Neurons expressing a novel rhodopsin can be activated by light; a complete story of protein structure and function follows.
Abstract
Generation of an electrochemical proton gradient is the first step of cell bioenergetics. In prokaryotes, the gradient is created by outward membrane protein proton pumps. Inward plasma membrane native proton pumps are yet unknown. We describe comprehensive functional studies of the representatives of the yet noncharacterized xenorhodopsins from Nanohaloarchaea family of microbial rhodopsins. They are inward proton pumps as we demonstrate in model membrane systems, Escherichia coli cells, human embryonic kidney cells, neuroblastoma cells, and rat hippocampal neuronal cells. We also solved the structure of a xenorhodopsin from the nanohalosarchaeon Nanosalina (NsXeR) and suggest a mechanism of inward proton pumping. We demonstrate that the NsXeR is a powerful pump, which is able to elicit action potentials in rat hippocampal neuronal cells up to their maximal intrinsic firing frequency. Hence, inwardly directed proton pumps are suitable for light-induced remote control of neurons, and they are an alternative to the well-known cation-selective channelrhodopsins.
INTRODUCTION
All cells maintain a particular concentration of ions H+, K+, Na+, and Cl− in the cytoplasm, which is crucial for life. Ion gradients across cell membranes are maintained by ion transporters, which are integral membrane proteins. In many archaea, bacteria, and unicellular eukaryotes, these gradients are created (among other mechanisms) by light-driven microbial rhodopsins, seven transmembrane α helix proteins comprising a cofactor chromophore named retinal (1). Proton gradients are maintained by outward proton pumps, and they play a crucial role in providing energy for most biochemical reactions (2). Although a light-driven proton-pumping rhodopsin [bacteriorhodopsin (BR)] was discovered long ago in archaea (3) and similar rhodopsins have now been found in other domains of life throughout soil, hypersaline, marine, and freshwater habitats, all known proton-pumping rhodopsins are outwardly directed (1). The same is true for non-rhodopsin proton pumps. Another inwardly directed cell plasma membrane rhodopsin proton pump was described very recently in a marine bacterium, Parvularcula oceani, during submission of the present manuscript (4). However, in that work (4), two major questions remained unanswered. First, the structure of xenorhodopsins (XeRs) and the true proton pathway and molecular mechanism of inward pumping are still not known. Second, their possible application in electrically excitable cells was not demonstrated (optogenetics). The question of whether these depolarizing ion pumps were powerful enough to achieve a similar remote control of neuronal cells as the well-known light-gated channelrhodopsins remained unanswered. In addition, the photocycle of XeR from P. oceani is about 10 times slower than that of XeR from Nanosalina (NsXeR), which also means that this protein cannot be used to elicit action potentials in neuronal cells at a high firing frequency.
In 2011, a new class of microbial rhodopsins, distinct from other rhodopsin types, was discovered (5). The authors of that work found several new homologs of Anabaena sensory rhodopsin (ASR) (6). The members of the class were named XeRs. Among these proteins were XeRs from a new major lineage of Archaea, specifically the nanohaloarchaeon Nanosalina sp. J07AB43 and Nanosalinarum sp. J07AB56 with 89% amino acid identity between them but with only 34% amino acid identity to ASR. By analyzing the amino acid sequence alignment, the authors concluded that XeRs do not share specific features common to proton pumps or halorhodopsin pumps. However, they are similar to ASR because they lack a common Asp at the donor position, as with the sensory rhodopsins known at that time.
We extended the analysis (figs. S1 and S2) and found that XeRs have remarkable differences from conservative amino acid residues usually found in other microbial rhodopsins. Arg82 (numbering is in accordance with BR amino acid sequence) is extremely conservative among rhodopsins, whereas in almost all XeRs from Nanohaloarchaea, this place is occupied by Trp. In XeR, we find a proline for the highly conserved Asp212. The positions of classical proton acceptors (Asp85) and donors (Asp96) in proton pumps are Asp and Ala, respectively, that is, XeRs have the WDSAPK motif, which is very different from that of conventional proton pumps such as BR (RDTDDK). Moreover, contrary to ASR where a proposed soluble transducer protein (ASR transducer) is encoded by a gene in the same operon, in nanohaloarchaeal genomes, there are only putative Na+/H+ antiporter genes adjacent to the XeR ones (5).
We characterized three proteins from the XeR family, namely NsXeR (5), HrvXeR (7), and AlkXeR (8), and found out that all these three proteins are inwardly directed proton pumps. All Nanohaloarchaea are extremely halophilic (live in saturated sodium chloride). NsXeR (5) was found in a metagenome from brines in Lake Tyrrell (Australia), whereas HrvXeR (7) was sequenced in a crystallizer pond of a solar saltern in Spain, and AlkXeR (8) comes from alkaline (pH >9) brines in Siberia. With one of these proteins, we performed a comprehensive function and structure study, and the results are described below. Moreover, we found that the light-induced depolarizing current by NsXeR is sufficient for reliable activation of neuronal cells with high temporal accuracy. Therefore, NsXeR as a proton pump is attractive for optogenetic studies because of the cation-independent activity and represents an alternative to the well-known cation-selective channelrhodopsins (9).
RESULTS AND DISCUSSION
Functional characterization of XeRs
pH changes in Escherichia coli suspensions
The protein NsXeR was heterologously expressed in E. coli C41 strain. The pH of the cell suspension increased upon illumination and decreased when the light was turned off (Fig. 1A). The effect of the pH change was completely abolished by the addition of carbonyl cyanide m-chlorophenylhydrazone (CCCP). Similar experiments performed with other proton pumps as BR gave the opposite behavior of pH upon illumination of the cells. Two other members of the XeR family, HrvXeR and AlkXeR, studied in the present work gave the same results as NsXeR (Fig. 1A). Thus, pH experiments provide evidence that Nanohaloarchaea rhodopsins are inwardly directed proton pumps.
Fig. 1. Electrogenic properties of XeR.
