Abstract
Paediatric sarcomas and brain tumours, remain cancers of significant unmet need, with a poor prognosis for patients with high risk disease or those who relapse, and significant morbidities from treatment for those that survive using standard treatment approaches. Novel treatment strategies, based on the underlying tumour biology, are needed to improve outcomes. Arginine is a semi-essential amino acid that is imported from the extracellular microenvironment or recycled from intracellular precursors through the combined expression of the enzymes ornithine transcarbamylase (OTC), argininosuccinate synthase (ASS) and argininosuccinate lyase (ASL) enzymes. The failure to express at least one of these recycling enzymes makes cells reliant on extracellular arginine – a state known as arginine auxotrophism. Here we show in large in silico patient cohorts that paediatric sarcomas and brain tumours express predominately the arginine transporter SLC7A1 and the arginine metabolising enzyme Arginase 2 (ARG2), but have low-absent expression of OTC. The arginine metabolic pathway correlated with the expression of genes associated with tumour pathogenesis, and overall survival in paediatric sarcomas. This gene signature of arginine auxotrophism indicates paediatric sarcomas and brain tumours are a viable target for therapeutic arginase drugs under current clinical trial development.
Keywords: paediatric, sarcomas, brain, arginine, auxotrophism
INTRODUCTION
Brain tumours and sarcomas account for around 50% of all paediatric tumours. Despite significant improvements in survival for paediatric cancer, these two groups of tumours have significantly inferior outcomes when compared with leukaemias and solid tumours such as Wilms’ and germ cell tumours [1].
Paediatric sarcomas are composed primarily of 3 major subtypes – osteosarcoma, Ewing's sarcoma, and rhabdomyosarcomas, whilst brain tumours are classified based on site, cell of origin and grade of tumour [2, 3].
For both groups of patients improvements in outcomes over the past 30 years have been achieved through the use of multi-drug cytotoxic chemotherapy, in conjunction with local control through surgery and radiotherapy. For patients that are cured, the morbidities of treatment may be significant not least due to surgical excision of limbs or organ tissue, and the long-term effects of chemotherapy. In particular local control measures for CNS tumours cause significant late effects in growth, endocrine regulation, as well as potential visual and hearing loss and significant neurocognitive and developmental effects [4]. For patients who present with high-risk sarcoma such as alveolar rhabdomyosarcoma, metastatic osteosarcoma, or those who relapse the prognosis remains dismal. Similarly, certain types of CNS tumours, such as embryonal tumours (atypical teratoid rhabdoid tumours, pineoblastoma) and the high grade tumours such as grade III and IV gliomas and medulloblastomas with metastases have an extremely poor prognosis. Diffuse intrinsic pontine gliomas (DIPG) is universally fatal with median survival of 9 months from diagnosis [5].
For both groups of patients, there has been a paucity of new and effective therapeutic strategies, often secondary to the limitations of accessing tumour tissue and modelling the underlying tumour biology. Therefore, there is a strong case for investigating new pathways in these tumours. Recently, targeting tumour amino acid metabolism has become a clinically-relevant therapeutic approach [6]. Arginine is a semi-essential amino acid which plays a key role in protein synthesis, polyamine synthesis, and the cell cycle. Arginine is transported from the extracellular microenvironment via the SLC7A family of transporters and catabolised through the tissue-specific expression of Arginase 1 (ARG1), Arginase 2 (ARG2), or Nitric Oxide Synthetase (iNOS) enzymes. Arginine can also be recycled from the precursors ornithine and citrulline through the intracellular expression of ornithine transcarbamylase (OTC), argininosuccinate synthase (ASS) and argininosuccinate lyase (ASL) enzymes. The failure to express at least one of these recycling enzymes makes cells reliant on extracellular arginine – a state known as arginine auxotrophism. It is increasingly recognised that a number of adult malignancies are deficient in one of more of these arginine-recycling enzymes, making them susceptible to a new class of therapeutic arginine depleting enzymes undergoing clinical investigation. However to date the status of the arginine metabolic pathway has never been reported in paediatric sarcomas and brain tumours despite the unmet clinical need. Using large in silico data sets from human tumours, we demonstrate that paediatric sarcomas and brain tumour subtypes are arginine auxotrophic, supporting the translation of arginine-depletion strategies for these patients.
RESULTS
Key components of arginine metabolism by cells include the ability to transport arginine intracellularly from the microenvironment, catabolise arginine into intermediates for downstream pathways, and the ability to re-synthesise arginine from precursor metabolites. We analysed the expression of the key arginine pathway genes in paediatric sarcomas (127 osteosarcoma samples, 117 Ewing's sarcoma samples, and 147 rhabdomyosarcoma samples) and paediatric CNS tumours (comprising 18 ATRT, 53 high grade glioma, 37 DIPG and 83 ependymoma, 73 medulloblastoma and 182 CNS-PNET) held within the R2: genomics analysis and visualization platform. This represents the largest analysis of paediatric tumours from patients, for arginine pathway alterations described in the literature. Data was revalidated against at least one other independent data set of the same tumour type (Supplementary Figure 1), confirming the same expression profiles.
Expression of arginine pathway genes in paediatric sarcomas and brain tumours
Within osteosarcoma (Figure 1a) we identified that cells have higher expression of the SLC7A1 and SLC7A2 amino acid transporters, compared to the SLC7A3 and SLC7A4 transporters (Figure 1a) consistent with the established role of these isoforms in transporting arginine and other key amino acids such as lysine and ornithine. In osteosarcoma, both ARG1 and ARG2 catabolic enzymes were moderately expressed. No data available for NOS2. Although osteosarcomas expressed ASS, expression of OTC and ASL were lower in comparison. Enzyme expression did not correlate with clinical features of the patient, histological subtype, anatomical location of the primary tumour, or response to chemotherapy.
Figure 1. Paediatric sarcomas have an arginine auxotrophic gene signature.
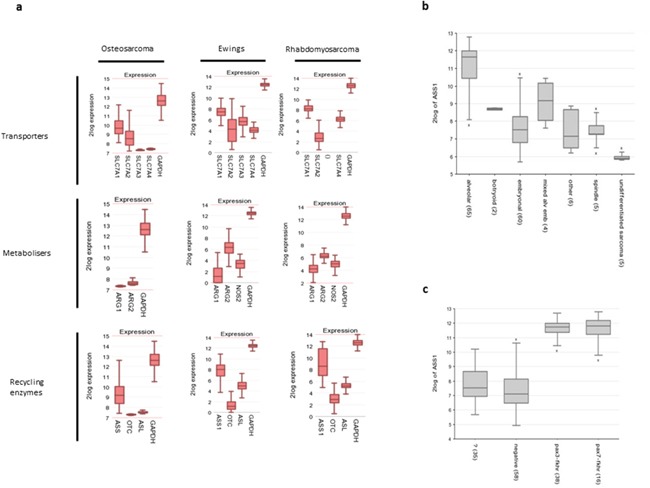
(a) Gene expression profiles of amino acid transporters, arginine catabolic enzymes, and arginine recycling enzymes, compared to GAPDH control. (b) ASS expression in rhabdomyosarcoma histological subgroups (c) ASS expression according to rhabdomyosarcoma cytogenetic status.
