Abstract
Chemoresistance remains a major drawback to osteosarcoma treatment. ZBTB7A, a member of the POK transcription repressor family, was shown to play an important role in tumorigenesis. However, the effect of ZBTB7A on osteosarcoma chemoresistance is completely unknown. In this study, we found that ZBTB7A is increased in cisplatin-resistant osteosarcoma cells and that elevated ZBTB7A inhibits cisplatin-induced apoptosis by repressing LINC00473 expression. Further mechanistic studies revealed that ZBTB7A directly binds to the promoter and suppresses the transcription of LINC00473. Additionally, our data indicate that LINC00473 interacts with the transcript factor C/EBPβ, facilitating its binding to the promoter of IL24, leading to decrease chemoresistance. Thus, these findings indicate that the ZBTB7A-mediated LINC00473-C/EBPβ-IL24 pathway is a promising novel target for overcoming cisplatin resistance in osteosarcoma.
Introduction
Osteosarcoma is the most common type of primary bone cancer that mainly arises in childhood and adolescence [1], [2], [3]. Use of chemotherapy along with surgery has improved the overall 5-year survival rate of osteosarcoma patients [4], [5], [6], [7]. Cisplatin is the most widely used platinum-based anticancer drug for osteosarcoma, which interacts with nucleophilic N7 sites of purine bases in DNA to induce DNA damage that leads to cell death [2], [8], [9]. Although this treatment strategy is effective, it is often limited by acquired or intrinsic resistance of cancer cells to the drug. Thus, understanding the molecular mechanisms that lead to chemoresistance is essential to developing more effective treatments against osteosarcoma.
ZBTB7A, also known as Pokemon, LRF, or FBI, is a member of the POK family of transcriptional repressors, which consists of an NH2-terminal POZ/BTB domain and 4 COOH-terminal krüppel-type zinc fingers. The POZ/BTB domain is involved in homodimerization or heterodimerization, and recruits some corepressors such as BcoR, NcoR, or SMRT, while the krüppel-type zinc finger domain mediates specific DNA recognition and binding [10], [11]. ZBTB7A was reported to increase in some human cancers, such as breast cancer, colorectal cancer, prostate cancer, bladder cancer, liver cancer, and lung cancer, and to play an important role in tumorigenesis [12], [13]. However, some studies have indicated that ZBTB7A acts as a tumor suppressor via repressing glycolysis and metastasis [14], [15]. Although different functions have been reported, the effect of ZBTB7A on chemoresistance in osteosarcoma is unclear.
Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides with no protein-coding capacity and are poorly conserved [16], [17]. Several functional lncRNAs were recently shown to play important regulatory roles in various biological processes, including embryonic development, cell migration, cell proliferation, apoptosis, and tumorigenesis [17], [18]. Chemoresistance of cancers remains a major reason leading to tumor recurrence. Recently, several lncRNAs were identified to regulate chemoresistance in many cancers, such as lncRNAs HOTAIR [19], MEG3 [20], LINC00161 [9], and AC023115.3 [21]. Although several lncRNAs have been indicated to involve in cancer chemoresistance, the lncRNAs regulated by ZBTB7A were still unknown.
In this study, we found that the expression level of ZBTB7A was increased in cisplatin-resistant osteosarcoma cells and that elevated ZBTB7A enhanced chemoresistance via transcriptionally repressing LINC00473 expression. Additionally, we found that LINC00473 promoted the activity of IL24 promoter and elevated IL24 expression. Further mechanistic studies revealed that LINC00473 interacted with C/EBPβ, thereby facilitating IL24 transcription. Thus, our data demonstrate that ZBTB7A is an essential regulator in cisplatin-induced apoptosis, and the ZBTB7A-LINC00473-IL24 signaling axis plays an important role in regulating osteosarcoma chemoresistance.
Materials and Methods
Cell Culture and Reagents
The human osteosarcoma cancer cell lines U2OS and MG63 were obtained from the American Type Culture Collection. U2OS cells were cultured in DMEM with 10% fetal bovine serum (FBS; ExCell Bio, Lot: FSP500). MG63 cells were cultured in EMEM medium with 10% fetal bovine serum (FBS; ExCell Bio, Lot: FSP500). The medium was renewed every day, and cells were passaged before reaching confluence. The following antibodies were used in this study: antibody against GAPDH (Santa Cruz Biotechnology, Dallas, TX; SC-25778); caspase 3 antibody (Cell Signaling Tech, 9662); PARP (Santa Cruz Biotechnology, SC-8007); ZBTB7A antibody (Santa Cruz Biotechnology, SC-33683); C/EBPβ antibody (Santa Cruz Biotechnology, SC-150); C/EBPα antibody (Santa Cruz Biotechnology, SC-9351); IL24 (Proteintech, 26,772–1-AP); and cisplatin (Sigma, P4394).
RNA Interference and Virus Infection
RNA interference was performed as previously described. The shRNA was purchased from Sigma. The sequences targeting ZBTB7A-1 were 5-CCACTGAGACACAAACCTATT-3 and ZBTB7A-2 5-GAACGTGTACGAGATCGACTT-3. The sequences targeting IL24 were 5-GCATACTTCCTAACAGAGGCT-3 and IL24–2 5-CTGTGAAAGACACTATGCAA-3. Human LINC00473 cDNA and shRNA were obtained as previously described [22], [23].
Real-Time RT-PCR and RT-PCR
Total RNA was isolated using Trizol (Invitrogen). One microgram of total RNA was used to synthesize cDNA using the PrimeScriptTM RT reagent kit (Takara, RR047A) according to the manufacturer's instructions. The primers were as follows: actin: F: 5-GACCTGACTGACTACCTCATGAAGAT-3 and R: 5-GTCACACTTCATGATGGAGTTGAAGG-3; IL24: F: 5-CATCGTGTCACAACTGCAAC-3 and R: 5-AATGTCCACTTCCCCAAGG-3; and LINC00473: F: 5-AAACGCGAACGTGAGCCCCG-3 and R: 5-CGCCATGCTCTGGCGCAGTT-3.
Dual-Luciferase Reporter Assay
Generation of Cisplatin-Resistant Osteosarcoma Cell Line
The cisplatin-resistant cells were obtained as previously described [9].
Cell Viability Assay
Cells were plated in 96-well plates at a density of 5000 cells in 100 ml of medium per well 24 hours before the experiment. The cells were treated with cisplatin as indicated in the figures, and the cell viability was determined by the CKK8 assay.