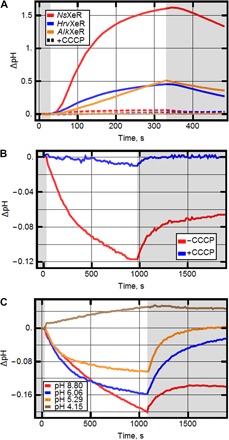
(A) pH changes upon illumination in E. coli cell suspensions expressing different XeRs. Graphs show the pH changes with and without the addition of CCCP. (B) pH changes upon illumination in liposome suspension with reconstructed NsXeR (with and without CCCP). (C) pH changes upon illumination in liposome suspension measured under different pH values.
pH changes in liposome suspension
Purified NsXeR was reconstituted in soybean liposomes (10). The pH changes upon illumination showed acidification of the solution outside the membrane (Fig. 1B). These pH changes were abolished when CCCP was added to the suspension. Because in similar experiments outwardly directed proton pumps [such as BR and proteorhodopsin (PR)] show the opposite pH behavior (11), we conclude that NsXeR is a real inwardly directed proton pump. What is interesting is that, in a wide range of pH values (between pH 5 and 9), our experiments still show inward proton pumping (Fig. 1C).
Absorption spectra, retinal isomer composition, and photocycle
The absorption maximum of NsXeR in solubilized form is 565 nm (Fig. 2A). It is similar for HrvXeR (562 nm), whereas the homolog AlkXeR is a red-shifted variant (577 nm; Fig. 2A). For NsXeR, the position of the maximum does not shift when the pH of the buffer is varied in the range from 4.5 to 9.0. NsXeR does not exhibit light and dark adaptation, as indicated by the absence of spectral changes. The same holds true for retinal extraction experiments (12) that show a predominant fraction of all-trans retinal independent of preillumination (Fig. 3).
Fig. 2. Spectroscopic characterization of NsXeR.
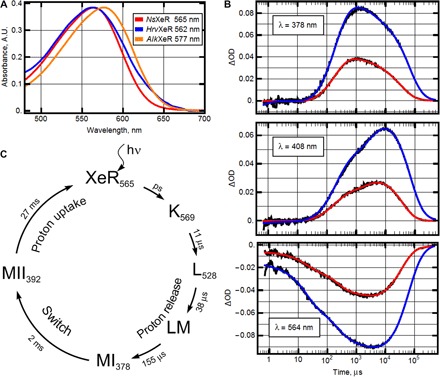
(A) Absorption spectra of representatives of XeR family solubilized in the detergent DDM (n-dodecyl-β-d-maltoside). The corresponding positions of absorption maximum are indicated in the legend. A.U., arbitrary units. (B) Transient absorption changes of NsXeR (pH 7.5, T = 20°C) at three representative wavelengths: 378, 408, and 564 nm. Black lines represent experimental data, and red and blue lines represent the result of global fit using five exponents. The photocycles were measured for the two preparations: NsXeR in nanodiscs (red) and in liposomes (blue). Note that the differences in amplitudes between the samples are due to the approximately two times higher concentration of NsXeR in liposomes than in nanodiscs (see fig. S3). ΔOD, change in absorbance. (C) Proposed model of NsXeR photocycle in nanodiscs.
Fig. 3. High-performance liquid chromatography measurements.
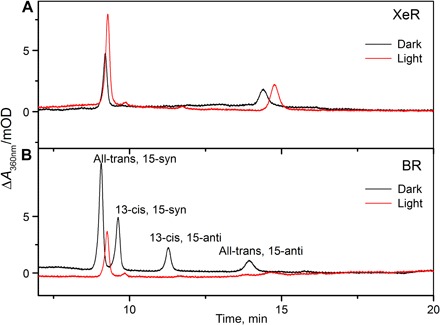
(A) Retinal extraction of light- and dark-adapted solubilized NsXeR reconstituted in liposomes and (B) of light- and dark-adapted solubilized BR (as reference). mOD, milli OD (optical density).
Transient absorption changes of NsXeR (pH 7.5, T = 20°C) are shown at three characteristic wavelengths—378, 408, and 564 nm—with NsXeR prepared in two different ways: in nanodiscs (red) and in single lipid vesicles (blue) (Fig. 2B). The results of global fit using five exponents are shown in fig. S3. The photocycle of NsXeR in nanodiscs is faster (27 ms) than that of NsXeR in lipid vesicles (50 ms). The photocycle of NsXeR in nanodiscs is shown in Fig. 2C.
The photocycle of NsXeR contains a microsecond-duration phase, which is usually assigned to the multistep reaction of release of the energized ion (the H+ in our case) and a millisecond-duration phase, consisting of relaxation and reuptake of the ion. However, the NsXeR photocycle reveals some distinct features, which, to our knowledge, have never been reported in the previous studies of retinal proteins (fig. S3). After the microsecond phase of the photocycle (P1, P2, and P3) that includes archetypal intermediates with the K- and L-like spectral shifts (P1, λmax = 570 nm; P2, λmax = 530 nm), in the millisecond time domain, we obtained two spectrally and kinetically different M intermediates (P4 and P5). The first M-form (P4) has a characteristic three-band absorption spectrum with the maximum at 360, 378, and 398 nm. This state with the half-time of 2 ms in nanodiscs (3 ms in lipid vesicles) converts to the state P5 with a single maximum at 392 nm. Both intermediates should correspond to the deprotonated state of the retinal Schiff base.
Protein structure, proton uptake and release regions, and active center
NsXeR crystals were grown using the in meso method (13). The structure was refined to a resolution of 2.5 Å. Crystallographic statistics is shown in table S1. P212121 space group crystals contain one trimer of NsXeR in the asymmetric unit (fig. S4).
The light-driven inward proton pump XeR has seven transmembrane α helices (A to G) and a cofactor retinal covalently bound to Lys213 via the Schiff base. The helix A is preceded with a small N-terminal α helix, which caps the protein on the extracellular side (fig. S5).
Comparison with BR structure (14) [Protein Data Bank (PDB): 1C3W] indicates considerable differences in helix localization and form, which are shown in Fig. 4A and fig. S6. A and G helices are significantly distorted. XeRs have a conservative residue Pro209 (position in NsXeR), which is located at the position of Asp212 in BR. Our experiments showed that its replacement with Asp makes the protein unstable. If changed to glycine, the pumping activity markedly decreases (table S2). Thus, Pro209 is crucial for proton pumping.
Fig. 4. NsXeR structure.
