D) Hprt, but reduced for (E) Fstl1 and (F) Il17rc, transcripts in FSTL-1 KO cells compared to WT * p<0.05, ** p<0.01, ***p<0.001, ****p<0.0001
To assess if FSTL-1 regulated de novo gene transcription, adherent WT and FSTL-1 KO cells were labeled with 5-ethynyl uridine, which incorporates only into newly synthesized (nascent) RNA. We then examined nascent RNA at various timepoints post-labeling (Fig 7 D–F). This approach revealed that Hprt, Fstl1 and Il17rc transcripts were actively synthesized in BMSCs within 6 hours showing similar nascent Hprt transcription between genotypes. However, FSTL-1 KO cells had decreased nascent Fstl1 and Il17rc mRNA identifying that newly synthesized l17rc transcripts were reduced in the absence of FSTL-1. Together, these studies suggest that FSTL-1 influences Il17rc transcript abundance by regulating gene transcription but not RNA stability.
Fstl1 expression correlates with Il17rc expression in the CD45-negative bone marrow population
To evaluate if these findings in vitro were also observed in vivo, we sought to evaluate Fstl1 and Il17rc transcription in a bone marrow stromal cell population. We harvested bone marrow cells from femur flushes of wild-type C57Bl//6 mice and FSTL-1 Hypomorphic mice. FACS-sorted CD45-negative cells (predominately bone marrow stromal cells) (CITE) underwent gene expression analysis for Fstl1, Il17rc and Il17ra (an IL-17 Receptor complex herterodimeric partner of IL-17RC). In this heterogenous CD45- population FSTL-1 Hypomorphs showed slightly reduced expression of (A) Fstl1, (B) Il17rc and (C) Il17ra compared to WT controls. However, Pearson’s correlation analysis demonstrated a strong positive correlation between (D) Fstl1 and Il17rc (r=0.9512, R2=0.9048, p<0.0001) but no correlation between (E) Fstl1 and Il17ra (r=−0.1982, R2=0.0393, p=0.5162), suggesting that the in vivo murine system likely reflects our in vitro observations.
Discussion
FSTL-1 is a molecule of emerging biological importance with recent studies identifying a role in tissue repair and development, novel mechanisms of transcriptional regulation and inflammation and host defense. While some of these pathways have begun to be defined, the role of FSTL-1 in IL-17 elicited gene production is unknown. Nonetheless, advances in understanding of IL-17/Th17 driven inflammation have provided targets for therapeutic intervention, including small molecule inhibitors and monoclonal antibodies aimed at IL-17 driven host responses, including autoimmune joint disease and psoariasis30–34. Therefore, understanding the mechanism underlying the influence of FSTL-1 on IL-17 inflammatory responses may spur development of novel therapies aimed at this disease pathway. Here, we identified the intersection of FSTL-1 and IL-17 using bone marrow stromal cells, a clinically relevant model system. Herein, we report that in vitro FSTL-1 modulates IL-17 mediated cytokine production by regulating Il17rc gene expression. Indeed, FSTL-1 appears to be necessary and sufficient to regulate Il17rc expression, and that complementation of IL-17RC rescues IL-17 driven cytokine production in the context of FSTL-1 deficiency.
This observation is the first to show that FSTL-1 regulates gene transcription, a cytokine receptor, opening a new avenue of investigation into the molecular mechanism by which FSTL-1 performs this function. Disco-interacting protein 2 homolog A (DIP2A) functions as an FSTL-1 receptor in mediating FSTL-1 dependent Akt phosphorylation in endothelial cells, cardiomyocytes and a rat ischemic stroke model11, 35. Other studies have suggested that FSTL-1 mediates multiple DAMP/PAMP stimulated inflammation pathways involving TLR4/CD14 and NF-κB perhaps via phosphorylating multiple intermediates36, 37. We have shown that FSTL-1 localizes to mitochondria in macrophages yielding increased NLRP3 inflammasome activation and IL-1β secretion15. FSTL-1 also plays a critical role in organogenesis as a TGFβ-inducible factor interacting with BMP signaling/Smad phosphorylation, as well as influencing cell cycling3, 8, 15, 29, 38, 39. It remains to be determined the common factors shared among these systems, as regulation of Il17rc gene expression fails to explain the diverse functions of FSTL-1 in mammalian biologic systems. Conversely, none of the above pathways have been implicated in Il1rc gene regulation, suggesting that additional cellular functions of FSTL-1 have yet to be identified.