For Ewing's sarcoma (Figure 1a) the expression levels of amino acid transporters are more equally distributed although SLC7A1 was again the most predominant. ARG2 expression was the main isoform expressed with low-absent ARG1, and low levels of NOS2. Of the arginine recycling enzymes, OTC expression was low-absent, with higher expression of ASS1 and ASL. Expression of these enzymes did not correlate with clinical features.
In rhabdomyosarcomas (Figure 1a) SLC7A1 and SLC7A4 were strongly expressed, with low SLC7A2 (SLC7A3 not available). Similar to the other sarcomas expression of ARG2 was highest, with modest expression of ARG1 and NOS2 genes. OTC expression was again significantly lower than ASS1, with comparable expression of ASL. Between histological subtypes of tumour ASS expression was significantly different (p=3.6e-19, ANOVA), and highest in the patient group with the worst prognosis - alveolar rhabdomyosarcoma (Figure 1b). ASS1 expression also corresponded with PAX3/PAX7-FKHR expression (p=4.6e-46), consistent with this finding (Figure 1c).
These findings show that these major paediatric sarcoma subtypes have the ability to transport arginine from the microenvironment and metabolise it. However, despite relatively preserved ASS and ASL expression they have a low or absent OTC indicating the potential of arginine auxotrophism.
Within CNS tumours the expression pattern of arginine pathway genes was consistent across all subtypes (Figure 2). SLC7A1 was the highest expressed transporter isoform with ARG2 as the main catabolic enzyme. OTC expression was very low or completely absent throughout all tumour subtypes with relatively preserved ASL and ASS expression. Thus all CNS tumour types share a predisposition to arginine auxotrophism.
Figure 2. Paediatric brain tumours have an arginine auxotrophic gene signature.
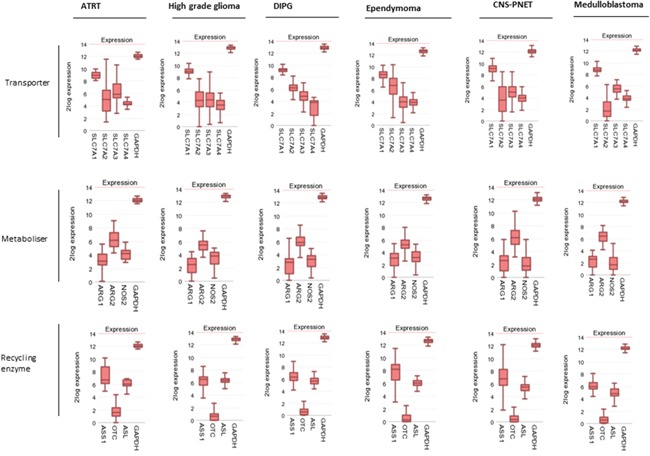
Gene expression profiles of amino acid transporters, arginine catabolic enzymes, and arginine recycling enzymes compared to. GAPDH control.
Impact of arginine pathway genes on overall survival
The analysis above indicates that the major paediatric sarcomas universally express SLC7A1, ARG2, and ASS, but have low OTC expression. We therefore assessed the impact of arginine pathway genes on overall survival in osteosarcoma and Ewing's sarcoma patients (Figure 3). Data on rhabdomyosarcoma was not available.
Figure 3. SLC7A1 and ASS expression correlate with survival in paediatric sarcomas.
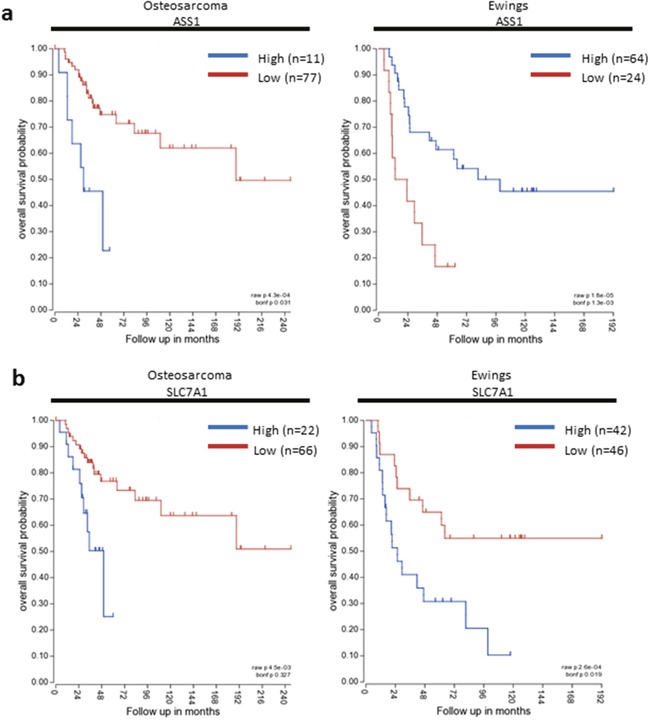
Kaplan-Meier survival curves based on SLC7A1 and ASS expression in osteosarcoma and Ewing's sarcoma. Blue – High gene expression. Red- Low gene expression.
In osteosarcoma, high ASS expression was associated with poorer overall survival probability (p=0.031). No significant difference in overall survival was detected based on expression of SLC7A1, ARG2 or OTC. In contrast for Ewing's sarcoma, low ASS expression was associated with poorer overall survival (p=0.0013), and high SLC7A1 expression was associated with poorer overall survival (p=0.019). Expression of ARG2 and OTC did not significantly correlate with overall survival. Differences in relative gene expression on survival for each tumour, is an indicator of the underlying varied tumour biology and the impact of other interacting cellular pathways.
Survival data was limited in the CNS tumours examined. Only survival data for high grade glioma was available of all the datasets used. There was no significant difference in overall survival in high grade glioma with SLC7A1, ARG2, ASS or OTC expression.
