Abstract
Methods to impute HLA alleles based on dense single nucleotide polymorphism (SNP) data provide a valuable resource to association studies and evolutionary investigation of the MHC region. The availability of appropriate training sets is critical to the accuracy of HLA imputation, and the inclusion of samples with various ancestries is an important pre-requisite in studies of admixed populations. We assess the accuracy of HLA imputation using 1000 Genomes Project data as a training set, applying it to a highly admixed Brazilian population, the Quilombos from the state of São Paulo. To assess accuracy, we compared imputed and experimentally determined genotypes for 146 samples at 4 HLA classical loci. We found imputation accuracies of 82.9%, 81.8%, 94.8% and 86.6% for HLA-A, -B, -C and -DRB1 respectively (two-field resolution). Accuracies were improved when we included a subset of Quilombo individuals in the training set. We conclude that the 1000 Genomes data is a valuable resource for construction of training sets due to the diversity of ancestries and the potential for a large overlap of SNPs with the target population. We also show that tailoring training sets to features of the target population substantially enhances imputation accuracy.
Keywords: HLA, Imputation, 1000 Genomes, Admixed populations, Relatedness
1. Introduction
Technological advances and the availability of large-scale genomic data have boosted the development of tools for the imputation of genotypes at both the genomic scale and in specific genomic regions of interest. Imputation methods combine training sets containing subjects genotyped for a high density of SNPs (single nucleotide polymorphisms) with samples of interest genotyped for only a subset of these markers. Based on population genetic models and allelic correlation measures (e.g. linkage disequilibrium), imputation methods predict unobserved genotypes from those present in the training set.
While high resolution HLA typing is still the gold standard in the field, imputation of HLA alleles is becoming increasingly used. The main advantage of HLA imputation is that it provides information on HLA variants for studies involving large samples, and for which HLA typing was not performed (e.g. many GWAS studies). The imputed HLA allele calls allow the GWAS hits to be interpreted with additional biological context [1]. For example, by analyzing GWAS SNPs with genomewide significance in the light of an individual’s HLA genotype, interactions can be tested for, and confounding effects can be controlled for (e.g. specific predisposing HLA alleles which are already known). Imputation can even provide, with a high reliability, the variant an individual carries at a specific amino-acid position, and this can be included in models testing for association between genotypes and disease phenotypes [2–4].
Given the complexity and costs associated with HLA genotyping and the increasing availability of genomewide SNP data, over the last years several methods have been developed with the goal of imputing the HLA alleles based on dense SNP data for the MHC region [3,5–7]. This is a challenging task, considering the large number of alleles of HLA genes, which makes methods more effective when: (a) the training set consists of a large number of samples [3,5]; (b) there is a suitable pairing among the population(s) that make up the training set and the sample of interest [8,9]; (c) the HLA alleles being queried in the target population are not rare. http://dx.doi.org/10.1016/j.humimm.2015.11.004
Choosing a suitable training set is critical to the success of the imputation methods. However, due to the high cost, it is not always possible to generate a training set tailored for the specific target population under study, so imputation is commonly made using public datasets as training sets such as the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) and the British 1958 birth cohort of Welcome Trust Case Control Consortium (http://www.ebi.ac.uk/ega/) [7]). The use of such resources can also be a challenge, since public datasets do not always have the populations related to those in the target sample, for which imputation is to be performed. This is especially critical for admixed populations, such as those from the Americas who carry Native American ancestry, which is underrepresented in public datasets. Because of the difficulties in obtaining Native American samples, an alternative is to use other admixed populations to make imputations for this ancestry component.
In recent years, one of the most widely used public resources for population genetic studies is the 1000 Genomes Project [10]. Phase I of the 1000 Genomes Project provided mainly low coverage sequencing data for two African, five European, three Asian, and four admixed populations from the Americas (African–Americans, Mexicans, Puerto Ricans and Colombians). These samples were recently genotyped at high resolution for the classical HLA genes [11], providing a valuable resource which integrates genomewide SNP data with HLA allele calls [12].
In the present study we examine the accuracy of HLA imputation in a highly admixed Brazilian population (with 40% African, 39% European and 21% Native American average ancestries [13]) using the 1000 Genomes HLA and SNP data as a training set. Our interest is motivated by the importance of admixed populations in studies with a focus on admixture mapping (e.g. [14]) and in understanding the role of introgression involving HLA genes (i.e. the observation that ancestry proportions in the HLA region deviate from genomewide averages for admixed populations, [15,16]).