Annexin V-FITC Staining and FACS
The staining protocol followed the manufacturer's instructions (BD). Generally, 5 × 105cells were harvested by a 5-minute centrifugation at 1000g and suspended in 195 μl of binding buffer followed by a 10-minute incubation with 5 μl of Annexin V-FITC at room temperature while avoiding light. After an additional centrifugation, the cells were resuspended in 190 μl of binding buffer, and 10 μl of PI staining reagent was added with slight shaking. The FASC (BD) analysis was employed for detecting cell apoptotic events.
Protein Identification and Quantitation
RIP Assay
RIP was performed using the EZ-Magna RIP kit (Millipore, USA) following the manufacturer's protocol. U2OS cells at 80% to 90% confluency were collected and lysed in complete RIPA buffer. The whole cell protein extract was then incubated with RIP wash buffer containing magnetic beads conjugated with human anti-C/EBPβ antibody or rabbit immunoglobulin G (IgG) control. The protein in the samples was digested with proteinase K, and the immunoprecipitated RNA was isolated. Finally, purified RNA was subjected to qRT-PCR analysis to demonstrate the presence of LINC00473.
Microarray and Computational Analysis
Arraystar Human LncRNA Microarray v3.0 is designed for the global profiling of human LncRNAs and protein-coding transcripts. The sample preparation and microarray hybridization were performed based on the manufacturer's standard protocols. Briefly, 10 quality mRNA samples from each group were purified from total RNA after the removal of rRNA using an mRNA-ONLY Eukaryotic mRNA Isolation Kit (Epicenter Biotechnologies, USA). Each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcript without 3′ bias utilizing a random priming method. The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-1000. Then, 1 μg of each labeled cRNA was fragmented by adding 5 μl of 10× blocking agent and 1 μl of 25× fragmentation buffer, heated to 60°C for 30 minutes, and diluted with 25 μl of 2× GE hybridization buffer. Fifty microliters of hybridization solution was dispensed into the gasket slide and assembled to the Human LncRNA Array v3.0 slide (8 × 60 K, Arraystar). The slides were incubated for 17 h at 65°C in an Agilent hybridization oven and then washed, fixed, and scanned using the Agilent DNA Microarray Scanner (part number G2505C). Approximately 30,586 lncRNAs and 26,109coding transcripts collected from the most authoritative databases, such as RefSeq (release 55), UCSC Human (GRCh37/hg19), GENCODE 13, and lncRNAdb (2.0), were detected using the microarray. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.0 software package (Agilent Technologies). Protein-coding genes were searched for differentially expressed lncRNAs using the UCSC Genome Browser. Genes transcribed within 300 kb were considered to represent nearby coding genes. The microarray work was performed by KangChen Bio-tech, Shanghai, China.
Statistics and Data Analyses
The data are expressed as the mean ± SEM, and statistical evaluation was performed using one-way analysis of variance. Values of P < .05 were considered statistically significant.
Results
ZBTB7A Inhibited Cisplatin-Induced Apoptosis in Osteosarcoma
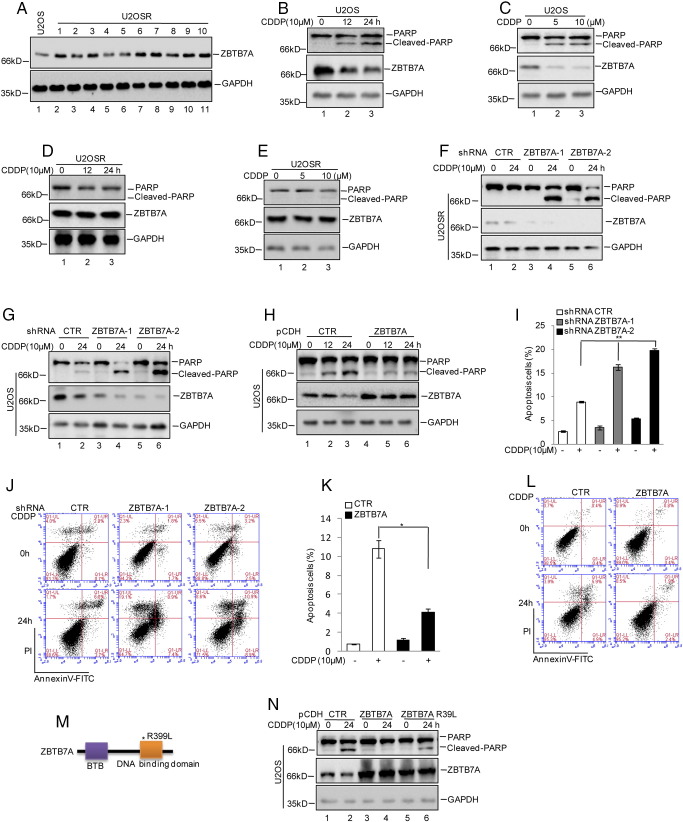
To assess the effect of ZBTB7A on cisplatin-induced apoptosis in osteosarcoma, we first analyzed the expression levels of ZBTB7A in cisplatin-resistant U2OS cells (U2OSR1-10). Compared with the control cells, the protein level of ZBTB7A was increased in most of the cisplatin-resistant cells, particularly in clone10 (Figure 1A). Thus, only U2OSR10 was used in subsequent functional studies. For convenience, U2OSR10 is hereafter referred to as U2OSR. After that, we detected the protein levels of ZBTB7A in response to cisplatin treatment in U2OS and U2OSR cells and found that the expression of ZBTB7A was reduced in U2OS cells under cisplatin treatment in a dose- and time-dependent manner (Figure 1, B and C). However, in U2OSR cells, the reduction of ZBTB7A was prevented (Figure 1, D and E). Then, the endogenous ZBTB7A was knocked down in U2OSR and U2OS cells. Compared with the control cells, inhibition of ZBTB7A significantly promoted cell apoptosis in U2OSR and U2OS cells, as indicated by PARP cleavage (Figure 1, F and G). However, overexpression of ZBTB7A inhibited cisplatin-induced apoptosis in U2OS cells (Figure 1H). To further confirm this result, the cell apoptosis was calculated by flow cytometric analyses with PI and Annexin V double staining. As shown in Figure 1, I and J, the apoptotic population was notably increased in ZBTB7A-suppressed cells during cisplatin treatment (approximately 20% versus 8% in the control group) and was decreased in ZBTB7A-overexpressed U2OS cells (approximately 4% versus 10% seen in the control group) (Figure 1, K and L).
Figure 1.
ZBTB7A inhibited cisplatin-induced apoptosis.