The arginine auxotrophic signature correlates with other intracellular targets for novel agents
Gene set enrichment was carried out for all sarcoma and CNS tumour subtypes, using pre-ranked gene lists correlating with ARG2 and OTC (Supplementary Figure 2). In all tumour types studied, except glioma, ARG2 and OTC ranked gene lists significantly correlated with KRAS signaling. MTOR signaling, a known downstream effector of amino acid metabolism and a target for novel agents in relapsed paediatric tumours, was enriched within osteosarcoma for both OTC and ARG2 (Supplementary Figure 2a). In medulloblastoma MYC (associated with a poor prognosis) had a positive correlation with FDR <0.05, for both the OTC and ARG2 oncogenic and hallmark gene lists (Supplementary Figure 2i). In ependymoma EGFR upregulation, a target of monoclonal antibodies and small molecule inhibitors, correlated with ARG2 (FDR 0.000) (Supplementary Figure 2g). ERB2 upregulation, targeted recently with small molecule inhibitors, also correlated with the OTC ranked gene list (FDR 0.000) for DIPG (Supplementary Figure 2f).
The interplay of the immune microenvironment and arginine metabolism as a contributor to tumour pathogenesis was also highlighted from our analysis. The continuing growth of immunotherapeutics makes targeting these pathways with anti-cytokine monoclonal antibodies of interest. In ependymomas and DIPG IL-6-JAK/STAT, IFN-γ, and TNF-α signaling were enriched (Supplementary Figure 2f and 2g). The NF-KB complex, a key player in immune and inflammatory signaling, was significantly enriched in both the hallmark and oncogenic data sets for gliomas, ependymomas, ATRT and Ewing's sarcoma. The above findings show that in both paediatric sarcoma and brain tumours, arginine metabolism influences a number of key pathways in cell and tumour development, and could stimulate novel therapeutic approaches in combination with arginine depletion therapy.
DISCUSSION
Cancer cell arginine auxotrophism is determined by 3 core cellular components: the ability to transport arginine from the extracellular microenvironment into the cell; the expression of arginine catabolising enzymes; and the absence of arginine recycling pathway enzymes [6]. Arginine is transported from the extracellular microenvironment into the cell cytoplasm predominantly through the system+ family of cationic amino acid transporters (SLC7A1, SLC7A2, SLC7A3, SLC7A4). 3 enzymes are principally involved in the catabolism of arginine. Arginase 1 (ARG1) and arginase 2 (ARG2) catabolise arginine in ornithine and urea, whilst nitric oxide synthetase (NOS2) converts arginine into citrulline and nitric oxide. In the majority of non-malignant cells arginine breakdown products are recycled back to arginine, through the expression of Ornithine transcarbamylase (OTC; converting ornithine into citrulline), Argininosuccinate Synthetase (ASS; converting citrulline into argininosuccinate), and finally Argininosuccinate Lyase (ASL; converting argininosuccinate back to arginine) (Supplementary Figure 3).
Here we identify, in large cohorts of paediatric tumours, that the arginine transporter SLC7A1 is highly expressed. ARG2 was the main catabolic enzyme found within the tissues, with lower expression of ARGI and iNOS enzymes. Although ASS was universally expressed, OTC was significantly underexpressed in all three subgroups of sarcomas and absent of very low in all CNS tumours. To date the status of the arginine metabolic pathway in human samples from paediatric sarcomas and brain tumours has never been reported.
The SLC7A family of transporters have been described in both non-malignant and adult malignant cells as the principle method of cellular arginine uptake from the microenvironment. We have shown that SLC7A1 is predominant in both adult and paediatric Acute Myeloid Leukaemia blasts which are dependent on arginine for survival [25]. Modulation of SLC7A1 expression in colorectal carcinoma and breast cancer cells decreases arginine uptake and reduces their proliferation and survival [26]. As SLC7A1 has an upstream open reading frame (uOR) in its proximal promoter which can be translated under conditions of arginine depletion, it suggests that this transporter is the cellular response to arginine concentrations in the microenvironment [27, 28]. Although the expression of other SLC7a family members has been reported to play a role in cancer associated myeloid-derived suppressor cells and in non-small cell lung cancer radiation resistance little is known about the unique functions of each SCL7A family member in the cancer pathogenesis, in particular in the paediatric setting [29, 30].
The paediatric tumours examined here show a predominance for Arginase II expression, over Arginase I and INOS. Both Arginase I and II catalyse the conversion of arginine into urea and ornithine, although they are reported to be localised in the cytoplasm and mitochondria respectively. The over-expression of arginase enzymes has been reported in adult soft tissue sarcomas with no correlation to histopathological parameters [31]. Hypoxia can drive Arginase II dependent proliferation of Osteosarcoma cell lines [32]. However the expression and function of Arginase enzymes in the malignant cells of human paediatric sarcomas and brain tumours has never been described to date. Thus the specific contribution of each of these enzymes in promoting tumour cell proliferation is still unclear although it is well established that a number of downstream pathways are dependent on arginine metabolism – such as mTOR regulation, ATF2 dependent regulation of cell cycle in leukaemia, and polyamine synthesis in neuroblastoma [33–55]. We have previously shown that Arginase II, is the main arginine catabolic enzyme in AML blasts and neuroblastoma tumour cells, and that its expression contributes to the creation of an immunosuppressive microenvironment [36, 37]. Indeed these tumour cells were found to be low or absent in Arginase I or iNOS enzymes. In contrast we have shown that cancer-induced myeloid-derived suppressor cells (MDSCs) can overexpress Arginase I and iNOS enzymes, to impair T cell proliferation and activation [38, 39]. Murine models of paediatric sarcomas, gliomas, and glioblastomas have been shown to induce Arginase 1+/iNOS+ MDSCs [40–43]. Our data supports that the tumour microenvironment in paediatric sarcomas and brain tumours is likely to be arginine deplete, and thus a suppressive environment for anti-cancer immunity.
The central tenant of arginine auxotrophy is that tumour cells have an incomplete complement of arginine resynthesis enzymes. Ornithine, produced by arginine breakdown, is converted first into citrulline through a reaction with carbamoyl phosphate by OTC. Here we show that paediatric sarcomas and brain tumours have low to absent expression of this enzyme, thus breaking the recycling pathway. In AML we showed that OTC has a range of expression in both adult and paediatric blasts. However in a range of adult solid cancer cell lines (keratinoctic carcinoma, lung adenocarcinoma, hepatocellular carcinoma and breast carcinoma) OTC was undetectable consistent with our findings here [44, 45]. An absence of OTC may suggest that tumours use ornithine for polyamine synthesis and not arginine regenertation [46]. Our data supports previous findings that no ASL negative tumours have been identified. The function of ASL can be regulated by fumarate hydratase expression in renal cell carcinoma and by epigenetic modulation in glioblastoma [47].