In this study we do not intend to compare the performance of different HLA imputation methods, as others have done before (eg. [7,8]). Rather, we assess the performance of the 1000 Genomes data as a training set for imputation of highly admixed populations, and explore how the quantity of SNPs and ancestry of the individuals in the training set impacts imputation accuracy. We perform imputation using HIBAG [7], an ensemble classifier that has been shown to provide accurate imputation, and for which imputation models can be built using training sets of choice.
We find that the 1000 Genomes data provides HLA imputation of 83–94% accuracy at the two-field level. We compare imputation accuracy to that obtained when other training sets are used, or when individuals which are related to the target sample are included in the training set. Finally, we discuss how SNP density and geographic origin of populations making up the target sample contribute to imputation accuracy, in the context of an admixed population.
2. Materials and methods
2.1. The Brazilian admixed sample
We imputed HLA genotypes for highly admixed samples from Brazilian communities known as “Quilombos” from Vale do Ribeira region, São Paulo State. These were founded by runaway, abandoned and free slaves in the 18th century, and established in remote areas in the Atlantic Rainforest of Southeastern Brazil, where they subsequently admixed with Native Americans, adding a third ancestry component, in addition to African and European (Table S1). A total of 365 samples (referred to as the “QUI dataset”) were genotyped using the Affymetrix Axiom Human Origins Array (600K SNPs), and a subset of 146 individuals were experimentally genotyped at HLA loci using PCR-SBT (Thermo Fisher) for HLA-A, -B. -C (exons 2, 3 and 4) and -DRB1 (exon 2). The ethics committee of the Instituto de Biociências da Universidade de São Paulo approved this study and informed consent was obtained from all participants.
2.2. Data for training set using 1000 Genomes Project samples (1000g)
We selected 931 samples from the 1000 Genomes Project for which SNP [10] and HLA genotypes [11] were available: 126 African, 317 European, 265 East Asian, and 223 admixed samples from the Americas (53 African-American, 60 Colombian, 55 Mexican and 55 Puerto Rican; Table S2). The SNP data was mainly of low coverage genotype calls ([10]; ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20110521/), and HLA typing was generated by sequence-based typing (PCR-SBT) for HLA-A, -B, -C and -DRB1 genes ([11]; data available at (http://www.ncbi.nlm.nih.gov/gv/mhc/xslcgi.fcgi?cmd=cellsearch).
2.3. Training set of Zheng et al. [7] (UW)
To place our result in the context of previous studies, we also used the multi-ethnic training set specific to the Affymetrix Axiom Human Origins Array platform assembled by Zheng et al. [7], which consists of 2 different datasets (HAPMAP Phase 2 and HLARES) and includes more than 3000 samples (details in [7]) (Table S3).
2.4. Data cleaning and SNP selection for imputation analysis
We filtered the Quilombo (QUI) SNP dataset for genotype quality using R Package GWASTools [17]. We selected a total of 1238 SNPs that flanked the HLA-A, -B, -C and -DRB1 genes in 500 kb windows. For the UW dataset, the 500 kb windows resulted in a set of 467 SNPs.
2.5. Building a multi-ethnic model for HLA allele imputation
HLA imputation was performed with Attribute Bagging, implemented in the HIBAG program, which averages over many classifiers (obtained by 100 bootstrap resamplings) to define HLA alleles with highest posterior probabilities [7]. This method has proven to be robust in a previous study with another admixed population [8]. We used HIBAG to build multi-ethnic models for HLA-A, -B, -C and -DRB1, with parameters to build the models used according to recommendations of the HIBAG authors [7]. We built three models (for both one and two field resolution), based on three different training sets: (a) 1000g; (b) 1000g with the inclusion of an additional set of 57 unrelated Quilombo samples (1000g+QUI); and (c) UW.
Details on how the unrelated individuals were selected to be added to the 1000g set are presented in Section 2.7. The models used in this study are available for download at www.ib.usp.br/genevol (1000g, 1000g+QUI) and www.biostat.washington.edu/~bsweir/HIBAG/ (UW model used in HIBAG).
