Abstract
Fragile X syndrome (FXS), a heritable intellectual and autism spectrum disorder, results from loss of Fragile X Mental Retardation Protein (FMRP). This neurodevelopmental disease state exhibits neural circuit hyperconnectivity and hyperexcitability. Canonically, FMRP functions as an mRNA-binding translation suppressor, but recent findings have enormously expanded proposed roles. Although connections between burgeoning FMRP functions remain unknown, recent advances have extended understanding of involvement in RNA-, channel- and protein-binding that modulates calcium signaling, activity-dependent critical period development and excitation-inhibition neural circuitry balance. This article contextualizes three years of FXS model research. Future directions extrapolated from recent advances focus on discovering links between FMRP roles; to determine whether FMRP has a multitude of unrelated functions, or combinatorial mechanisms can explain its multifaceted existence.
Keywords: Fragile X Syndrome (FXS), Autism Spectrum Disorder (ASD), Synapse, Translation Regulation, Activity-Dependent Critical Period
Overview of Expanding FMRP Functions
Fragile X syndrome (FXS), a common genetic root of both intellectual and autism spectrum disorders, is usually caused by a 5’UTR trinucleotide repeat expansion in the FMR1 gene, resulting in loss of the Fragile X Mental Retardation Protein (FMRP). FMRP functions as a master regulator of activity-dependent neurodevelopment, with null mutants manifesting hyperexcitability and reduced activity-dependent modulation of synapse maturation, refinement and plasticity [1]. FMRP is canonically defined as an mRNA-binding translational repressor, with a broad but largely indeterminate range of transcript targets [2], but the scope of FMRP genetic functions continues to explode (Table 1). From the cytosol, FMRP is classically described to shuttle to/from the nucleus, with a recent Drosophila study mapping a novel C-terminus mutation to this nuclear export function [3]. In the nucleus, recent work indicates that FMRP binds chromatin through tandem Tudor (Agenet) domains during the DNA damage response (DDR) to regulate genome stability in mice (Table 1) [4]. The involvement with DDR machinery is shown to be important during spermatogenesis, but a requirement in neurodevelopment has not been established. In the Drosophila FXS model, FMRP chromatin-binding function mediates replication stress induced H2av phosphorylation, one of the earliest DDR responses to double strand breaks and replication stress [5], but a requirement in neurodevelopment has also not been shown. Downstream of DNA interactions, but prior to canonical translational regulation roles, FMRP is proposed to act at multiple RNA life stages: in mRNA editing, pre-mRNA splicing, and in the microRNA pathway (Table 1). Recent work from zebrafish and mouse FXS models shows FMRP alters RNA-editing via interaction with the adenosine deaminase ADAR, supporting earlier Drosophila studies (Table 1) [6,7]. In the mouse FXS model, FMRP also works with RNA-binding protein 14 (RBM14) in pre-mRNA alternative splicing (Table 1) [8]. FMRP has long been associated with the microRNA pathway, and recent studies suggest key interactions in circuit plasticity and behavioral output in Drosophila (Table 1) [9,10]. In both canonical and newly discovered RNA-binding functions of FMRP, the mechanism of FMRP mRNA binding specificity has long been a conundrum, but recent insights into mRNA diversification provide further possible means for FMRP to recognize and regulate target transcripts [11,12].
Table 1. Recently defined FMRP genetic functions.
The table lists recently validated roles of FMRP (column 1), the FXS model system of the work (column 2), and the primary reference(s) discussed in the text (column 3).
| Proposed FMRP Roles | FXS Model | References |
|---|---|---|
| mRNA-binding translational regulator; canonically suppressing translation | Mouse and Drosophila | [2,33,34,46] |
| Chromatin-binding; regulates genome stability, required for early DDR | Drosophila and Mouse | [4,5] |
| ADAR-binding RNA editing regulator; alters RNA editing of neural genes | Drosophila, Zebrafish, Mouse | [6,7] |
| RBM14-binding in pre-mRNA splicing; promotes mRNA target binding | Mouse | [8] |
| microRNA pathway regulation; neural circuit plasticity and behavioral output | Drosophila | [9,10] |
| Regulation of cell differentiation kinetics; effects both neurons and glia | Mouse | [14] |
| Ion channel-binding to regulate gating; circuit excitability and plasticity | Mouse | [16,17] |
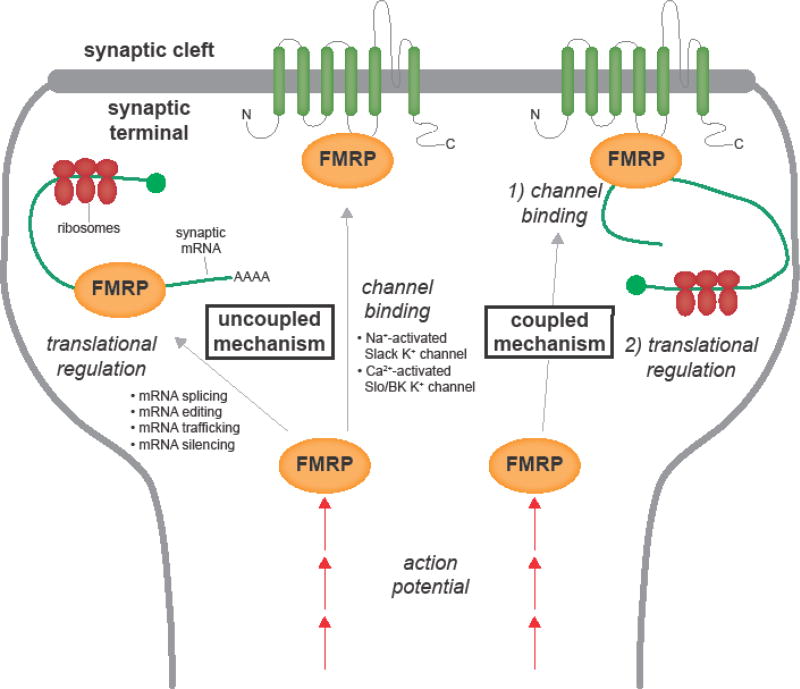
The FXS field is rife with debates about FMRP roles, including cellular locations, temporal timing and FMRP functions beyond DNA/RNA regulation. A long-term question concerns roles in neurons versus glia. A recent study of human patients shows FMR1 epigenetic alterations silence FMRP specifically in neurons, but not glia or neurons obtained from reprogrammed pluripotent stem cells [13]. In the mouse FXS model, recent work shows FMRP loss changes cell differentiation kinetics for both neurons and glia (Table 1) [14], and astrocyte-specific FMRP knockout in mice increases neuronal dendritic spine density similar to the global FXS condition [15]. Within neurons in all model systems, the soma contains the vast majority of FMRP, yet the lion’s share of research and discussion focuses on local FMRP functions at the synapse [2,16–19]. Most study postsynaptic mechanisms, but there appears to be at least as many presynaptic mRNA targets and presynaptic defects in mutants [2,16,20,21]. Yet another long-term debate concerns the timing of FMRP requirements [2]. Although FXS defects persist throughout life in both patients and animal models, this does not necessarily require continuous, maintained FMRP function [22–24]. Indeed, peak FMRP levels in both mouse and Drosophila FXS models occur during development, and prominent defects restricted to critical period neural circuit refinement may drive mature dysfunction [1,9,25–27]. Finally, the central role of FMRP mRNA-binding itself has been strongly challenged. In addition to the wide range of newly-discovered FMRP functions discussed above, including nuclear roles such as chromatin-binding, mRNA splicing and mRNA editing (Table 1) [9,28], FMRP shows direct ion channel binding modulating pore conductivity properties, and it is argued that many core FXS neurological defects may be explained by channel-binding alone [20]. These two widely separated biological roles could represent completely divergent FMRP functions in neurons (Fig. 1). Alternatively, FMRP RNA- and channel-binding functions could be linked in a common mechanism controlling activity-dependent protein synthesis (Fig. 1).
