Abstract
Hsp90 is a conserved molecular chaperone that facilitates the folding and function of client proteins. Hsp90 function is dynamically regulated by interactions with co-chaperones and by post-translational modifications. In the fungal pathogen Candida albicans, Hsp90 enables drug resistance and virulence by stabilizing diverse signal transducers. Here, we review studies that have unveiled regulators of Hsp90 function, as well as downstream effectors that govern the key virulence traits of morphogenesis and drug resistance. We highlight recent work mapping the Hsp90 genetic network in C. albicans under diverse environmental conditions, and how these interactions provide insight into circuitry important for drug resistance, morphogenesis, and virulence. Ultimately, elucidating the Hsp90 chaperone network will aid in the development of therapeutics to treat fungal disease.
Keywords: Candida albicans, stress response, Hsp90, virulence, development, drug resistance
Hsp90 enables temperature-dependent morphogenesis, drug resistance and virulence in fungal pathogens
For fungal pathogens, the ability to grow at human physiological temperatures is a requirement for successful colonization and infection [1-4]. Of the estimated 1.5 million fungal species, only approximately 300 can cause disease in humans and only a handful are common human pathogens [5]. The requirement for high temperature growth has been proposed as the reason for the paucity of successful human fungal pathogens [6], and for the increased number of fungal infections in tropical regions [7]. Species of Aspergillus, Candida, Cryptococcus and Pneumocystis are the predominant causal agents of systemic fungal infections, accounting for approximately 90% of human mortality due to fungal infections [8]. Of these, Candida albicans, which can exist as a normal member of the human mucosal microbiota, is the primary cause of systemic candidiasis, infecting ∼400,000 individuals per year with mortality rates approaching 40%, even with current treatments [8].
Of the successful human fungal pathogens, elevated temperatures are often associated with entry into the host. Many fungi use elevated temperature as the cue for the initiation of developmental programs that allow for expression of virulence traits, including morphological transitions [9]. The thermally dimorphic fungi use high temperature as a signal to transition from filaments to the virulent yeast form [10,11]. In contrast, C. albicans transitions from yeast to the filamentous form upon exposure to elevated temperatures, such as those experienced during febrile episodes [12,13]. This ability to transition between morphotypes is a key trait that underpins the virulence of this pathogen. A second form of morphological plasticity in C. albicans involves epigenetic switching between the standard white cells and opaque, grey, and GUT cellular forms in response to environmental cues [14-17]. For example, in response to lower temperatures, such as those found on the skin, C. albicans can switch from the white to opaque state; although C. albicans white cells filament in response to elevated temperatures, opaque cell filamentation occurs only at the lower temperatures [14,15].
Temperature fluctuations can be sensed by these organisms via changes in many aspects of cellular homeostasis. One mechanism is through changes in membrane fluidity; extreme temperatures can change membrane properties, thereby allowing membrane dynamics to act as a thermal signal [18]. In fungi, temperature can also be sensed by histidine kinases, which are analogous to the sensor component of two-component signalling systems in bacteria [19], and these histidine kinase sensors can be used by the thermal dimorphs to trigger morphogenetic transitions [20]. Fungi can also respond to temperature fluctuations through the induction of a diverse set of heat shock proteins. These include the small heat shock proteins, which can shift from a low to high-affinity chaperone state upon heat stress [21,22], Hsp70 proteins, which are required for maintaining protein homeostasis [23], and the molecular chaperone Hsp90. Hsp90 is a highly conserved ATP-dependent molecular chaperone that acts to stabilize components of signal transduction cascades, especially those involved in adaptation to stress [24-26]. Hsp90 promotes the folding of hundreds of client proteins (see Glossary) in a highly regulated fashion [27]. Hsp90's unique position as a central hub in cellular circuitry enables it to exert a powerful influence on the phenotypic consequences of genetic variation in response to environmental change [28-30].
In the fungal pathogen C. albicans, Hsp90 is a key regulator of virulence traits. Impairment of C. albicans Hsp90 function abrogates drug resistance, reduces tolerance to a myriad of stresses, induces a morphological transition from yeast to filamentous growth, and attenuates virulence [12,28,31-33]. Targeting Hsp90 is a powerful strategy to treat fungal infections; however, the antifungal utility of current Hsp90 inhibitors is compromised by toxicity due to the effects of concurrent inhibition of host Hsp90. Thus, it is critical to identify Hsp90 interacting proteins that control drug resistance, stress response, and virulence, and that are sufficiently divergent to enable selective targeting in the pathogen. This includes interacting proteins that regulate Hsp90 function, those that rely on Hsp90 for proper folding and function, and those that work in concert with Hsp90 to mediate important cellular processes. This review will focus on two fundamental questions: 1) how is Hsp90 regulated in C. albicans, and 2) what are the effectors downstream of Hsp90 in C. albicans that modulate the key virulence traits of drug resistance and morphogenesis? By exploring these questions, we are poised to gain further insight into the Hsp90 interaction network and reveal novel therapeutic targets for treating fungal infections.
