Abstract
The neural crest is a transient, multipotent population of cells that arises at the border of the developing nervous system. After closure of the neural tube, these cells undergo an epithelial to mesenchymal transition to delaminate and migrate, often to distinct locations in the embryo. Neural crest cells give rise to a diverse array of derivatives including neurons and glia of the peripheral nervous system, melanocytes, and bone and cartilage of the face. A gene regulatory network (GRN) controls specification, delamination, migration, and differentiation of this fascinating cell type. With increasing technological advances, direct linkages within the neural crest GRN are being uncovered. The underlying circuitry is useful for understanding important topics such as reprogramming, evolution, and disease.
Introduction to the Neural Crest
Neural crest formation occurs during early vertebrate development. This multipotent stem cell-like population forms within the future central nervous system (CNS) but then migrates away, undergoing some of the longest migrations of any embryonic cell type. These cells then contribute to a wide variety of derivatives including craniofacial cartilage and bone, neurons and glia of the peripheral nervous system, and melanocytes. As a vertebrate apomorphy, the neural crest is an innovation thought to be necessary for the origin, expansion, and acquisition of predation of vertebrates (the new head hypothesis) [1]. As a consequence, it has often been called “the fourth germ layer” [2,3]. Due to its developmental plasticity, migratory ability, and importance in vertebrate evolution, studies of the neural crest have had a profound impact on the fields of both embryology and evolutionary biology for over the last century.
Wilhelm His (1868) first identified the neural crest or “Zwischenstrang” (translation: intermediate cord) as a band of cells between the neural tube and non-neural ectoderm that gives rise to the spinal and cranial ganglia of chicken embryos. Later, grafting experiments revealed the developmental potential of neural crest cells in amphibians and birds, the latter using quail/chick chimera [4,5]. Tracking cells over time revealed distinct migratory pathways and a diverse array of neural crest derivatives. More recently, the neural crest field has expanded to reveal the cellular and molecular components underlying neural crest development.
With continuing advances in systems-level approaches and genomic analyses, Gene Regulatory Networks (GRNs) describing the formation and diversification of various stages of neural crest development are being uncovered at high-resolution (see Box 1). Here, we briefly describe how connections have been established within the neural crest GRN with emphasis on the most recent findings. Space limitation precludes description of the entire literature underlying building of each node, but please see Simoes-Costa and Bronner (2015) for further details [5].
Box 1. Increasing the resolution of GRN architecture with technological advances.
Investigating gene regulatory networks requires tools for interrogating each node and regulatory linkage in detail. Most linkages in the GRN were initially established using simple perturbation analyses where a genetic knockdown was performed and gene expression changes were assayed by in situ hybridization. Cis-regulatory analyses to uncover transcription factor occupancy were done on a gene by gene basis and took years to examine each node. To achieve the goal of elucidating each node and establishing direct linkages in the GRN, high throughput examinations are a crucial. Genomic tools such as CRISPR/Cas9, RNA-seq, ATAC-seq, and ChIP-seq have become a standard for GRN analysis.
Tissue-specific transcriptomes have been used to uncover temporal and spatially specific regulatory states, even at the single cell level [62,95]. Fluorescent activated cell sorting (FACS) allows isolation of distinct cell populations even within an intermingled, heterogeneous population such as the neural crest. Using tissue-specific transgenic lines or tissue-specific enhancers driving fluorescent reporter expression, one can isolate a pure population of neural crest cells or derivatives at any time during their development. Establishing a comprehensive neural crest GRN depends upon establishing complete regulatory states at each event of neural crest development. Once the suite of genes involved in each regulatory module is uncovered, it is possible to establish direct regulatory linkages therein.
Recent advances in molecular biology enable uncovering the whole regulatory landscape such that direct linkages can be easily established computationally and tested in vivo. Tissues that were previously inaccessible to tissue-specific isolation can now be purified using a technique called “biotagging” which takes advantage of the strong non-covalent nature of in vivo biotinylation to specifically label cells in a tissue-specific manner, enabling study of their in vivo chromatin landscape [103]. Assay for transposase accessible chromatin using sequencing (ATAC-seq) surveys regions of open chromatin and transcription factor occupancy and has opened new opportunities for probing direct linkages within the GRN. Methods such as ChIP-seq, which investigates DNA-binding, and Hi-C, which uncovers chromatin conformation are also avenues for neural crest researchers to enhance each node of the GRN. Future studies will likely allow GRN investigation at even higher resolution by more rapidly and efficiently establishing the whole hierarchy.