Strikingly, there is very little information regarding the molecular mechanisms regulating Il17rc gene transcription. The methylation status of Il17rc has been associated with macular degeneration in human studies, though the precise role of Il17rc epigenetic regulation in this disease remains an area of active investigation40, 41. Additionally, multiple transcription factor binding sites have been identified within Il17rc exons suggesting that several independent pathways may influence its expression. Furthermore, while IL-17RC is highly expressed in bone, joint, prostate, liver and lung, IL-17RC is selectively and minimally detected in the leukocyte compartment suggesting that cell specific expression is tightly regulated42–47. Notably, several splice variants of Il17rc are expressed in a tissue specific manner, and factors influencing their regulation are unknown47, 48. Thus, further study of the FSTL-1 regulation is likely to yield enhanced understanding of pathways influencing in IL17RC expression and possibly function.
These studies are currently limited by the lack of knowledge regarding molecular interactions between FSTL-1 and transcription factors or epigenetic regulators capable of modulating Il17rc gene expression, and studies to identify these aspects are needed to inform the cellular function(s) of FSTL-1. Additionally, as our work examines in vitro mechanisms of FSTL-1/IL-17 interaction, further work exploring the role of FSTL-1 in in vivo IL-17/Th17 models of inflammation and clinical disease processes is warranted to clarify the significance of our findings. While FSTL-1 is expressed in heart, lung, neuron and bone amongst others tissues, the cell and tissue specific expression of FSTL-1 and the relationship to complex inflammatory diseases has yet to be explored.
In summary, this study identified a new function for FSTL-1, gene transcription modulation. Specifically FSTL-1 mediated Il17rc regulation is a mechanism by which FSTL-1 mediates IL-17 driven cytokine production in vitro in bone marrow stromal cells. This discovery represents a significant advance in our understanding of IL-17 biology, and ongoing mechanistic investigation of FSTL-1 function in this pathway may prove valuable for the design of therapies aimed at modulating Th17/IL-17 driven diseases. Additionally, this work suggests that further FSTL-1 focused investigation into innate and adaptive immune responses may yield new therapeutic targets, as the precise function of FSTL-1 in different inflammatory processes remains elusive.
Methods
Bone marrow stromal cell culture
The murine ST2 bone marrow stromal cell line has been utilized extensively for the in vitro study of IL-17 signaling18, 49. Cells were stably transduced with murine FSTL1 short hairpin RNA (shRNA) or a control lentivirus shRNA (Santa Cruz Biotechnology) according to the manufacturer’s protocol and clonally selected, as previously described13. Primary bone marrow stromal cells (BMSCs) were isolated from FSTL-1 KO embryos as previously described29. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with heat-inactivated 10% fetal bovine serum (FBS) for ST2 cells and 15% FBS for BMSCs. In 12 wells, we plated 100,000 cells/well followed by stimulation with either 8ng/ml IL-17A (R&D 7956-ML) and 2ng/ml TNFα (R&D 410-MT-010) or media alone in duplicate; for microarray studies each group contained n=3. Cells were then harvested for RNA at 6 hours post-stimulation or collection of supernatant 24 hours post-stimulation. Randomization was included in treatment design prior to each experiment. Each experiment depicted was performed at least two times to ensure reproducibility.
RNA and microarray analysis
Total RNA was isolated from cells using the RNeasy kit (Qaigen), and cDNA was synthesized using iScrip 8t (Bio-Rad) according to the manufacturers instructions. Gene expression was analyzed by qPCR performed using primer/probe pairs for Hprt, Fstl1, Il6, Csf3, Il17rc (Applied Biosystems) in the C1000 Touch/CFX96 Real-Time System (Bio-Rad). RNA quality was verified using the 2100 Bioanalyzer (Agilent) before undergoing microarray analysis performed on ST2 cells using Mouse WG8.0 and BMSCs using Mouse WG6 2.0 Expression Bead Chip (Affymetrix) at the Genomics and Proteomics Core Laboratories, University of Pittsburgh. We defined differential expression as significant by setting threshold levels of ≥1.1 fold-change and p<0.05 by t-test with Bonferroni correction. Gene lists meeting these criteria were cross-analyzed for common FSTL-1 dependent differentially expressed genes.
Protein secretion analysis
Supernatant protein concentrations were determined for FSTL-1 by ELISA, as previously described13, and for IL-6 and G-CSF using the xMAP multiplex system (Millipore) according to the manufacturer’s instructions.