Mapping Hsp90 Chemical Genetic Interaction Networks
A powerful strategy to map interaction networks is based on genetic interactions, which are defined as an unexpected double mutant phenotype when alleles of two genes are combined (See Box 1). Identifying genetic interactions can facilitate assigning molecular function to genes based on shared genetic interactions [34], and reviewed in [35]. Genetic interaction studies are complementary to protein-protein interaction studies, which can be hampered by transience or low abundance. One approach for mapping genetic interactions is chemical genomic screening, where instead of constructing double mutants, a gene product is inhibited by a specific chemical and a mutant library is screened for hypersensitivity to that compound [36]. This is especially useful for C. albicans Hsp90 given the diploid nature of the pathogen, the lack of a complete sexual cycle, and the essential cellular function of Hsp90. There are potent and highly specific inhibitors of Hsp90 function, including the natural product geldanamycin [37]. Performing chemical genomic screens under multiple conditions also provides insight into the functional relationship of the genes involved, as those genes displaying an interaction under multiple conditions are more likely to act upstream and regulate expression or function of the target gene product, whereas those genes displaying an interaction only under a specific environmental condition are more likely to act downstream of the target gene [38,39]. These principles have been applied to map the C. albicans Hsp90 interaction network, revealing new mechanisms for the regulation of this molecular chaperone and the function of downstream regulators that impact key virulence traits.
Box 1. Chemical genomic screening in C. albicans.
Genetic interactions are defined as an unexpected double mutant phenotype. An extreme example of a negative genetic interaction is when two mutations that have little effect on their own cause synthetic lethality. A positive genetic interaction is when the double mutant has a less than expected defect in fitness. In general, negative genetic interactions can help define functionally related processes, while positive genetic interactions can aid in the identification of regulatory processes [34].
A genetic interaction is determined by examining the observed fitness or phenotype of a double mutant strain compared to the expected phenotype, which is calculated as the product of the fitness defect of each single mutant [95]. In principle, any combination where the observed fitness of the double mutant is significantly different than the expected fitness would be considered a genetic interaction. In practice, a cut-off of 50% greater than expected decrease or increase in fitness provides a robust indicator of a genetic interaction.
For chemical genomic screening, instead of creating double mutants, single mutants are subjected to growth in the presence of a small molecule inhibitor of a specific gene product. Here, the fitness of the single mutant in the presence of drug is compared to the product of the fitness of the wild-type strain in the presence of drug and the mutant strain in the absence of drug. The clearest interaction maps will result from chemical probes with high specificity for a single cellular target. To map the effect of environment on chemical genetic interactions, fitness is quantified in multiple environmental stress conditions.
Chemical Genetic Interactors
Chemical genomic screening of ∼20% of the C. albicans genome under distinct environmental conditions revealed 352 distinct HSP90 genetic interactors, and established that the Hsp90 genetic network is environmentally contingent [38,39]. Many of the Hsp90 genetic interactors play crucial roles in drug resistance, virulence, or both, as demonstrated in Figure 1 (Key Figure). These screens also identified a subset of genes that, when deleted, resulted in hypersensitivity to Hsp90 inhibition under multiple environmental cues. These were hypothesized, and several subsequently validated, to be upstream regulators of Hsp90 function. Consistent with a role for Hsp90 in regulating drug resistance and virulence, many of these high-connectivity Hsp90 interactors have roles in both of these phenotypes (Figure 1).
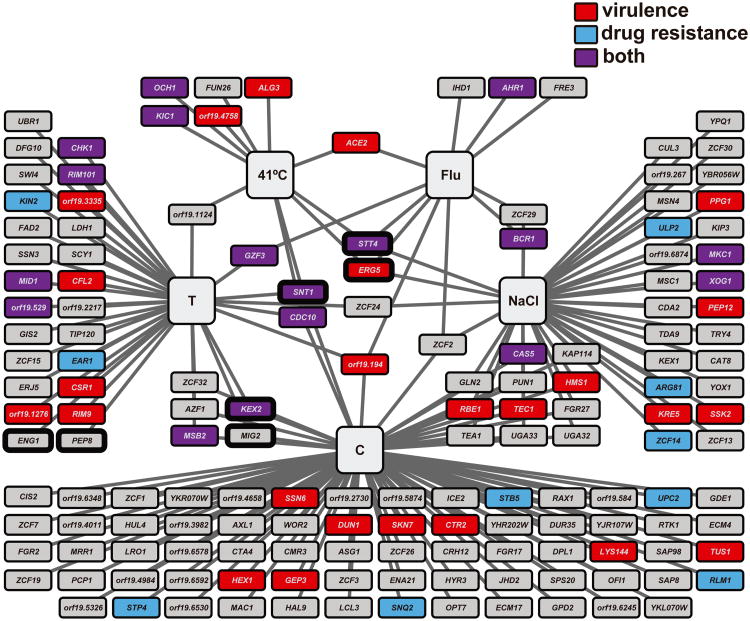
Figure 1. Key Figure. Hsp90 genetic interactors play important roles in drug resistance and virulence.
A recent Hsp90 chemical genetic network [39] was re-colored to emphasize the role of each interactor in drug resistance (blue), virulence (red), or both (purple). The network is composed of 158 genetic interactors identified in five different growth conditions (large grey boxes). T = tunicamycin, Flu = fluconazole, C = caspofungin. Each HSP90 genetic interactor is indicated by a rectangle, with edges connecting it to the environmental conditions in which it interacts with HSP90. Interactors with a thick black outline were identified as hypersensitive to Hsp90 inhibition in the absence of stress.