Figure 1. FMRP roles in RNA- and channel-binding at the neuronal synapse.
Activity-dependent functions of FMRP in RNA-binding translation regulation and direct channel-binding activity regulation. In the uncoupled mechanism (left), RNA- and channel-binding roles are unrelated, representing two evolutionarily divergent functions. In the coupled mechanism (right), channel-binding is an integral activity-sensing step in the translational regulation of FMRP-bound transcripts.
This article provides an update on major advances in the FXS field over the past three years, and has been divided into five parts grouped by FMRP biological functions. While FXS phenotypes present similarly in different genetic models, we carefully define the animal system used in each body of work, and note when any contradictory findings have been found between models or with human patients. Part I addresses recent progress on FMRP RNA-binding roles, predominantly in the suppression of protein translation, although translational activation has also been documented in some cases. We discuss all the newly identified FMRP mRNA targets, involved in a diverse range of biological functions including neuronal signal transduction, internal cellular architecture and intercellular signaling mechanisms. New FMRP roles are summarized in Table 1, new mRNA targets of FMRP canonical function in translationally regulation are shown in Table 2, and protein partners recently identified to interact with FMRP in translation control are shown in Table 3. We also briefly discuss new advances in non-mRNA binding translational regulation mediated by FMRP. Part II addresses recent progress on FMRP channel-binding roles, primarily in the control of neuronal excitability. We also discuss calcium-signaling mechanisms, downstream of direct channel-binding as well as indirect FMRP regulation. Calcium signaling functions highlight the role of FMRP as a translation activator in some contexts. Part III addresses new insights into FMRP synaptic development roles, including signaling, structural and functional requirements. We also discuss the activity-dependent regulation of critical period synaptic remodeling during the refinement of neural circuitry. Part IV briefly addresses excitatory/inhibitory (E/I) imbalance in the FXS disease state. We discuss recent advances in understanding FMRP roles regulating the balance between strengthened excitatory and repressed inhibitory connections in model neural circuits. Finally, in part V, we highlight important future directions for FXS research. We discuss logical extensions of the above ongoing work, and also emphasize key objectives that fall outside the scope of these areas.
Table 2. Recently defined FMRP direct RNA-binding targets.
The table lists recently validated mRNA targets bound by FMRP (column 1), encoded protein function (column 2), impacted signaling pathway (column 3), disease state effect (column 4), and the FXS model system of the work (column 5). Primary reference(s) discussed in text are listed in the rightmost column (column 6).
| RNA transcript | Related Protein Function | Related Signal Pathway | Disease State Impact | FXS | References |
|---|---|---|---|---|---|
| ARC | Synaptic ARC facilitates removal of AMPA receptors | Depends on MAPK signal cascade for activation | Spine overgrowth, Ca2+ signaling misregulation | Human cell lines | [29,38,39] |
| CAMKII reporter construct | Translational reporter binds Ca2+-Calmodulin | Activity-dependent Ca2+ signaling at the synapse | Elevated activity underlying LTH | Drosophila | [10] |
| DAG Kinase | Converts DAG to PA for lipid signaling and homeostasis | Lipid signaling during vesicle trafficking | Postsynaptic spine deformation | Mouse | [33,44] |
| Rac1, MAPB1, GluR1 | Rho GTPase, microtubule stability, AMPA-R subunit | Wide range of central signaling pathways | Defective hippocampal spinogenesis | Mouse and human | [34] |
| ADCY1 | Adenylyl cyclase produces cAMP second messenger | cAMP upregulates ERK & PI3K, which activate RSK | Hyperphosphorylates s6, spine overgrowth | Mouse | [47] |
| DSCAM | Cell adhesion and repulsion during synaptic growth | Relies on FMRP binding with Abelson kinase (Ab-1) | Unregulated presynaptic growth | Drosophila | [49] |
| EF1α | Overexpression of hyperphosphorylated Mdm2 | MEF2 transcription factor resistant Mdm2 | Eliminates excitatory synapse scaffold | Drosophila | [50] |
| BMPR2 | Receptor S/T kinase in growth and differentiation | Activates LIMK1, inhibiting cofilin, an actin destabilizer | Actin reorganization for dendritic overgrowth | Mouse and Drosophila | [35] |
| Kv3.1b mRNA | Voltage-gated K+ channels in brainstem | K+ influx to regulate auditory circuit processing | Disrupts processing of auditory information | Mouse | [52] |
| Kv4.2 mRNA | Voltage-gated K+ channels in hippocampus | Major K+ channel regulating hippocampal excitability | Channel expression repression/activation | Mouse | [54,55] |
| L-type Ca2+ channel | Voltage-dependent, slow L-type Ca2+ channel | Ca2+ influx to modulate gene transcription | Defective synaptic architecture | Human | [53] |
| Calmodulin, Calbindin | Ca2+-binding proteins act as second messengers | Ca2+ signaling activates many signaling pathways | Impaired sequestration, Impaired E/I Balance | Drosophila | [58] |
| NCS-1, Ric8a | Ca2+ signaling to regulate synaptic mechanisms | Coregulate synapse activity and synapse number | Increased activity and synapse number | Drosophila | [56] |
Table 3. Recently defined FMRP direct protein-binding partners.
The table lists recently validated direct protein partners bound by FMRP (column 1), mechanisms involved in FMRP-binding functions (column 2), and the FXS model system of the work (column 3). Primary reference(s) discussed in text are listed in the rightmost column (column 4).