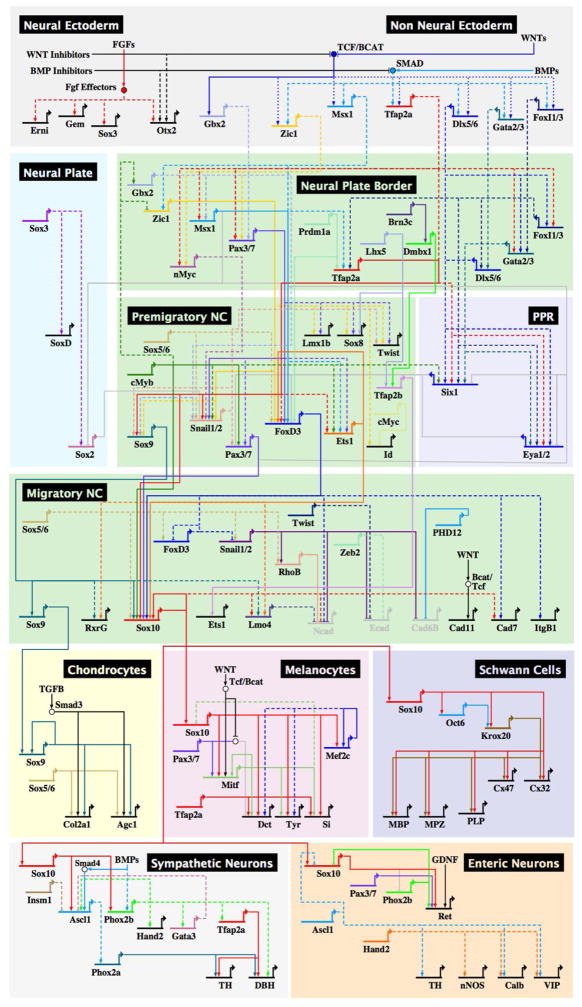
Gene Networks Overview
GRN circuitry provides a view of development from a logical perspective. Formulated via a systems-level integration of data accumulated in many vertebrate model systems, the neural crest GRN consists of sequential, separate regulatory modules, each comprised of a suite of transcription factors and signaling molecules (Figure 2) that explain the process of neural crest formation and differentiation into the vast array of derivatives. Underlying each regulatory event is a stable module embedded within a larger “neural crest gene regulatory network”, that combines signaling and transcriptional interactions. Through a series of feedback loops, each dynamic state within the network stabilizes, so that drivers of downstream gene batteries can be activated [6]. Also embedded within this GRN are circuits that confer multipotency and migratory ability.
Figure 2.
The GRN underlying neural crest development. Constructed from a systems-level amalgamation of literature in many vertebrate model systems, the neural crest GRN is comprised of sequential regulatory modules, focusing on transcription factors and signaling molecules for each stage of neural crest formation: induction, specification, delamination, migration, and differentiation. Differentiation gene batteries for five representative neural crest derivatives are placed downstream in the GRN hierarchy. Direct regulatory interactions within the neural crest GRN are represented by a solid line, while interactions that have yet to be verified as direct are represented with a dashed line. The GRN is largely conserved across vertebrates with some variation between species. For example, Twist1 is part of the neural crest specification module in frog and zebrafish, but missing in amniotes.
Initial regulatory interactions within the neural crest GRN were largely established by perturbation analyses in mouse, zebrafish, frog, chick, and the sea lamprey, with focus on the role of transcription factors and signaling molecules. Recent advances in technology have increased the resolution of each interaction by enabling analysis of direct regulatory interactions (Box 1). Thus, we are now beginning to understand, in detail, the genetic control of the sequential events of neural crest formation from a multipotent progenitor to a differentiated state.