2.6. Quantification of imputation accuracy
To assess the accuracy of imputation at each locus, we quantified the number of chromosomes with correctly called HLA alleles over the total number of imputations made (corresponding to 292 chromosomes for which experimentally generated HLA calls were available). We did not require a minimum posterior probability (implying a call threshold of 0%). For 1000g+QUI we adopted the same strategy, but used only 89 of the 146 samples for comparisons, since 57 QUI samples were used in the training sample.
2.7. Effect of relatedness on imputation
The Quilombo communities have a small population size (between 100 and 450 inhabitants per community) and are geographically close to one another. As a consequence, there is a substantial degree of relatedness between individuals [18]. Using approximately 500.000 genomewide SNPs from the Affymetrix Axiom Human Origins Array platform, we estimated the kinship coefficient between all pairs of Quilombo samples based on the Moment Method implemented in SNPRelate R Package [19]. We defined an “unrelated group” as a set of individuals among which there was no pair for which the kinship coefficient exceeded 1/32. This “unrelated group”, comprising 57 samples, was added to 1000g to define the 1000g+QUI model. In addition, the kinship information was used to evaluate if relatedness between individuals in the target populations and the training sets influenced the accuracy of the analyses.
3. Results
Using three training sets (1000g, 1000g+QUI and UW), we created multi-ethnic models to impute alleles for HLA-A, -B, -C and -DRB1 for the admixed Quilombo population. Tables 1 and S4 describe each training set (numbers of training samples, HLA alleles present, and SNPs flanking the HLA genes in an 500 kb window), as well their imputation accuracies, estimated as the proportion of correct allele calls over the 292 chromosomes with experimentally defined alleles for the QUI sample.
Table 1.
Imputation accuracy for five training sets.
| Training set | Target sample size | HLA-A (%) | HLA-B (%) | HLA-C (%) | HLA-DRB1 (%) |
|---|---|---|---|---|---|
| One-field | |||||
| 1000g | 146 | 92.1 | 87.3 | 98.6 | 93.5 |
| 1000g+QUI | 89 | 92.7 | 93.8 | 99.4 | 94.4 |
| UW | 146 | 89.0 | 72.9 | 94.4 | 84.6 |
| 1000g_UW_shared_SNPs | 146 | 92.1 | 76.0 | 95.5 | 88.7 |
| 1000g_89_samples | 89 | 88.8 | 86.5 | 94.5 | 92.1 |
| Two-fields | |||||
| 1000g | 146 | 82.9 | 81.8 | 94.8 | 86.6 |
| 1000g+QUI | 89 | 86.5 | 88.8 | 97.7 | 89.3 |
| UW | 146 | 83.2 | 72.6 | 94.1 | 84.6 |
| 1000g_UW_shared_SNPs | 146 | 84.2 | 65.7 | 86.8 | 77.7 |
| 1000g_89_samples | 89 | 80.3 | 85.4 | 96.1 | 87.1 |
When using the 1000g data as a training set, we found that the imputation accuracy at the one-field level of HLA resolution was above 92% for all loci, except for HLA-B (87%) (Table 1). At the two-field level imputation accuracies were lower, ranging from 82% to 94%, with the lowest being for HLA-B and the highest for HLA-C (Table 1). These values are generally higher (with the exception of HLA-A) than those obtained using the UW model (Table 1), a finding that maybe influenced by the lower number of SNPs in the UW training set (467 SNPs) (Table S4), when compared to the number present in the 1000g data (1238 SNPs).
We next developed a training set in which the 1000g samples were supplemented with a subset of 57 unrelated individuals from the Quilombo population (1000g+QUI model). For this analysis, we removed individuals which were related to the 57 QUI samples included in the training set from the target population, resulting in a set of 89 individuals. For this dataset, we found that imputation accuracies were higher when the training set was supplemented with QUI samples (compare Table 1, rows B and D; median increase in accuracy of 4.6% over all loci).
To better evaluate how relatedness among target population and training set influence imputation accuracy, we divided the of 89 samples for which imputation accuracies were estimated into two groups: one consisting of individuals with relatedness above 1/32 with at least one individual in the training set (resulting in a set of 60 individuals, which we refer to as the “related set”), and another consisting of individuals with no cases of relatedness above 1/32 in the training set (resulting in a set of 29 samples, which we refer to as “unrelated set”; Table 2 and S5). Because sample sizes for accuracy estimation are small, measures of accuracy were taken over all loci and chromosomes (totaling 360 and 174 allele calls for the “related” and “unrelated” sets, respectively). For analyses at the two-field resolution, the overall imputation accuracies were 92.5% and 86.6% for the “related” and “unrelated”, respectively, confirming the contribution of related samples to imputation accuracy (Table 2). The exception was HLA-C for which the “unrelated” sample had a small improvement, although the “related” sample was also highly accurate. We note that even among individuals which were not related to those in the training set, there was an increase in imputation accuracy (from imputation accuracy of 82.7% when using the 1000g without the QUI, to 86.6% when they were included).