Strikingly, the vast majority of genetic interactions were only observed under a single environmental stressor. Clustering of interactors provides a powerful approach to identify the function of uncharacterized genes based on correlation of genetic interactions with genes of known function, as has been done to great effect in Saccharomyces cerevisiae [34,35,40]. For example, we observe multiple members of the C. albicans Rim101 signaling cascade as genetic interactors with Hsp90 in response to tunicamycin [38,39,41-44]. This suggests that additional Hsp90 genetic interactors identified in the tunicamycin stress conditions may function with the Rim101 cascade. As another example, orf19.194 was identified as a genetic interactor with Hsp90 in response to three antifungal drugs: tunicamycin, fluconazole, and caspofungin [39]. Although it has been previously implicated in virulence in a systemic model of candidiasis [45], it has no annotated function, no ortholog in S. cerevisiae, and the only predicted protein domain is a putative nuclear pore complex domain. Given the genetic interactions identified under multiple environmental conditions, this protein may function upstream of Hsp90.
Hsp90 Regulation
Although Hsp90 is a conserved molecular chaperone in all eukaryotes, a comprehensive understanding of its regulation remains elusive. This is in part due to the dynamic and transient nature of the complexes Hsp90 forms and the variable participation of co-chaperones that contribute to its distinct functions [25]. Here, we discuss recent advances in understanding of the complex regulation governing Hsp90 abundance and function and highlight examples where genetic interaction mapping has unveiled novel regulators of Hsp90 function (Figure 2).
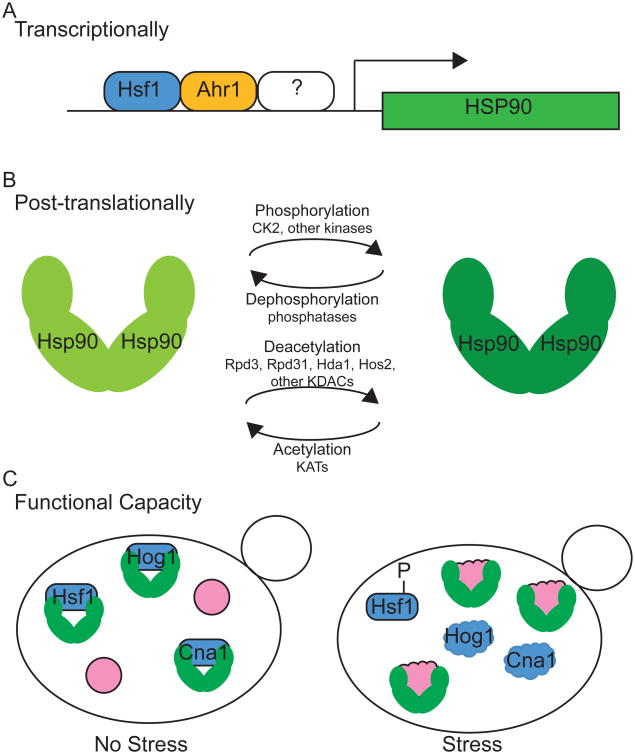
Figure 2. Models of Hsp90 regulation in C. albicans.
Hsp90 has been found to be regulated at the transcriptional and post-translational levels in response to environmental signals, with Hsp90 functional capacity modulated by cellular stress. A. HSP90 transcription can be regulated by Hsf1 and Ahr1, in addition to other factors. B. Hsp90 can be post-translationally modified by phosphorylation and acetylation. The phosphorylated and deacetylated form is required for functions in client stabilization and drug resistance. The phosphatase and lysine acetyltransferases (KATs) involved have not been elucidated. C. Increased cellular stress can overwhelm the functional capacity of Hsp90, impairing its ability to chaperone specific client proteins. Unfolded proteins are represented by ‘cloud’ shapes.
Transcriptional Regulation
Although Hsp90 is among the most abundant proteins in the eukaryotic cell [46], its expression is further induced in response to diverse environmental stresses [46-48]. In most eukaryotes, including the fungal pathogen C. albicans, inducible transcription of HSP90 is controlled by the essential transcription factor heat shock factor 1 (Hsf1) [13,49]. Under basal conditions, Hsf1 is a client of Hsp90, where Hsp90 exerts a repressive effect on Hsf1 activity [50]. When Hsp90 function is compromised, including during stress conditions, it can no longer repress Hsf1, allowing for Hsf1 activation via phosphorylation and the induction of Hsf1 targets, including HSP90 [50,51]. In C. albicans, heat shock induces profound transcriptional changes, resulting in transcriptional induction of approximately 18% of the genome, controlled in large part by Hsf1 acting in concert with Hsp90 [51]. Depletion of HSP90 impairs the rapid global transcriptional response to heat shock, which may be attributable to the increase in nucleosome density at promoters of Hsf1 target genes with Hsp90-dependent expression [51]. Thus, the functional relationship between Hsf1 and Hsp90 is complex and dynamic.
Although Hsf1 plays a vital role in HSP90 expression, there are other transcription factors that also contribute to the transcriptional regulation of HSP90. For human HSP90α, multiple transcriptional activators govern HSP90 expression in response to different stimuli, including the nuclear factor-κB subunit p65, NF-Il6β (also known as C/EBPδ), signal transducer and activator of transcription 3 (STAT3), and STAT1 [25]. Similar regulation is observed in fungi. Ahr1 is a zinc finger transcription factor identified as a genetic interactor with C. albicans Hsp90 under multiple environmental conditions [38]. Deletion of AHR1 reduces HSP90 transcript levels, resulting in filamentous growth in the absence of inducing cues [38]. There are likely to be additional transcription factors that can co-ordinate with Hsf1 to regulate HSP90 levels in response to distinct environmental insults.