Neural Crest Formation along the Body Axis
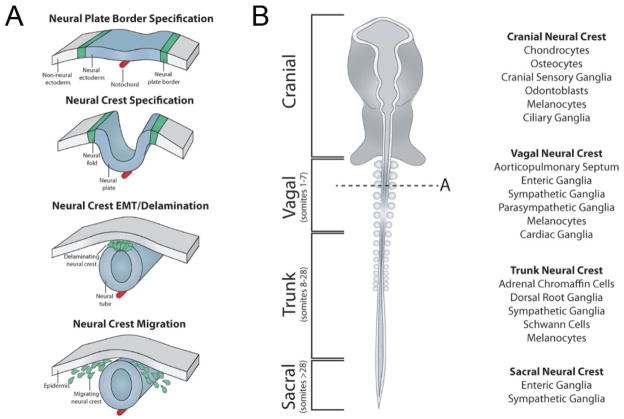
The neural crest forms by a series of sequential regulatory steps: induction, specification, delamination, migration, and differentiation (Figure 1A). During gastrulation, many signaling events influence the region at the border between neural plate and non-neural ectoderm to promote neural crest formation. Initial specification of the neural crest from the neural plate border is complete when the neural folds become elevated during neurulation. Around the time of neural tube closure, premigratory neural crest cells initially reside within the neural folds/dorsal neural tube and then undergo an epithelial-to-mesenchymal transition (EMT) to emigrate. This occurs after neural tube closure in chick and zebrafish, but prior to tube closure in frog and mouse. Following EMT, neural crest cells then migrate extensively along distinct, stereotypical pathways often to distant locations where they differentiate into a variety of derivatives (Figure 1A, 1B).
Figure 1.
Neural crest formation and its derivatives. (A) The neural crest forms by a series of important regulatory events: induction at the neural plate border, specification, delamination from the neural tube, migration throughout the embryo, and differentiation into many derivatives. During gastrulation, coordinated signaling events specify the region at the border between neural plate and non-neural ectoderm to promote neural crest formation. Initial specification of the neural crest from the neural plate border is complete when the neural folds become elevated during neurulation. In the chick, premigratory neural crest cells undergo an epithelial-to-mesenchymal transition (EMT) after neural tube closure to migrate extensively along distinct, stereotypical pathways to differentiate into a variety of derivatives. (B) Along the anterior-posterior length of the developing vertebrate embryo (left), different derivatives will form depending on the axial level (cranial, vagal, trunk, or sacral) from which they have originated. Neural crest cells at all axial levels contribute to melanocytes, the cranial neural crest gives rise to craniofacial skeleton, connective tissue, and cranial ganglia. The vagal neural crest cells contribute to the cardiac outflow tract and enteric nervous system. Trunk neural crest cells form dorsal root and sympathetic ganglia. Sacral neural crest cells will give rise to parts of the enteric and sympathetic nervous systems.
Along the anterior-posterior body axis, there are distinct neural crest populations, (cranial, vagal, trunk, and sacral) with differential abilities to form different derivatives (Figure1B). Each is controlled by unique downstream gene batteries that are influenced by extrinsic environmental factors that direct migration and activate downstream differentiation genes. Whereas neural crest cells at all axial levels contribute to melanocytes, there are differences in neural crest derivatives from different axial levels: the cranial neural crest gives rise to craniofacial skeleton, connective tissue, and cranial ganglia [7]; vagal neural crest cells contribute to the cardiac outflow tract and enteric nervous system [4,8]; trunk neural crest cells form dorsal root and sympathetic ganglia [9]; and sacral neural crest cells will give rise to components of the sympathetic and enteric nervous systems [10]. While each population is multipotent, only the cranial neural crest can give rise to cartilage [11–13] (see Box 2).
Box 2. Axial reprogramming by re-wiring GRNs.
Along the body axis of the embryo, distinct neural crest populations have different developmental potentials and migratory behaviors. Whereas individual neural crest cells at all axial levels can contribute to multiple fates, the potential to form bone and cartilage is restricted to the cranial neural crest. When trunk neural crest is grafted in vivo to the head, it fails to give rise to cartilage. While the GRNs at different axial levels (cranial, trunk, vagal, and sacral) are similar in that they each drive neural crest formation, maintain multipotency, and bestow migratory ability, each axial level is endowed with slightly regulatory differences leading to formation of different derivatives. Axial level differences in cis-regulatory activity and regulatory circuitry confer differential ability to respond to environmental cues during later development and, ultimately, lead to differences in cell fate.
Different enhancers control the activity of neural crest specifier genes Foxd3 and Sox10 in the cranial neural crest versus the trunk neural crest [29,45,59]. For instance, two enhancers for Foxd3, NC1 and NC2, are activated differentially in the cranial and trunk crest, respectively. In the cranial crest, NC1 is activated by Ets1 along with Msx1 and Pax3/7 to drive expression; in the trunk, Msx1 and Pax3/7 are also important for activation of NC2 in the trunk, but work together with Zic1 rather than Ets1 [29]. Such axial level differences likely occur throughout the GRN, though these remain to be completely understood.