Table 2.
Imputaiton accuracy of the 1000g+QUI model in “related” and “unrelated” sets (two-field HLA resolution).
| Dataset | HLA-A (%) | HLA-B (%) | HLA-C (%) | HLA-DRB1 (%) | Overall (%) |
|---|---|---|---|---|---|
| Related (60 samples, 1000g+QUI as training set) | 90.0 | 92.5 | 96.6 | 90.8 | 92.5 |
| Unrelated (29 samples, 1000g+QUI as training set) | 79.3 | 81.0 | 100.0 | 86.2 | 86.6 |
| Unrelated (29 samples, 1000g without QUI as training set) | 72.4 | 77.6 | 96.5 | 84.5 | 82.7 |
To better understand the differences between the imputation models we quantified the effect of training set identity while controlling for the number of SNPs. We compared the accuracies for 1000g and UW models when only the SNPs shared by both models were used. We found that even when identical SNPs were present in both training sets, the 1000g consistently had higher accuracies than the UW training set (compare rows C and E of Table 1; median increase in accuracy of 2.9% over all loci).
Next, we evaluated if the number of SNPs had an effect on imputation accuracy. We did this by comparing the accuracy obtained using the 1000g training set with either all available SNPs or only with the SNPs also present in the UW dataset. We found that increasing the number of SNPs improved accuracy at all loci (compare rows A and C of Table 1; median increase in accuracy of 7.6% over all loci).
Overall, our results show that all three attributes analyzed (SNP number, training set identity, presence of individuals related to target population in training set) affect the imputation accuracy.
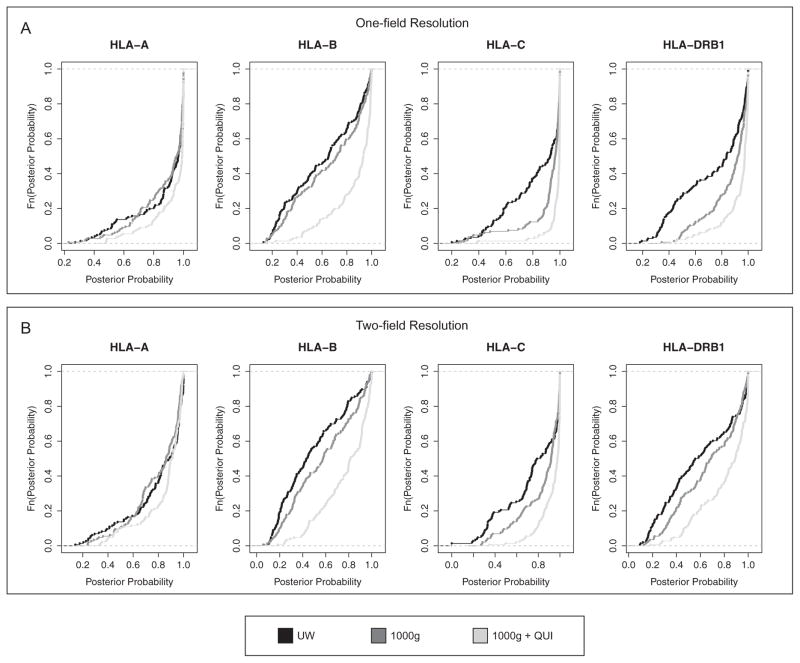
Next, we examined how informative are the posterior probabilities for genotype calls, reported by the imputation programs. First, we examined the posterior probability distribution associated with the different training sets (Fig. 1), and found that the 1000g+QUI model had the highest posterior probabilities. Additionally, we investigated the relationship between accuracy and posterior probabilities of our models. For all HLA loci, imputation accuracy exceeded 85% at a posterior probability threshold >50%, suggesting that this threshold is associated with high imputation accuracy in our population (Table S6).
Fig. 1.