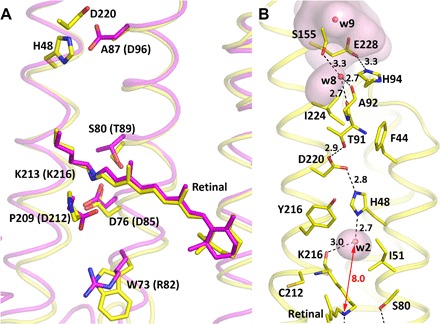
(A) Comparison of NsXeR (yellow) and BR (magenta) motifs. Residues are shown as an NsXeR motif (WDSAPK) and a BR motif (RDTDDK). Two residues, H48 and D220, in NsXeR are shown as an analog of the D96 residue in BR. (B) Putative proton acceptor region in detail. The distance between the Schiff base and water molecule 2 is shown with a double-arrow red line (8.0 Å). Distances between D76 (D85) and the Schiff base in NsXeR (BR) are 4.9 Å (3.8 Å), respectively. Cavities inside the protein calculated by HOLLOW1.2 are shown transparent pink.
Proton uptake region and active center
NsXeR has a proton uptake cavity (Fig. 5), which is accessible from the bulk through the residue Gln122. The cavity is filled with water molecules. The putative proton donor Asp76 is available from that cavity through the residue Tyr59. Mutations of Asp76 to Glu, Ser, Thr, and Asn do not allow the protein to fold correctly (mutants were not colored; table S2). This is evidence of not only functional but also significant structural role of these amino acids. Ser55 is located close to Asp76, and it may stabilize this residue. Substitution of Ser55 with alanine (Ala53 in BR) also breaks protein folding. Asp76 is connected to the Schiff base through the water molecule 1 (the Asp76-w1 distance is 2.9 Å, and the w1–Schiff base distance is 3.2 Å in the ground state).
Fig. 5. Putative NsXeR ion translocation pathway.
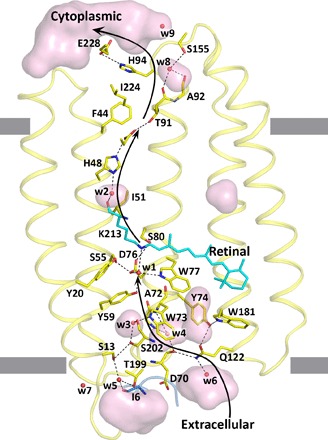
Cavities (transparent pink) and putative key residues inside protein are shown. Black arrows show the putative proton path. Helices F and G are not shown. The hydrophobic membrane core boundaries were calculated using the PPM (Positioning of Proteins in Membrane) server and are shown by gray lines.
Residue Trp73, which replaces the amino acid Arg82 (position in BR) that is highly conserved in bacteriorhodopsins, separates the active center from the “proton uptake pathway cavities” (Fig. 5). Substitution of Trp73 with Arg was fatal for protein folding (W73R mutant is not colored). The W73A mutant binds retinal and has the color of the wild-type protein but demonstrates no pumping activity, which means that this residue is critical for proton translocation.
The unique feature of NsXeR structure is the presence of the chain of strong hydrogen bonds Ile6-Thr199-w5-Ser13-Ser202-w3-Trp73, which connects the bulk on the extracellular side with the active center of the protein (Fig. 5). These interactions between helices A and G and N-terminal helix play important roles in protein structure stabilization.
Proton release region
Another major difference of NsXeR from other known microbial retinal proton pumps is that it has no ionizable amino acid at the position equivalent to Asp96 in BR (in NsXeR, it is Ala71). However, the residues His48 (10 Å from the Schiff base in the ground state) and Asp220 (12 Å), which are connected via a hydrogen bond, are located close to the expected proton acceptor position (Fig. 4B). Substitution of Asp220 with Asn completely disrupts proton pumping.
His48 is a unique residue, which is not present at a similar place in other known microbial rhodopsins. Our experiments showed that substitution of His48 with any other amino acid prevents binding of retinal (all mutants are not colored), which indicated its crucial role in protein architecture. We suggest that the pair His48-Asp220 is a proton acceptor, and the protonation processes from the Schiff base through the water molecule 2 and His48 residue, more precisely, through the pair His48-Asp220. Remarkably, it is exactly the same proton acceptor pair as in PRs (15, 16). However, contrary to PRs in XeRs, the pair is placed at the cytoplasmic side of the protein close to the Schiff base and serves as a Schiff base proton acceptor.
Putative mechanism of inwardly directed proton transport
The structure and experiments with mutated amino acids provide insights into the mechanism of inwardly directed proton transport. A putative pathway of proton translocation is shown in Fig. 5, and more details are shown in fig. S7 (C to H). Upon illumination, retinal isomerizes, the Schiff base deprotonates, and the proton is translocated to the deprotonated His48-Asp220 pair. This happens in MI and MII intermediate states because both intermediates correspond to the deprotonated state of the retinal Schiff base. It is known that Asp-His interaction in rhodopsins can substantially lower the pKa (where Ka is the acid dissociation constant) of Asp by stabilizing its deprotonated state (16). A key role of the Asp-His pair in proton translocation is supported by our experiments with the mutated Asp and His. We suggest that after reisomerization of the retinal, the protonated Asp-His pair releases proton through the Glu228-His94 pair directly to the cytoplasm (Figs. 4B and 5). We also suggest that after isomerization of the retinal, Asp76 is protonated through the hydrophilic cavities (proton uptake pathway cavities; Fig. 5). Reisomerization of the retinal also results in reprotonation of the Schiff base from Asp76.
Optogenetic implications
Experiments with human embryonic kidney and neuroblastoma glioma cells
The human embryonic kidney (HEK) 293 and neuroblastoma glioma (NG) 108-15 cells were transfected with the pcDNA3.1(−) vector bearing NsXeR gene with membrane trafficking signal (17), P2A self-cleaving peptide (18), and a green fluorescent protein (GFP) variant (19) at the C terminus. Whole-cell voltage-clamped experiments were conducted, and Fig. 6A shows photocurrents generated by NsXeR in the HEK293 cell. Typical photocurrent values vary from 40 to 150 pA at −60-mV applied potential, whereas the currents normalized to the capacitance (meaning the size) of the cell are about 1.4 ± 0.5 pA/pF (measured for n = 7 cells). An additional control experiment in NG108-15 cells was performed. To exclude the transport of Cl− ions (which may account for apparent “inward” current), we replaced chloride salts in buffers with sulfate. To exclude monovalent ion transport into the cell, we replaced Na+ in the bath solution by larger molecule, N-methyl-d-glucosamine. The pH of the solutions was symmetric (pH 7.4). However, similar photocurrents were recorded in this experimental configuration (Fig. 6B), convincing us that the experiments with HEK and NG cells confirm that NsXeR is an inwardly directed proton pump.