(A) The protein levels of ZBTB7A were analyzed in U2OS and U2OSR cells. (B-E) U2OS and U2OSR cells were treated with cisplatin as indicated, and cell lysates were then subjected to Western blotting analysis using the indicated antibodies. (F and G) U2OSR and U2OS cells with or without knocked down ZBTB7A were treated with cisplatin as indicated, and the cell lysates were analyzed by Western blotting with the indicated antibodies. (H) U2OS cells with or without overexpressing ZBTB7A were treated with cisplatin as indicated, and the cell lysates were then subjected to Western blotting analysis using the indicated antibodies. (I and J) U2OS cells with or without knocked down ZBTB7A were treated with cisplatin as indicated. The percentage of cell apoptosis was analyzed by flow cytometry. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR. (K and L) U2OS cells with or without overexpressedZBTB7A were treated with cisplatin as indicated. Cell apoptosis was analyzed by flow cytometry. The data represent the mean ± SD of three independent experiments. *P < .05 versus CTR. (M and N) The ZBTB7A zinc finger mutant was constructed (M). U2OS cells with or without overexpressed ZBTB7A or ZBTB7A R399L were treated with cisplatin as indicated, and the cell lysates were then subjected to Western blotting analysis using the indicated antibodies (N).
Additionally, to test whether ZBTB7A enhanced cisplatin resistance by its transcriptional activity, we generated a zinc finger (R399L) mutant of ZBTB7A, which was shown to be defective in DNA binding [15]. As shown in Figure 1, M and N, unlike the wild-type ZBTB7A, the R399L mutant failed to suppress the cisplatin-induced apoptosis.
ZBTB7A Transcriptionally Represses lncRNA LINC00473 in Response to Cisplatin Treatment
To explore the molecular mechanism whereby ZBTB7A contributed to osteosarcoma chemoresistance, we used microarray analysis to compare lncRNA expression profiles between ZBTB7A knockdown U2OS cells and control cells after cisplatin treatment. Among the changes in lncRNAs, we found that lncRNA LINC00473 was dramatically increased in ZBTB7A-suppressed cells under cisplatin treatment, which was confirmed by qRT-PCR (Figure 2, A and B). Consistently, inhibition of ZBTB7A expression promoted LINC00473 increase in U2OSR cells (Figure 2C). In contrast, ZBTB7A overexpression suppressed LINC00473 expression. However, the suppression was abolished by the R399L mutant, indicating that the effect of ZBTB7A on LINC00473 was dependent on its transcriptional activity (Figure 2D).
Figure 2.
ZBTB7A repressed LINC00473 expression.
(A) U2OS cells with or without knocked down ZBTB7A were treated with 10 μM cisplatin for the indicated times. The filtered lncRNA array data were subjected to unsupervised hierarchical clustering analysis. The metric was set as the Euclidean distance. (B and C) U2OS and U2OSR cells with or without knocked down ZBTB7A were treated with cisplatin as indicated and the mRNA levels of LINC00473 were analyzed by qRT-PCR. The protein levels of ZBTB7A were detected by Western blotting. The data represent the mean ± SD of three independent experiments. **P < .01, ***P < .001 versus CTR. (D) U2OS cells with or without overexpressed ZBTB7A or theR399L mutant were treated with cisplatin as indicated. The mRNA level of LINC00473 was analyzed by qRT-PCR. The data represent the mean ± SD of three independent experiments. **P < .01versus CTR. (E) Schematic illustration of pGL3-based reporter constructs used in luciferase assays to examine the transcriptional activity of LINC00473. (F) The promoter of LINC00473 (F1) was transfected into U2OSR cells with or without ZBTB7A knockdown. The cells were then treated with cisplatin. Luciferase activity was measured by the luciferase assay. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR. (G) The promoters of LINC00473 (F, F2, F3) were individually transfected into U2OSR cells with or without ZBTB7A knockdown. The cells were then treated with cisplatin. Luciferase activity was measured. The data represent the mean ± SD of three independent experiments. ***P < .001 versus CTR. (H) ChIP analysis showed the binding of ZBTB7A to the promoter of LINC00473. An isotype-matched IgG was used as a negative control. (I) Schematic illustration of pGL3-based reported constructs used in luciferase assays to examine the transcriptional activity of LINC00473. The dots indicated the ZBTB7A binding sites. (J) The promoters of LINC00473 WT and mutants (MUT1, 2, 3) were transfected into U2OSR cells with or without knockdown ZBTB7A, and then the cells were treated with cisplatin. Luciferase activity was measured. The data represent the mean ± SD of three independent experiments. ***P < .001 versus CTR.
To investigate whether ZBTB7A can directly repress LINC00473 transcription, we first cloned the upstream sequence of LINC00473 and different truncations by PCR and inserted them into the pGL3-based luciferase reporter plasmids. These truncations were named F1 to F3 (Figure 2E). As shown in Figure 2F, the luciferase activity of the LINC00473 promoter was increased in ZBTB7A knockdown U2OSR cells, and the sequence of F3 (0 ~ −383 bp) was essential for the increase (Figure 2G). To further confirm this result, the chromatin immunoprecipitation (ChIP) assay was performed. As shown in Figure 2H, the fragment of F3 was specifically detected in ZBTB7A immunoprecipitates, and the bond was increased after cisplatin treatment.
A previous report has shown that ZBTB7A binds to GC-rich DNA sequences [25]. To further verify the potential sites bound by ZBTB7A, we inspected the sequence of F3 using JASPAR software. Three positive binding sits of ZBTB7A were identified. As show in Figure 2I, these binding sites were named 1 to 3 based on the distance from the transcription start site, and then a series of pGL3-based luciferase reporter plasmids containing the wild-type ZBTB7A binding regions or different mutants was constructed. As shown in Figure 2J, specific mutations of predicated ZBTB7A binding site 2 attenuated the luciferase activity, indicating that binding site 2 was a potential binding site of ZBTB7A.