A number of genes correlated with the expression of arginase enzymes in paediatric tumours, in particular related to transcription factors and intracellular signal transduction. Although no single pathway was prevalent, corresponding with the different underlying biology of these tumours, these findings do suggest arginine metabolism has a wide influence on cellular functions. The most significant signalling hallmarks and genes enriched in the gene set enrichment analysis for the paeditatric tumours analysed here, was KRAS and KRAS signalling pathway. The pathway plays an established role in cell proliferation and differentiation. A number of human genetic syndromes have emerged that are caused by germline mutations in components of the Ras/mitogen activated protein kinase (MAPK) pathway, and are associated with an increased risk of developing cancer [48]. Transgenic expression of KRAS in neural stem and progenitor cells induces brain tumorigenesis in a zebra fish model [49]. Dysregulated KRAS signaling has also been implicated in rhabdomyosarcoma and Ewing's sarcoma pathogenesis [50, 51].
As such targeting arginine metabolism could provide a new therapeutic approach for these childhood cancers of significant unmet need. Until recently, drugs targeting arginine metabolism in a clinically relevant manner did not exist. Small molecules which inhibit arginine transport or arginase enzymes are only available for in vitro research use (NG-hydroxy-L-arginine: NOHA and L-NG-monomethyl arginine: L-NMMA) and suffer from a number of issues. Inhibitors of SLC7 transporters are likely to lead to toxicity from inhibiting uptake of other amino acids by non-malignant tissues as well as the tumour cells, and due to the structural homology between different SLC7 family members. The arginase I and iNOS enzyme inhibitors are commonly arginine analogues which are reversible in binding to the enzymes, and due to the close interplay between iNOS and Arginase pathways require concurrent administration in order to fully inhibit cellular use of arginine. New arginase I inhibitors, such as CB-1158 (Calithera, Inc) are at various stages of preclinical and early clinical development (NCT02903914).
The most clinical relevant approach to targeting tumour arginine metabolism is through therapeutic arginine depletion. Normal human arginase has limited clinical value because of a short plasma-half life. We have shown that the addition of a 5000 MW polyethylene glycol molecule (PEG) significantly increases the plasma half-life of arginase I with minimal loss of enzyme activity. BCT-100 is a pegylated recombinant human arginase which we have demonstrated to have significant pre-clinical activity against leukaemias, hepatocellular carcinoma, melanoma, and prostate cancer [52, 53]. Phase I and II clinical trials have been completed in adults with relapsed/refractory hepatocellular carcinoma, demonstrating that BCT-100 can lead to a sustained depletion of plasma arginine to undetectable levels (<8uM) for 5 days or more with weekly dosing [54, 55]. Patients experienced a significant improvement in overall survival with an excellent toxicity profile. Pegyated mycoplasma derived arginine demininase (PEG-ADI) is an alternative strategy that catabolises arginine into citrulline and ammonia [56]. The immunogenicity of the molecule has significantly limited sustained arginine depletion and anti-tumour efficacy of this molecule.
Our findings suggest future clinical studies could rationally include a combination of arginase therapy in combination with molecules targeting other intersecting intracellular pathways. A number of early phase clinical trials are ongoing in the field of relapsed paediatric malignancies, including those targeting the ERBB2, EGFR, MTOR, and Aurora Kinase pathways, we identified as correlating with arginine metabolism [57–59]. Sarcomas remain a disease of significant unmet clinical need, with a lack of new agents in upfront or relapsed treatment protocols. In vitro cell line studies have shown that paediatric sarcomas including osteosarcoma, Ewing's sarcoma, synovial sarcoma and malignant peripheral nerves sheath tumours, overexpress arginine metabolising enzymes consistent with a dependence on arginine for growth. Resistance to doxorubicin, one of the principal chemotherapy agents used in sarcoma treatment, is associated with ASS1 downregulation, suggesting relapsed sarcomas are more likely to be arginine auxotrophic [60]. Immunohistochemical analysis of sarcomas shows they may have reduced or absent ASS1 expression. Arginine deprivation induces cell death of osteosarcoma, Ewing's sarcoma, and synovial sarcoma cell lines by cell cycle arrest and subsequent autophagy or necroptosis in vitro, and inhibition of tumour growth in murine xenografts [61, 62]. For High Grade Gliomas disease representative preclinical models are still in early stages of development, contributing to a paucity of new agents for this group of patients. The immediate downstream metabolite of arginine - polyamine, plays a critical role in medulloblastoma and glioblastoma cell pathogenesis [63, 64]. Glioblastoma cells have been shown to undergo alterations in invasiveness in low arginine conditions [65]. Cell lines undergo caspase-independent cell death in vitro and have a significant reduction in in xenograft growth when treated with arginine depleting enzymes [66, 67].
Our study is limited to the patient cohorts within the R2: Genomics Analysis and Visualization platform and generates a number of hypotheses of the role of arginine in tumour cells. Although this provides data on large datasets from primary patient samples, further in vitro and in vivo studies is necessary to further investigate how arginine metabolism can be functionally linked to other biological pathways and the impact of therapeutic approaches. Overall this study highlights an arginine auxotrophic signature in paediatric sarcomas and brain tumours and provides an important rationale for investigating clinically relevant arginine depletion strategies in these children.
MATERIALS AND METHODS
Arginine pathway gene expression profiles
Using the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl), we analysed the expression of argininosuccinate synthetase 1 (ASS1); ornithine transcarbamylase (OTC); argininosuccinate lyase (ASL), nitric oxide synthase 2 (NOS2), Arginase 1 and 2 (ARG1 and ARG2) isoforms and the solute carrier family/cationic amino acid transporters SLC7A1, SLC7A2, SLC7A3, and SLC7A4, and the housekeeping gene GAPDH in the range of published sarcoma and CNS tumour datasets as described in Table 1 [8–24]. Arginine auxotrophy was defined as the low or absent expression of ASS alone or in combination with low or absent expression of OTC. The relative expression of the genes are shown, including GAPDH and TUBB as housekeeping genes.