| Protein Partner | FMRP-binding Function | FXS Model | References |
|---|---|---|---|
| Adenosine deaminase acting on RNA (ADAR) | RNA editing; binds double stranded RNA to convert adenosine to inosine by deamination | Drosophila, Zebrafish, Mouse | [6] |
| RNA-binding protein 14 (RBM14) | Alternative splicing; binds pre-mRNA for splicing, results in differential mRNA expression | Mouse | [8] |
| L5 protein of 80s ribosome | FMRP blocks tRNAs and elongation factors from binding 80s ribosome, suppressing translation | Drosophila | [36] |
| Telomere repeat-binding factor 2 (TRF2-S) | FMRP antagonizes TRF2-S target mRNA binding to limit axonal mRNA localization and function | Mouse | [37] |
| Staufen, Pumilio, Lark and other translational regulators | Translation regulation via RNA-binding in direct or indirect partnership with FMRP function | Human, Mouse, & Drosophila | [40–43] |
| Ataxin-2 mRNA-binding translational regulator | RNA-binding: microRNA pathway-induced RNA silencing for regulating & dampening LTH | Drosophila | [10,42] |
| Cytoplasmic FMR1-interacting protein 1 (CYFIP1) | Actin cytoskeleton and mTor signaling; binds UG/GU RNA with TDR-43 and Staufen | Human, Mouse, & Drosophila | [32,45–47] |
| Janus kinase and microtubule interacting protein 1 (JAKMIP1) | Microtubule regulation and GABAB receptor expression regulation in inhibitory neurons | Human | [32] |
| TAR DNA-binding protein of 43kDa (TDP-43) | Complex with FMRP, Staufen and CYFIP1 binds mRNA to suppress hippocampal spinogenesis | Mouse and human | [34,40] |
| Slack and Slowpoke (Slo) BK K+ channels | FMRP regulates gating to shape AP kinetics, modulate Ca2+ influx and neurotransmitter release | Mouse and Xenopus | [16,17,20] |
Part I: New Progress in RNA-Binding/Translation Suppression Mechanisms
The canonical FMRP role is direct mRNA-binding translation suppression (Fig. 1), although FMRP has long been associated with mRNA throughout its lifecycle, including splicing, editing, trafficking and stability [6,11,12,29,30]. FMRP selectively associates with a subset of mRNAs: the candidate target list is long, but altered protein levels have been established for only a tiny handful of proteins (Table 2) [29,31]. Indeed, proteomic screens suggest that the number of protein changes in both mouse and Drosophila FXS models is surprisingly small, and many of these changes may be indirect, since they don’t align well with mRNA-binding data [32]. Given synaptic defects in FXS patients and models, there is a particular bias towards mRNA targets encoding synaptic proteins [33]. However, it is clear that FMRP also binds many non-synaptic transcripts, and that translational regulation of most synaptic transcripts does not occur exclusively (or even predominantly) locally at synapses [5,12,30]. Although many mechanisms have been proposed to explain the RNA-binding specificity of FMRP, including both secondary structure (G-quartet) and consensus nucleotide sequences [34], there is little compelling evidence for any universal mechanism [35]. Therefore, it remains unclear by which mechanism(s) FMRP specifically binds target mRNAs, or which mRNAs contribute predominantly to FXS neuropathology. The basis of inherent RNA-binding specificity, or perhaps lack thereof, is an outstanding question in need of continued investigation. Here, we focus only on new FMRP mRNA targets and translational control mechanisms identified over the past three years (Tables 2,3).
FMRP represses translation via interaction with translation machinery, including RNA-binding proteins and microRNA components. A newly-defined FMRP partner is the ribosomal L5 protein (Table 3) [36]. In Drosophila, FMRP-L5 binding prevents tRNA and elongation factor ribosome association to suppress translation. A recent study in human FXS patient cells suggests translation repression also arises from FMR1 5'UTR CGG binding to activity-regulated cytoskeleton associated protein (ARC) transcripts in mobile ribosome granules, inhibiting mRNA localization/translation (Table 2) [29,37,38]. In mice, telomere repeat-binding factor 2 (TRF2-S) binding to target mRNAs facilitates axonal delivery, which is antagonized by FMRP binding the TRF2-S GAR domain [39]. FMRP knockdown promotes mRNA localization to enhance growth and presynaptic transmitter release (Table 3). FMRP has long been known to partner with multiple other RNA-binding proteins (e.g. Staufen, Pumilio, Lark and many others; Table 3), which may provide mRNA-binding specificity and/or coupled regulation of joint targets [40–43]. This co-regulation has striking effects on neural circuit plasticity. For example, it was recently shown in Drosophila that FMRP partners with mRNA-binding Ataxin-2 in the microRNA-dependent plasticity mechanism of long-term olfactory habituation (LTH) [10]. FMRP, Ataxin-2 or microRNA component knockdown elevates CAMKII translation reporter activity during LTH. FMRP and Ataxin-2 both display transdominant genetic interactions with two microRNA-mediated translational suppression proteins, RNA-induced silencing complex (RISC) component Ago1 and deadbox helicase me31B, required for long-term central adaptation (LTCA) from odorant exposure [9,10].
FMRP has long been known to regulate core signal transduction pathways through multiple mechanisms. A recent mouse study establishes a newly-defined FMRP mRNA target for diacylglycerol (DAG) kinase, which converts DAG to phosphatidic acid (PA) during lipid signaling (Table 2). Loss of DAG kinase translation repression by FMRP of causes postsynaptic signaling and spine defects [33,44]. FMRP also binds signaling proteins, including Janus kinase and microtubule interacting protein 1 (JAKMIP1) and cytoplasmic FMR1 interaction protein 1 (CYFIP1; Table 3). A recent gene expression study shows JAKMIP1 modulates microtubule transport and GABABR levels [32]. WAVE complex CYFIP1 links FMRP to actin regulation, and mechanistic target of rapamycin (mTor) signaling in both mice and Drosophila [45]. A FMRP/CYFIP1/Staufen complex recruited by TAR DNA binding protein 43kDa (TDP-43) represses the translation of Ras-related C3 botulinum toxin substrate 1 (Rac1), microtubule-associated protein 1B (MAP1B) and glutamate receptor 1 (GluR1) to limit spinogenesis in mice and human cell lines (Table 2) [34,40]. Downstream of CYFIP1, the mTor target ribosomal protein s6 is hyperphosphorylated by p90-ribosomal S6 kinase (RSK), downstream of both extracellular signal-regulated kinase (ERK) and phosphatidylinositide 3 kinase (PI3K) signaling elevated in FXS model mice [46,47]. A recent study found Acamprosate decreases elevated ERK signaling to attenuate functional and behavioral defects [48]. Finally, in mice FMRP also binds adenylyl cyclase type 1 (ADCY1) mRNA to suppress translation, and elevated ADCY1 expression in the FXS condition also activates the above pathway to promote postsynaptic spine overgrowth (Table 2) [47].
In addition to core signal transduction and cytoskeletal regulation within the cell, FMRP also binds mRNA targets encoding proteins involved in intercellular interactions. Recent work shows FMRP binds Down syndrome cell adhesion molecule (DSCAM) mRNA to suppress translation in Drosophila (Table 2) [49]. In the Drosophila FXS model, enlarged synaptic termini are associated with DSCAM misregulation via an Abelson kinase 1 (Ab1) mechanism, and removal of just one Ab1 gene copy restores synapse structure in the disease state [49]. In the same Drosophila model, FMRP binds eukaryotic elongation factor 1 (EF1α) mRNA to suppress translation (Table 2) [50]. Increased EF1α levels in the FXS condition causes elevated nuclear localization of hyperphosphorylated ubiquitin ligase murine double minute-2 (Mdm2), which is resistant to myocyte enhancer factor 2 (MEF2) control (Table 1). When activated by MEF2, Mdm2 normally disassembles synaptic scaffolds to limit excitatory synapse number, so this defect drives FXS hyperexcitability [50]. In both Drosophila and mouse models, FMRP binds mRNA encoding bone morphogenic protein receptor type II (BMPR2), which activates LIM-domain kinase 1 (LIMK1) to inhibit actin-binding Cofilin and cause actin reorganization that promotes synaptogenesis in the FXS condition (Table 2) [35]. Genetic or pharmacological correction of BMPR2-LIMK signaling rectifies synaptic and movement defects in the Drosophila FXS model [51]. The FMRP C-terminal domain (CTD) binds BMPR2 mRNA, highlighting the question of which RNA-binding domains mediate different interactions, and whether they act alone or in combination.