ZBTB7A Inhibits Cisplatin-Induced Apoptosis via Regulating LINC00473 Expression
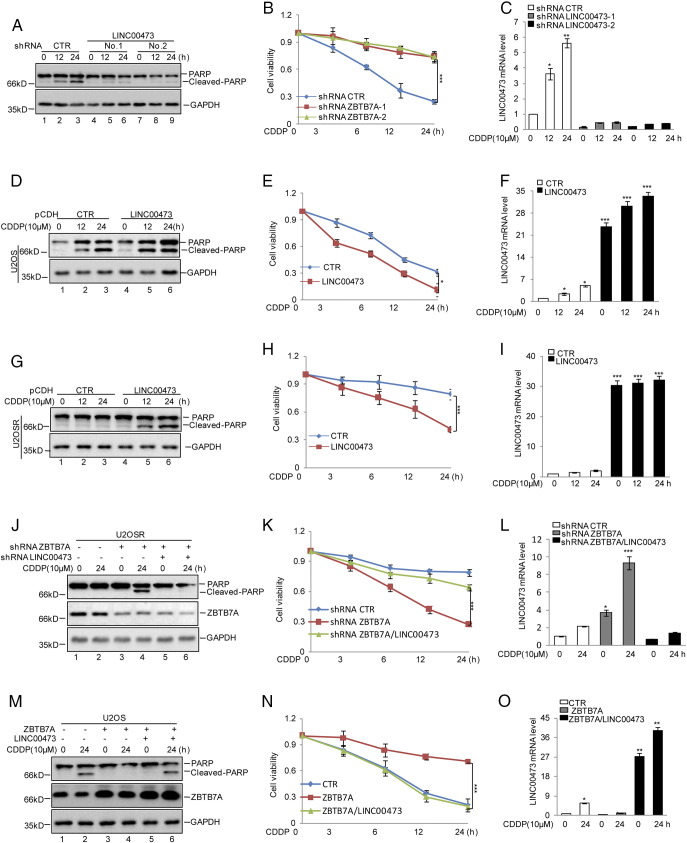
To evaluate whether the effect of ZBTB7A on osteosarcoma chemoresistance was regulated by LINC00473, we first knocked down LINC00473 in U2OS cells and found that inhibition of endogenous LINC00473decreased cisplatin-induced apoptosis and increased cell viability (Figure 3, A-C). Conversely, overexpression of LINC00473 in U2OS and U2OSR cells accelerated cell apoptosis and decreased cell viability (Figure 3, D-I). Then, we decreased LINC00473 in ZBTB7A suppressed-U2OSR cells. As shown in Figure 3, J, K, and L, we found that the inhibition of ZBTB7A promoted cisplatin-induced apoptosis and reduced cell viability, but the effect was diminished when LINC00473 was knocked down. Similarly, overexpression of LINC00473 decreased ZBTB7A-enhanced cisplatin resistance (Figure 3, M-O).
Figure 3.
ZBTB7A enhanced cisplatin resistance by repressing LINC00473.
(A-C) U2OS cells with or without knockdown LINC00473 were treated with cisplatin as indicated. Cell lysates were then subjected to Western blotting analysis using the indicated antibodies (A). Cell viability was analyzed by the CKK8 assay (B). LINC00473 RNA level was detected by qRT-PCR. The data represent the mean ± SD of three independent experiments. *P < .05, **P < .01, ***P < .001 versus CTR. (D-I) U2OS and U2OSR cells with or without LINC00473 overexpression were treated with cisplatin as indicated. Cell lysates were then subjected to Western blotting analysis using the indicated antibodies (D and G). Cell viability was analyzed by the CKK8 assay (E, H).The LINC00473 RNA level was detected by qRT-PCR (F and I). The data represent the mean ± SD of three independent experiments. *P < .05, ***P < .001 versus CTR. (J-L) LINC00473 was inhibited using shRNA in U2OSR cells with or without knocked down ZBTB7A, and then the cells were treated with cisplatin as indicated. Cell lysates were subjected to Western blotting analysis using the indicated antibodies (J). Cell viability was analyzed by the CKK8 assay (K). The LINC00473 RNA level was detected by qRT-PCR (L). The data represent the mean ± SD of three independent experiments. *P < .05, ***P < .001 versus CTR. (M-O) LINC00473 was overexpressed in U2OS cells with or without ZBTB7A overexpression, and then the cells were treated with cisplatin as indicated. Cell lysates were subjected to Western blotting analysis using the indicated antibodies (M). Cell viability was analyzed by the CKK8 assay (N).The LINC00473 RNA level was detected by qRT-PCR (O). The data represent the mean ± SD of three independent experiments. *P < .05, **P < .01, ***P < .001 versus CTR.
ZBTB7A Inhibits IL24 Expression Relying on LINC00473
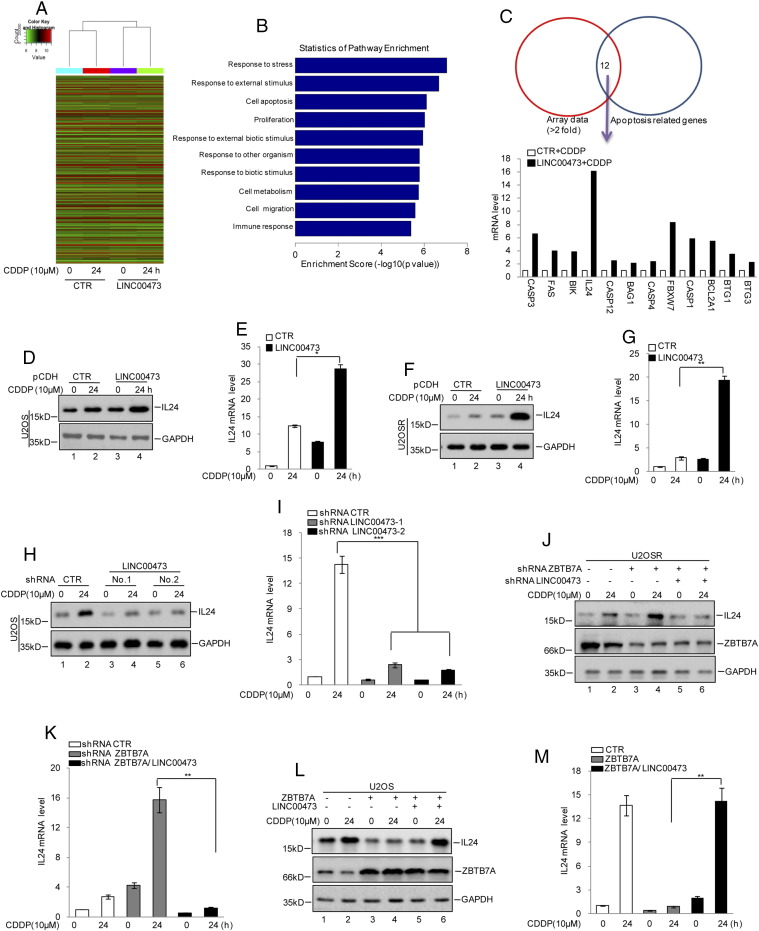
To determine the molecular mechanism underlying the LINC00473 promotion of cisplatin-induced apoptosis, gene expression profiles in U2OS cells with or without LINC00473 overexpression were analyzed using mRNA microarray analysis (Figure 4A). As shown in Figure 4, B and C, we focused on the altered genes (more than two-fold) involved in regulation of cell apoptosis. Among the changed genes, IL24 was remarkably upregulated in LINC00473-overexpressing cells under cisplatin treatment, which was subsequently confirmed by Western blotting and qRT-PCR (Figure 4, D and E). Similar results were obtained in U2OSR cells (Figure 4, F and G). Furthermore, the expression levels of IL24 were analyzed in U2OS cells with or without LINC00473 knockdown. Compared with the control cells, IL24 was significantly downregulated when LINC00473was knocked down (Figure 4, H and I).