FACS Analysis
WT and FSTL-1 KO cells were stained for IL-17RC surface expression using APC-conjugated anti-IL-17RC (R&D FAB2270A) or APC-IgG control (R&D IC108A). Cells were trypsinized, washed in 0.5%FBS in PBS, centrifuged at low speed, Fc blocked with CD16/32 and stained with APC-conjugated anti-mouse IL-17RC or APC-conjugated isotype control. Cells were sorted using an LSR II flow cytometer, and data was analyzed using FlowJo software (TreeStar).
Il17rc and Fstl1 plasmid transfection
Murine Il17rc (Addgene 46864), Fstl1 (Origene MR204305) and empty vector pCMV6-Entry control (Origene PS 100001) plasmids were transformed in and grown in Mix and Go E. coli (Zymo Research) in 5ml Luria-Bertani broth containing selection antibiotic and prepared using the QIAprep Spin Miniprep kit (Qiagen 27106), per manufacturer’s instructions. FSTL-1 KO BMSCs were transfected with plasmid by electroporation using the Neon Transfection System (ThermoFischer Scientific), per manufacturer’s instructions, with transfection settings of 30ms at 1400V for 1 pulse. Cells were cultured in triplicate in complete DMEM + 15% FBS and 12–18 hours post-transfection treated identically as described above.
mRNA regulation studies
To assess mRNA stability, WT and FSTL-1 KO cells were incubated with 5ug/ml ActinomycinD, to inhibit gene transcription, followed by RNA isolation at various timepoints post-ActinomycinD treatment. At each time, untreated cells were used as a normalizing control to assess rate of decay. Gene expression was assessed as described above.
To assess nascent mRNA systhesis, WT and FSTL-1 KO BMSCs were cultured for 18hours to allow adherence. Using the Click-iT Nascent RNA Capture Kit (ThermoFisher Scientific Cat# C10365), cells were labeled for various lengths of time (0, 1, 2, 4, and 6 hours) with 150uM 5-ethyl Uridine (EU). Cells were harvested using TRIzol reagent and half of this volume was used to isolate total RNA, while the remainder was used as input for the nascent RNA capture, which was followed according to the manufacturer’s protocol to recover the labeled, newly-synthesized mRNA. Labeled mRNA underwent reverse transcription using iScript in addition to total cell RNA as described above. Nascent mRNA for each transcript was normalized to total Hprt for relative abundance analysis.
Bone marrow population analysis
C57Bl/6 and FSTL-1 Hypomorphic (on C57Bl/6 background) mice were housed in accordance with the approved University of Pittsburgh School of Medicine Institutional Animal Care and Use Committee protocol12. The bone marrow compartment of male and female wild-type and FSTL-1 Hypomorhic mice was isolated by femur flush protocol50. Isolated bone marrow cells were resuspended in sterile phosphate-buffered saline and counted using trypan blue stain and a hemocytometer. The cells were first blocked using anti-mouse CD16/CD32 (eBioscience) and subsequently surfaced stained in the presence of anti-mouse CD45 antibody (clone 30-F11, BD Bioscience). CD45-positive and CD45-negative cell populations were then sorted on a FACSAria via FACSDiva software (Becton-Dickinson) into phosphate-buffered saline. The sorted cell populations were pelleted and resuspended in RNA Lysis Buffer (Zymo Research). RNA was processed for gene expression analysis as previously described. These experiments were performed twice with n=3 or n=4 per group each time. Data shown include cells from all animals in the two experiments.
Statistical analysis
Investigators were not blinded to treatment, but were blinded to individual/group during data analysis. All statistics were performed using GraphPad Prism 6. Briefly, all data are presented with mean ± SEM. Studies comparing two groups were analyzed by two-sided student’s t-test. Studies comparing more than two groups were analyzed by ordinary one-way ANOVA with Tukey’s multiple comparisons. Ex vivo gene expression correlation was determined using Pearson’s correlation coefficient analysis. All statistical analyses considered p<0.05 significant.
Figure 8. Fstl1 expression correlates with Il17rc expression in the CD45 negative bone marrow population.

CD45 negative cells from wild-type and FSTL-1 Hypomorphic mouse bone marrow had slightly reduced (A) Fstl1, (B) Il17rc and (C) Il17ra expression. Correlation analysis revealed highly significant association between (D) Fstl1 and Il17rc (r=0.9512, R2=0.9048, p<0.0001) but not (E) Fstl1 and Il17ra (r=−0.1982, R2=0.0393, p=0.5162).