Part II: New Progress in Channel-Binding and Calcium Signaling Mechanisms
In addition to translation roles, FMRP directly binds ion channels (Fig. 1), and modulates Ca2+ signaling to control neural activity [1]. FMRP binds multiple classes of K+ channels, including Na+-activated Slack [16] and Ca2+-activated Slowpoke (Slo) BK channels [17]. FMRP-Slack binding regulates gating to shape action potential kinetics, especially during high-frequency activity. FMRP binding to presynaptic BK channels modulates Ca2+ influx and neurotransmitter release (Fig. 1). In addition to direct channel binding, FMRP controls channel levels by translational regulation, including repression of Kv3.1b K+ channels in mice and activation of L-type Ca2+ channels in human cells (Table 2) [52,53]. Contradictory studies report Kv4.2 K+ channels are translationally repressed [54] or activated [55] by FMRP in mice (Table 2). FMRP indirectly controls presynaptic N-type Ca2+ channel levels (Cav2.2) via proteasomal degradation in human cells [18]. Downstream of channels, FMRP also regulates calcium binding protein (CaBP) function, through both RNA- and protein-binding mechanisms [10,29,56,57]. Moreover, the Drosophila FXS model shows reduced Calmodulin and Calbindin CaBP mRNA levels, resulting in the impaired sequestration of cytosolic Ca2+ (Table 2) [58]. This transcript change could be the result of mRNA-binding mechanisms or impaired transcriptional regulation. Thus, FMRP controls channel/CaBP expression and function via multiple overlapping direct and indirect avenues, providing a rich and confusing tapestry of activity misregulation in the FXS disease state.
Recent work continues to identify mechanisms by which FMRP controls channels via both RNA- and protein-binding mechanisms (Fig. 1). A human patient-derived FMR1 missense mutation (R138Q) has helped to discriminate these two FMRP functions by blocking channel-binding while preserving mRNA-binding translational control [20]. In the Drosophila FXS model, this mutant variant fails to rescue synaptic structural defects, suggesting growth phenotypes are separable from channel functions directly modulating presynaptic activity [20]. A caveat to this conclusion is that the R138Q mutation blocks FMRP interaction with Ca2+-gated BK channels, but may not impair other channel binding. Moreover, Drosophila FMRP has not yet been shown to bind ion channels, and it remains critical to determine whether this is an evolutionarily conserved mechanism, as suggested by the binding of FMRP to Slack K+ channels in Aplysia [59]. Furthermore, it is unknown whether channel-binding activity regulation and RNA-binding translational suppression can actually be separated, as they may form two parts of a coupled activity-dependent regulatory mechanism (Fig. 1). Recent studies into activity regulation via channel expression may result in new therapeutic treatments. In the Drosophila FXS model, a recent study shows that overexpression of N-type Ca2+ channels can be corrected by lowering channel activity with chronic magnetic nanoparticle stimulation [60]. This modulation also rescues action potential duration, intensity and frequency, and may enhance GABAA receptor expression in the disease state [60].
Recent studies show calcium signaling is misregulated at multiple levels to alter FXS model synapse and circuit excitability. As referenced above, Drosophila FMRP binds CAMKII mRNA, and FMRP knockdown in the Drosophila FXS model activates a CAMKII Ca2+-dependent translational reporter [10]. Drosophila FMRP also binds to the transcripts of neuronal calcium sensor 1 (NCS-1) and the associated guanine exchange factor (GEF) Ric8a, which operate in another Ca2+-sensing mechanism. NCS-1 and Ric8a antagonistically regulate neuronal activity and synapse number downstream of voltage-gated Ca2+ channels (Table 2) [56]. In the Drosophila FXS model, the drug phenothiazine inhibits the NCS-1/Ric8a interaction to prevent the assembly of this Ca2+ sensing complex and restore normal synapse architecture [57]. FMRP also modulates calcium homeostasis in non-canonical mechanisms. As referenced above, in human cells the FMR1 5'UTR CGG expansion interacts with ARC mRNA in mobile ribosomal granules, which also disrupts Ca2+ homeostasis by misregulating CaBP mRNAs that localize to these granules in a similar translation misregulation mechanism [29]. Using a drug that destabilizes the FMR1 CGG repeats (TMPyP4), normal Ca2+ homeostasis can be restored, indicating some relationship between the FXS trinucleotide expansion and neural Ca2+ dynamics [29]. Added to this complexity, FMRP-dependent Ca2+ signaling is altered differentially in excitatory versus inhibitory neurons as they mature during the early-use critical period in the Drosophila FXS model [1].
Part III: New Progress in Activity-Dependent Synapse Development Mechanisms
The critical question of FMRP roles during neurodevelopment versus maturity is centrally important for determining optimal FXS treatment strategies [1,2,21,25,26]. Recent FXS clinical trials have been greatly disappointing, but this may be in large part due to targeting adult patients [21]. Moreover, many mouse FXS model interpretations may be complicated by “adult” studies initiated in animals at ~4-weeks, which overlaps the neurodevelopmental period [61]. In the Drosophila FXS model, studies over the last decade have revealed clear, selective FMRP roles restricted to early-use critical periods [1,25,26]. This is not to say that there are not maintained roles at maturity, but only that FMRP clearly has specialized functions during neurodevelopment. Recent work shows that depleting FMRP in mouse embryonic stem cells alters the normal kinetics of both neuronal and glial differentiation [14]. Later on in the Drosophila model, multiple aspects of sensory- and activity-dependent synaptic remodeling and calcium signaling refinement are limited to FMRP functions during early-use critical periods, without any apparent persisting roles at adult maturity [1]. Indeed, FMRP expression has been suggested to functionally delineate temporal windows of activity-dependent neural circuitry optimization, with sharply dropping FMRP levels marking the end of the critical period in Drosophila [1]. FMRP has been proposed to act as a core component of the critical period activity-sensing mechanism during this developmental window, but much more investigation is needed to rigorously test this hypothesis.
Recent work in the Drosophila FXS model shows FMRP is absolutely required in activity-dependent remodeling of synaptic connectivity during an early-use critical period [25,26]. Although FMRP is similarly required in both excitatory and inhibitory neurons, the direction of activity-dependent changes appears opposite in these neuronal classes, including both synaptic architecture and calcium signaling refinement [1]. Optogenetic manipulations in Drosophila result in opposing changes in dendritic arborization in these two neuron classes, but either directional change absolutely requires FMRP during the developmental critical period [25]. In the Drosophila FXS model, glial phagocytosis is delayed in this same circuitry, suggesting a possible synaptic pruning mechanism [62]. However, FMRP may not work in glia but rather within neurons, which may signal glial phagocytosis through extracellular protein expression. Interestingly, neuronal FMRP regulates synaptic matrix metalloproteinase (MMP) classes in Drosophila, which sculpt intercellular signaling via proteolytic cleavage of extracellular proteins [63,64,65]. In the Drosophila FXS model, MMP levels are elevated, resulting in altered intercellular signaling, including trans-synaptic Wnt signaling [66,67]. This mechanism depends on regulation of the cell surface Wnt co-receptor Heparan Sulfate Proteoglycan (HSPG) Dally-like Protein (Dlp). Future studies will be needed to test whether activity-dependent MMP/Dlp intersections underlie FXS synaptogenic defects, especially in the context of excitatory versus inhibitory neuron classes.