Empirical cumulative distribution of posterior probabilities for 3 models: UW (black), 1000g (dark gray) and 1000g+QUI (light gray). (A) One-field resolution. (B) Two-field resolution.
Throughout our analyses we have evaluated the imputation accuracy of all loci independently. However, the close proximity among certain loci may result in non-independence in accuracy estimates. To address this, we created a contingency table with estimation accuracies for locus pairs and compared this to the expectation under the null hypothesis of independence in imputation accuracy among markers. The only locus pair for which a significant association was found was for HLA-B and -C (Table S7). The likely basis for this non-random association of accuracies is that the models for imputing alleles at these loci share many SNPs, due to their close proximity. As a consequence, underrepresented flanking sequences in the reference panel can cause inaccurate imputation at both loci.
Finally, we examined in greater detail the identity of the alleles for which imputation accuracy was particularly low. As previously reported by Zheng et al. [7], we found that alleles with low accuracy tend to have lower frequencies, and in most instances where an allele is miscalled, it is replaced with the same incorrect allele over many individuals (Tables S8–S10). Additionally, we examined the ancestry of the miscalled alleles. We focus on HLA-B alleles, for which there is extensive geographic information allowing us to classify alleles with respect to a likely continent of origin (http://igdawg.org/software/browser-beta.html). As shown in Table S11, the HLA-B alleles with low sensitivity in 1000g and UW models were frequently from Native American and African ancestry. This set of alleles represents 12.6% of HLA-B allele frequency in the target population, a value close to the error rate when using the 1000g and UW models (13.5% and 16.38%, respectively). When the 57 QUI individuals were added to the training set (1000g+QUI), some of the previously incorrectly imputed alleles were correctly imputed. The low sensitivity, mainly for the Native American and African alleles, reflects their absence or rarity in the 1000g and UW training sets.
4. Discussion
Imputation of HLA genotypes is a methodology that has grown in recent years, driven by the availability of large samples of dense genotype data, primarily generated by GWAS studies. It has proven a valuable resource through its potential to enrich the degree of information about HLA polymorphism in association studies [1–4].
Our study was motivated by the need to impute HLA alleles for highly admixed populations, such as the Quilombo communities of Southeastern Brazil. This population is representative of other highly admixed populations, and also contains individuals with varying degrees of relatedness, an additional factor whose impact on imputation accuracy we explore.
Reliable imputation requires genotype data for a target population and an appropriate training set, for which HLA and SNP data must be available. In the case of admixed populations, training sets with samples from multiple ancestries are particularly important, since the underrepresentation of ancestries in the training set can result in poor imputation accuracy. In the present study we evaluate the accuracy of HLA imputation using the 1000 Genomes Phase I data as a training set.
The 1000 Genomes data is of interest for imputation for several reasons. First, it contains populations from various continents, as well as admixed populations, enhancing the coverage of distinct ancestries. Secondly, the 1000 Genomes data was generated by genome sequencing, which implies that genotype calls are available for a large number of SNPs. Thus, regardless of the SNP array chosen for the target population, the overlap with those in the 1000 Genomes data is expected to be high. Third, the 1000 Genomes data have publicly available HLA typing at high resolution [11].
In our study of highly admixed Brazilian individuals, imputation using the 1000g as a training sample resulted in accuracies of 82.9%, 81.8%, 94.8% and 86.6% for HLA-A, -B, -C and -DRB1 alleles respectively (at the two fields level of HLA resolution). We note that the 1000g dataset is comparatively small with respect to other training sets (e.g. from 2000 to 3000 samples used by Zheng et al. [7]), and its accuracy was slightly higher than Zheng et al. [7] for the case of admixed or African-ancestry populations. Two main features of the 1000g data as a training set contribute to its accuracy.
First, the 1000g data contains samples from various continents, spanning a broad array of human diversity, whereas other datasets such as HLARES (used by Zheng et al. [7]) were aggregated from multiple GlaxoSmithKline clinical trials including samples from 34 countries, and these clinical trials did not necessarily prioritize genetic background or geographic variation. It is well documented that imputation works best when the training samples and the test samples come from similar populations or share ancestral populations [20]. As discussed by Levin et al. [8], the dataset used by Zheng et al. [7] contains South African samples, which contributed little to the formation of the admixed populations of the Americas and therefore may be less informative for imputing HLA in these populations than the 126 sub-Saharan Africans present in the 1000g data.