Recently, a cranial specific subcircuit reflecting an axial level specific regulatory modulate was show to be missing from the trunk population [13]. A tissue-specific transcriptomics approach, used to isolate pure populations of cranial versus trunk neural crest, led to identification of regulatory state differences in the cranial and trunk neural crest. Just 15 transcription factors were found to be differentially expressed within the cranial neural crest compared to the trunk GRN. Of those 15, only 6 were expressed throughout the cranial neural crest. Focusing on these six transcription factors enabled building of a regulatory circuit with direct linkages. Surprisingly, ectopically driving 3 downstream factors (Sox8, Tfap2b, and Ets1), of this small gene circuit gave the trunk neural crest the ability to form cartilage in an ectopic location. By rewiring the trunk NC GRN by the ectopic expression of this cranial-specific subcircuit, trunk neural crest cell identity was changed to cranial-like. The ability to rewire GRNs, in vivo, has powerful implications on therapeutic approaches.
While different neural crest GRN regulatory programs are deployed at each axial level, most experiments to date have focused on the cranial neural crest. Due to the extensive literature on this cell type, here we focus on the cranial crest, but highlight axial level differences where known. Interestingly, GRN subcircuits unique to one axial level can be ectopically expressed at other axial levels to change their identity; for example, transplantation of a cranial subcircuit comprised of three transcription factors is sufficient to reprogram trunk neural crest cells and imbue them with the ability to make cartilage, a derivative they normally cannot make [13] (Box 2).
Induction and Specification of the Neural Plate Border
During gastrulation, restrictive boundaries are established between the neural plate (CNS) and non-neural ectoderm (epidermis) to form the neural plate border [14] (Figure 1A). Coordinated signaling events initiate a cascade of transcriptional activation to give rise to neural crest and placodal progenitors within the neural plate border domain [14]. While these domains become refined throughout gastrulation, many neural plate border cells can contribute to neural, neural crest, and placodal ectodermal fates, such that the fates of each cell type are only established after neural tube closure [15].
Inductive signals from neighboring tissues activate a cascade of transcription factors that specify the neural plate border from which the neural crest will arise. Three major signaling pathways, BMP, WNT, and FGF, work combinatorially to drive expression of neural plate border specifier genes [14,16–18]. Crosstalk between signaling pathways result in the formation of the neural plate border; for example, Schille and colleagues in frog, found that WNT/PCP receptor, Ror2, upregulates Gdf6 at the neural plate border [19], thus, linking Wnt/PCP signaling to BMP signaling.
The initial signaling inputs that restrict the neural plate border also regulate activation of a transcriptional circuit, involving Msx1, Dlx5/6, Pax3/7, and Gbx2, to further refine the expression domain [20]. Also activated at the border are Zic1, Tfap2, Gata2/3, Foxi1/2, and Hairy2 [14,21,22]. The components of the circuit are largely conserved across vertebrates though there are often paralogue switches between species. Positive feedback interactions within this hierarchical gene network, e.g. between Tfap2a and Msx1, ensure expression of important neural crest specifier genes Msx1 and Pax3/7. These specifier genes are critical players in activation of the downstream specification module (see below). Also important at the neural plate border are epigenetic factors like DNA methyltransferase 3a (Dnmt3a), which directly represses neural genes [23].
While signaling events initially activate neural plate border specifier genes, understanding of how these genes are directly activated by each signaling pathway remains incomplete. In frog, Gbx is directly activated by β–catenin through a proximal enhancer [24]. In zebrafish, Zic3 and Pax3a expression depend on multiple enhancers that respond differentially to BMP, WNT, and FGFs [25]. Sharpening of neural plate boundaries is achieved by reciprocal transcriptional repression, or mutual exclusion across regulatory states, which stabilizes each state (i.e. by excluding neural crest from placodal fate) (Figure 2).
Does neural crest induction occur via a gain of developmental potential or by maintenance of early embryonic stem cell properties? Recently, it has been proposed in frog that neural crest cells retain pluripotency from the blastula stage [26]. Accordingly, a parsimonious explanation may be that neural crest multipotency occurs via maintenance of a pluripotent state from the early blastula rather than induction of a new regulatory state. Thus, the shared molecular underpinnings of neural crest and blastula cells may reflect the possibility that transcription factors like Pax7 and Snai1/2 are simply pluripotency maintenance factors. To gain insight into whether neural crest formation occurs via induction or pluripotency maintenance, it will be important to better resolve direct linkages in the neural plate border regulatory module. These circuits can then be compared with other pluripotency programs.