Part IV: New Progress in Excitatory/Inhibitory (E/I) Balance Mechanisms
The metabotropic glutamate receptor (mGluR) theory of FXS hyperexcitability [68,69] is developmentally restricted, with early defects later corrected [22,24]. In mice, FMRP loss causes mGluR type 1 and/or 5 (mGluR1/5) pathway activation. In the Drosophila FXS model, elevated phosphatidylinositide 3-kinase enhancer (PIKE) levels exaggerate mGluR1/5 signaling to both impair synaptic plasticity and cause hyperexcitability [70]. One result of this mGluR hyperexcitability is the well-documented decrease in cAMP levels in both mouse and Drosophila FXS models [71]. Interestingly, phosphodiesterase 4 (PDE4) inhibition in adult Drosophila reportedly restores mGluR-dependent learning/memory defects, independent of correcting brain learning/memory circuitry defects, showing therapies administered after the developmental critical period can still have efficacy [71]. FMRP and mGluR5 also may modulate excitability by acting together with amyloid precursor protein (APP) to alter signaling pathways, including mTor and ERK discussed above [14,68]. Drosophila FMRP binds APP mRNA, and APP translation is elevated in the Drosophila FXS model. Increased APP levels are proposed to mediate hyperexcitation via a variety of mechanisms, including mGluR5 redistribution at synapses via Αβ oligomers metabolites of APP. In the mouse FXS model, genetically reducing APP levels prevents hyperexcitability and corrects behavioral, postsynaptic dendritic spine and glutamate signaling defects [14,68]. Dual dysregulation of APP and α-secretase ADAM10 in the mouse FXS model during the critical period activates the MAP kinase pathway to cause synaptogenic and behavioral deficits [72].
In addition to the mGluR hyperexcitability theory, GABAergic hypoinhibition may underlie FXS symptoms [27]. Earlier work in both mouse and Drosophila FXS models showed impaired GABAergic circuit assembly and function. Recent mouse work shows FMRP binds multiple GABAAR mRNAs (although this is debated [73]), resulting in delay of the developmental GABA inhibitory switch in FMR1 knockouts [68]. In FXS mice, underdevelopment and reduction of many components of the GABAergic system occurs at an early age [73]. For example, a GABAbR subunit (R1a) mediating presynaptic inhibition is reduced in FMR1 knockout mice [73]. Similarly, the Drosophila FXS model shows deficits in GABAbR suppression of presynaptic glutamate release [19]. This early delay in GABA signaling is followed by later overelaboration of GABAergic neurons in a Drosophila brain learning/memory center [27]. In the mouse FXS model, recent work shows that both pre- and postsynaptic alterations cause abnormal GABAAR-mediated phasic inhibition [73], although GABAergic defects vary widely across brain regions [73]. In the Drosophila FXS model, GABAergic neuron structure and Ca2+ signaling are both impaired, although correction of hypoinhibition is not sufficient to restore learning [27]. A recent Drosophila study shows altered GABAergic signaling reduces lateral inhibition across olfactory projection neurons [74]. However, in this case, GABAergic neurons are actually more active in the FXS condition, although postsynaptic neurons are less sensitive to inhibitory signaling. A recent mouse study shows glycinergic inhibitory signaling is unaffected but excitatory input elevated in the lateral superior olive [75], suggesting E/I imbalance also underlies FXS auditory processing defects.
Part V: Future Directions
Canonically, FMRP is an mRNA-binding translational suppressor. However, at a foundational level, we still fail to grasp the binding mechanism to specific mRNA targets. This question is complicated by our ever-increasing grasp of mRNA complexity [11,12], as well as by examples of FMRP acting as a translational activator [55]. FMRP also has multiple mRNA-binding domains [35], but it is not clear whether they work together, separably or sequentially. Moreover, FMRP interacts with multiple other RNA-binding translational regulators [40–43], microRNA [10,42] and ribosomal components [36], highlighting unanswered questions about FMRP mechanisms of translation regulation. Partnerships with other mRNA-binding proteins could provide the long-sought specificity for coupled control of transcript targets in neuron class-specific mechanisms [40–43]. FMRP roles in glia may also contribute to FXS [13–15], and this remains an important question. Intercellular glial-neuron signaling, as well as trans-synaptic signaling, may have central roles in FXS neuropathology, which are just coming to light [63–67]. FMRP roles in the extracellular space, particularly in MMP regulation, may regulate intercellular signaling. Studies in this area may help address the historical divide in the FXS field between pre- and postsynaptic mechanisms. Although the bulk of research remains focused on postsynaptic roles, there are at least as many FMRP targets encoding presynaptic proteins [2,16,20,21]. Future work needs to build a consensus view of FMRP roles on both sides of the synapse, as well as within the synaptic cleft itself.
As we begin to bridge the gap between pre- and postsynaptic mechanisms, there remains a wide divide between FMRP roles in development vs. maturity. At the earliest time point, some work suggests FMRP roles in neuron/glia differentiation [14]. However, the majority of developmental studies focus on early-use critical periods of peak FMRP expression, theorizing that critical period defects may set the stage for dysfunction at maturity [1,9,25–27]. It is vital to define how critical periods are delineated, possibly by FMRP itself, and test FMRP roles in activity-dependent synaptic and circuit refinement [1,22,24–26]. On the other hand, FMRP function is maintained at maturity. In both mouse and Drosophila FXS models, genetic and pharmacological interventions in mature animals have proven efficacious in some cases [68,71]. For example, PDE4 inhibitors in Drosophila adults rescue FXS learning/memory deficits, despite persistent malformation of underlying brain circuitry [71]. In the mouse FXS model, many argue that persistent changes in mGluR signaling must be taken into account in therapies [22]. E/I imbalance theories, including mGluR hyperexcitability and GABA hypoinhibition, are entangled in this ongoing question of development versus mature functions. Although debate continues over the relative importance of hyperexcitability and hypoinhibition in the FXS condition [27], the two theories have recently been framed more compatibly [67,68]. Future work needs to build a consensus view of FMRP roles in different neuron classes and in different temporal windows of requirement.
The biggest recent change in the FXS field has been the challenge to the predominant function of FMRP as an mRNA-binding translational suppressor. While most recent studies continue to identify new FMRP mRNA targets (Table 2), other work reveals new FMRP protein partners in an ever-broadening arena of biology (Table 3). Although some interactions support canonical roles in translation control, others have expanded FMRP biology to a dizzying scale spanning from chromatin-binding to channel-binding (Table 1) [3,4,6,7,8]. Considerable work has shown the importance of channel-binding interactions in FXS, especially for disease state hyperexcitability. There is a critical need to address whether channel-binding is a wholly separate FMRP function, or rather part of an integrated mechanism driving activity-dependent translation control (Fig. 1). How the other, more diverse FMRP functions may contribute to FXS neuropathology remains largely unknown. Future work on the downstream effects of these broad changes, and identifying key alterations in the FXS disease state, will be crucial to deciphering unfolding non-canonical roles. We should also question whether FMRP is truly a multifarious protein with diverse, unrelated functions adopted through the long course of evolution, or rather a central link in an integrated mechanism linking activity input to appropriate protein translation. Occam’s razor supports a combinatorial mechanism of FMRP requirement at the heart of the sensory input- and activity-dependent optimization of neural circuitry.
Trends Box.
Fragile X syndrome (FXS), a leading heritable autism, is caused by a 5’UTR trinucleotide repeat expansion in the gene encoding Fragile X Mental Retardation Protein (FMRP)
The disease presents with stereotypical hyperexcitability and synaptic overelaboration, which are well replicated across a broad range of neural circuits in genetic models
Canonically, FMRP is a mRNA-binding translational suppressor, but genetic roles range from chromatin-binding to mRNA splicing/editing to other forms of translation control
Additional FMRP roles include direct ion channel-binding to regulate neural excitability, which may be an independent function or linked to activity-dependent translation control
FXS cell-type specific defects in neurons and glia, include altered calcium signaling, critical period synapse refinement and excitation/inhibition balance in neural circuitry
Outstanding Questions.