Post-translational Regulation
In addition to the complex transcriptional regulatory networks that orchestrate changes in HSP90 expression, there is a remarkable complexity in the code of posttranslational modifications that control distinct aspects of Hsp90 function. Hsp90 function is modified by numerous posttranslational modifications, including acetylation, phosphorylation, and S-nitrosylation [52]. In the model yeast S. cerevisiae, Hsp90 is phosphorylated on at least 11 residues [53], including on threonine 22 by the casein kinase 2 (CK2) complex [53]. The ability to cycle between phosphorylation states is key for proper Hsp90 function [53]. Similarly, in C. albicans, CK2 phosphorylates Hsp90, and compromised CK2 function reduces the functional capacity of Hsp90 [38]. Hsp90 is also regulated by acetylation on numerous lysine residues [54-57]. In S. cerevisiae, Hsp90 is acetylated on lysine 27 and 270, as well as on other residues that remain enigmatic [56]. In C. albicans, lysine 30 and 271 are the analogous critical acetylation sites, and are important for Hsp90 function [57]. Hsp90 acetylation also regulates drug resistance and virulence in the fungal pathogen Aspergillus fumigatus [58,59]. In S. cerevisiae, the lysine deacetylases (KDACs) important for regulating Hsp90 are Hda1 and Rpd3 [56]. Intriguingly, in C. albicans, there is a high level of functional redundancy among KDACs, with Hos2, Hda1, Rpd3, and Rpd31 mediating azole resistance and morphogenesis [57]. Multiple members of the Hos2 complex, including HOS2 and SNT1, are genetic interactors with Hsp90 under diverse environmental stresses, implicating this complex in regulation of Hsp90 function [38,39]. Notably, the acetyltransferase required for adding the acetyl moiety to Hsp90 has not been identified in C. albicans.
Co-chaperones
Hsp90 function can also modulated by co-chaperones, which are thought to mediate recognition of client proteins and aid in ATP cycling and conformational changes of the Hsp90 complex [25,52,60]. These co-chaperones bind to different domains on Hsp90 and have distinct effects on the chaperone cycle. However, the identity, function, and regulation of most of the C. albicans co-chaperones have not been fully elucidated. One Hsp90 co-chaperone, Sgt1, regulates C. albicans morphogenesis and drug resistance [61], in part through interactions with the adenylyl cyclase Cyr1. The Hsp90 co-chaperones Cdc37 and Sti1 interact with the Crk1 kinase in C. albicans and are required for Crk1 stability [62]. Crk1 is a kinase that is required for morphogenesis and virulence [63]. To add to the complexity in regulation, the activity of co-chaperones is also subject to posttranslational modifications. For example, CK2 phosphorylates not only Hsp90, but also the co-chaperone Cdc37 in both S. cerevisiae and C. albicans, and this phosphorylation is critical for proper chaperone activity [38,64]. By guiding client protein recognition, these co-chaperones can have profound impacts on Hsp90 function.
Alterations in Cellular Environment
Hsp90 function can also be regulated by changes in the cellular environment. The functional capacity of Hsp90 can be overwhelmed in response to stress, as the chaperone is diverted from its set of select client proteins to deal with more global problems of protein misfolding. A classic example is with heat stress conditions, including febrile temperatures, which increase the cellular load of misfolded proteins that engage with Hsp90, resulting in relief of Hsp90-mediated repression of Hsf1, thereby activating the heat shock response [25,50,65]. Recently, other cellular stresses have been identified that similarly overwhelm Hsp90 function. The ergosterol biosynthesis gene ERG5 and the phosphatidylinositol-4-kinase (PI4K) gene STT4 were identified as HSP90 genetic interactors under multiple conditions [39]. Systematic analysis of the ergosterol biosynthetic cascade and the phosphatidylinositol pathway demonstrated that defects in ergosterol biosynthesis and actin remodeling can induce cellular stresses that overwhelm Hsp90's functional capacity [39]. Perturbation of ergosterol biosynthesis genes induces the expression of stress response genes, consistent with a model that defects in ergosterol biosynthesis result in cellular stress [66]. Hence, the function of Hsp90 displays substantial environmental contingency with profound biological consequences.
Hsp90 Physical Interactors
The biochemical function of Hsp90 is to act as a chaperone and aid in the folding of client proteins, many of which are important for C. albicans survival and virulence. Currently, there are limited methods to accurately predict Hsp90 client proteins, emphasizing the need for experimental approaches to define the Hsp90 network. One approach is to perform affinity purification of Hsp90 coupled to mass spectrometry [67]; purification of Hsp90 co-chaperones in parallel has been an effective strategy to aid in the identification of transient interactions [24]. Studies in mammalian systems have adapted and developed LUMIER (luminescence-based mammalian interactome mapping) to systematically measure Hsp90 physical interactions with the majority of human protein kinases, transcription factors and ubiquitin ligases in vivo [68].
