Abstract
Diffusion-weighted magnetic resonance imaging (MRI) provides a unique approach to understand the geometric structure of brain fiber bundles and to delineate the diffusion properties across subjects and time. It can be used to identify structural connectivity abnormalities and helps to diagnose brain-related disorders. The aim of this paper is to develop a novel, robust, and efficient dimensional reduction and regression framework, called hierarchical functional principal regression model (HFPRM), to effectively correlate high-dimensional fiber bundle statistics with a set of predictors of interest, such as age, diagnosis status, and genetic markers. The three key novelties of HFPRM include the simultaneous analysis of a large number of fiber bundles, the disentanglement of global and individual latent factors that characterizes between-tract correlation patterns, and a bi-level analysis on the predictor effects. Simulations are conducted to evaluate the finite sample performance of HFPRM. We have also applied HFPRM to a genome-wide association study to explore important genetic variants in neonatal white matter development.
Keywords: Fiber Bundle Statistics, Varying Coefficient Model, Functional Principal Component Analysis, Factor Analysis, Imaging Genetics
1 Introduction
Scientifically, investigation in the connectional organization of human brain and its variation across subjects is a critical step to understand the pathology of many neuro-related disorders. Diffusion-weighted MRI offers a non-invasive approach to study the tissue structure of white matter fiber bundles in vivo, including both the geometric shape and the diffusion properties [2,6,9,12,17,24,27]. Delineating diffusion statistics along fiber bundles may help identify structural connectivity abnormalities across different spatial-temporal scales. It could eventually inspire new approaches for disease preventions, diagnoses and clinical treatments.
Group analysis of fiber bundle statistics poses remarkable computational and mathematical challenges to existing statistical methods. The first challenge is to efficiently and simultaneously study multiple fiber bundles with heterogeneous geometric structures and variation patterns. The second challenge is to correlate fiber bundle statistics with a large number of covariates, such as millions of genetic markers. This challenge is motivated by the demand to carry out a genome-wide association study on fiber bundle statistics. Voxel-wise methods [21] and single tract analysis [8, 26, 28] suffer from performing massive multiple comparison adjustments, which would severely reduce detection power. The third challenge is to properly handle the potential correlation among multiple tracts and to disentangle tract-specific information from global information shared by a large portion of fiber bundles.
The aim of this paper is to develop a hierarchical functional principal regression model (HFPRM) framework to address the three challenges discussed above. HFPRM consists of three statistical models, including a varying coefficient model (VCM), a latent factor analysis (LFA) procedure, and a multivariate regression model (MRM). The path diagram of HFPRM is presented in Fig. 1. The VCM not only captures the functional structure of fiber bundle statistics for each single tract, but also maps the heterogeneous geometric structure of multiple fiber bundles onto a common coordinate system. The LFA is applied to characterize potential inter-tract correlation across multiple bundles. It allows us to explicitly identify both tract-specific and global latent signals. The integration of VCM and LFA dramatically reduces the dimension of fiber bundle statistics. Finally, using MRM, we are able to examine the effect of selected predictors on both global level and individual level.
Fig. 1. A Schematic Overview of HFPRM.
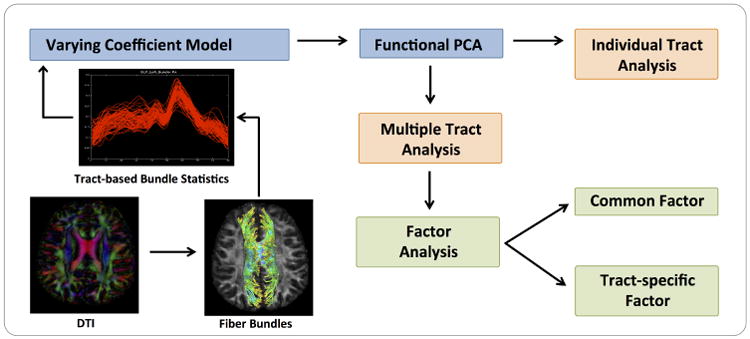
In Section 2, we introduce the general framework of HFPRM and propose a two stage estimation procedure to study both global effect and individual tract effect. In Sections 3 and 4, we use numerical simulations and a real data example to examine the finite sample performance of HFPRM. Section 5 concludes with some remarks.