Fig. 6. Photocurrents in HEK293 and NG108-15 cells.
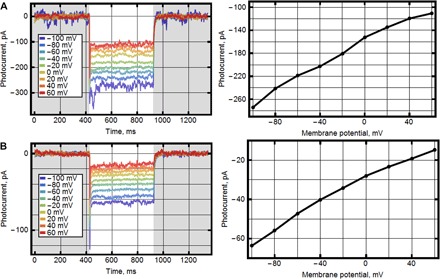
Photocurrents in cells expressing NsXeR at the membrane potentials changed in 20-mV steps from −100 mV and corresponding current-voltage curves. (A) HEK293 cells with pipette solution [110 mM NaCl, 2 mM MgCl2, 10 mM EGTA, and 10 mM Hepes (pH 7.4)] and bath solution [140 mM NaCl, 2 mM MgCl2, and 10 mM Hepes (pH 7.4)]. (B) NG108-15 cells with pipette solution [110 mM Na2SO4, 4 mM MgSO4, 10 mM EGTA, and 10 mM Hepes (pH 7.4)] and bath solution [140 mM N-methyl-d-glucosamine, 4 mM MgSO4, and 10 mM Hepes (pH 7.4)] (control measurements to confirm that protons are responsible for inwardly directed current).
For the measurement of the action spectra (fig. S8), ultrashort nanosecond light pulses were generated by the Opolette 355 tunable laser system (Optoprim). The pulse energies at the different wavelengths were set to values that corresponded to equal photon counts of 1019 photons/m2. To gain additional kinetic information, we recorded time-resolved photocurrents in response to ultrashort nanosecond light pulses at different membrane potentials (fig. S9). The time-resolved photocurrents are characterized by two relaxation times. At 0 mV, which corresponds to the membrane potential in the flash photolysis experiments, a biexponential relaxation with τ1 = 1.1 ± 0.2 ms (n = 5) and τ2 = 11.6 ± 1.9 ms (n = 5) was found. By comparing it to the photocycle (Fig. 2), we assigned τ1 (1.1 ms versus 2 ms) to the MI/MII transition and τ2 (11.6 ms versus 27 ms) to the M2 decay. The minor difference between the time constants derived from the photocycle and the photocurrent might be due to the different environments of the protein (nanodiscs versus NG cells). At a physiological membrane potential of −60 mV, a relaxation constant of τ2 = 6.2 ± 2.8 ms (n = 5) was found for the rate-limiting step, which corresponds to a turnover rate of ~160 s−1.
Light-triggered spiking in rat hippocampal neurons
We heterologously expressed NsXeR, C-terminally fused to the Kir2.1 membrane trafficking signal (17), in rat hippocampal neurons by means of adeno-associated virus–mediated gene transfer. For the electrophysiological characterization of the transduced cells, we performed whole-cell patch-clamp experiments. Thereby, NsXeR-expressing neurons showed normal morphology and membrane properties, which indicates that heterologous expression had no detectable toxic effects. The membrane capacitance and the resting membrane potential were 32.5 ± 12.9 pF (n = 6) and −60 ± 6 mV (n = 6), respectively. The mean photocurrent at a membrane potential of −70 mV, measured upon green light illumination at saturating intensity (fig. S10), was −189 ± 80 pA (n = 6), which corresponded to a mean current density of 6.7 ± 4.4 pA/pF (n = 6).
As expected from the current densities of the photocurrents and as proven in current clamp experiments, light-triggered spiking in rat hippocampal neurons was possible (Fig. 7). NsXeR enabled a fast, neural photostimulation with a firing success rate of 100% up to a frequency of 40 Hz. Spike failures at higher stimulation frequencies can be explained by intrinsic properties of the rat hippocampal neurons because a vast majority of rat hippocampal neurons have a maximal firing frequency of 40 to 60 Hz (20).
Fig. 7. Spiking traces at different light-pulse frequencies.
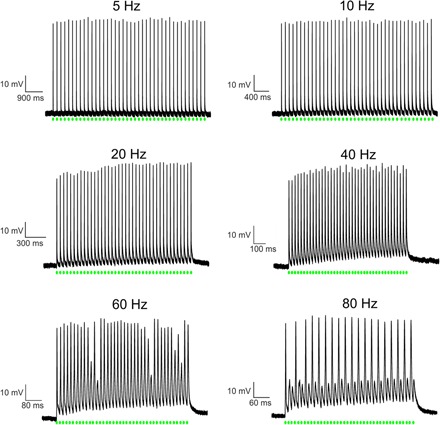
Rat hippocampal neurons heterologously expressing NsXeR, C-terminally fused to the Kir2.1 membrane trafficking signal, investigated by current clamp measurements in the whole-cell configuration. Action potentials were triggered by 40 light pulses at indicated frequencies. The light pulses had a pulse width of 3 ms, a wavelength of λ = 532 nm, and an intensity of 23 mW/mm2.
An important observation is that light-triggered spiking could be achieved with a pulse width of only 3 ms (fig. S11A), which is shorter than the turnover time of the pump (6.2 ± 2.8 ms). Hence, the extent of depolarization due to the transport of a single proton by each NsXeR is sufficient to successfully trigger action potentials. A similar behavior was observed in hippocampal neurons transduced with the inhibitory light-driven Cl− pump NpHR (21). However, a variability of the spike latencies was observed (fig. S11), which, in some cases, required longer pulse widths for the light-triggered spiking (fig. S11B). Longer spike latencies could be explained by a comparatively lower expression of NsXeR in those neurons.
CONCLUSIONS
We performed comprehensive functional studies of the representatives of the yet noncharacterized XeRs from the Nanohaloarchaea family of microbial rhodopsins and showed that they are inwardly directed proton pumps. A rigorous study of the pumping activity of NsXeR in model membrane systems, E. coli cells, HEK cells, NG cells, and rat hippocampal neuronal cells confirmed that, in all these cells, NsXeR works as an inwardly directed proton pump. Its biological function is rather obscure. The functionally similar pump PoXeR found in the marine bacterium P. oceani has been proposed to have a role in signal transduction (4), but this is highly unlikely in extremely simplified and small cells as the Nanohaloarchaea with a single rhodopsin gene in their genomes (P. oceani has three). Given that there are no extant cultures of these microbes, determining the biological function of this inwardly directed proton pump remains elusive.