Acknowledgments
This work was supported by NIH grants K08HL128809 (B.T.C.), F30AI114146 (T.E), R01AI107825 (S.L.G), R01AI073556, T32AR052282 (R.H) and R37HL079142 (J.K.K.) and in part by the Children’s Hospital of Pittsburgh of UPMC Health System.
Footnotes
Competing Interests
The University of Pittsburgh has a patent “Immunomodulation of inflammatory conditions utilizing Follistatin-like Protein-1 and agents that bind thereto” (No 8334274) listing RH as an inventor.
References
- 1.Shibanuma M, Mashimo J, Mita A, Kuroki T, Nose K. Cloning from a mouse osteoblastic cell line of a set of transforming-growth-factor-beta 1-regulated genes, one of which seems to encode a follistatin-related polypeptide. Eur J Biochem. 1993;217(1):13–9. doi: 10.1111/j.1432-1033.1993.tb18212.x. [DOI] [PubMed] [Google Scholar]
- 2.Dong Y, Geng Y, Li L, Li X, Yan X, Fang Y, et al. Blocking follistatin-like 1 attenuates bleomycin-induced pulmonary fibrosis in mice. The Journal of experimental medicine. 2015;212(2):235–52. doi: 10.1084/jem.20121878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geng Y, Dong Y, Yu M, Zhang L, Yan X, Sun J, et al. Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(17):7058–63. doi: 10.1073/pnas.1007293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sylva M, Li VS, Buffing AA, van Es JH, van den Born M, van der Velden S, et al. The BMP antagonist follistatin-like 1 is required for skeletal and lung organogenesis. PloS one. 2011;6(8):e22616. doi: 10.1371/journal.pone.0022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sylva M, Moorman AF, van den Hoff MJ. Follistatin-like 1 in vertebrate development. Birth defects research Part C, Embryo today : reviews. 2013;99(1):61–9. doi: 10.1002/bdrc.21030. [DOI] [PubMed] [Google Scholar]
- 6.Umezu T, Yamanouchi H, Iida Y, Miura M, Tomooka Y. Follistatin-like-1, a diffusible mesenchymal factor determines the fate of epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(10):4601–6. doi: 10.1073/pnas.0909501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams D, Larman B, Oxburgh L. Developmental expression of mouse Follistatin-like 1 (Fstl1): Dynamic regulation during organogenesis of the kidney and lung. Gene expression patterns: GEP. 2007;7(4):491–500. doi: 10.1016/j.modgep.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei K, Serpooshan V, Hurtado C, Diez-Cunado M, Zhao M, Maruyama S, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525(7570):479–85. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson DC, Marinov AD, Blair HC, Bushnell DS, Thompson SD, Chaly Y, et al. Follistatin-like protein 1 is a mesenchyme-derived inflammatory protein and may represent a biomarker for systemic-onset juvenile rheumatoid arthritis. Arthritis and rheumatism. 2010;62(8):2510–6. doi: 10.1002/art.27485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clutter SD, Wilson DC, Marinov AD, Hirsch R. Follistatin-like protein 1 promotes arthritis by up-regulating IFN-gamma. Journal of immunology. 2009;182(1):234–9. doi: 10.4049/jimmunol.182.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang X, Hu Q, Li B, McBride D, Bian H, Spagnoli P, et al. Follistatin-like 1 attenuates apoptosis via disco-interacting protein 2 homolog A/Akt pathway after middle cerebral artery occlusion in rats. Stroke. 2014;45(10):3048–54. doi: 10.1161/STROKEAHA.114.006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campfield BT, Nolder CL, Marinov A, Bushnell D, Davis A, Spychala C, et al. Follistatin-like protein 1 is a critical mediator of experimental Lyme arthritis and the humoral response to Borrelia burgdorferi infection. Microbial pathogenesis. 2014;73:70–9. doi: 10.1016/j.micpath.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaly Y, Marinov AD, Oxburgh L, Bushnell DS, Hirsch R. FSTL1 promotes arthritis in mice by enhancing inflammatory cytokine/chemokine expression. Arthritis and rheumatism. 2012;64(4):1082–8. doi: 10.1002/art.33422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamae T, Marinov AD, Sowders D, Wilson DC, Devlin J, Boudreau R, et al. Follistatin-like protein-1 is a novel proinflammatory molecule. Journal of immunology. 2006;177(7):4758–62. doi: 10.4049/jimmunol.177.7.4758. [DOI] [PubMed] [Google Scholar]