Second, whole genome sequencing provides information on the majority of SNPs contained in our target population. For example, the SNPs contained in the Zheng et al. [7] training set were an intersection of Illumina 1M, Illumina 1M Duo and Affymetrix Axiom Human Origins Array, and as a consequence covered a smaller number of SNPs than those we were able to use based on the 1000g sample.
We also found that accuracies were improved when we included a set of 57 Quilombo individuals in the training set, and that this improvement was higher for the individuals which were more closely related to those in the training set, as expected (Tables 1 and 2). However, the inclusion of the 57 samples also improved, although more modestly, the accuracy of imputation for individuals which are not related to any sample in the training set (Table 2).
Our study allows some general guidelines to be proposed. First, the coupling of high resolution HLA genotyping with genomewide resequencing, as is the case for the 1000g data, provides a valuable resource of HLA imputation. In particular, the availability of genotypes for all variable sites (as opposed to a subset included in genotyping arrays) increases the utility of such data in training sets, since a greater number of SNPs in the target population can be used. Secondly, we find that for studies of relatively isolated populations such as the Quilombos (e.g. [21,22]), the imputation approach is particularly powerful when a subset of target samples are include in the training set, due to the high degree of sharing of MHC region haplotypes between training set and target samples. We have shown that in the case of the Quilombo samples the addition of as few as 57 samples to the training set can bring about substantial improvements to the accuracy of estimates.
The present study illustrates how training set size, identity, and SNP composition will influence the accuracy of HLA imputation in a highly admixed population. Adding a subset of individuals from the target population to the training set increases accuracy, as does increasing its size by including underrepresented ancestries. In light of this, we predict that the expansion of training sets will be an important development, allowing imputation to be performed increasingly more accurately. Fortunately, existing imputation methods are well placed to be enhanced by the acquisition of new SNP and HLA data. For example, commonly used programs such as HiBAG [7], MAGpredition [23] and SNP2HLA [2] are easily accessible and allow models to be formulated by the user, with whatever data is chosen. With the sharing and merging of these training sets, HLA imputation has the potential to play an increasingly important role in characterizing the HLA diversity of human populations, including those which are highly admixed.
Supplementary Material
Acknowledgments
We thank Bruce Weir for helpful discussions.
K.N. wrote the paper, designed, performed the experiments and analyzed the data. X.Z contributed with analysis tools and data analysis. M.T, M.E.M, B.Z.P and G.N.P. genotyped HLA genes on the samples. L.K and J.E.P.C prepared DNA samples. R.C.M.N. contributed with reagents/material. D.M wrote the paper, designed the experiments and contributed with reagents.
We thank 3 anonymous reviewers for an extremely useful set of comments.
This research was financially supported by grants from São Paulo Research Foundation (FAPESP), The Brazilian National Council for Scientific and Technological Development (CNPq) and United States – National Institutes of Health (NIH). K.N was funded by FAPESP Grant #2012/09950-9 and CEPID-FAPESP Grant #13/08028-1; D.M. has a FAPESP research Grant #12/18010-0 and a CNPq productivity Grant #308167/2012-0; R.C.M.N., L.K. and J.E.P.C. were funded by CEPID-FAPESP – Grant #13/08028-1. X.Z were supported by United States NIH Grant #GM07591.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.humimm.2015.11.004.