We also demonstrated that the NsXeR is a powerful pump with a turnover rate of 160 s−1, which is able to elicit action potentials in rat hippocampal neuronal cells up to their maximal intrinsic firing frequency. Because of its intrinsic properties as a proton pump, NsXeR is completely independent of the ion conditions, which makes this rhodopsin an attractive alternative for light-induced remote control of neurons as the well-known cation-selective channelrhodopsins.
The crystallographic structure of NsXeR reveals the ion translocation pathway that is very different from that of the known rhodopsins. The structure and our data on the function of the protein mutants allowed us to propose a mechanism of inwardly directed proton pumping.
MATERIALS AND METHODS
Phylogenetic analysis and sequence alignment
For the phylogenetic tree of microbial rhodopsin proteins, 52 rhodopsin sequences were aligned using MUSCLE. Phylogenetic reconstruction was conducted by maximum likelihood using MEGA6 with the following parameters: Jones-Taylor-Thornton model, 100 bootstraps, and γ distribution with five discrete categories. Positions with less than 80% site coverage were eliminated. Only bootstraps greater than 50% are shown. GenBank accession numbers were written in parentheses for each protein.
Cloning
NsXeR (UniProt ID: G0QG75), HrvXeR (7), and AlkXeR (8) coding DNAs were synthesized commercially (Eurofins). The nucleotide sequences were optimized for E. coli expression using the GeneOptimizer software (Life Technologies). The genes, together with the 5′ ribosome-binding sites and the 3′ extensions coding additional LEHHHHHH* tags, were introduced into the pET15b expression vector (Novagen) via Xba I and Bam HI restriction sites. The mutants were prepared by site-directed mutagenesis and verified by sequencing.
E. coli expression, solubilization, and purification
The protein was expressed as described previously (22) with modifications. E. coli cells of strain C41 (DE3) (Lucigen) were transformed with the expression plasmids. Transformed cells were grown at 37°C in shaking baffled flasks in an autoinducing medium, ZYP-5052 (23), containing ampicillin (100 mg/liter), and were induced at an OD600 (OD at 600 nm) of 0.6 to 0.7 with 1 mM isopropyl-β-d-thiogalactopyranoside and supplemented with 10 μM all-trans retinal. Three hours after induction, the cells were collected by centrifugation at 3000g for 30 min. The collected cells were disrupted in M-110P Lab Homogenizer (Microfluidics) at 25,000 psi in a buffer containing 20 mM tris-HCl (pH 8.0), 5% glycerol, 0.5% Triton X-100 (Sigma-Aldrich), and deoxyribonuclease I (50 mg/liter) (Sigma-Aldrich). The membrane fraction of cell lysate was isolated by ultracentrifugation at 90,000g for 1 hour at 4°C. The pellet was resuspended in a buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 8.0), 0.1 M NaCl, and 1% DDM (Anatrace, Affymetrix) and stirred overnight for solubilization. The insoluble fraction was removed by ultracentrifugation at 90,000g for 1 hour at 4°C. The supernatant was loaded on a Ni-NTA column (Qiagen), and XeRs were eluted in a buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 7.5), 0.1 M NaCl, 0.3 M imidazole, and 0.2% DDM. The eluate was dialyzed against 100 volumes of 50 mM NaH2PO4/Na2HPO4 (pH 7.5) and 0.1 M NaCl buffer twice for 2 hours to dispose imidazole. Finally, proteins were concentrated to 70 mg/ml for crystallization.
Liposome preparation
Phospholipids (asolectin from soybean, Sigma-Aldrich) were dissolved in CHCl3 (chloroform ultrapure, PanReac AppliChem) and dried under a stream of N2 in a glass vial. Residual solvent was removed using a vacuum pump overnight. The dried lipids were resuspended at a final concentration of 1% (w/v) in 0.15 M NaCl supplemented with 2% (w/v) sodium cholate. The mixture was clarified by sonication at 4°C, and XeR was added at a protein/lipid ratio of 7:100 (w/w). The detergent was removed by overnight stirring with detergent-absorbing beads (Amberlite XAD-2, Supelco). The mixture was dialyzed against 0.15 M NaCl (adjusted to a desired pH) at 4°C for 1 day (four 200-ml changes) to obtain a certain pH.
Incorporation in nanodiscs
The proteonanodiscs were assembled using standard protocol (24). DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) (Avanti Polar Lipids) was used as lipid. An elongated MSP1E3 version of apolipoprotein-1 was used. The molar ratio during assembly was DMPC/MSP1E3/NsXeR = 100:2:3.
Measurements of pump activity in E. coli suspensions
The protein was expressed as described above. The cells were collected by centrifugation at 3000g for 10 min and were washed three times with an unbuffered salt solution (100 mM NaCl and 10 mM MgCl2), with 30-min intervals between the washes to allow exchange of the ions inside the cells with the bulk. After that, the cells were resuspended in 100 mM NaCl solution and adjusted to an OD600 of 8.5. The measurements were performed in 3-ml aliquots of stirred cell suspension kept at 1°C. The cells were illuminated for 5 min using a halogen lamp (Intralux 5000-1, Volpi), and the light-induced pH changes were monitored with a pH meter (Lab 850, SCHOTT Instruments). Measurements were repeated under the same conditions after the addition of 30 μM CCCP.
Measurement of pump activity in the liposomes
The measurements were performed on 2 ml of stirred proteoliposome suspension at 0°C. Proteoliposomes were illuminated for 18 min using a halogen lamp (Intralux 5000-1, Volpi) and then were kept in the dark for another 18 min. Changes in pH were monitored using a pH meter (Lab 850, SCHOTT Instruments). Measurements were repeated for different starting pH and in the presence of 40 μM CCCP under the same conditions.