2 Methods
2.1 Data Structure
Suppose that we obtain a data set with clinical, genetic variables as well as DTI statistics along M fiber bundles from n subjects. For the m-th fiber bundle, m = 1, …, M, we use sm ∈ [0, Sm] to denote the arc length of any point relative to a fixed end point, where Sm is the longest arc length on the tract. For the i-th subject where i = 1, …, n, yi,m(sm) denotes a specific diffusion statistics observed at arc-length sm along the m-th tract, and xi is a q × 1 vector of covariates.
2.2 HFPRM
HFRPM is proposed to study the association between diffusion properties (e.g., FA, MD or RD) along M fiber bundles with a set of covariates, such as age, gender, and genetic markers. It consists of three key components, a varying coefficient model (VCM), a latent factor analysis (LFA) procedure, and a multivariate regression model (MRM).
The VCM describes the functional association between {yi,m(sm) : sm ∈ [0, Sm]} and xi for a single tract. It admits the following form,
| (1) |
where μm(sm) is the function of population mean, ηi,m(sm) is an individual function characterizing subject-specific spatial variations along the m-th tract, and ei,m(sm) is the measurement error. Let SP(0, Σ) represent a stochastic process with mean zero and covariance operator . It is assumed that ηi,m(sm) and ei,m(sm) are mutually independent and identical copies of stochastic processes SP(0, Σηm) and SP(0, Σem) respectively, in which and 1(·) is an indicator function.
The major challenge to simultaneously study M fiber bundles is the heterogenuity in their geometric structures. It is necessary to find a common coordinate system for . Specifically, we use functional principal component analysis (fPCA) to extract the key features in ηi,m(sm). Based on Mercer's theorem, admits a spectral decomposition as follows:
| (2) |
where {λmd ≥ 0} are eigenvalues in descending order with and {ϕmd(sm)} are the corresponding orthonormal eigenfunctions. Using Karhunen-Loeve expansion [13, 16], ηim(sm) can be expressed as
| (3) |
Individual function ηi,m(sm) can then be equivalently represented by a set of functional principal component (fPC) scores {zi,md : d = 1, …, ∞}. In practice, a relatively small number of fPC scores would account for the majority of variation in ηi,m(s). Therefore, we can approximate ηi,m(sm) by a finite vector zi,m = (zi,m1, …, zi,mD)T of dimension D. For notational simplicity, it is assumed that D is the same across all M bundles. Now we use zi,m to integrate information across M bundles and denote zi as a p × 1 long vector that concatenates all zi,ms together, where p = DM.
A LFA is then proposed to account for potential inter-tract correlation across multiple bundles. Specifically, zi is assumed to have the following latent factor structure,
| (4) |
where Λ is a p × L loading matrix and fi and ui, respectively, represent global and individual latent factors. When there exist homogeneous signal patterns across multiple fiber bundles, L is expected to be much smaller than p. Global factor fi thus allows us to study the shared pattern in a low dimensional space. And tract-specific pattern can also be captured by each component in ui = (ui,1, …, ui,m)T.
Finally, a MLM is introduced to correlate the global and individual latent factors with covariate xi,
| (5) |
where Bf and Bum are, respectively, q × L and q × D coefficient matrices and ∊f,i and ∊um,i are residual terms. Using (5), we are able to perform a hierarchical analysis on both global level and individual level.
2.3 Estimation and Inference Procedure
In practice, diffusion statistics are observed on discrete grid points along each tract. For the m-th tract, assume yi,m(sm) is observed on sample point set Sm = {sm,1, …, sm,k, …, sm, Km} ⊂ [0, Sm], we use the following two-stage procedure to estimate fPC scores Z = {zi}1≤i≤n, global factors F = {fi}1≤i≤n and individual factors U = {ui}1≤i≤n.
– Stage I: For each tract, μm(sm) and ηi,m(sm) are estimated from (1) and functional principal component analysis is applied to calculate ϕ̂md(sm) and ẑi,
– Stage II: Perform factor analysis on ẑi to extract global factor f̂i and individual factor ûi. Regression and hypothesis testing can then be applied on f̂i and ûi respectively.
Details of the two stages are given below.
In Stage I, to estimate the mean curve from model (1), we apply the local linear kernel smoothing technique. μm(sm) is first approximated by the following taylor expansion,
| (6) |
Let K(s) be a predetermined smoothing kernel and denote as the rescaled function with bandwidth h, μ̂m(sm) and dμ̂m(sm) can be estimated as the minimizers of the following weighted least square function,
| (7) |
and solution μ̂m(sm) is smooth curve with local linearity. More complicated polynomial structure can be applied using higher order expansion if necessary.