References
- 1.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. http://dx.doi.org/10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bakker PI, Raychaudhuri S. Interrogating the major histocompatibility complex with high-throughput genomics. Hum Mol Genet. 2012;21:R29–36. doi: 10.1093/hmg/dds384. http://dx.doi.org/10.1093/hmg/dds384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. http://dx.doi.org/10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Mazas A, Meyer D. The relevance of HLA sequencing in population genetics studies. J Immunol Res. 2014;2014:971818. doi: 10.1155/2014/971818. http://dx.doi.org/10.1155/2014/971818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. http://dx.doi.org/10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dilthey A, Leslie S, Moutsianas L, Shen J, Cox C, Nelson MR, et al. Multi-population classical HLA type imputation. PLoS Comput Biol. 2013;9:e1002877. doi: 10.1371/journal.pcbi.1002877. http://dx.doi.org/10.1371/journal.pcbi.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14:192–200. doi: 10.1038/tpj.2013.18. http://dx.doi.org/10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin AM, Adrianto I, Datta I, Iannuzzi MC, Trudeau S, McKeigue P, et al. Performance of HLA allele prediction methods in African Americans for class II genes HLA-DRB1, -DQB1, and -DPB1. BMC Genet. 2014;15:72. doi: 10.1186/1471-2156-15-72. http://dx.doi.org/10.1186/1471-2156-15-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khor SS, Yang W, Kawashima M, Kamitsuji S, Zheng X, Nishida N, et al. High-accuracy imputation for HLA class I and II genes based on high-resolution SNP data of population-specific references. Pharmacogenomics J. 2015 doi: 10.1038/tpj.2015.4. http://dx.doi.org/10.1038/tpj.2015.4. [DOI] [PMC free article] [PubMed]
- 10.1000 Genomes Project consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. http://dx.doi.org/10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gourraud PA, Khankhanian P, Cereb N, Yang SY, Feolo M, Maiers M, et al. HLA diversity in the 1000 genomes dataset. PLoS One. 2014;9:e97282. doi: 10.1371/journal.pone.0097282. http://dx.doi.org/10.1371/journal.pone.0097282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappas D, Paunic V, Lizee AM, Taylor K, Leslie S, Meller J, et al. The immpute project: evaluating SNP imputation methodologies for HLA-A,-B,-C,- DRB1 and-DQB1 genotypes. Tissue Antigens. 2014;1:12–13. [Google Scholar]
- 13.Kimura L, Ribeiro-Rodrigues EM, De Mello Auricchio MT, Vicente JP, Batista Santos SE, Mingroni-Netto RC. Genomic ancestry of rural African-derived populations from Southeastern Brazil. Am J Hum Biol. 2013;25:35–41. doi: 10.1002/ajhb.22335. http://dx.doi.org/10.1002/ajhb.22335. [DOI] [PubMed] [Google Scholar]
- 14.Shriner D, Adeyemo A, Ramos E, Chen G, Rotimi CN. Mapping of disease-associated variants in admixed populations. Genome Biol. 2011;12:223. doi: 10.1186/gb-2011-12-5-223. http://dx.doi.org/10.1186/gb-2011-12-5-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang H, Choudhry S, Mei R, Morgan M, Rodriguez-Cintron W, Burchard EG, et al. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am J Hum Genet. 2007;81:626–633. doi: 10.1086/520769. http://dx.doi.org/10.1086/520769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J, et al. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 2012;84:343–364. doi: 10.3378/027.084.0401. http://dx.doi.org/10.3378/027.084.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gogarten SM, Bhangale T, Conomos MP, Laurie CA, McHugh CP, Painter I, et al. GWASTools: an R/bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–3331. doi: 10.1093/bioinformatics/bts610. http://dx.doi.org/10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemes RB, Nunes K, Meyer D, Mingroni-Netto RC, Otto PA. Estimation of inbreeding and substructure levels in african-derived Brazilian Quilombo populations. Hum Biol. 2014;86:276–288. doi: 10.13110/humanbiology.86.4.0276. [DOI] [PubMed] [Google Scholar]
- 19.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. http://dx.doi.org/10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erlich HA. HLA typing using next generation sequencing: an overview. Hum Immunol. 2015;76(12):887–890. doi: 10.1016/j.humimm.2015.03.001. http://dx.doi.org/10.1016/j.humimm.2015.03.001.S0198-8859(15)00093-2. [DOI] [PubMed] [Google Scholar]
- 21.Garagnani P, Laayouni H, González-Neira A, Sikora M, Luiselli D, Bertranpetit J, et al. Isolated populations as treasure troves in genetic epidemiology: the case of the Basques. Eur J Hum Genet. 2009;17:1490– 1494. doi: 10.1038/ejhg.2009.69. http://dx.doi.org/10.1038/ejhg.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeggini E. Using genetically isolated populations to understand the genomic basis of disease. Genome Med. 2014;6:83. doi: 10.1186/s13073-014-0083-5. http://dx.doi.org/10.1186/s13073-014-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li SS, Wang H, Smith A, Zhang B, Zhang XC, Schoch G, Geraghty D, Hansen JA, Zhao LP. Predicting multiallelic genes using unphased and flanking single nucleotide polymorphisms. Genet Epidemiol. 2011;35(2):85–92. doi: 10.1002/gepi.20549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.