Spectroscopic characterization and time-resolved absorption spectroscopy
The absorption spectra were recorded using the Shimadzu UV-2401PC spectrophotometer. The laser flash photolysis setup was similar to that described by Chizhov and co-workers (25). The excitation/detection systems were composed as such: a Surelite II-10 Nd:YAG laser (Continuum Inc.) was used, providing pulses of 5-ns duration at 532-nm wavelength and an energy of 3 mJ per pulse. Samples (5 × 5–mm spectroscopic quartz cuvette; Hellma GmbH & Co.) were placed in a thermostated house between two collimated and mechanically coupled monochromators (1/8 m model 77250, Oriel Corp.). The probing light (xenon arc lamp, 75 W, Osram) passed the first monochromator sample and arrived after a second monochromator at a photomultiplier tube (PMT) detector (R3896, Hamamatsu). The current-to-voltage converter of the PMT determines the time resolution of the measurement system of ca. 50 ns (measured as an apparent pulse width of the 5-ns laser pulse). Two digital oscilloscopes (LeCroy 9361 and 9400A, 25 and 32 kilobytes of buffer memory per channel, respectively) were used to record the traces of transient transmission changes in two overlapping time windows. The maximal digitizing rate was 10 ns per data point. Transient absorption changes were recorded from 10 ns after the laser pulses until full completion of the phototransformation. At each wavelength, 25 laser pulses were averaged to improve the signal-to-noise ratio. The quasi-logarithmic data compression reduced the initial number of data points per trace (~50,000) to ~600 points evenly distributed in a log time scale giving ~100 points per time decade. The wavelengths were varied from 300 to 730 nm in steps of 2 nm (altogether, 216 spectral points) using a computer-controlled step motor. Absorption spectra of the samples were measured before and after each experiment on a standard spectrophotometer (DU-800, Beckman Coulter). Here, we report results of the analysis of two data sets: the XeR protein reconstituted in nanodiscs and in liposomes.
Data treatment and global fit analysis
Each data set was independently analyzed using the global multiexponential nonlinear least-squares fitting program MEXFIT (25). The number of exponential components was incremented until the SD of weighted residuals did not further improve. After establishing the apparent rate constants and their assignment to the internal irreversible transitions of a single chain of relaxation processes, the amplitude spectra of exponents were transformed to the difference spectra of the corresponding intermediates in respect to the spectrum of the final state. Subsequently, the absolute absorption spectra of states were determined by adding the difference spectra divided by the fraction of converted molecules to the spectra of the final states. Criteria for the determination of the fraction value were the absence of negative absorbencies and contributions from the initial state to the calculated spectra of final state. For further details of the methods, see the study by Chizhov et al. (25).
Retinal extraction
Retinal extraction was performed on samples that have been dark-adapted for 16 hours or after preillumination with a light from a 75-W halogen light longpass-filtered at 480 nm for 1 min. The conditions were similar to those in the study by Inoue et al. (4). Briefly, 500 mM hydroxylamine was added to the samples just before denaturation with ice-cold methanol. Retinal oximes were extracted with hexane and loaded on a ProntoSIL-OH 120-3 column. The flow rate was 1 ml/min of 12% (v/v) ethyl acetate and 0.12% (v/v) in hexane at room temperature.
Crystallization
The crystals were grown using the in meso approach (13), similar to that used in our previous work (26). The solubilized protein in the crystallization buffer was mixed with monoundecenoin (Nu-Chek Prep) (premelted at 50°C) in a 1:1 ratio to form a lipidic mesophase. Aliquots (150 nl) of a protein-mesophase mixture were spotted on a 96-well lipidic cubic phase glass sandwich plate (Marienfeld) and overlaid with 600 nl of precipitant solution by means of the NT8 crystallization robot (Formulatrix). The best crystals were obtained with a protein concentration of 30 mg/ml and 2.8 to 3.4 M sodium malonate (pH 8.0) (Hampton Research). The crystals were grown at 22°C and appeared in 1 to 4 weeks. Note that the mesophase mixture was spotted for crystallization directly after preparation, because the XeR protein embedded into the bilayer of the mesophase (at 22°C) and not overlaid with precipitant solution was losing its purple color (significantly within several hours) likely because of denaturation processes and loss of retinal.
Data collection
X-ray diffraction data (wavelengths, 0.969 and 0.972 Å) were collected at ID23-1, ID29, and ID30B beamlines of the European Synchrotron Radiation Facility at 100 K using a PILATUS 6M detector. Diffraction images were processed with XDS software (27). The reflection intensities were scaled with Scala from the CCP4 suite (28). The crystallographic data statistics are shown in table S1.
Structure refinement
Reference model (archaerhodopsin-2, PDB 2EI4) for molecular replacement was chosen with the MoRDa pipeline (29). Initial phases were successfully obtained in the P212121 space group by an Automated Model Building and Rebuilding using AutoBuild (30). The initial model was iteratively refined using REFMAC5 (31), PHENIX, and Coot (32).
NsXeR expression in HEK and NG cells
The human codon–optimized NsXeR gene was synthesized commercially (Eurofins). The gene was cloned into the pcDNA3.1(−) vector bearing an additional membrane trafficking signal (17), a P2A self-cleaving peptide (18), and a GFP variant (19). The gene was cloned under the cytomegalovirus promoter. The sequence was verified by sequencing. The HEK293 and NG108-15 cells at a confluency of 80% were transfected with the plasmid and Lipofectamine LTX according to the manufacturer’s protocol (Thermo Fisher Scientific). The cells were incubated under 5% CO2 at 37°C for 2 days before measurements.
Hippocampal neuron culture and adeno-associated virus (AAV2/1) transduction
Hippocampi were isolated from postnatal day 1 (P1) Sprague-Dawley rats (by the laboratory of E. Schuman, Max Planck Institute for Brain Research, Frankfurt) and treated with papain (20 U ml−1) for 20 min at 37°C. The hippocampi were washed with Dulbecco’s modified Eagle’s medium (high glucose, Invitrogen/Gibco) supplemented with 10% fetal bovine serum and titrated in a small volume of this solution. Approximately 96,000 cells were plated on poly-d-lysine/laminin–coated glass coverslips in 24-well plates. After 3 hours, the plating medium was replaced by culture medium (Neurobasal A containing 2% B-27 supplement and 2 mM GlutaMAX-I).
rAAV2/1 virus was prepared using a pAAV2 vector with a human synapsin promoter containing the humanized DNA sequence of NsXeR, C-terminally fused to the Kir2.1 membrane trafficking signal (17), a P2A self-cleaving peptide (18), and a GFP variant (19). Briefly, rAAV2/1 virus (5 × 109 genome copies/ml) was added to each well 4 to 9 days after plating. The electrophysiological recordings were performed 19 to 23 days after transduction. No neurotoxicity was observed for the lifetime of the culture (~5 weeks).