Figure 4.
ZBTB7A inhibited IL24 expression relying on LINC00473.
(A) An unbiased genome mRNA expression profiling heat map between CTR and LINC00473 overexpression under cisplatin treatment as indicated. (B) The numbers of changed genes involved in different biological processes were statistically analyzed. (C) Comparison of the array data (more than two-fold) with apoptosis-related genes. The related genes are listed. (D-G) U2OS and U2OSR cells with or without overexpressing LINC00473 were treated with cisplatin as indicated. The protein and mRNA levels of IL24 were detected by Western blotting and qRT-PCR. The data represent the mean ± SD of three independent experiments. *P < .05, **P < .01 versus CTR. (H and I) U2OS cells with or without LINC00473 knockdown were treated with cisplatin as indicated. The protein and mRNA levels of IL24 were detected by Western blotting and qRT-PCR. The data represent the mean ± SD of three independent experiments. ***P < .001 versus CTR. (J and K) LINC00473 was inhibited in U2OSR cells with or without knocked down ZBTB7A. The expression of IL24 was analyzed by Western blotting and qRT-PCR. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR. (L and M) LINC00473 was overexpressed in U2OS cells with or without ZBTB7A overexpression. The expression of IL24 was analyzed by Western blotting and qRT-PCR. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR.
Based on the observation that ZBTB7A repressed LINC00473 expression, we asked whether ZBTB7A could mediate IL24 expression by LINC00473. As shown in Figure 4, J and K, inhibition of ZBTB7A in U2OSR cells promoted IL24 expression. However, the increase was abolished when LINC00473 was knocked down. Otherwise, overexpression of ZBTB7A suppressed IL24 expression, which was reversed by LINC00473 (Figure 4, L and M).
LINC00473 Promotes IL24 Transcription via Interacting with C/EBPβ
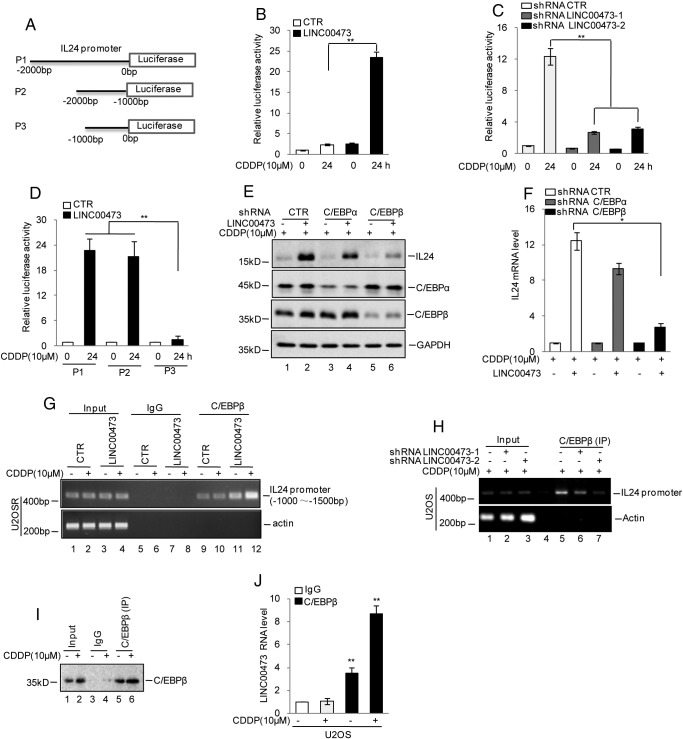
To further investigate the mechanism underlying the LINC00473 regulation of IL24 expression, we first examined whether LINC00473 could regulate IL24 expression transcription. We first cloned the promoter of IL24 and inserted it into the luciferase reporter vector (Figure 5A). As shown in Figure 5B, the firefly luciferase activity was higher after LINC00473 overexpression in U2OSR cells. Conversely, inhibition of LINC0047 prevented the upregulation of luciferase activity (Figure 5C).
Figure 5.
LINC00473 upregulated IL24 expression via interacting with C/EBPβ.
(A) Schematic illustration of pGL3-based reported constructs used in luciferase assays to examine the transcriptional activity of IL24. (B) The promoter of IL24 (P1) was transfected into U2OSR cells with or without LINC00473 overexpression. Luciferase activity was measured. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR. (C) The promoter of IL24 (P1) was transfected into U2OS cells with or without LINC00473 knockdown. Luciferase activity was measured. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR. (D) The promoter of IL24 (P1, P2, P3) was transfected into U2OSR cells with or without LINC00473 overexpression. Luciferase activity was measured. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR. (E and F) C/EBPα and C/EBPβ were individually knocked down in U2OS cells with or without LINC00473 overexpression. The protein and mRNA levels of IL24 were analyzed by Western blotting and qRT-PCR. The data represent the mean ± SD of three independent experiments. *P < .05 versus CTR. (G and H) ChIP analysis showed the binding of C/EBPβ to the promoter of IL24 when LINC00473 was overexpressed or knocked down. An isotype-matched IgG was used as a negative control. (I and J) RIP analysis showed the interaction of C/EBPβ with LINC00473. The data represent the mean ± SD of three independent experiments. **P < .01 versus CTR.
To determine which part of the IL24 promoter was essential for LINC00473, we separated the promoter into two parts named P2 and P3 (Figure 5A). We transfected them into U2OSR cells with or without overexpressing LINC00473. As shown in Figure 5D, we found that the firefly luciferase activity of P1 and P2 was increased in LINC00473-overexpressing cells; however, the increase was abolished when P3 was transfected, indicating that the region of P2 was essential for the regulation of LINC00473.
Previous reports have shown that the transcription factors C/EBPα and C/EBPβ could bind the region of P2 and promote IL24 expression [26]. Therefore, we asked whether LINC00473 promoted the upregulation of IL24 via these transcription factors. We first knocked down these transcription factors using shRNA and found that the effect of LINC00473 on IL24 disappeared with C/EBPβ knockdown, suggesting that the upregulation of IL24 by LINC00473 relied on C/EBPβ (Figure 5, E and F). To further confirm this result, we next evaluated whether binding of C/EBPβ to the promoter of IL24 was increased by LINC00473. As shown in Figure 5G, the chromatin fragments corresponding to C/EBPβ were increased when LINC00473 was overexpressed. LINC00473 knockdown decreased the binding of C/EBPβ to the promoter of IL24 (Figure 5H). In addition, RNA immunoprecipitation (RIP) was performed to validate the interaction between C/EBPβ and LINC00473. As expected, we observed a significant enrichment of LINC00473 in the C/EBPβ RIP relative to the IgG control RIP (Figure 5, I and J).