Table 1. Table of patient cohorts analysed.
| Tumour type | Database name | Data features and Source Pubmed reference |
|---|---|---|
| DIPG | Glioma DIPG - Paugh - 37 - MAS5.0 - u133p2 | n= 43 Affymetrix Human Genome U133 Plus 2.0 arrays. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3209696/ [8] |
| Glioma | Glioma - French - 284 - MAS5.0 - u133p2 Glioma pediatric - Paugh - 53 - MAS5.0 - u133p2 | n=28 Affymetrix https://www.ncbi.nlm.nih.gov/pubmed/19920198 [9] n=53 Affymetrix Human Genome U133 Plus 2.0 arrays https://www.ncbi.nlm.nih.gov/pubmed?term=20479398 [10] |
| ATRT | ATRT - Kool - 49 - MAS5.0 - u133p2 ATRT - Birks - 18 - MAS5.0 - u133p2 | n=49 Affymetrix GeneChip Human Genome U133. https://www.ncbi.nlm.nih.gov/pubmed/26923874 [11] n=18 Affymetrix HG-U133 Plus 2 GeneChip microarrays. https://www.ncbi.nlm.nih.gov/pubmed?term=21946044 [12] |
| Ependymoma | Ependymoma - Donson - 19 - MAS5.0 - u133p2 Ependymoma - Gilbertson - 83 - MAS5.0 - u133p2 | n=19 Affymetrix HG-U133 Plus 2 microarray chips https://www.ncbi.nlm.nih.gov/pubmed?term=19917695 [13] n=83 Affymetrix U133 Plusv2 and Agilent miRNA arrays https://www.ncbi.nlm.nih.gov/pubmed?term=20639864 [14] |
| PNET | CNS-PNET - Kool - 182 - MAS5.0 - u133p2 CNS/PNET - Grundy - 24 - MAS5.0 - u133p2 | n=182 Affymetrix GeneChip Human Genome U133 Plus 2.0 Array. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5139621/ [15] n=24 Affymetrix U133 plus 2.0 arrays. https://www.ncbi.nlm.nih.gov/pubmed/21798848 [16] |
| Medulloblastoma | Medulloblastoma - Thompson - 46 - MAS5.0 - u133a Medulloblastoma PLoS One - Kool - 62 - MAS5.0 - u133p2 | n=46 U133av2 Affymetrix oligonucleotide array. https://www.ncbi.nlm.nih.gov/pubmed/16567768 [17] n=62 Affymetrix HG-U133 plus 2.0. https://www.ncbi.nlm.nih.gov/pubmed?term=18769486 [18] |
| Osteosarcoma | Osteosarcoma - Kobayashi - 27 - MAS5.0 - u133p2 Osteosarcoma - Buddingh - 53 - vst - ilmnhwg6v2 Osteosarcoma - Kuijjer - 127 - vst - ilmnhwg6v2 | n=27 Affymetrix Human Genome U133 Plus 2.0 array http://www.ncbi.nlm.nih.gov/pubmed?term=20159990 [19] n=53 Illumina Human-6 v2.0 Expression BeadChips http://www.ncbi.nlm.nih.gov/pubmed?term=21372215 [20] n=84 Illumina Human-6 v2.0 arrays/ http://www.ncbi.nlm.nih.gov/pubmed?term=22454324 [21] |
| Ewings sarcoma | Ewing Sarcoma - Delattre - 117 - MAS5.0 - u133p2 Ewing Sarcoma - Francesconi - 37 - MAS5.0 - u133p2 | n=117 Affymetrix hgu133Plus2 arrays. http://www.ncbi.nlm.nih.gov/pubmed?term=22327514 [22] n=37 Affymetrix https://www.ncbi.nlm.nih.gov/pubmed/19307502 [23] |
| Rhabdomyosarcoma | Rhabdomyosarcoma - Davicioni - 147 - MAS5.0 - u133a | n=147 Affymetrix GeneChip Human U133A http://www.ncbi.nlm.nih.gov/pubmed?term=16849537 [24] |
Databases used in the analysis are included in Table 1. To ensure there was no selection bias we validated findings using other databases with the same tumour type in the R2 visualisation platform. According the primary literature for each study Affymetrix and Illumina technologies were used and sample library preparation, hybridization, and quality control were performed according to manufacturer's protocol.
Kaplan-Meier survival plots
Kaplan-Meier overall survival plots were generated using the R2: Kaplan Meier tool for the key SLC7A1, OTC, ASS and ARG2 genes, where data was available. Bonferroni corrected p-values were used.
Gene set enrichment analysis
Gene set enrichment analysis was carried out, using ranked gene lists generated from the R2 platform (Table 1). Genes correlating with ARG2 and OTC were retrieved and ranked according to the correlation p-coefficient., An initial threshold of p<2 was used to generate the ranked genes list before enrichment analysis was carried out using Broad Institute software (http://software.broadinstitute.org/gsea/index.jsp). We used datasets from MsigDB (http://software.broadinstitute.org/gsea/msigdb/index.jsp) including ‘hallmark’ and ‘oncogenic signatures’. Oncogenic signatures were generated from microarray gene expression data from cancer gene perturbations. Hallmark gene sets were coherently expressed signatures derived by aggregating many MSigDB gene sets to represent well-defined biological states or processes. We used 1000 permutations with weighted enrichment statistics in the analysis. Hallmarks and oncogenic genes were selected on the basis of the 5 most significant or those with an FDR q value less than 25% (0.25).
SUPPLEMENTARY MATERIALS FIGURES
Acknowledgments
We thank G. Papatzikas (University of Birmingham Centre for Computational Biology), N Jude Davies (Institute for Immunology and Immunotherapy) and A Beggs (Institute for Cancer and Genomics) at the University of Birmingham for their assistance with the genomic analyses in the study. The work was supported by the Amber Phillpott Trust, Niayah's Fund, Team Isobel, the Birmingham Children's Hospital Research Fund, Treating Children with Cancer, and Cancer Research UK.
Abbreviations
- ARG1
arginase 1
- ARG2
arginase 2
- ASL
argininosuccinate lyase
- ASS
argininosuccinate synthase
- DIPG
diffuse intrinsic pontine gliomas
- MDSC
myeloid-derived suppressor cells
- iNOS
nitric oxide synthetase
- OTC
ornithine transcarbamylase
Footnotes
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
FUNDING
We are grateful for funding from Cancer Research UK, Amber Phillpott Trust, Treating Children with Cancer, Niayah's Fund, Birmingham Children's Hospital Research Fund and Neuroblastoma UK, for supporting the work presented in this manuscript.
Author contributions
Contribution: F.M. and C.D.S designed and led the study, performed research, analysed data and wrote the manuscript. A.V, D.C, M.D, P.C designed and performed research and reviewed the manuscript.