- 15.Chaly Y, Fu Y, Marinov A, Hostager B, Yan W, Campfield B, et al. Follistatin-like protein 1 enhances NLRP3 inflammasome-mediated IL-1beta secretion from monocytes and macrophages. European journal of immunology. 2014;44(5):1467–79. doi: 10.1002/eji.201344063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K, McAleer JP, Lin Y, Paterson DL, Zheng M, Alcorn JF, et al. Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity. 2011;35(6):997–1009. doi: 10.1016/j.immuni.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conti HR, Peterson AC, Brane L, Huppler AR, Hernandez-Santos N, Whibley N, et al. Oral-resident natural Th17 cells and gammadelta T cells control opportunistic Candida albicans infections. The Journal of experimental medicine. 2014;211(10):2075–84. doi: 10.1084/jem.20130877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. The Journal of experimental medicine. 2009;206(2):299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nature reviews Immunology. 2009;9(8):556–67. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAleer JP, Nguyen NL, Chen K, Kumar P, Ricks DM, Binnie M, et al. Pulmonary Th17 Antifungal Immunity Is Regulated by the Gut Microbiome. Journal of immunology. 2016;197(1):97–107. doi: 10.4049/jimmunol.1502566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11(10):763–76. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 22.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. The Journal of experimental medicine. 2001;194(4):519–27. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baeten D, Sieper J, Braun J, Baraliakos X, Dougados M, Emery P, et al. Secukinumab, an Interleukin-17A Inhibitor, in Ankylosing Spondylitis. The New England journal of medicine. 2015;373(26):2534–48. doi: 10.1056/NEJMoa1505066. [DOI] [PubMed] [Google Scholar]
- 24.Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, et al. Secukinumab in plaque psoriasis–results of two phase 3 trials. The New England journal of medicine. 2014;371(4):326–38. doi: 10.1056/NEJMoa1314258. [DOI] [PubMed] [Google Scholar]
- 25.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nature reviews Immunology. 2014;14(9):585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Rosa F. T-lymphocyte interaction with stromal, bone and hematopoietic cells in the bone marrow. Immunol Cell Biol. 2009;87(1):20–9. doi: 10.1038/icb.2008.84. [DOI] [PubMed] [Google Scholar]
- 27.Onishi RM, Park SJ, Hanel W, Ho AW, Maitra A, Gaffen SL. SEF/IL-17R (SEFIR) is not enough: an extended SEFIR domain is required for il-17RA-mediated signal transduction. The Journal of biological chemistry. 2010;285(43):32751–9. doi: 10.1074/jbc.M110.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garg AV, Ahmed M, Vallejo AN, Ma A, Gaffen SL. The deubiquitinase A20 mediates feedback inhibition of interleukin-17 receptor signaling. Sci Signal. 2013;6(278):ra44. doi: 10.1126/scisignal.2003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaly Y, Blair HC, Smith SM, Bushnell DS, Marinov AD, Campfield BT, et al. Follistatin-like protein 1 regulates chondrocyte proliferation and chondrogenic differentiation of mesenchymal stem cells. Annals of the rheumatic diseases. 2015;74(7):1467–73. doi: 10.1136/annrheumdis-2013-204822. [DOI] [PubMed] [Google Scholar]
- 30.Huh JR, Littman DR. Small molecule inhibitors of RORgammat: targeting Th17 cells and other applications. European journal of immunology. 2012;42(9):2232–7. doi: 10.1002/eji.201242740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu T, Wang X, Zhong B, Nurieva RI, Ding S, Dong C. Ursolic acid suppresses interleukin-17 (IL-17) production by selectively antagonizing the function of RORgamma t protein. The Journal of biological chemistry. 2011;286(26):22707–10. doi: 10.1074/jbc.C111.250407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472(7344):491–4. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonardi C, Matheson R, Zachariae C, Cameron G, Li L, Edson-Heredia E, et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. The New England journal of medicine. 2012;366(13):1190–9. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 34.Papp KA, Langley RG, Lebwohl M, Krueger GG, Szapary P, Yeilding N, et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 52-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 2) Lancet. 2008;371(9625):1675–84. doi: 10.1016/S0140-6736(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 35.Ouchi N, Asaumi Y, Ohashi K, Higuchi A, Sono-Romanelli S, Oshima Y, et al. DIP2A functions as a FSTL1 receptor. The Journal of biological chemistry. 2010;285(10):7127–34. doi: 10.1074/jbc.M109.069468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J, Liang W, Li J, Long J. Knockdown of FSTL1 inhibits oxLDL-induced inflammation responses through the TLR4/MyD88/NF-kappaB and MAPK pathway. Biochem Biophys Res Commun. 2016;478(4):1528–33. doi: 10.1016/j.bbrc.2016.08.138. [DOI] [PubMed] [Google Scholar]