ZBTB7A Enhances Cisplatin Resistance by Suppressing LINC00473-IL24 Pathway
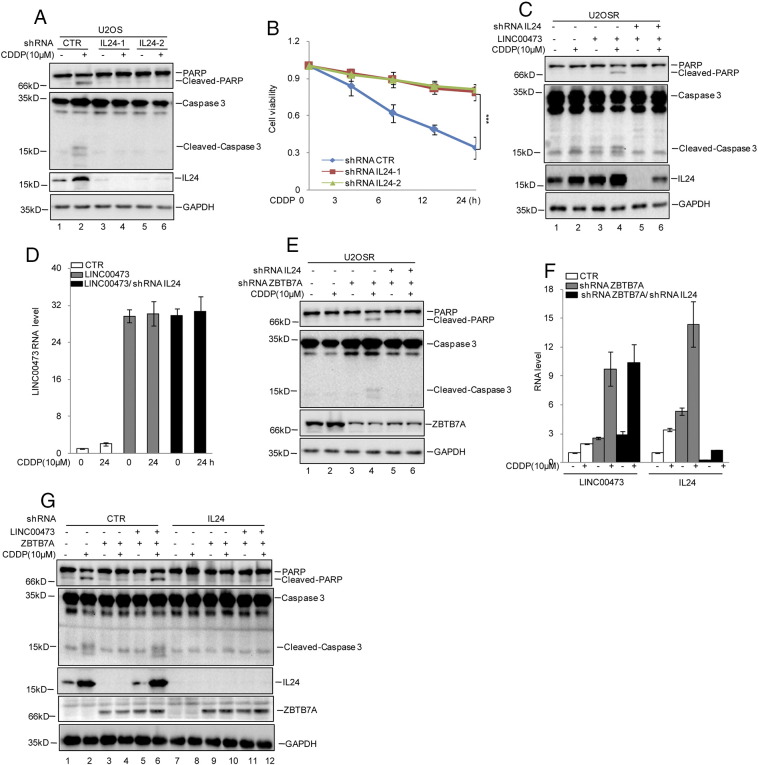
To assess whether ZBTB7A enhanced cisplatin-resistance via regulation of the LINC00473-IL24 axis, we first knocked down IL24 expression using shRNA. Compared with control cells, IL24 knockdown inhibited cisplatin-induced apoptosis and increased cell viability in U2OS cells (Figure 6, A and B). Subsequently, we found that the promotion byLINC00473 of cisplatin-induced apoptosis was abolished when IL24 was knocked down in U2OSR cells (Figure 6, C and D). A similar result was obtained in ZBTB7A-suppressed U2OS cells, indicating that the effects of ZBTB7A and LINC00473 were dependent on IL24 (Figure 6, E and F). To confirm that result, we knocked down IL24 in ZBTB7A and LINC00473 overexpressed U2OS cells. Compared with control cells, we found that the effects of ZBTB7A or LINC00473 on cisplatin resistance were diminished when IL24 was knocked down (Figure 6G).
Figure 6.
ZBTB7A repressed cisplatin-induced apoptosis via regulating the LINC00473-IL24 axis.
(A and B) U2OS cells with or without IL24 knockdown were treated with cisplatin as indicated. Cell lysates were then subjected to Western blotting analysis using the indicated antibodies (A). Cell viability was analyzed by CKK8 assay (B). The data represent the mean ± SD of three independent experiments. ***P < .001 versus CTR. (C and D) IL24 was knocked down in U2OSR cells with or without LINC00473 overexpression. The cells were treated with cisplatin as indicated. Cell lysates were then subjected to Western blotting analysis using the indicated antibodies. The mRNA level of LINC00473 was analyzed by qRT-PCR. (E and F) IL24 was inhibited in U2OSR cells with or without ZBTB7A knockdown. The cells were treated with cisplatin as indicated. Cell lysates were then subjected to Western blotting analysis using the indicated antibodies (E). The mRNA levels of LINC00473 and IL24 were analyzed by qRT-PCR (F). (G) IL24 was knocked down in U2OS cells with or without ZBTB7A overexpression/LINC00473 knockdown. The cells were treated with cisplatin as indicated. Cell lysates were then subjected to Western blotting analysis using the indicated antibodies.
Discussion
In this study, we report that ZBTB7A is an important regulator of osteosarcoma chemoresistance. ZBTB7A was increased in cisplatin-resistant cells, and elevation of ZBTB7A inhibited cisplatin-induced apoptosis via transcriptionally repressing LINC00473 expression. In addition, we found that LINC00473 could interact with the transcription factor C/EBPβ and facilitated its binding to the IL24 promoter, leading to IL24 increase and decreasing chemoresistance. Therefore, our data indicated that ZBTB7A-mediated LINC00473-C/EBPβ-IL24 plays an important role in osteosarcoma chemoresistance.
ZBTB7A is a POK family transcription repressor that is best known for its pro-oncogenic role in various cancers. Maeda et al. reported that ZBTB7A promoted cellular transformation by suppressing ARF and that ZBTB7A also promoted cell proliferation and lymphoma formation in transgenic mice [12]. Overexpressed ZBTB7A was capable of promoting cell proliferation by regulating p53, p21, and p27 expression [27]. Consistently, we observed that ZBTB7A was increased in cisplatin-resistant cells and elevated ZBTB7A inhibited cisplatin-induced apoptosis.
Recent reports have shown that lncRNAs play an important role in chemoresistance in many cancers [17], [28], [29], [30], [31], [32], [33], [34]. However, ZBTB7A-mediated lncRNAs have not been reported. Thus, to identify the lncRNAs that were regulated by ZBTB7A in osteosarcoma cells under cisplatin treatment, the cells with or without knocked down ZBTB7A were analyzed using lncRNAs microarray. Long noncoding RNA LINC00473 was identified as a ZBTB7A-regulated lncRNA.
LINC00473, also known as C6orf176, encodes an intergenic lncRNA from the chromosome 6q27 locus. The previous reports have shown that LINC00473 was in response to cAMP signaling and mediates decidualization of human endometrial stromal cells and was highly induced in LKB-inactivated lung cancer cells. Elevated LINC00473 promoted cell growth and survival [1]. However, the role of LINC00473 in chemoresistance of cancer cells has not been explored. Here, we found that LINC00473 was induced by cisplatin and the increase in LINC00473 under cisplatin was transcriptionally inhibited by ZBTB7A, leading to a cisplatin-induced apoptosis decrease.