REFERENCES
- 1.Gatta G, Botta L, Rossi S, Aareleid T, Bielska-Lasota M, Clavel J, Dimitrova N, Jakab Z, Kaatsch P, Lacour B, Mallone S, Marcos-Gragera R, Minicozzi P, et al. Childhood cancer survival in Europe 1999-2007: results of EUROCARE-5–a population-based study. Lancet Oncol. 2014;15:35–47. doi: 10.1016/S1470-2045(13)70548-5. [DOI] [PubMed] [Google Scholar]
- 2.Johnson KJ, Cullen J, Barnholtz-Sloan JS, Ostrom QT, Langer CE, Turner MC, McKean-Cowdin R, Fisher JL, Lupo PJ, Partap S, Schwartzbaum JA, Scheurer ME. Childhood brain tumor epidemiology: a brain tumor epidemiology consortium review. Cancer Epidemiol Biomarkers Prev. 2014;23:2716–2736. doi: 10.1158/1055-9965.EPI-14-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alaggio R, Coffin CM. The evolution of Pediatric Soft Tissue Sarcoma Classification in the LAst 50 Years. Pediatr Dev Pathol. 2015;18:481–94. doi: 10.2350/15-07-1666-MISC.1. [DOI] [PubMed] [Google Scholar]
- 4.Fidler MM, Reulen RC, Winter DL, Kelly J, Jenkinson HC, Skinner R, Frobisher C, Hawkins MM. Long term cause specific mortalit among 34489 five year survivors of childhood cancer in Great Britain: population based cohort study. BMJ. 2016;354:i4351. doi: 10.1136/bmj.i4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol. 2006;7:241–248. doi: 10.1016/S1470-2045(06)70615-5. [DOI] [PubMed] [Google Scholar]
- 6.Fultang L, Vardon A, De Santo C, Mussai F. Molecular basis and current strategies of therapeutic arginine depletion for cancer. Int J Cancer. 2016;139:501–509. doi: 10.1002/ijc.30051. [DOI] [PubMed] [Google Scholar]
- 7.R2: microarray analysis and visualization platform’. http://r2.amc.nl
- 8.Paugh BS, Broniscer A, Qu C, Miller CP, Zhang J, Tatevossian RG, Olson JM, Geyer JR, Chi SN, da Silva NS, Onar-Thomas A, Baker JN, Gajjar A, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gravendeel LA, Kouwenhoven MC, Gevaert O, de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB, Kloosterhof NK, De Moor B, Eilers PH, van der Spek PJ, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 10.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, Bax DA, Coyle B, Barrow J, Hargrave D, Lowe J, Gajjar A, Zhao W, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johann PD, Erkek S, Zapatka M, Kerl K, Buchhalter I, Hovestadt V, Jones DT, Sturm D, Hermann C, Segura Wang M, Korshunov A, Rhyzova M, Gröbner S, et al. Atypical teratoid/rhabdoid tumors are comprised of three epigenetic subgroups with distinct enhancer landscapes. Cancer Cell. 2016;29:379–393. doi: 10.1016/j.ccell.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Birks DK, Donson AM, Patel PR, Dunham C, Muscat A, Algar EM, Ashley DM, Kleinschmidt-Demasters BK, Vibhakar R, Handler MH, Foreman NK. High expression of BMP pathway genes distinguishes a subset of atypical teratoid/rhabdoid tumors associated with shorter survival. Neuro Oncol. 2011;13:1296–1307. doi: 10.1093/neuonc/nor140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donson AM, Birks DK, Barton VN, Wei Q, Kleinschmidt-Demasters BK, Handler MH, Waziri AE, Wang M, Foreman NK. Immune gene and cell enrichment is associated with a good prognosis in ependymoma. J Immunol. 2009;183:7428–7440. doi: 10.4049/jimmunol.0902811. [DOI] [PubMed] [Google Scholar]
- 14.Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, Rand V, Leary SE, White E, Eden C, Hogg T, Northcott P, Mack S, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, Ryzhova M, Koelsche C, Pfaff E, Allen SJ, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller S, Rogers HA, Lyon P, Rand V, Adamowicz-Brice M, Clifford SC, Hayden JT, Dyer S, Pfister S, Korshunov A, Brundler MA, Lowe J, Coyle B, et al. Genome-wide molecular characterization of central nervous system primitive neuroectodermal tumor and pineoblastoma. Neuro Oncol. 2011;13:866–879. doi: 10.1093/neuonc/nor070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, Taylor MD, Curran T, Gajjar A. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 18.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, Mrsić A, Ylstra B, Grajkowska W. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;28(3):e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N, Morioka H, Toyama Y, Hirohashi S, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535–44. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 20.Buddingh EP, Kuijjer ML, Duim RA, Bürger H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PC, Lankester AC, Cleton-Jansen AM. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17:2110–19. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 21.Kuijjer ML, Rydbeck H, Kresse SH, Buddingh EP, Lid AB, Roelofs H, Bürger H, Myklebost O, Hogendoorn PC, Meza-Zepeda LA, Cleton-Jansen AM. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer. 2012;51:696–706. doi: 10.1002/gcc.21956. [DOI] [PubMed] [Google Scholar]
- 22.Postel-Vinay S, Véron AS, Tirode F, Pierron G, Reynaud S, Kovar H, Oberlin O, Lapouble E, Ballet S, Lucchesi C, Kontny U, González-Neira A, Picci P. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44:323–7. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 23.Scotlandi K, Remondini D, Castellani G, Manara MC, Nardi F, Cantiani L, Francesconi M, Mercuri M, Caccuri AM, Serra M, Knuutila S, Picci P. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J Clin Oncol. 2009;27:2209–16. doi: 10.1200/JCO.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- 24.Davicioni E1, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–46. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 25.Mussai F, Egan S, Higginbotham-Jones J, Perry T, Beggs A, Odintsova E, Loke J, Pratt G, U KP, Lo A, Ng M, Kearns P, Cheng P, De Santo C. Arginine dependence of acute myeloid leukemia blast proliferation: a novel therapeutic target. Blood. 2015;125:2386–96. doi: 10.1182/blood-2014-09-600643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Wang W, Wang J, Yang C, Mao H, Fu X, Wu Y, Cai J, Han J, Xu Z, Zhuang Z, Liu Z, Hu H, Chen B. Overexpression of arginine transporter CAT-1 is associated with accumulation of L-arginine and cell growth in human colorectal cancer tissue. PLoS One. 2013;8:e73866. doi: 10.1371/journal.pone.0073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez J, Lopez AB, Wang C, Mishra R, Zhou L, Yaman I, Snider MD, Hatzoglou M. Transcriptional control of the argrinine/lysine transporter, cat-1, by physiological stress. J Biol Chem. 2003;278:50000–9. doi: 10.1074/jbc.M305903200. [DOI] [PubMed] [Google Scholar]