While global analyses of Hsp90 physical interactors have not been performed in fungal pathogens, there are a select number of known Hsp90 client proteins in C. albicans (Table 1). Many of these play important roles in C. albicans drug resistance, morphogenesis, or virulence. For example, Hsp90 chaperones key components of the protein kinase C (PKC) signalling cascade. The PKC mitogen activated protein kinase (MAPK) cascade is required for tolerance to both the echinocandins [69] and azoles [70], which are the most widely deployed classes of antifungal drugs. Pkc1 is also a global regulator of C. albicans morphogenesis [71], and is required for virulence in a murine model of systemic infection [70]. Hsp90 is required for the stability of the terminal MAPK Mkc1 in both the basal and activated forms [50,70]. Mkk2, which is the MAPK that phosphorylates Mkc1, also depends on Hsp90 for stability [38]. Another key cellular regulator that depends on Hsp90 for its function is the protein phosphatase calcineurin. Calcineurin is required for tolerance to ergosterol biosynthesis inhibitors [70] and cell wall inhibitors [32], and is an Hsp90 client protein in C. albicans [32], S. cerevisiae [72] and A. fumigatus [73]. Finally, Hsp90 is required for the stability of Cdr1, a major drug pump in C. albicans [38], whose overexpression is implicated in drug-resistant clinical isolates.
Table 1. Hsp90 client proteins in C. albicans.
| Protein | Evidence | Reference |
|---|---|---|
| Mkc1 | Destabilized upon Hsp90 depletion | [50,70] |
| Cna1 | Physical interaction, destabilized upon Hsp90 depletion, activation depends on Hsp90 | [32,70] |
| Hog1 | Activation depends on Hsp90, destabilized upon Hsp90 depletion | [38,50] |
| Cdr1 | Destabilized upon Hsp90 depletion | [38] |
| Mkk2 | Destabilized upon Hsp90 depletion | [38] |
| Crk1 | Destabilized upon Cdc37 or Sti1 depletion | [62] |
| Cdc28 | Physical interaction, destabilized upon Hsp90 depletion | [88] |
| Clb4 | Destabilized upon Hsp90 depletion and inhibition | [88] |
| Tup1 | Physical interaction | [93] |
| Hsf1 | Physical interaction | [50] |
In addition to stabilizing client proteins important for antifungal drug resistance, Hsp90 also regulates proteins with important roles in host immune responses. Hog1 is a MAPK involved in a signal transduction cascade that controls responses to diverse environmental stresses, including osmotic stress [74]. Hsp90 is required for the accumulation of the activated form of Hog1, the stability of Hog1 protein, and the transcription of HOG1 [38,50]. In addition to the role of Hog1 in tolerance of many cellular stresses [75-77], it was recently implicated in regulating cell wall remodelling in response to neutrophil extracellular traps (NETs) [78]. Hsp90 is also required for activation of the Cek1 MAP kinase [50], which has been implicated in cell wall remodelling that modulates immune cell recognition [79-81]. Consistent with a role for Hsp90 in regulating cell wall remodelling and host immune responses, the Hsp90 genetic interactors Stt4, Snt1, and Erg5 are all required for induction of host cell pyroptosis [39].
Circuitry Through Which Hsp90 Governs Morphogenesis
Thus far, we have described key Hsp90 circuitry important for drug resistance, stress responses, and regulation of host immune responses. In addition, Hsp90 governs temperature-dependent C. albicans morphogenesis, a key virulence trait [45,82]. Chemical genomic screening, looking for alterations in morphogenesis instead of decreased growth, allows for the identification of genes required for filamentation in response to Hsp90 inhibition. This approach implicated genes in the Ras1-PKA signalling cascade, which is required for filamentation in response to diverse cues [83,84]. Hsp90 physically interacts with the adenylyl cyclase Cyr1 via the co-chaperone Sgt1 to repress PKA/cAMP signaling [61]. This is in contrast to S. cerevisiae, where Sgt1 is required to activate, not repress, Cyr1 signaling [85]. Notably, filamentation in response to Hsp90 inhibition does not require Efg1, the transcription factor downstream of PKA that is required for filamentation in response to serum and other cues [86]. Recent work implicated components of the cell wall integrity pathway in filamentation in response to diverse cues, including Hsp90 inhibition; key components include the GTPase Rho1 [87], the GTPase activator Lrg1, and the kinase Pkc1 [71]. Notably, downstream MAPK components are not required for filamentation in response to Hsp90 inhibition, implicating alternate downstream effectors [71]. In contrast to core circuitry required for filamentation in response to diverse cues, other pathways have more specific roles in filamentation in response to Hsp90 inhibition. For example, signalling through the cyclin Pcl1, cyclin-dependent kinase Pho85, and transcription factor Hms1 is required specifically for filamentation in response to Hsp90 inhibition and elevated temperature [12,88,89]. Additional transcription factors required for filamentation in response to Hsp90 inhibition include Cph2, Hap5, and Stp2 [12,88,89], and additional cell cycle regulators include the cyclin-dependent kinase Cdc28, the GTPase activator Bub2, and the cyclin Clb4 [88]. The engagement of such a large portfolio of diverse regulators in morphogenetic responses to Hsp90 inhibition is remarkable, and we expect screens with enhanced genomic coverage to reveal additional circuitry through which Hsp90 governs this important virulence trait.
Distinct Hsp90 Genetic Interactions in Planktonic and Biofilm Cellular States
The Hsp90 interaction network described thus far has been defined under planktonic or free floating conditions. However, C. albicans is also capable of forming biofilms [90]. Fungal biofilms are composed of communities of cells with distinct morphology that are surrounded by an extra-cellular matrix [90,91]. Striking differences in Hsp90 interactions have been observed between biofilm and planktonic states. For example, Hsp90 is required to stabilize calcineurin and Mkc1 in planktonic but not biofilm growth, suggesting that Hsp90 regulates drug resistance through different mechanisms in these distinct cellular states [92]. Hsp90 genetic interactors are also divergent in biofilm conditions. The transcription factor genes BCR1, MIG1, TEC1, TUP1, and UPC2 are Hsp90 genetic interactors in planktonic conditions [38], but not in biofilms [93]. This highlights the need for a systematic analysis of Hsp90 interactions in biofilms to establish how the chaperone network is rewired in distinct cellular states. Hsp90 function may also be different in other cellular states, including those that show distinct temperature-dependent responses, such as white versus opaque cells [14]. Distinct Hsp90 interaction networks are likely to be uncovered in other cellular states that differ in gene expression, cellular architecture and virulence [17,94].