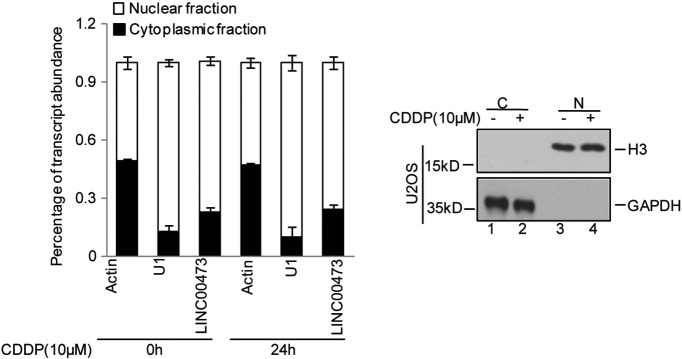
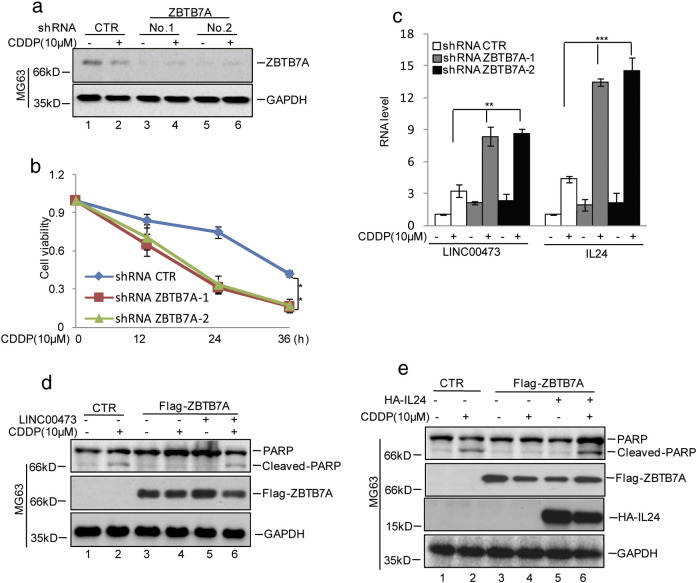
Studies have indicated that lncRNAs may be involved in various processes, including transcription, splicing, posttranscriptional regulation, organization of protein complexes, cell-cell signaling, and allosteric regulation of proteins [35], [36], [37], [38], [39], [40], [41]. LINC00473 was reported to locate to the nucleus in lung cancer cells. Consistently, LINC00473 was also located in the nucleus in osteosarcoma cells, and the treatment with CDDP did not alter the sublocalization of LINC00473 (Supplementary Figure 1). Subsequently, we found that LINC00473 promoted IL24 expression via interacting with the transcription factor C/EBPβ under cisplatin treatment. The interaction between LINC00473 and C/EBPβ facilitated its binding to the promoter of IL24, leading to an increase in IL24 and a decrease in chemoresistance. Furthermore, we found that ZBTB7A enhanced osteosarcoma chemoresistance via repressing the LINC00473-IL24 pathway, which was subsequent proved in another osteosarcoma cell line, MG63 (Supplementary Figure 2). Thus, our data provide an effective therapeutic strategy for use in osteosarcoma treatment.
Supplementary Figure 1.
Fractionation of U2OS cells with or without treatment with cisplatin followed by qRT-PCR (left panel) and fractionation controls by Western blot (right panel). U1 RNA served as a positive control for nuclear gene expression. N, nuclear fraction; C, cytoplasmic fraction. The data are representative of at least three independent experiments.
Supplementary Figure 2.
(A-C) MG63 cells with or without knockdown ZBTB7A were treated with cisplatin as indicated. Cell lysates were then subjected to Western blotting analysis using the indicated antibodies (A). Cell viability was analyzed by CKK8 assay (B). The mRNA levels of LINC00473 and IL24 were analyzed by qRT-PCR (C). The data represent the mean ± SD of three independent experiments. **P < .01, ***P < .001 versus CTR. (D and E) LINC00473 or HA-IL24 was transfected into MG63 with or without overexpressing Flag-ZBTB7A. The cells were treated with cisplatin as indicated, and cell lysates were then subjected to Western blotting analysis using the indicated antibodies.
The following are the supplementary data in this article.
lncRNA Expression Profiles between ZBTB7A Knockdown U2OS Cells and Control Cells after Cisplatin Treatment
mRNA Expression Profiles between LINC00473 Overpressed U2OS Cells and Control Cells after Cisplatin Treatment
Conflict of Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgements
This research was supported by the National Natural Science Foundation of China (no. 31270999 to Wenzhi Zhao, 81402260 to Chuanchun Han) and the Liaoning Provincial Natural Science Foundation of China (no. 20170540298) to Lu Zhang and (no. 2015020309) to Chuanchun Han.
Contributor Information
Chuanchun Han, Email: hanchuanchun@163.com.
Wenzhi Zhao, Email: drzhaowenzhi@sina.com.