- 28.Lopez Ab, Wang C, Huang CC, Yaman I, Li Y, Chakravarty K, Johnson PF, Chiang CM, Snider MD, Wek RC, Hatzoglou M. A feedback transcriptional mechanism controls the levels of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem J. 2007;402:163–73. doi: 10.1042/BJ20060941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cimen Bozkus C, Elzey BD, Crist SA, Ellies LG, Ratliff TL. Expression of cationic amino acid transporter 2 is required for myeloid-derived suppressor cell-mediated control of T cell immunity. J Immunol. 2015:5237–50. doi: 10.4049/jimmunol.1500959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie L, Song X, Yu J, Guo W, Wei L, Liu Y, Wang X. Solute carrier protein family may involve in radiation-induced radioresistance of non-small cell lung cancer. J Cancer Res Clin Oncol. 2011;137:1739–47. doi: 10.1007/s00432-011-1050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan X, Takahara M, Xie L, Gondo C, Setsu N, Oda Y, Takeuchi S, Tu Y, Moroi Y, Furue M. Arginine metabolism in soft tissue sarcoma. J Dermatol Sci. 2011;61:211–5. doi: 10.1016/j.jdermsci.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Setty BA, Jin Y, Houghton PJ, Yeager ND, Gross TG, Nelin LD. Hypoxic prolifration of osteosarcoma cells depends on arginase II. Cell Physiol Biochem. 2016;39:802–13. doi: 10.1159/000447790. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, Wang T, Bar-Peled L, Zoncu R, et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–94. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An JJ, Shi KJ, Wei W, Hua FY, Ci YL, Jiang Q, Li F, Wu P, Hui KY, Yang Y, Xu CM. The ROS/JNK/ ATF2 pathway mediates selenite-induced leukemia NB4 cell cycle arrest and apoptosis in vitro and in vivo. Cell Death Dis. 2013;4:e973. doi: 10.1038/cddis.2013.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evageliou NF, Haber M, Vu A, Laetsch TW, Murray J, Gamble LD, Cheng NC, Liu K, Reese M, Corrigan KA, Ziegler DS, Webber H, Hayes CS, et al. Polyamine antagonist therapies inhibit neuroblastoma initiation and progression. Clin Cancer Res. 2016;22:4391–404. doi: 10.1158/1078-0432.CCR-15-2539. [DOI] [PubMed] [Google Scholar]
- 36.Mussai F, De Santo C, Abu-Dayyeh I, Booth S, Quek L, McEwen-Smith RM, Qureshi A, Dazzi F, Vyas P, Cerundolo V. Acute myeloid leukemia creates an arginase-dependent immunosuppressive microenvironment. Blood. 2013;122:749–58. doi: 10.1182/blood-2013-01-480129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mussai F, Egan S, Hunter S, Webber H, Fisher J, Wheat R, McConville C, Sbirkov Y, Wheeler K, Bendle G, Petrie K, Anderson J, Chesler L, De Santo C. Neuroblastoma arginase activity creates and immunosuppressive microenvironment that impairs autologous and engineered immunity. Cancer Res. 2015;75:3043–53. doi: 10.1158/0008-5472.CAN-14-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Santo C, Arscott R, Booth S, Karydis I, Jones M, Asher R, et al. Invariant NKT cells modulate the suppressive activity of Il-10-escreting neutrophils differentiated with serum amyloid A. Nat Immunol. 2010;11:1039–46. doi: 10.1038/ni.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Santo C, Serafini P, Marigo I, Dolcetti L, Bolla M, Del Soldato P, Melani C, Guiducci C, Colombo MP, Iezzi M, Musiani P, Zanovello P, Bronte V. Nitroaspirin corrects immune dysfunction in tumor-bearing hosts and promotes tumor eradication by cancer vaccination. Proc Natl Acad Sci U S A. 2005;102:4185–90. doi: 10.1073/pnas.0409783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long AH, Highfill SL, Cui Y, Smith JP, Walker AJ, Ramakrishna S, El-Etriby R, Galli S, Tsokos MG, Orentas RJ, Mackall CL. Reduction of MDSCs with all-trans-retinoic acid improves CAR therapy efficacy for sarcomas. Cancer Immunol Res. 2016;4:869–80. doi: 10.1158/2326-6066.CIR-15-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, Kaplan RN, Mackall CL. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med. 2014;6:237ra67. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohanbash G, McKaveney K, Skakaki M, Ueda R, Mintz AH, Amankulor N, Fujita M, Ohlfest JR, Okada H. GM-CSF promotes the immunosuppressive activity of glioma-infiltrating myeloid cells through interlukin-4 receptor-α. Cancer Res. 2013;3:6413–23. doi: 10.1158/0008-5472.CAN-12-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otvos B, Silver DJ, Mulkearns-Hubert EE, Alvarado AG, Turaga SM, Sorensen MD, Rayman P, Flavahan WA, Hale JS, Stoltz K, Sinyuk M, Wu Q, Jarrar A. Cancer stem cell-secreted macrophages migration inhibitory factor stimulates myeloid derived suppressor cell function and facilitates glioblastoma immune evasion. Stem Cells. 2016;34:2026–39. doi: 10.1002/stem.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bobak YP, Vynnytska BO, Kurlishchuk YV, Sibirny AA, Stasyk OV. Cancer cell sensitivity to arginine deprivation in vitro is not determined by endogenous levels of arginine metabolic enzymes. Cell Biol Int. 2010;34:1085–9. doi: 10.1042/CBI20100451. [DOI] [PubMed] [Google Scholar]
- 45.Selamnia M, Robert V, Mayeru C, Delpal S, Blachier F. De novo synthesis of arginine and ornithine from citrulline in human colon carcinoma cells: meabolic fate of L-ornithine. Biochim Biophys Acta. 1998;1425:93–102. doi: 10.1016/s0304-4165(98)00056-7. [DOI] [PubMed] [Google Scholar]