Electrophysiological recordings
For the electrophysiological characterization of NsXeR, whole-cell patch-clamp recordings were performed (Axopatch 200B interface, Axon Instruments). Patch pipettes with resistances of 2 to 5 megohms were fabricated from thin-walled borosilicate glass (GB150F-8P) on a horizontal puller (Model P-1000, Sutter Instruments). For experiments in HEK293 cells, the pipette solution contained 110 mM NaCl, 2 mM MgCl2, 10 mM EGTA, and 10 mM Hepes (pH 7.4) (pipette solution 1), and the bath solution contained 140 mM NaCl, 2 mM MgCl2, and 10 mM Hepes (pH 7.4) (bath solution 1). For the experiments in NG108-15 cells, the pipette solution contained 110 mM Na2SO4, 4 mM MgSO4, 10 mM EGTA, and 10 mM Hepes (pH 7.4) (with H2SO4) (pipette solution 2), and the bath solution contained 140 mM N-methyl-d-glucosamine, 4 mM MgSO4, and 10 mM Hepes (pH 7.4) (with H2SO4) (bath solution 2).
Photocurrents were measured in response to light pulses with a saturating intensity of 23 mW/mm2 using diode-pumped solid-state lasers (λ = 532 nm) focused into a 400-μm optic fiber. Light pulses were applied by a fast computer-controlled shutter (Uniblitz LS6ZM2, Vincent Associates). Ultrashort nanosecond light pulses were generated by the Opolette 355 tunable laser system (Optoprim). For the measurement of the action spectra, the pulse energies at the different wavelengths were set to values that corresponded to equal photon counts of 1019 photons/m2.
For whole-cell recordings in cultured hippocampal neurons, patch pipettes with resistances of 3 to 8 megohms were filled with 129 mM potassium gluconate, 10 mM Hepes, 10 mM KCl, 4 mM MgATP, and 0.3 mM Na3GTP, titrated to pH 7.3. The extracellular solution contained 125 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 30 mM glucose, and 25 mM Hepes, titrated to pH 7.3. Electrophysiological signals were filtered at 10 kHz, digitized with an Axon Digidata 1322A (50 kHz), and acquired and analyzed using pCLAMP9 software (Axon Instruments).
Supplementary Material
Acknowledgments
We acknowledge the Structural Biology Group of the European Synchrotron Radiation Facility for granting access to the synchrotron beamlines. We thank T. Wießalla for additional time-resolved current measurements. Funding: This work was supported by the common program of Agence Nationale de la Recherche (ANR) France and Deutsche Forschungsgemeinschaft, Germany (ANR-15-CE11-0029-02); by funding from Frankfurt: Cluster of Excellence Frankfurt Macromolecular Complexes to (E.B. and C.B.) by the Max Planck Society (to E.B.): and by the Commissariat à l’Energie Atomique et aux Energies Alternatives (Institut de Biologie Structurale)–Helmholtz-Gemeinschaft Deutscher Forschungszentren (Forschungszentrum Jülich) Special Terms and Conditions 5.1 specific agreement. This work was also supported by the Russian Science Foundation research project 14-14-00995. This work used the platforms of the Grenoble Instruct Centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from French Infrastructure for Integrated Structural Biology (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. This work was also supported by the ERA.Net RUS Plus program (project RFMEFI58715X0011). R.G. was supported by the research grant 17-04828S from the Grant Agency of the Czech Republic. F.R.-V. acknowledges the following funding: projects MEDIMAX BFPU2013-48007-P, VIREVO CGL2016-76273-P, and CGL2015-71523-REDC from the Ministerio Español de Economía y Competitividad and project AQUAMET PROMETEO II/2014/012 from Generalitat Valenciana. Author contributions: V.G. defined the targets of research with input from F.R.-V., C.V., and R.G. V.G. designed the project and directed and supervised all the research. E.B. supervised the optogenetic part of the project. V.G., E.B., V.S., and T.M. analyzed the results with contribution from K.K. and G.B. V.G. and V.S. prepared the manuscript with major contributions from E.B. and additional contributions from F.R.-V., V.P., T.M., I.G., A.A., and K.K. V.S., K.K., and A.A. expressed and purified the proteins with contribution from T.B. V.S., T.M., K.K., and A.A. performed the function studies. I.C. and V.S. did the flash photolysis with contribution from D.J.M. C.B. performed the retinal isomer composition experiments. V.S. and T.M. performed the optogenetic studies with contribution from J.J. V.P. crystallized the protein and collected the crystallographic data with strong contribution from A.P. K.K. solved the structure with contribution from V.B., I.M., A.P., I.G., and A.R. Competing interests: E.B., T.M., V.G., and V.S. are authors on a pending European patent application related to this work, filed by Max-Planck-Gesellschaft zur Förderung der Wissenschaften e.V. (application no. 17 166 302.4; priority date, 12 April 2017). All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/9/e1603187/DC1
fig. S1. Phylogenetic tree of microbial rhodopsin proteins.
fig. S2. Sequence alignment of microbial rhodopsins.
fig. S3. Photocycles of the NsXeR in nanodisc (ND, upper row) and liposome (LIP, lower row) preparations (20°C, pH 7.5).
fig. S4. Crystal packing of NsXeR.
fig. S5. Overall architecture of NsXeR.
fig. S6. Comparison of NsXeR structure with structures of other microbial rhodopsins.
fig. S7. Details of the NsXeR proton-translocation pathway.
fig. S8. Action spectrum of photocurrents in NG108-15 cells expressing NsXeR (black) and NsXeR absorption spectrum (red).
fig. S9. NsXeR photocurrents in response to ultrashort light pulses.
fig. S10. Light intensity dependence of NsXeR.
fig. S11. Variability of spike latency.
table S1. Crystallographic data collection and refinement statistics.
table S2. List of mutations to NsXeR and their implications on proton pumping.