Similarly, we expand individual function ηi,m(sm) for subject i as follows,
| (8) |
The corresponding weighted least square function is given by,
| (9) |
When smoothed individual functions are obtained as , we can calculate the empirical covariance function . And eigenbases {ϕ̂md(sm)} can be estimated from spectral decomposition,
| (10) |
Then individual random effect η̂i,m(sm) is projected onto basis functions {ϕ̂md(sm)} to get functional PC scores,
| (11) |
There are several strategies to determine the number of fPCs to be extracted. For example, the analog of some model selection techniques have been generalized for this purpose, such as Akaike information criterion (AIC), Bayesian information criterion (BIC) [25] and cross-validation (CV) [20]. Alternatively, the percentage of explained variation has been widely used to give an appropriate cut-off in practice. Here, we choose D as the minimum number of fPCs that incorporates at least V% of total variation in each tract. When the optimal D = Dm is different across tracts, the largest Dm will be used for all tracts.
In Stage II, a PCA-based factor analysis is performed. Let ξ̂1, …, ξ̂L be the first L eigenvectors of sample covariance matrix . The loading matrix, the global factors and the individual factors are estimated as,
| (12) |
Finally, the MLM (5) is used to estimate regression coefficients. Standard test statistics, such as wald and score statistics, can be applied subsequently for inference purpose.
3 Simulations
In this section, numerical simulations are conducted to evaluate the proposed method. Particularly, we examine the performance of HFPRM to detect covariate effect in hypothesis testing.
3.1 Setup
11 fiber tracts with FA measure shown in Table 1 were selected from diffusion tensor tractography in UNC Early Human Brain Development Studies [7]. Functional responses were simulated from a vary coefficient model with fixed covariate effects,
Table 1. List of Fiber Tracts in Simulation and Real Data Experiment.
| Bundle Group | Tract Segments |
|---|---|
| Arcuate Fasciculus | left fronto-parietal, right fronto-parietal, left fronto-temporal*, right fronto-temporal*, right temporo-parietal |
| Corpus Callosum | motor body*, occipital splenium, parietal body*, premotor body, rostrum*, genu*, temporal tapetum* |
| Cingulum | left premotor, left cingulate gyrus, right cingulate gyrus, right hippocampal, right prefrontal cortex |
| Corticothalamic | left motor, right motor,left premotor, right premotor, left parietal, right parietal, left prefrontal, right prefrontal |
| CorticoFugal | left motor, right motor, left parietal, right parietal, left prefrontal cortex, |
| Others | left fornix, right fornix, left inferior fronto-occipital fasciculi, right inferior fronto-occipital fasciculi, left inferior longitudinal fasciculi*, right inferior longitudinal fasciculi*, left medial lemniscus, right medial lemniscus, left optic, right optic, left superior longitudinal fasciculus, right superior longitudinal fasciculus, left uncinate fasciculus*, right uncinate fasciculus* |
Selected tracts for simulation study
| (13) |
where i = 1, …, n and m = 1, …, 11, β(sm) was a q × 1 vector of coefficient functions along the m—th tract, covariates xi = (xi1, …, xiq)T were generated from N(0, 1) for continuous variables or from multinomial distribution with equal probabilities for categorical variables, ηi,m(sm) followed gaussian process GP{0, Σηm} and ei,m(sm) followed GP{0, Σem}. Compared to model (1), the above equation directly specified the covariates as fixed effect. Sample size n was set to be 100 and true parameters (β(sm), Σηm, Σem) were estimated from real data using FADTTS [28].
To examine our method, the following two scenarios on β(sm)Txi were simulated. In case I, the aim is to study shared effect of multiple tracts. Gender (G) and gestational age at birth (Gage) were included as covariates for all the 11 tracts,
in which we assumed c = 0, 0.2, 0.4, 0.6 and Gage effect was tested.
In case II, we want to examine a tract-specific effect. Birth weight (BW) was added as covariate to one particular tract, right uncinate fasciculus (m = 11), in addition to case I,
where effect size c was set to take values 0, 0.5, 1, 1.5 and the effect of BW was tested.
We applied HFPRM to the simulated dataset. The varying coefficient model (1) was first fitted to estimate individual functions. Functional principal components were then extracted such that at least 85% of total variation is included for each tract. In factor analysis, the first elbow point in the scree plot was taken as a cut-off to determine the number of global factors. In testing step, type I error and statistical power were calculated at significance level α = 0.05 based on 1000 simulation replications. FADTTS was also applied on each single tract and the results were compared.