- 37.Murakami K, Tanaka M, Usui T, Kawabata D, Shiomi A, Iguchi-Hashimoto M, et al. Follistatin-related protein/follistatin-like 1 evokes an innate immune response via CD14 and toll-like receptor 4. FEBS letters. 2012;586(4):319–24. doi: 10.1016/j.febslet.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Ouchi N, Oshima Y, Ohashi K, Higuchi A, Ikegami C, Izumiya Y, et al. Follistatin-like 1, a secreted muscle protein, promotes endothelial cell function and revascularization in ischemic tissue through a nitric-oxide synthase-dependent mechanism. The Journal of biological chemistry. 2008;283(47):32802–11. doi: 10.1074/jbc.M803440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Fang Y, Li X, Liang J, Jiang D, Geng Y, et al. Apical Secretion of FSTL1 in the Respiratory Epithelium for Normal Lung Development. PloS one. 2016;11(6):e0158385. doi: 10.1371/journal.pone.0158385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei L, Liu B, Tuo J, Shen D, Chen P, Li Z, et al. Hypomethylation of the IL17RC promoter associates with age-related macular degeneration. Cell Rep. 2012;2(5):1151–8. doi: 10.1016/j.celrep.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliver VF, Franchina M, Jaffe AE, Branham KE, Othman M, Heckenlively JR, et al. Hypomethylation of the IL17RC promoter in peripheral blood leukocytes is not a hallmark of age-related macular degeneration. Cell Rep. 2013;5(6):1527–35. doi: 10.1016/j.celrep.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor PR, Roy S, Leal SM, Jr, Sun Y, Howell SJ, Cobb BA, et al. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORgammat and dectin-2. Nature immunology. 2014;15(2):143–51. doi: 10.1038/ni.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You Z, Shi XB, DuRaine G, Haudenschild D, Tepper CG, Lo SH, et al. Interleukin-17 receptor-like gene is a novel antiapoptotic gene highly expressed in androgen-independent prostate cancer. Cancer research. 2006;66(1):175–83. doi: 10.1158/0008-5472.CAN-05-1130. [DOI] [PubMed] [Google Scholar]
- 44.Trevejo-Nunez G, Chen K, Dufour JP, Bagby GJ, Horne WT, Nelson S, et al. Ethanol impairs mucosal immunity against Streptococcus pneumoniae infection by disrupting interleukin 17 gene expression. Infection and immunity. 2015;83(5):2082–8. doi: 10.1128/IAI.02869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, et al. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. Journal of immunology. 2007;179(8):5462–73. doi: 10.4049/jimmunol.179.8.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Baarsen LG, Lebre MC, van der Coelen D, Aarrass S, Tang MW, Ramwadhdoebe TH, et al. Heterogeneous expression pattern of interleukin 17A (IL-17A), IL-17F and their receptors in synovium of rheumatoid arthritis, psoriatic arthritis and osteoarthritis: possible explanation for nonresponse to anti-IL-17 therapy? Arthritis Res Ther. 2014;16(4):426. doi: 10.1186/s13075-014-0426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haudenschild D, Moseley T, Rose L, Reddi AH. Soluble and transmembrane isoforms of novel interleukin-17 receptor-like protein by RNA splicing and expression in prostate cancer. The Journal of biological chemistry. 2002;277(6):4309–16. doi: 10.1074/jbc.M109372200. [DOI] [PubMed] [Google Scholar]
- 48.You Z, Dong Y, Kong X, Zhang Y, Vessella RL, Melamed J. Differential expression of IL-17RC isoforms in androgen-dependent and androgen-independent prostate cancers. Neoplasia. 2007;9(6):464–70. doi: 10.1593/neo.07109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramer JM, Yi L, Shen F, Maitra A, Jiao X, Jin T, et al. Evidence for ligand-independent multimerization of the IL-17 receptor. Journal of immunology. 2006;176(2):711–5. doi: 10.4049/jimmunol.176.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemeth K, Mayer B, Sworder BJ, Kuznetsov SA, Mezey E. A practical guide to culturing mouse and human bone marrow stromal cells. Current protocols in immunology/edited by John E Coligan … [et al] 2013;102 doi: 10.1002/0471142735.im22f12s102. Unit 22F 12. [DOI] [PubMed] [Google Scholar]