References
- 1.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- 2.Han XG, Du L, Qiao H, Tu B, Wang YG, Qin A, Dai KR, Fan QM, Tang TT. CXCR1 knockdown improves the sensitivity of osteosarcoma to cisplatin. Cancer Lett. 2015;369:405–415. doi: 10.1016/j.canlet.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Yan GN, Lv YF, Guo QN. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016;370:268–274. doi: 10.1016/j.canlet.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari S, Mercuri M, Bacci G. Comment on "Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols". J Clin Oncol. 2002;20:2910. doi: 10.1200/JCO.2002.20.12.2910. [author reply 2910-2911] [DOI] [PubMed] [Google Scholar]
- 5.Fiorini C, Cordani M, Gotte G, Picone D, Donadelli M. Onconase induces autophagy sensitizing pancreatic cancer cells to gemcitabine and activates Akt/mTOR pathway in a ROS-dependent manner. Biochim Biophys Acta. 2015;1853:549–560. doi: 10.1016/j.bbamcr.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Li G, Ma D, Chen Y. Cellular functions of programmed cell death 5. Biochim Biophys Acta. 2016;1863:572–580. doi: 10.1016/j.bbamcr.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 7.Wen H, Xu WJ, Jin X, Oh S, Phan CH, Song J, Lee SK, Park S. The roles of IP3 receptor in energy metabolic pathways and reactive oxygen species homeostasis revealed by metabolomic and biochemical studies. Biochim Biophys Acta. 2015;1853:2937–2944. doi: 10.1016/j.bbamcr.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Zhang L, Zheng X, Zhong W, Tian X, Yin B, Tian K, Zhang W. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 2016;382:137–146. doi: 10.1016/j.canlet.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 10.Costoya JA. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic. 2007;6:8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 11.Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, Di Cesare PE. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J Biol Chem. 2004;279:47081–47091. doi: 10.1074/jbc.M405288200. [DOI] [PubMed] [Google Scholar]
- 12.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 13.Shi DB, Wang YW, Xing AY, Gao JW, Zhang H, Guo XY, Gao P. C/EBPalpha-induced miR-100 expression suppresses tumor metastasis and growth by targeting ZBTB7A in gastric cancer. Cancer Lett. 2015;369:376–385. doi: 10.1016/j.canlet.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 14.Liu XS, Genet MD, Haines JE, Mehanna EK, Wu S, Chen HI, Chen Y, Qureshi AA, Han J, Chen X. ZBTB7A suppresses melanoma metastasis by transcriptionally repressing MCAM. Mol Cancer Res. 2015;13:1206–1217. doi: 10.1158/1541-7786.MCR-15-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu XS, Haines JE, Mehanna EK, Genet MD, Ben-Sahra I, Asara JM, Manning BD, Yuan ZM. ZBTB7A acts as a tumor suppressor through the transcriptional repression of glycolysis. Genes Dev. 2014;28:1917–1928. doi: 10.1101/gad.245910.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia. 2015;17:79–88. doi: 10.1016/j.neo.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalevee S, Feil R. Long noncoding RNAs in human disease: emerging mechanisms and therapeutic strategies. Epigenomics. 2015;7:877–879. doi: 10.2217/epi.15.55. [DOI] [PubMed] [Google Scholar]
- 18.Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 19.Liu MY, Li XQ, Gao TH, Cui Y, Ma N, Zhou Y, Zhang GJ. Elevated HOTAIR expression associated with cisplatin resistance in non–small cell lung cancer patients. J Thorac Dis. 2016;8:3314–3322. doi: 10.21037/jtd.2016.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma B, Gao Z, Lou J, Zhang H, Yuan Z, Wu Q, Li X, Zhang B. Long noncoding RNA MEG3 contributes to cisplatininduced apoptosis via inhibition of autophagy in human glioma cells. Mol Med Rep. 2017;16:2946–2952. doi: 10.3892/mmr.2017.6897. [DOI] [PubMed] [Google Scholar]
- 21.Ma B, Yuan Z, Zhang L, Lv P, Yang T, Gao J, Pan N, Wu Q, Lou J, Han C. Long non-coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta. 2017;1864:1393–1404. doi: 10.1016/j.bbamcr.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126:2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han C, Gu H, Wang J, Lu W, Mei Y, Wu M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells. 2013;31:953–965. doi: 10.1002/stem.1335. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Du T, Wang X, Zhang Y, Hu W, Du X, Miao L, Han C. IDH1, a CHOP and C/EBPbeta-responsive gene under ER stress, sensitizes human melanoma cells to hypoxia-induced apoptosis. Cancer Lett. 2015;365:201–210. doi: 10.1016/j.canlet.2015.05.027. [DOI] [PubMed] [Google Scholar]
- 25.Tsuji-Takechi K, Negishi-Koga T, Sumiya E, Kukita A, Kato S, Maeda T, Pandolfi PP, Moriyama K, Takayanagi H. Stage-specific functions of leukemia/lymphoma-related factor (LRF) in the transcriptional control of osteoclast development. Proc Natl Acad Sci U S A. 2012;109:2561–2566. doi: 10.1073/pnas.1116042109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madireddi MT, Dent P, Fisher PB. AP-1 and C/EBP transcription factors contribute to mda-7 gene promoter activity during human melanoma differentiation. J Cell Physiol. 2000;185:36–46. doi: 10.1002/1097-4652(200010)185:1<36::AID-JCP3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 27.Choi WI, Jeon BN, Yun CO, Kim PH, Kim SE, Choi KY, Kim SH, Hur MW. Proto-oncogene FBI-1 represses transcription of p21CIP1 by inhibition of transcription activation by p53 and Sp1. J Biol Chem. 2009;284:12633–12644. doi: 10.1074/jbc.M809794200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chitambar CR. Gallium and its competing roles with iron in biological systems. Biochim Biophys Acta. 2016;1863:2044–2053. doi: 10.1016/j.bbamcr.2016.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanduri C. Long noncoding RNA and epigenomics. Adv Exp Med Biol. 2011;722:174–195. doi: 10.1007/978-1-4614-0332-6_11. [DOI] [PubMed] [Google Scholar]
- 31.Lee E, Wang J, Yumoto K, Jung Y, Cackowski FC, Decker AM, Li Y, Franceschi RT, Pienta KJ, Taichman RS. DNMT1 regulates epithelial-mesenchymal transition and cancer stem cells, which promotes prostate cancer metastasis. Neoplasia. 2016;18:553–566. doi: 10.1016/j.neo.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lizardo MM, Morrow JJ, Miller TE, Hong ES, Ren L, Mendoza A, Halsey CH, Scacheri PC, Helman LJ, Khanna C. Upregulation of glucose-regulated protein 78 in metastatic cancer cells is necessary for lung metastasis progression. Neoplasia. 2016;18:699–710. doi: 10.1016/j.neo.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenza GL. The hypoxic tumor microenvironment: a driving force for breast cancer progression. Biochim Biophys Acta. 2016;1863:382–391. doi: 10.1016/j.bbamcr.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Guo X, Li G, Shi Y, Li L. Long noncoding RNAs as potential biomarkers in gastric cancer: opportunities and challenges. Cancer Lett. 2016;371:62–70. doi: 10.1016/j.canlet.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Mammucari C, Raffaello A, Vecellio Reane D, Rizzuto R. Molecular structure and pathophysiological roles of the mitochondrial calcium uniporter. Biochim Biophys Acta. 2016;1863:2457–2464. doi: 10.1016/j.bbamcr.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Nicholas NS, Apollonio B, Ramsay AG. Tumor microenvironment (TME)–driven immune suppression in B cell malignancy. Biochim Biophys Acta. 2016;1863:471–482. doi: 10.1016/j.bbamcr.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Palmieri F, Monne M. Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim Biophys Acta. 2016;1863:2362–2378. doi: 10.1016/j.bbamcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan M, Jurasz P. The role of platelets in the tumor microenvironment: from solid tumors to leukemia. Biochim Biophys Acta. 2016;1863:392–400. doi: 10.1016/j.bbamcr.2015.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lncRNA Expression Profiles between ZBTB7A Knockdown U2OS Cells and Control Cells after Cisplatin Treatment
mRNA Expression Profiles between LINC00473 Overpressed U2OS Cells and Control Cells after Cisplatin Treatment