REFERENCES AND NOTES
- 1.Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., Kandori H., Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell P., Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol. Rev. 41, 445–501 (1966). [DOI] [PubMed] [Google Scholar]
- 3.Oesterhelt D., Stoeckenius W., Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233, 149–152 (1971). [DOI] [PubMed] [Google Scholar]
- 4.Inoue K., Ito S., Kato Y., Nomura Y., Shibata M., Uchihashi T., Tsunoda S. P., Kandori H., A natural light-driven inward proton pump. Nat. Commun. 7, 13415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ugalde J. A., Podell S., Narasingarao P., Allen E. E., Xenorhodopsins, an enigmatic new class of microbial rhodopsins horizontally transferred between archaea and bacteria. Biol. Direct 6, 52 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogeley L., Sineshchekov O. A., Trivedi V. D., Sasaki J., Spudich J. L., Luecke H., Anabaena sensory rhodopsin: A photochromic color sensor at 2.0 Å. Science 306, 1390–1393 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghai R., Pašić L., Fernández A. B., Martin-Cuadrado A.-B., Mizuno C. M., McMahon K. D., Papke R. T., Stepanauskas R., Rodriguez-Brito B., Rohwer F., Sánchez-Porro C., Ventosa A., Rodríguez-Valera F., New abundant microbial groups in aquatic hypersaline environments. Sci. Rep. 1, 135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vavourakis C. D., Ghai R., Rodriguez-Valera F., Sorokin D. Y., Tringe S. G., Hugenholtz P., Muyzer G., Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline soda lake brines. Front. Microbiol. 7, 211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., Ollig D., Hegemann P., Bamberg E., Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang K. S., Bayley H., Khorana H. G., Delipidation of bacteriorhodopsin and reconstitution with exogenous phospholipid. Proc. Natl. Acad. Sci. U.S.A. 77, 323–327 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Racker E., Stoeckenius W., Reconstitution of purple membrane vesicles catalyzing light-driven proton uptake and adenosine triphosphate formation. J. Biol. Chem. 249, 662–663 (1974). [PubMed] [Google Scholar]
- 12.Moise A. R., Kuksa V., Imanishi Y., Palczewski K., Identification of all-trans-retinol:All-trans-13,14-dihydroretinol saturase. J. Biol. Chem. 279, 50230–50242 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.V. I. Gordeliy, R. Schlesinger, R. Efremov, G. Büldt, J. Heberle, in Membrane Protein Protocols, B. S. Selinsky, Ed. (Humana Press, 2003), vol. 228, pp. 305–316. [Google Scholar]
- 14.Luecke H., Schobert B., Richter H.-T., Cartailler J.-P., Lanyi J. K., Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291, 899–911 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Gushchin I., Chervakov P., Kuzmichev P., Popov A. N., Round E., Borshchevskiy V., Ishchenko A., Petrovskaya L., Chupin V., Dolgikh D. A., Arseniev A. S., Kirpichnikov M., Gordeliy V., Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 12631–12636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balashov S. P., Petrovskaya L. E., Lukashev E. P., Imasheva E. S., Dioumaev A. K., Wang J. M., Sychev S. V., Dolgikh D. A., Rubin A. B., Kirpichnikov M. P., Lanyi J. K., Aspartate–histidine interaction in the retinal schiff base counterion of the light-driven proton pump of Exiguobacterium sibiricum. Biochemistry 51, 5748–5762 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gradinaru V., Zhang F., Ramakrishnan C., Mattis J., Prakash R., Diester I., Goshen I., Thompson K. R., Deisseroth K., Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzmich A. I., Vvedenskii A. V., Kopantzev E. P., Vinogradova T. V., Quantitative comparison of gene co-expression in a bicistronic vector harboring IRES or coding sequence of porcine teschovirus 2A peptide. Russ. J. Bioorg. Chem. 39, 406–416 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Shcherbo D., Merzlyak E. M., Chepurnykh T. V., Fradkov A. F., Ermakova G. V., Solovieva E. A., Lukyanov K. A., Bogdanova E. A., Zaraisky A. G., Lukyanov S., Chudakov D. M., Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 4, 741–746 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Gunaydin L. A., Yizhar O., Berndt A., Sohal V. S., Deisseroth K., Hegemann P., Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Kleinlogel S., Terpitz U., Legrum B., Gökbuget D., Boyden E. S., Bamann C., Wood P. G., Bamberg E., A gene-fusion strategy for stoichiometric and co-localized expression of light-gated membrane proteins. Nat. Methods 8, 1083–1088 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Gushchin I., Shevchenko V., Polovinkin V., Kovalev K., Alekseev A., Round E., Borshchevskiy V., Balandin T., Popov A., Gensch T., Fahlke C., Bamann C., Willbold D., Büldt G., Bamberg E., Gordeliy V., Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 22, 390–395 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Studier F. W., Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Ritchie T. K., Grinkova Y. V., Bayburt T. H., Denisov I. G., Zolnerciks J. K., Atkins W. M., Sligar S. G., Chapter 11–Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 464, 211–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chizhov I., Chernavskii D. S., Engelhard M., Mueller K.-H., Zubov B. V., Hess B., Spectrally silent transitions in the bacteriorhodopsin photocycle. Biophys. J. 71, 2329–2345 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordeliy V. I., Labahn J., Moukhametzianov R., Efremov R., Granzin J., Schlesinger R., Büldt G., Savopol T., Scheidig A. J., Klare J. P., Engelhard M., Molecular basis of transmembrane signalling by sensory rhodopsin II–transducer complex. Nature 419, 484–487 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin A., Lebedev A., MoRDa, an automatic molecular replacement pipeline. Acta Crystallogr. A Found. Adv. 71, s19 (2015). [Google Scholar]
- 30.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX : A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K., Coot : Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/9/e1603187/DC1
fig. S1. Phylogenetic tree of microbial rhodopsin proteins.
fig. S2. Sequence alignment of microbial rhodopsins.
fig. S3. Photocycles of the NsXeR in nanodisc (ND, upper row) and liposome (LIP, lower row) preparations (20°C, pH 7.5).
fig. S4. Crystal packing of NsXeR.
fig. S5. Overall architecture of NsXeR.
fig. S6. Comparison of NsXeR structure with structures of other microbial rhodopsins.
fig. S7. Details of the NsXeR proton-translocation pathway.
fig. S8. Action spectrum of photocurrents in NG108-15 cells expressing NsXeR (black) and NsXeR absorption spectrum (red).
fig. S9. NsXeR photocurrents in response to ultrashort light pulses.
fig. S10. Light intensity dependence of NsXeR.
fig. S11. Variability of spike latency.
table S1. Crystallographic data collection and refinement statistics.
table S2. List of mutations to NsXeR and their implications on proton pumping.