- 46.Zheng L, Mackenzie ED, Karim SA, Hedley A, Blyth K, Kalna G, Watson DG, Szlosarek P, Frezza C, Gottlieb E. Reversed argininosuccinae lyase activity in fumarate hydratase-deficient cancer cells. Cancer Metabo. 2013;1:12. doi: 10.1186/2049-3002-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Syed N, Langer J, Janczar K, Singh P, Lo Nigro C, Lattanzio L, Coley HM, Hatzimichael E, Bomalaski J, Szlosarek P, Awad M, O'Neil K, Roncaroli F, Crook T. Epigenetic status of argininosuccinate synthetase and argininosuccinate lyase modulates autophagy and cell death in glioblasoma. Cell Death Dis. 2013;4:e458. doi: 10.1038/cddis.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–6. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ju B, Chen W, Orr BA, Spitsbergen JM, Jia S, Eden CJ, Henson HE, Taylor MR. Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol Cancer. 2015;14:18. doi: 10.1186/s12943-015-0288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinelli S, McDowell HP, Vigne SD, Kokai G, Uccini S, Tartaglia M, Dominici C. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer. 2009;48:975–82. doi: 10.1002/gcc.20702. [DOI] [PubMed] [Google Scholar]
- 51.Jiang Y, Subbiah V, Janku F, Ludwig JA, Naing A, Benjamin RS, Brown RE, Anderson P, Kurzrock R. Novel secondary somatic mutations in Ewing's sarcoma and desmoplastic small round cell tumors. PLoS One. 2014;9:e93676. doi: 10.1371/journal.pone.0093676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lam TL, Wong GK, Chow HY, Cong HC, Chow TL, Kwok SY, Cheng PN, Wheatley DN, Lo WH, Leung YC. Recombinant human arginase inhibits the in vitro and in vivo proliferation of human melanoma by inducing cell cycle arrest and apoptosis. Pigment Cell Melanoma Res. 2011;24:366–76. doi: 10.1111/j.1755-148X.2010.00798.x. [DOI] [PubMed] [Google Scholar]
- 53.Hsueh EC, Knebel SM, Lo WH, Leung YC, Cheng PN, Sueh CT. Deprivation of arginine by recombinant human arginase in prostate cancer cells. J Hematol Oncol. 2012;5:17. doi: 10.1186/1756-8722-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yau T, Cheng PN, Chan P, Chan W, Chen L, Yuen J, Pang R, Fan ST, Poon RT. A phase 1 dose-escalating study of pegylated recombinant human arginase 1 in patients with advanced hepatocellular carcinoma. Invest New Drugs. 2013;31:99–107. doi: 10.1007/s10637-012-9807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yau T, Cheng PN, Chan P, Chen L, Yuen J, Pang R, Fan ST, Wheatley DN, Poon RT. Preliminary efficacy, safety, pharmacokinetics, pharmacodynamics and quality of life study of pegylated recombinant human arginase 2 in patients with advanced hepatocellular carcinoma. Invest New Drugs. 2015:496–504. doi: 10.1007/s10637-014-0200-8. [DOI] [PubMed] [Google Scholar]
- 56.Tomlinson BK, Thomson JA, Bomalaski JS, Diaz M, Akande T, Mahaffey N, Li T, Dutia MP, Kelly K, Gong IY, Semrad T, Gandara DR, Pan CX, Lara PN., Jr Phase I trial of arginine deprivation therapy with ADI-PEG 20 plus docetaxel in patients with advanced malignant solid tumours. Clin Cancer Res. 2015;21:2480–6. doi: 10.1158/1078-0432.CCR-14-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markant SL, Esparza LA, Sun J, Barton KL, McCoig LM, Grant GA, Crawford JR, Levy ML, Northcott PA, Shih D, Remke M, Taylor MD, Wechsler-Reya RJ. Targeting sonic hedgehog-associated medulloblastoma through inhibition of Aurora and Polo-like kinases. Cancer Res. 2015;73:6310–22. doi: 10.1158/0008-5472.CAN-12-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bautista F, Van den Lugt J, Kearns PR, Mussai FJ, Zwaan CM, Moreno L. The development of targeted new agents to improve the outcome for children with leukemia. Expert Opin Drug Discov. 2016;11:1111–1122. doi: 10.1080/17460441.2016.1237939. [DOI] [PubMed] [Google Scholar]
- 59.Saletta F, Wadham C, Ziegler DS, Marshall GM, Haber M, McCowage G, Norris MD, Byrne JA. Moleuclar profiling of childhood cancer: Biomarkers and novel therapies. BBA Clin. 2014;1:59–77. doi: 10.1016/j.bbacli.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N, Morioka H, Toyama Y, Hirohashi S, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastaseis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535–44. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 61.Kim Y, Kobayashi E, Kubota D, Suehara Y, Mukaihara K, Akaike K, Ito A, Kaneko K, Chuman H, Kawai A, Kitano S. Reduced argininosuccinate synthetase expression in refractory sarcomas: impacts on therapeutic potential and drug resistance. Oncotarget. 2016;7:70832–44. doi: 10.18632/oncotarget.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bean GR, Kremer JC, Prudner BC, Schenone AD, Yao JC, Schultze MB, Chen DY, Tanas MR, Adkins DR, Bomalaski J, Rubin BP, Michel LS, Van Tine BA. A metabolic synthetic lethal strategy with arginine deprivation and chloroquine leads to cell death in ASS1-deficient sarcomas. Cell Death Dis. 2016;7:e2406. doi: 10.1038/cddis.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D'Amico D, Antonucci L, Di Magno L, Coni S, Sdruscia G, Macone A, Miele E, Infante P, Di Marcotullio L, De Smaele E, Ferretti E, Ciapponi L, Giangaspero F. Non-canonical Hedgehog/AMPK-mediated control of polyamine metabolism supports neuronal and medulloblastoma cell growth. Dev Cell. 2015;35:21–35. doi: 10.1016/j.devcel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brett-Morris A, Wright BM, Seo Y, Pasupuleti V, Zhang J, Lu J, Spina R, Bar EE, Gujrati M, Schur R, Lu ZR, Welford SM. The polyamine catabolic enzyme SAT1 modulates tumorigenesis and radiation response in GBM. Cancer Res. 2014;74:6925–34. doi: 10.1158/0008-5472.CAN-14-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pavlyk I, Rzhepetskyy Y, Jagielski AK, Drozak J, Wasik A, Pereverzieva G, Olchowik M, Kunz-Schugart LA, Stasyk O, Redowicz MJ. Arginine deprivation affects glioblastoma cell adhesion, invasiveness and actin cytoskeleton organisation by impairment of b-actin arginylation. Amino Acids. 2015;47:199–212. doi: 10.1007/s00726-014-1857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khoury O, Ghazale N, Stone E, El-Sibai M, Frankel AE, Abi-Habib RJ. Human recombinant arginase I (C0)-PEG5000 [HuArgI (Co)-PEG5000]-induced arginine depletion is selectively cytotoxic to human glioblastoma cells. J Neurooncol. 2015;122:75–85. doi: 10.1007/s11060-014-1698-5. [DOI] [PubMed] [Google Scholar]
- 67.Fielder T, Strauss M, Hering S, Redanz U, William D, Rosche Y, Classen CF, Kreikemeyer B, Linnebacher M, Maletzki C. Arginine deprivation by arginine deiminase of Streptococcus pyogenes controls primary glioblastoma growth in vitro and in vivo. Cancer Biol Ther. 2015;16:1047–55. doi: 10.1080/15384047.2015.1026478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