Concluding Remarks
For the fungal pathogen C. albicans, Hsp90 is a powerful therapeutic target given its function in orchestrating morphogenesis, drug resistance, and virulence. However, problems with host toxicity demand the development of fungal-selective Hsp90 inhibitors, or the development of inhibitors targeting other components of the Hsp90 interaction network. The complex network of environmentally contingent Hsp90 genetic interactors in C. albicans highlights the vast number of synergistic relationships possible among compounds targeting different nodes within the network. Many important discoveries have been made with genomic resources limited to a fraction of the genome, but continued efforts to expand C. albicans mutant libraries to full genome scale will be vital to unleash the full potential of this approach. Further, deciphering Hsp90 interacting partners in different C. albicans cellular states and in different host-relevant environments will provide fascinating biological insight into the function of Hsp90 in responses to different stresses (see Outstanding Questions). A global appreciation of the Hsp90 chaperone network in C. albicans and other fungal pathogens will illuminate new targets and accelerate the development of much needed antifungal drugs.
Outstanding Questions.
How is C. albicans Hsp90 regulated? What are the transcription factors, in addition to Hsf1 and Ahr1, that regulate its expression in response to environmental perturbation? What are the additional residues on C. albicans Hsp90 regulated by posttranslational modifications and what complexes are involved? What are the identity and function of the C. albicans co-chaperones and how to they dictate Hsp90 function?
What is the complete Hsp90 interactome in C. albicans? What methods are optimal to systematically evaluate Hsp90 physical interactors in C. albicans given the often low expression of unstable client proteins? How will this interactome change in response to stress?
Once complete genome-scale mutant libraries in C. albicans are available, what other Hsp90 genetic interactors will be identified? Which genetic interactors will be involved in virulence? What are the best environmental stresses to uncover genetic interactors with roles in C. albicans virulence? Which interactors will we be able to target for the development of novel antifungal therapies?
How do cellular states, including biofilms and white and opaque cells, impact the Hsp90 interactome? How do they influence Hsp90 genetic interactions?
Trends.
The Candia albicans Hsp90 chaperone network is environmentally contingent, with a vast majority of genetic interactors important for growth only under a specific environmental condition.
Gene products displaying a genetic interaction with Hsp90 in diverse conditions are more likely to regulate Hsp90 levels or function.
Hsp90 function can be modulated transcriptionally, post-translationally, by co-chaperones, and by overwhelming its functional capacity in response to diverse cellular stressors.
Chemical genomic screens mapping Hsp90 interactors unveil novel circuitry governing morphogenesis, a key virulence trait.
Distinct cellular forms, including the formation of C. albicans biofilms, dramatically alter the Hsp90 interaction network.
Acknowledgments
LEC is supported by the Canadian Institutes of Health Research Operating Grants (MOP-86452 and MOP-119520), the Natural Sciences and Engineering Council (NSERC) of Canada Discovery Grants (06261 and 462167), and an NSERC E.W.R. Steacie Memorial Fellowship (477598). TRO is supported by a National Institutes of Health (NIH) Ruth L. Kirschstein National Research Service Award (AI115947-01) from the NIAID. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Glossary
- Hsp90 Client Proteins
are those that 1) physically interact with Hsp90, and 2) depend on Hsp90 for stabilization, maturation, or activation. Hsp90 client proteins are enriched for kinases and transcription factors
- Co-chaperones
are proteins that act as cofactors for Hsp90 function. Co-chaperones can assist in the recognition of client proteins, regulating the rate of client protein release, and altering the rate of Hsp90 ATP cycling
- Genetic interactors
are two gene products that, when deleted together, have an unexpected double mutant phenotype
- Geldanamycin
is a natural product that is a highly specific Hsp90 inhibitor. It acts by binding in the unusual ADP/ATP binding pocket of Hsp90 and competitively inhibiting ATP
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nichols CB, et al. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in beythe fungal pathogen Cryptococcus neoformans. Molecular Microbiology. 2007;63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- 2.Cooney NM, Klein BS. Fungal adaptation to the mammalian host: it is a new world, after all. Current Opinion in Microbiology. 2008;11:511–516. doi: 10.1016/j.mib.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall A. Determinants of virulence in the pathogenic fungi. Fungal Biology Reviews. 2007;21:130–132. doi: 10.1016/j.fbr.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhodes JC. Aspergillus fumigatus: Growth and virulence. Med Mycol. 2006;44:77–81. doi: 10.1080/13693780600779419. [DOI] [PubMed] [Google Scholar]
- 5.Taylor LH, et al. Risk factors for human disease emergence. Philosophical Transactions of the Royal Society B: Biological Sciences. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergman A, Casadevall A. Mammalian endothermy optimally restricts fungi and metabolic costs. mBio. 2010;1:e00212–10–e00212–11. doi: 10.1128/mBio.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Solache MA, Casadevall A. Global warming will bring new fungal diseases for mammals. mBio. 2010;1:e00061–10–e00061–10. doi: 10.1128/mBio.00061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown GD, et al. Hidden killers: human fungal infections. Science Translational Medicine. 2012;4:165rv13–165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 9.Klein BS, Tebbets B. Dimorphism and virulence in fungi. Current Opinion in Microbiology. 2007;10:314–319. doi: 10.1016/j.mib.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beyhan S, et al. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. Plos Biol. 2013;11:e1001614. doi: 10.1371/journal.pbio.1001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Nat Acad Sci. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro RS, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 2009;19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholls S, et al. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm‐ blooded animals. Molecular Microbiology. 2009;74:844–861. doi: 10.1111/j.1365-2958.2009.06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si H, et al. Candida albicans white and opaque cells undergo distinct programs of filamentous growth. PLoS Pathog. 2013;9:e1003210. doi: 10.1371/journal.ppat.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JM, Soll DR. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. Journal of Bacteriology. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao L, et al. Discovery of a “white-gray-opaque” tristable phenotypic switching system in Candida albicans: roles of non-genetic diversity in host adaptation. Plos Biol. 2014;12:e1001830. doi: 10.1371/journal.pbio.1001830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pande K, et al. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–1091. doi: 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Los DA, Murata N. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta. 2004;1666:142–157. doi: 10.1016/j.bbamem.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Santos JL, Shiozaki K. Fungal histidine kinases. Sci STKE. 2001;2001:re1. doi: 10.1126/stke.2001.98.re1. [DOI] [PubMed] [Google Scholar]
- 20.Nemecek JC, et al. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 21.Mayer FL, et al. Small but crucial: the novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS ONE. 2012;7:e38584. doi: 10.1371/journal.pone.0038584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzmann TM, et al. Activation of the chaperone Hsp26 is controlled by the rearrangement of its thermosensor domain. Mol Cell. 2008;29:207–216. doi: 10.1016/j.molcel.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Zuiderweg ERP, et al. The remarkable multivalency of the Hsp70 chaperones. Cell Stress and Chaperones. 2017;22:173–189. doi: 10.1007/s12192-017-0776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taipale M, et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taipale M, et al. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 26.Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. BBA - Molecular Cell Research. 2012;1823:674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 29.Lachowiec J, et al. Hsp90 promotes kinase evolution. Mol Biol Evol. 2015;32:91–99. doi: 10.1093/molbev/msu270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohner N, et al. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowen LE, Steinbach WJ. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Euk Cell. 2008;7:747–764. doi: 10.1128/EC.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh SD, et al. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowen LE, et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci USA. 2009;106:2818–2823. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420–aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boone C, et al. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 36.Roemer T, et al. Bugs, drugs and chemical genomics. Nat Chem Biol. 2012;8:46–56. doi: 10.1038/nchembio.744. [DOI] [PubMed] [Google Scholar]
- 37.Whitesell L, et al. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Nat Acad Sci. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diezmann S, et al. Mapping the Hsp90 genetic interaction network in Candida albicans reveals environmental contingency and rewired circuitry. PLoS Genet. 2012;8:e1002562–15. doi: 10.1371/journal.pgen.1002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Meara TR, et al. Mapping the HSP90 genetic network reveals ergosterol biosynthesis and phosphatidylinositol-4-kinase signaling as core circuitry governing cellular stress. PLoS Genet. 2016;12:e1006142. doi: 10.1371/journal.pgen.1006142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaut M, et al. Protein complexes are central in the yeast genetic landscape. PLoS Comput Biol. 2011;7:e1001092. doi: 10.1371/journal.pcbi.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kullas AL, et al. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Euk Cell. 2004;3:1609–1618. doi: 10.1128/EC.3.6.1609-1618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornet M, et al. The homologue of the Saccharomyces cerevisiae RIM9 gene is required for ambient pH signalling in Candida albicans. Res Microbiol. 2009;160:219–223. doi: 10.1016/j.resmic.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Davis D, et al. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infection and Immunity. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornet M, et al. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infection and Immunity. 2005;73:7977–7987. doi: 10.1128/IAI.73.12.7977-7987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noble SM, et al. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borkovich KA, et al. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Molecular and Cellular Biology. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leach MD, et al. Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs. Nat Rev Micro. 2012;10:693–704. doi: 10.1038/nrmicro2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981;293:311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- 49.Sorger PK. Heat shock factor and the heat shock response. Cell. 1991;65:363–366. doi: 10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- 50.Leach MD, et al. Hsp90 orchestrates transcriptional regulation by Hsf1 and cell wall remodelling by MAPK signalling during thermal adaptation in a pathogenic yeast. PLoS Pathog. 2012;8:e1003069. doi: 10.1371/journal.ppat.1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leach MD, et al. Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat Comm. 2016;7:11704. doi: 10.1038/ncomms11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prodromou C. Mechanisms of Hsp90 regulation. Biochem J. 2016;473:2439–2452. doi: 10.1042/BCJ20160005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mollapour M, et al. Casein kinase 2 phosphorylation of Hsp90 threonine 22 modulates chaperone function and drug sensitivity. Oncotarget. 2011;2:407–417. doi: 10.18632/oncotarget.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bali P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 55.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 56.Robbins N, et al. Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. CellReports. 2012;2:878–888. doi: 10.1016/j.celrep.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, et al. Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Molecular Microbiology. 2017;103:635–656. doi: 10.1111/mmi.