3.2 Results
In case I, the first five functional principal components were extracted for each tract and the first factor was identified as global factor. The rejection rates for global factor analysis and FADTTS on testing Gage effect are presented by Fig. 2(a). The global factor analysis is substantially more powerful than the single tract analysis when detecting commonly shared effect. Such results are expected since common effect tends to be accumulated in the global factor.
Fig. 2. Simulation Result.
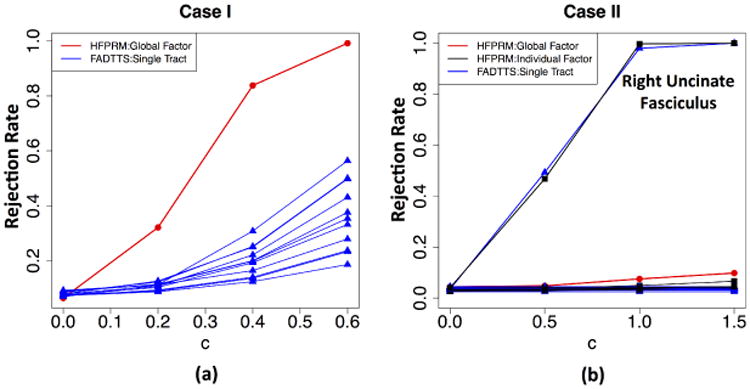
In case II, the first five functional principal components were extracted for each tract and the first two factors were identified as global factors. Fig. 2(b) shows the rejection rates for global factor analysis, individual factor analysis and FADTTS on testing BW effect. As can be seen, individual factor analysis in HFPRM achieves comparable power to single tract analysis for detecting tract-specific effect.
4 Early Human Brain Development Study
To investigate how genetic factors influence brain structure in prenatal and early postnatal stage, we conducted a genome-wide association study on the fiber bundle statistics in a unique cohort of infants. A total number of 662 neonatal twin subjects were taken from the UNC Early Brain Development Studies [7].
4.1 Data Acquisition and Preprocessing
MRI scans were acquired either on a 3T Siemens Allegra head-only scanner (N = 566) or on a 3T Siemens TIM Trio 3T scanner (N = 96). For the Allegra model, 339 diffusion weighted images were acquired by a single shot EPI DTI sequence with the following parameters: TR/TE = 5200/73 ms, voxel resolution = 2 × 2 × 2 mm3, 6 non-collinear directions with b = 1000 s/mm2 and 1 baseline image with b = 0. To improve the signal-to-noise ratio, five scans were repeated and averaged. For the remaining subjects scanned on Allegra, DWI was acquired with the following parameters: TR/ TE = 7680/82 ms, voxel resolution = 2 × 2 × 2 mm3, 42 non-collinear directions with diffusion gradients of b = 1000 s/mm2 in addition to 7 baseline images. For the Trio model, DWIs were acquired using a similar protocol to that of the 42 direction Allegra model with TR/TE = 7200/83 ms. Quality control was applied on raw DWIs using DTIPrep [18], and FSL [11,22] was performed for skull stripping and brain masking. We used a weighted least squares method [8] to estimate diffusion tensors and followed the UNC-Utah NA-MIC framework [23] to create a study-specific atlas. Subsequently, a total number of 44 fiber tracts listed in Table 1 were reconstructed in the atlas space using a streamline algorithm [5]. For each subject, four scalar diffusion properties, FA, MD, AD and RD, were then calculated at each location along each tract using neighboring diffusion tensors.
Genotyping of single nucleotide polymorphisms (SNPs) was conducted on Affymetrix Axiom genome-wide LAT Array. Samples with call rates less than 95%, outliers for homozygosity, ancestry outliers and unexpected relatedness were excluded from the study. We also removed genetic markers with Hardy-Weinberg equilibrium p-value less than 10−8, call rate less than 95% and Mendelian error rate larger than 10%. Population stratification was assessed using PCA [19]. Imputation was performed with MaCH-Admix [15] using 1000G reference panel [3]. To evaluate the quality of imputed SNPs, we computed the mean R2 under varying minor allele frequency (MAF) categories and selected R2 cutoffs as described in [14]. SNPs with MAF less than 0.01 were excluded from imputed dataset. Eventually, 472 twin subjects (32 MZ pairs, 75 DZ pairs and 259 singletons or unpaired twin subjects) and 8,538,562 genetic markers were retained for further analysis.