How does FMRP manifest RNA-binding specificity to repress the translation of a select subset of transcripts?
Do FMRP RNA-binding domains work together, sequentially or separably to mediate translation control?
Why does FMRP interact with the microRNA pathway, and what role does this play in translation control?
How does FMRP work with other RNA-binding proteins in translation control; together, sequentially or separably?
What percentage of FMRP translation control occurs locally at the synapse as compared to the soma?
Does FMRP involved in RNA- vs. channel-binding represent a single protein pool for both binding functions?
Is channel-binding a separate function of FMRP, or is it mechanistically tied to RNA-binding functions?
How does lack of FMRP in glia contribute to the FXS disease state, and what processes are impacted?
Does FMRP in neurons and/or glia regulate intercellular signaling required for synaptic pruning/refinement?
How does trans-synaptic signaling depend on FMRP, and defects in this process contribute to the FXS state?
Are FMRP-dependent pre/postsynaptic changes independent or coupled to affect synaptic connectivity?
What MMP roles in the extracellular space are regulated by FMRP, and is this a direct translation mechanism?
How are activity-dependent critical periods delineated, and does FMRP play a role in this temporal restriction?
Do FXS developmental defects cause defects at maturity, or are these separable stages of requirement?
How do FMRP roles differ by neuron classes and within different temporal windows of requirements?
What is the role of FMRP in establishing or modulating excitation/inhibition balance in neural circuits?
Are the hyperexcitation and hypoinhibition characterizing FXS mechanistically linked or separable defects?
How does the apparently diverse range of FMRP functions contribute to FXS neurological phenotypes?
Does FMRP have many unrelated functions, or is there an integrated mechanism of FMRP requirement?
Acknowledgments
We thank Caleb Doll for help making Figure 1. We are grateful to Dominic Vita, Jim Sears, Randy Golovin and Tyler Kennedy for critical input on this article. This work is supported by R01 MH084989 to K.B.
Glossary Box
- Activity-Regulated Cytoskeleton-Associated Protein (ARC)
An immediate-early gene (IEG) displaying activity-dependent mRNA localization to the synapse, where local translation is involved in synaptic plasticity, learning and memory.
- Adenosine Deaminase Acting on RNA (ADAR)
A class of RNA-editing enzymes that bind double stranded RNA to convert adenosine to inosine by direct deamination.
- Adenylyl Cyclase (ADCY1)
Converts adenosine triphosphate (ATP) into the second messenger cyclic adenosine monophosphate (cAMP).
- Amyloid Precursor Protein (APP)
An integral membrane protein concentrated at neuronal synapses.
- Bone Morphogenetic Protein Receptor Type 2 (BMPR2)
A serine-threonine receptor kinase of the transforming growth factor-β superfamily.
- Calcium/Calmodulin-Dependent Protein Kinase II (CAMKII)
A serine-threonine protein kinase regulated by calcium-calmodulin complexes, involved in many signaling cascades at synapses.
- Cyclic Adenosine Monophosphate (cAMP)
Derivative of adenosine triphosphate (ATP) and a common second messenger in intracellular signaling at synapses.
- Cytoplasmic FMR1-interacting Protein 1 (CYFIP1)
Serves as a Rac-1 (an activating Ras GTPase) binding site in the WAVE Complex to facilitate actin nucleation through ARP2/3 complex activation.
- Diacylglycerol Kinase (DAG Kinase)
Converts Diacylglycerol (DAG) into Phosphatidic Acid (PA) during lipid signaling.
- Down Syndrome Cell Adhesion Molecule (DSCAM)
A transmembrane protein regulating synaptic growth. In Down Syndrome patients, Dscam is over-expressed owing to chromosome 21 trisomy.
- DNA Damage Response (DDR)
Phosphatidylinositol 3 kinases are activated by double-stranded DNA breaks, and open replication forks to enable histone phosphorylation driving chromatin folding changes.
- Extracellular Signal-Regulated Kinases (ERK)
A core component of the Ras-Raf-MEK-ERK signaling pathway that regulates many functions; also known as Mitogen-Activated Protein Kinase (MAPK).
- Fragile X Mental Retardation Protein (FMRP)
Classically defined as an mRNA-binding translation regulator, FMRP also binds multiple cytosolic proteins and ion channels to regulate neural excitation, calcium signaling, excitatory/inhibitory balance, and has a wide range of other proposed mechanistic functions discussed in this article.
- Fragile X Syndrome (FXS)
A leading heritable cause of intellectual disability and autism spectrum disorders. Typically caused by a trinucleotide repeat in the 5’ untranslated region (UTR) of the fragile X mental retardation 1 (FMR1) gene, leading to hypermethylation and transcriptional silencing.
- Long-Term Central Adaptation (LTCA) / Long-Term Habituation (LTH)
Two terms both describing sensory input-dependent plasticity in Drosophila olfactory neural circuits.
- Matrix Metalloproteinase (MMP)
Class of extracellular proteases responsible for proteolytic cleavage of a wide range of extracellular protein targets.
- Metabotropic Glutamate Receptor (mGluR)
A class of G-protein coupled receptor (GPCR) that binds the excitatory neurotransmitter glutamate.
- MicroRNA-Mediated Translational Suppression
MicroRNAs bind Argonaute proteins to form RNA-Induced Silencing Complexes (RISC) that associate with complementary mRNAs to silence expression.
- Phosphodiesterase 4 (PDE4)
An enzyme that hydrolyzes and degrades the second messenger cyclic adenosine monophosphate (cAMP), terminating cAMP signaling.
- Phosphatidylinositide 3 Kinase Enhancer (PIKE)
Enhances activity of the phosphatidylinositide 3 kinase (PI3K) enzyme family, which phosphorylates the 3-hydroxyl group on the phosphatidylinositol inositol ring.
- Pre-mRNA Alternative Splicing
The process by which transcripts are spliced into different mRNAs by the differential inclusion and exclusion of exon transcript sequences.