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lamoth F, et al. Histone deacetylase inhibition as an alternative strategy against invasive aspergillosis. Front Microbiol. 2015;6:96. doi: 10.3389/fmicb.2015.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamoth F, et al. Identification of a key lysine residue in heat shock protein 90 required for azole and echinocandin resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 2014;58:1889–1896. doi: 10.1128/AAC.02286-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prodromou C. The “active life” of Hsp90 complexes. BBA - Molecular Cell Research. 2012;1823:614–623. doi: 10.1016/j.bbamcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shapiro RS, et al. The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PLoS ONE. 2012;7:e44734. doi: 10.1371/journal.pone.0044734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ni J, et al. Candida albicans Cdc37 interacts with the Crk1 kinase and is required for Crk1 production. FEBS Letters. 2004;561:223–230. doi: 10.1016/S0014-5793(04)00172-3. [DOI] [PubMed] [Google Scholar]
- 63.Chen J, et al. Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans. Molecular and Cellular Biology. 2000;20:8696–8708. doi: 10.1128/mcb.20.23.8696-8708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bandhakavi S, et al. A positive feedback loop between protein kinase CKII and Cdc37 promotes the activity of multiple protein kinases. J Biol Chem. 2003;278:2829–2836. doi: 10.1074/jbc.M206662200. [DOI] [PubMed] [Google Scholar]
- 65.Zou J, et al. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 66.Vincent BM, et al. Fitness trade-offs restrict the evolution of resistance to amphotericin B. Plos Biol. 2013;11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 68.Taipale M, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sussman A, et al. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Euk Cell. 2004;3:932–943. doi: 10.1128/EC.3.4.932-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.LaFayette SL, et al. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 2010;6:e1001069. doi: 10.1371/journal.ppat.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie JL, et al. Signaling through Lrg1, Rho1 and Pkc1 governs Candida albicans morphogenesis in response to diverse cues. PLoS Genet. 2016;12:e1006405. doi: 10.1371/journal.pgen.1006405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imai J, Yahara I. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Molecular and Cellular Biology. 2000;20:9262–9270. doi: 10.1128/mcb.20.24.9262-9270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lamoth F, et al. In vitro activity of calcineurin and heat shock protein 90 inhibitors against Aspergillus fumigatus azole- and echinocandin-resistant strains. Antimicrob Agents Chemother. 2013;57:1035–1039. doi: 10.1128/AAC.01857-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith DA, et al. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munro CA, et al. The PKC, HOG and Ca 2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Molecular Microbiology. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bahn YS. Master and Commander in Fungal Pathogens: the Two-Component System and the HOG Signaling Pathway. Euk Cell. 2008;7:2017–2036. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monge RA. The MAP kinase signal transduction network in Candida albicans. Mic78. robiology. 2006;152:905–912. doi: 10.1099/mic.0.28616-0. [DOI] [PubMed] [Google Scholar]
- 78.Hopke A, et al. Neutrophil attack triggers extracellular trap-dependent Candida cell wall remodeling and altered immune recognition. PLoS Pathog. 2016;12:e1005644. doi: 10.1371/journal.ppat.1005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roman E, et al. The Cek1-mediated MAP kinase pathway regulates exposure of α-1,2 and β-1,2-mannosides in the cell wall of Candida albicans modulating immune recognition. Virulence. 2016;7:558–577. doi: 10.1080/21505594.2016.1163458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Galán-Díez M, et al. Candida albicans beta-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infection and Immunity. 2010;78:1426–1436. doi: 10.1128/IAI.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Wijlick L, et al. Candida albicans responds to glycostructure damage by Ace2-mediated feedback regulation of Cek1 signaling. Molecular Microbiology. 2016;102:827–849. doi: 10.1111/mmi.13494. [DOI] [PubMed] [Google Scholar]
- 82.Lo HJ, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 83.Feng Q, et al. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. Journal of Bacteriology. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rocha CRC, et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dubacq C, et al. Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Euk Cell. 2002;1:568–582. doi: 10.1128/EC.1.4.568-582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bockmühl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157:1523–1530. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Corvest V, et al. Spatiotemporal regulation of Rho1 and Cdc42 activity during Candida albicans filamentous growth. Molecular Microbiology. 2013;89:626–648. doi: 10.1111/mmi.12302. [DOI] [PubMed] [Google Scholar]
- 88.Senn H, et al. Cdc28 provides a molecular link between Hsp90, morphogenesis, and cell cycle progression in Candida albicans. Mol. Biol. Cell. 2012;23:268–283. doi: 10.1091/mbc.E11-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shapiro RS, et al. Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr Biol. 2012;22:461–470. doi: 10.1016/j.cub.2012.01.062. [DOI] [PubMed] [Google Scholar]
- 90.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Micro. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nobile CJ, Mitchell AP. Genetics and genomics of Candida albicans biofilm formation. Cellular Microbiology. 2006;8:1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- 92.Robbins N, et al. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 2011;7:e1002257. doi: 10.1371/journal.ppat.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diezmann S, et al. Functional divergence of Hsp90 genetic interactions in biofilm and planktonic cellular states. PLoS ONE. 2015;10:e0137947. doi: 10.1371/journal.pone.0137947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lohse MB, Johnson AD. White-opaque switching in Candida albicans. Current Opinion in Microbiology. 2009;12:650–654. doi: 10.1016/j.mib.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mani R, et al. Defining genetic interaction. Proc Natl Acad Sci USA. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]