4.2 Data Analysis
In this experiment, we chose to focus on the fractional anisotropy (FA) measure. FA quantifies the extent of local directional water diffusion and partially reflects the degree of bundle maturation in premature brains [4]. To eliminate the heterogeneity in variance among different tracts, yi,m(sm) was rescaled by the total standard deviation along the tract. For the twin study, ACE model was fitted in (5) to account for correlation within twin pairs. Seven variables were added as covariates, including gestational age at birth, gender, DTI direction, scanner type and the first three genetic principal component to adjust for population stratification.
4.3 Results
In functional PCA, the first 5 functional principal components were extracted for each tract to include at least 70% of variation. Fig. 3(a) shows the scree plot in factor analysis and the elbow point is located at factor 2. Therefore, the first factor is identified as the global factor. We then performed GWAS on the global factor. The result is visualized by Fig. 3(b). In the Manhattan plot, we observed a significant region in anaplastic lymphoma kinase (ALK) gene on chromosome 2. The ALK gene is a neuronal orphan receptor tyrosine kinase that plays an important role in the nervous system development [1], and is highly expressed in the neonatal brain [10]. As a comparison, we also performed association analysis for top hit rs66556850 on each single tract. The result is presented by Fig. 3(c). A number of tracts have relatively small pvalue yet not small enough to be detected by a single tract GWAS. It indicates that the global factor analysis is more powerful to detect commonly shared genetic effect than single tract analysis.
Fig. 3.
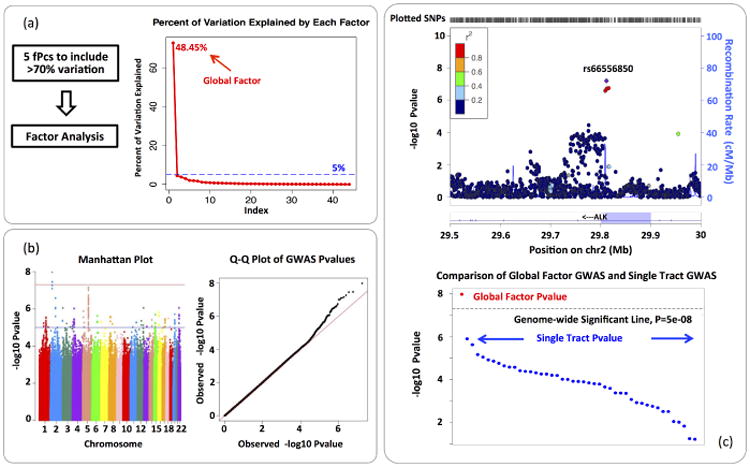
Real Data Analysis Result: (a) Functional PCA and Factor Analysis. (b) Visualization of GWAS result of the global factor. (c) A comparison between global factor analysis and single tract analysis on marker rs66556850, the −log10p value in the association test is plotted. The majority of pvalues in single tract analysis are around 10−2 ∼ 10−6.
5 Conclusion
We have developed a hierarchical functional principal regression model (HF-PRM) to efficiently conduct joint analysis on diffusion statistics from multiple neurofiber bundles. A varying coefficient model is introduced and functional PCA is applied to capture major tract variation. Factor analysis is then adopted to extract key features at both global level and individual level. Finally, standard estimation and testing procedures can be applied to study global effect and tract-specific effect. Simulation results demonstrated that HFPRM is powerful to detect common effect shared by multiple tracts. HFPRM has also been successfully applied to a genome-wide association study on neonatal twins. We are able to identify some important genetic variants related to early childhood brain development that were ignored by single tract analysis.
References
- 1.National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/gene/238.