- TAR DNA Binding Protein, 43 Kilodaltons (TDP-43)
An mRNA-binding protein functioning during development; involved in cell cycle progression, apoptosis, RNA processing and alternative splicing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doll CA, Broadie K. Neuron class-specific requirements for Fragile X Mental Retardation Protein in critical period development of calcium signaling in learning and memory circuitry. Neurobiol. Dis. 2016;89:76–87. doi: 10.1016/j.nbd.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weisz ED, et al. Deciphering discord: How Drosophila research has enhanced our understanding of the importance of FMRP in different spatial and temporal contexts. Exp. Neurol. 2015;274:14–24. doi: 10.1016/j.expneurol.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Okray Z, et al. A novel fragile X syndrome mutation reveals a conserved role for the carboxy-terminus in FMRP localization and function. EMBO Mol. Med. 2015;7(4):423–37. doi: 10.15252/emmm.201404576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alpatov R, et al. A chromatin-dependent role of the Fragile X Mental Retardation Protein FMRP in the DNA damage response. Cell. 2014;157(4):869–881. doi: 10.1016/j.cell.2014.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, et al. A feed-forward mechanism involving Drosophila Fragile X Mental Retardation Protein triggers a replication stress-induced DNA damage response. Hum. Mol. Genet. 2014;23(19):5188–96. doi: 10.1093/hmg/ddu241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamay-Ramot A, et al. FMRP Interacts with Adar and Regulates RNA Editing, Synaptic Density and Locomotor Activity in Zebrafish. PLOS Genet. 2015;11(12):e1005702. doi: 10.1371/journal.pgen.1005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippini A, et al. Absence of the Fragile X Mental Retardation Protein results in defects of RNA editing of neuronal mRNAs in mouse. RNA Biol. 2017 doi: 10.1080/15476286.2017.1338232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou LT, et al. A novel role of Fragile X Mental Retardation Protein in pre-mRNA alternative splicing through RNA-binding protein 14. Neuroscience. 2017;349:64–75. doi: 10.1016/j.neuroscience.2017.02.044. [DOI] [PubMed] [Google Scholar]
- 9.Golovin RM, Broadie K. Developmental experience-dependent plasticity in the first synapse of the Drosophila olfactory circuit. J. Neurophysiol. 2016;116:2730–2738. doi: 10.1152/jn.00616.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudhakaran IP, et al. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. PNAS. 2014;111(1):E99–108. doi: 10.1073/pnas.1309543111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue Y, et al. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2017;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao BS, et al. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2016;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esanov R, et al. The FMR1 promoter is selectively hydroxymethylated in primary neurons of Fragile X Syndrome patients. Hum. Mol. Genet. 2016;25(22):4870–4880. doi: 10.1093/hmg/ddw311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalfallah O, et al. Depletion of the Fragile X Mental Retardation Protein in Embryonic Stem Cells Alters the Kinetics of Neurogenesis. Stem Cells. 2017;35:374–385. doi: 10.1002/stem.2505. [DOI] [PubMed] [Google Scholar]
- 15.Hodges JL, et al. Astrocytic Contributions to Synaptic and Learning Abnormalities in a Mouse Model of Fragile X Syndrome. Biol. Psychiatry. 2016;S0006-3223(16):32779–2. doi: 10.1016/j.biopsych.2016.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown MR, et al. Fragile X Mental Retardation Protein controls gating of the sodium-activated potassium channel Slack. Nat. Neurosci. 2010;13(7):819–21. doi: 10.1038/nn.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng PY, et al. FMRP regulates neurotransmitter release and synaptic information transmission by modulating action potential duration via BK channels. Neuron. 2013;77(4):696–711. doi: 10.1016/j.neuron.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferron L, et al. Fragile X Mental Retardation Protein controls synaptic vesicle exocytosis by modulating N-type calcium channel density. Nat. Commun. 2014;5:3628. doi: 10.1038/ncomms4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang JY, et al. Deficits in the activity of presynaptic γ-aminobutyric acid type B receptors contribute to altered neuronal excitability in Fragile X Syndrome. J. Biol. Chem. 2017 doi: 10.1074/jbc.M116.772541. jbc.M116.772541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myrick LK, et al. Independent role for presynaptic FMRP revealed by an FMR1 missense mutation associated with intellectual disability and seizures. PNAS. 2015;112(4):949–956. doi: 10.1073/pnas.1423094112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo SY, et al. Molecular medicine of Fragile X Syndrome: based on known molecular mechanisms. World Journal of Pediatrics. 2016;12(1):19–27. doi: 10.1007/s12519-015-0052-0. [DOI] [PubMed] [Google Scholar]
- 22.Martin HG, et al. Differential Adulthood Onset mGlu5 Signaling Saves Prefrontal Function in the Fragile X Mouse. Cereb. Cortex. 2016:1–11. doi: 10.1093/cercor/bhw328. [DOI] [PubMed] [Google Scholar]
- 23.Scharkowski F, et al. Altered Connectivity and Synapse Maturation of the Hippocampal Mossy Fiber Pathway in a Mouse Model of the Fragile X Syndrome. Cereb. Cortex. 2017:1–16. doi: 10.1093/cercor/bhw408. [DOI] [PubMed] [Google Scholar]
- 24.Berzhanskaya J, et al. Sensory hypo-excitability in a rat model of fetal development in Fragile X Syndrome. Sci. Rep. 2016;6:30769. doi: 10.1038/srep30769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doll CA, Broadie K. Activity-dependent FMRP requirements in development of the neural circuitry of learning and memory. Development. 2015;142:1346–1356. doi: 10.1242/dev.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doll CA, Broadie K. Impaired activity-dependent neural circuit assembly and refinement in autism spectrum disorder genetic models. Front. Cell. Neurosci. 2014;8:30. doi: 10.3389/fncel.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatto CL, et al. GABAergic circuit dysfunction in the Drosophila Fragile X Syndrome model. Neurobiol. Dis. 2014;65:142–159. doi: 10.1016/j.nbd.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Q, Ge W. FMRP: a new chapter with chromatin. Protein Cell. 2014;5(12):885–888. doi: 10.1007/s13238-014-0105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rovozzo R, et al. CGG Repeats in the 5’UTR of FMR1 RNA Regulate Translation of Other RNAs Localized in the Same RNA Granules. PLoS One. 2016;11(12):e0168204. doi: 10.1371/journal.pone.0168204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang E, et al. Dysregulation of mRNA Localization and Translation in Genetic Disease. J. Neurosci. 2016;36(45):11418–11426. doi: 10.1523/JNEUROSCI.2352-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boland M, et al. Molecular analyses of neurogenic defects in a human pluripotent stem cell model of Fragile X Syndrome. Brain. 2017;140:582–598. doi: 10.1093/brain/aww357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ansel A, et al. Variation in Gene Expression in Autism Spectrum Disorders: An Extensive Review of Transcriptomic Studies. Frontiers in Neuroscience. 2017;10:601. doi: 10.3389/fnins.2016.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabet R, et al. Fragile X Mental Retardation Protein (FMRP) controls diacylglycerol kinase activity in neurons. PNAS. 2016;113(26):E3619–28. doi: 10.1073/pnas.1522631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumder P, et al. Co-regulation of mRNA translation by TDP-43 and Fragile X Syndrome protein FMRP. Acta Neuropathol. 2016;132:721–738. doi: 10.1007/s00401-016-1603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashima R, et al. Augmented noncanonical BMP type II receptor signaling mediates the synaptic abnormality of Fragile X Syndrome. Sci. Signal. 2016;9(431):ra58. doi: 10.1126/scisignal.aaf6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen E, et al. Fragile X Mental Retardation Protein regulates translation by binding directly to the ribosome. Mol. Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waltereit R, et al. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J. Neurosci. 2001;21(15):5484–93. doi: 10.1523/JNEUROSCI.21-15-05484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chowdhury S, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52(3):445–59. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang P, et al. Novel RNA- and FMRP-binding protein TRF2-S regulates axonal mRNA transport and presynaptic plasticity. Nat. Commun. 2015;6:8888. doi: 10.1038/ncomms9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu Z, et al. Neurodegeneration-associated TDP-43 interacts with Fragile X Mental Retardation Protein (FMRP)/Staufen (STAU1) and regulates SIRT1 expression in neuronal cells. J. Biol. Chem. 2012;287(27):22560–22572. doi: 10.1074/jbc.M112.357582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siemen H, et al. Pumilio-2 Function in the Mouse Nervous System. PLOS One. 2011;6(10):e25932. doi: 10.1371/journal.pone.0025932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kenny P, Ceman S. RNA Secondary Structure Modulates FMRP's Bi-Functional Role in the MicroRNA Pathway. Int. J. Mol. Sci. 2016;17:985. doi: 10.3390/ijms17060985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sofola O, et al. The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J. Neurosci. 2008;28(41):10200–10205. doi: 10.1523/JNEUROSCI.2786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabet R, et al. Fragile X Syndrome: Are signaling lipids the missing culprits? Biochimie. 2016;130:188–194. doi: 10.1016/j.biochi.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Abekhoukh S, et al. New insights into the regulatory function of CYFIP1 in the context of WAVE- and FMRP-containing complexes. Dis. Model. Mech. 2017;10(4):463–474. doi: 10.1242/dmm.025809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawicka K, et al. Elevated ERK/p90 ribosomal S6 kinase activity underlies audiogenic seizure susceptibility in Fragile X mice. PNAS. 2016:E6290–E6297. doi: 10.1073/pnas.1610812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sethna F, et al. Enhanced expression of ADCY1 underlies aberrant neuronal signalling and behaviour in a syndromic autism model. Nat. Commun. 2016;8:14359. doi: 10.1038/ncomms14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer TL, et al. Acamprosate in a mouse model of Fragile X Syndrome: modulation of spontaneous cortical activity, ERK1/2 activation, locomotor behavior, and anxiety. J. Neurodev. Disord. 2017;9:6. doi: 10.1186/s11689-017-9184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterne GR, et al. Dysregulated Dscam levels act through Abelson tyrosine kinase to enlarge presynaptic arbors. eLIFE. 2015;4:e05196. doi: 10.7554/eLife.05196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai NP, et al. FMRP-dependent Mdm2 dephosphorylation is required for MEF2-induced synapse elimination. Hum. Mol. Genet. 2016;26(2):293–304. doi: 10.1093/hmg/ddw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kashima R, et al. Hyperactive locomotion in a Drosophila model is a functional readout for the synaptic abnormalities underlying Fragile X Syndrome. Sci. Signal. 2017;10(477) doi: 10.1126/scisignal.aai8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strumbos JG, et al. Fragile X Mental Retardation Protein is required for rapid experience-dependent regulation of the potassium channel Kv3.1b. J. Neurosci. 2010;30(31):10263. doi: 10.1523/JNEUROSCI.1125-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L, et al. The Fragile X Mental Retardation Protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- 54.Lee HY, et al. Bidirectional regulation of dendritic voltage-gated potassium channels by the Fragile X Mental Retardation Protein. Neuron. 2011;72(4):630. doi: 10.1016/j.neuron.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gross C, et al. Fragile X Mental Retardation Protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J. Neurosci. 2011;31(15):5693. doi: 10.1523/JNEUROSCI.6661-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dason JS, et al. Frequenin/NCS-1 and the Ca2+-channel α1-subunit co-regulate synaptic transmission and nerve-terminal growth. J. Cell Sci. 2009;122:4109–4121. doi: 10.1242/jcs.055095. [DOI] [PubMed] [Google Scholar]
- 57.Mansilla A, et al. Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for Fragile X Syndrome. PNAS. 2016;114(6):E999–E1008. doi: 10.1073/pnas.1611089114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tessier CR, Broadie K. The Fragile X Mental Retardation Protein developmentally regulates the strength and fidelity of calcium signaling in Drosophila mushroom body neurons. Neurobiol. Dis. 2011;41(1):147–159. doi: 10.1016/j.nbd.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. Regulation of neuronal excitability by interaction of Fragile X Mental Retardation Protein with slack potassium channels. J. Neurosci. 2012;32(44):15318. doi: 10.1523/JNEUROSCI.2162-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tay A, Di Carlo D. Magnetic Nanoparticle-Based Mechanical Stimulation for Restoration of Mechano-Sensitive Ion Channel Equilibrium in Neural Networks. Nano Lett. 2017;17:886–892. doi: 10.1021/acs.nanolett.6b04200. [DOI] [PubMed] [Google Scholar]
- 61.Santos AR, et al. Learning and behavioral deficits associated with the absence of the Fragile X Mental Retardation Protein: what a fly and mouse model can teach us. Learn. Mem. 2014;21:543–555. doi: 10.1101/lm.035956.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Connor RM, et al. A Drosophila model of Fragile X Syndrome exhibits defects in phagocytosis by innate immune cells. J. Cell Biol. 2017;216(3):595–605. doi: 10.1083/jcb.201607093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lepeta K, et al. A normal genetic variation modulates synaptic MMP-9 protein levels and the severity of schizophrenia symptoms. EMBO Mol. Med. 2017 doi: 10.15252/emmm.201707723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siller SS, Broadie K. Matrix Metalloproteinases and Minocycline: Therapeutic Avenues for Fragile X Syndrome. Neural Plast. 2012;5:124548. doi: 10.1155/2012/124548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janusz A, et al. The Fragile X mental Retardation Protein regulates matrix metalloproteinase 9 mRNA at synapses. J. Neurosci. 2013;33(46):18234–18241. doi: 10.1523/JNEUROSCI.2207-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parkinson WM, et al. Synaptic roles for phosphomannomutase type 2 in a new Drosophila congenital disorder of glycosylation disease model. Dis. Model. Mech. 2016;9:513–527. doi: 10.1242/dmm.022939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dear ML, et al. Two classes of matrix metalloproteinases reciprocally regulate synaptogenesis. Development. 2016;143:75–87. doi: 10.1242/dev.124461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westmark CJ, et al. APP Causes Hyperexcitability in Fragile X Mice. Front. Mol. Neurosci. 2016;9:147. doi: 10.3389/fnmol.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hays SA, et al. Altered Neocortical Rhythmic Activity States in FMR1 KO mice are Due to Enhanced mGluR5 Signaling and Involve Changes in Excitatory Circuitry. J. Neurosci. 2011;31(40):14223–14234. doi: 10.1523/JNEUROSCI.3157-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gross C, et al. Increased expression of the PI3K enhancer PIKE mediates deficits in synaptic plasticity and behavior in fragile X syndrome. Cell Reports. 2015;11:727–736. doi: 10.1016/j.celrep.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi CH, et al. PDE-4 inhibition rescues aberrant synaptic plasticity in Drosophila and mouse models of Fragile X Syndrome. J. Neurosci. 2015;35(1):396–408. doi: 10.1523/JNEUROSCI.1356-12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pasciuto E, et al. Dysregulated ADAM10-Mediated Processing of APP during a Critical Time Window Leads to Synaptic Deficits in Fragile X Syndrome. Neuron. 2015;87(2):382–98. doi: 10.1016/j.neuron.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 73.Sabanov V, et al. Impaired GABAergic inhibition in the hippocampus of FMR1 knockout mice. Neuropharmacol. 2016;116:71–81. doi: 10.1016/j.neuropharm.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 74.Franco LM, et al. Reduced Lateral Inhibition Impairs Olfactory Computations and Behaviors in a Drosophila Model of Fragile X Syndrome. Curr. Biol. 2017;27(8):1111–23. doi: 10.1016/j.cub.2017.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia-Pino E, et al. Enhanced Excitatory Connectivity and Disturbed Sound Processing in the Auditory Brainstem of Fragile X Mice. J. Neurosci. 2017 doi: 10.1523/JNEUROSCI.2310-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]