- 2.Bach M, Laun FB, Leemans A, Tax CM, Biessels GJ, Stieltjes B, Maier-Hein KH. Methodological considerations on tract-based spatial statistics (tbss) Neuroimage. 2014;100:358–369. doi: 10.1016/j.neuroimage.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 3.Consortium GP, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubois J, Hertz-Pannier L, Dehaene-Lambertz G, Cointepas Y, Le Bihan D. Assessment of the early organization and maturation of infants' cerebral white matter fiber bundles: a feasibility study using quantitative diffusion tensor imaging and tractography. Neuroimage. 2006;30(4):1121–1132. doi: 10.1016/j.neuroimage.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 5.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, et al. 3d slicer as an image computing platform for the quantitative imaging network. Magnetic resonance imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garyfallidis E, Ocegueda O, Wassermann D, Descoteaux M. Robust and efficient linear registration of white-matter fascicles in the space of streamlines. NeuroImage. 2015;117:124–140. doi: 10.1016/j.neuroimage.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Gilmore JH, Schmitt JE, Knickmeyer RC, Smith JK, Lin W, Styner M, Gerig G, Neale MC. Genetic and environmental contributions to neonatal brain structure: a twin study. Human brain mapping. 2010;31(8):1174–1182. doi: 10.1002/hbm.20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodlett CB, Fletcher PT, Gilmore JH, Gerig G. Group analysis of DTI fiber tract statistics with application to neurodevelopment. NeuroImage. 2009;45:S133–S142. doi: 10.1016/j.neuroimage.2008.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guevara P, Poupon C, Rivi`ere D, Cointepas Y, Descoteaux M, Thirion B, Mangin J. Robust clustering of massive tractography datasets. NeuroImage. 2011;54(3):1975–1993. doi: 10.1016/j.neuroimage.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Iwahara T, Fujimoto J, Wen D, Cupples R, Bucay N, Arakawa T, Mori S, Ratzkin B, Yamamoto T. Molecular characterization of alk, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14(4):439–449. doi: 10.1038/sj.onc.1200849. [DOI] [PubMed] [Google Scholar]
- 11.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Jin Y, Shi Y, Zhan L, Gutman BA, de Zubicaray GI, McMahon KL, Wright MJ, Toga AW, Thompson PM. Automatic clustering of white matter fibers in brain diffusion mri with an application to genetics. NeuroImage. 2014;100:75–90. doi: 10.1016/j.neuroimage.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karhunen K. Zur spektraltheorie stochastischer prozesse. 1946 [Google Scholar]
- 14.Liu EY, Buyske S, Aragaki AK, Peters U, Boerwinkle E, Carlson C, Carty C, Crawford DC, Haessler J, Hindorff LA, et al. Genotype imputation of metabochipsnps using a study-specific reference panel of 4,000 haplotypes in african americans from the women's health initiative. Genetic epidemiology. 2012;36(2):107–117. doi: 10.1002/gepi.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu EY, Li M, Wang W, Li Y. Mach-admix: genotype imputation for admixed populations. Genetic epidemiology. 2013;37(1):25–37. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loève M. Fonctions aléatoires à décomposition orthogonale exponentielle. La Revue Scientifique. 1946;84:159–162. [Google Scholar]
- 17.O'Donnell LJ, Westin CF, Golby AJ. Tract-based morphometry for white matter group analysis. NeuroImage. 2009;45:832–844. doi: 10.1016/j.neuroimage.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oguz I, Farzinfar M, Matsui J, Budin F, Liu Z, Gerig G, Johnson HJ, Styner MA. Dtiprep: quality control of diffusion-weighted images. Frontiers in neuroinformatics. 2014;8:4. doi: 10.3389/fninf.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Rice JA, Silverman BW. Estimating the mean and covariance structure non-parametrically when the data are curves. Journal of the Royal Statistical Society Series B (Methodological) 1991:233–243. [Google Scholar]
- 21.Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31(4):1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 22.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, et al. Advances in functional and structural mr image analysis and implementation as fsl. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 23.Verde AR, Budin F, Berger JB, Gupta A, Farzinfar M, Kaiser A, Ahn M, Johnson HJ, Matsui J, Hazlett HC, et al. Unc-utah na-mic framework for dti fiber tract analysis. Frontiers in neuroinformatics. 2014;7:51. doi: 10.3389/fninf.2013.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng WYI. The geometric structure of the brain fiber pathways. Science. 2012;335(6076):1628–1634. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao F, Müler HG, Wang JL. Functional data analysis for sparse longitudinal data. Journal of the American Statistical Association. 2005;100(470):577–590. [Google Scholar]
- 26.Yuan Y, Gilmore JH, Geng X, Martin S, Chen K, Wang Jl, Zhu H. Fmem: Functional mixed effects modeling for the analysis of longitudinal white matter tract data. NeuroImage. 2014;84:753–764. doi: 10.1016/j.neuroimage.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yushkevich PA, Zhang H, Simon TJ, Gee JC. Structure-specific statistical mapping of white matter tracts. NeuroImage. 2008;41:448–461. doi: 10.1016/j.neuroimage.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Kong L, Li R, Styner M, Gerig G, Lin W, Gilmore JH. Fadtts: functional analysis of diffusion tensor tract statistics. NeuroImage. 2011;56(3):1412–1425. doi: 10.1016/j.neuroimage.2011.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]