Neural Crest Specification
After formation of the neural plate border, signaling inputs are closely linked to upregulation of neural crest specifier genes. For example, Wnt signaling is upstream of Snai2 and Foxd3 in chick and Zic1 and Pax3 in frog neural crest specification [17] [27]. In chick, the initial neural crest specifier circuit is controlled by upstream neural plate border specifiers Pax3/7 and Msx1, which form a transcriptional complex with Wnt effector, Axud1, to directly bind to and activate neural crest specifier Foxd3 [28,29], thus connecting Wnt signaling at the border with the downstream specification modules. Further integrating upstream Wnt signaling to downstream modules is a proximal enhancer of Snai2, which is directly activated by TCF/Lef and β–catenin [30].
In chick, Pax7 and Msx1 are also required for activation of neural crest specifier, Ets1 [31], which also directly regulates Foxd3 in the cranial neural crest; in the trunk, Zic1 works together with Msx1 and Pax3/7 to directly regulate Foxd3 [29]. In frog, Zic1 and Pax3/7 bind directly to Snai1/2 promoters [32,33]. Epigenetic modifier JmjD2A, a histone demethylase, is responsible for removing repressive methylation marks from the Snai2 promoter [34]. In turn, Snai1/2 in frog positively regulates Twist, Foxd3, and Sox9 [35].
Transcription factor ap2a (Tfap2a) is required for neural crest specification in fish, frog, chick, and mouse [36–39] and has a role in differentiation of melanocytes and sympathetic ganglia (as discussed below) [37,40,41]. In addition, transcription factors Myb, Myc, and Prdm1 are also required for specification [42–44]. Myb directly interacts with Pax3/7 as well as Sox10 in chick, and Prdm1 directly interacts with Tfap2a and Foxd3 in fish [43–46].
In addition to specifying the neural crest, these transcription factors are also important for activating the downstream EMT programs and stabilizing the specification regulatory state through a series of coherent feed-forward and feedback loops (Figure 2). This robust circuitry will further delineate the neural crest domain at the neural plate border from the pre-placodal domain. Below, we will discuss how this stable state initiates downstream of EMT effector modules and how environmental cues then promote many differentiation gene batteries.
Transcriptional control of morphogenesis
EMT and delamination from the neural tube
Once specified, the neural crest undergoes an epithelial to mesenchymal transition and delaminates from the dorsal neural tube to migrate extensively throughout the embryo (Figure 1A). EMT coordinates signaling and transcriptional regulation to trigger major structural changes, including de-adhesion, cytoskeletal rearrangements, and gain of motility. Comprehension of the molecular underpinnings of this complex morphogenesis is incomplete; our current understanding largely reflects the effector gene modules controlling de-adhesion.
Changes in adhesion involve dissolution of cadherin-mediated adherens junctions. In neural crest delamination, there is a “Cadherin switch” between type I Cadherins (including Ecad and Ncad) that are more important for epithelial adhesion, and more mesenchymal type II Cadherins (including Cad7 and Cad11). The transcriptional regulation of Cadherins is crucial for EMT. Snail transcription factors downregulate type I Cadherins by directly binding the Ecad promoter [47]. Snai1/2 also has been implicated, along with Lmo4, in downregulating Ncad (Type I) and Cad6b (a Type II Cadherin that is downregulated during delamination) [48]. Epigenetically, PHD12, an adapter protein, complexes Snai2 with Sin3A/HDAC to bind the Cad6b promoter that will, in turn, control repression of Snail2 on Cad6b[49]. Similarly, the Polycomb repressive complex 2 (PRC2) catalytic subunit EZH2 interacts directly with Snai2 to repress Ecad by binding its promoter[50].
Also important for downregulating Ecad are transcription factors Zeb2, FoxD3 [51,52] and Twist, which is downstream of Hif-1α [53] Transcription factor Sox9 also directly interacts with Snai1/2 to drive EMT [54,55]. While type I Cadherins (and Cad6b) are being repressed, type II Cadherins, such as Cadherin-7, are upregulated by upstream specifiers FoxD3 and Sox10 [52]. This switch in expression, along with activation of Integrin-β1 initiates downstream dispersal and migratory mechanisms.
Wnt signaling also influences EMT by transcriptionally regulating Snai1/2 [30,56]. However, Wnt/β–catenin signaling is transiently downregulated at the onset of delamination [57] by the scaffold protein DACT1/2 that regulates the subcellular distribution of β–catenin[57].
Acquisition of a Migratory ability
After delamination, neural crest cells acquire a regulatory state that enables long distance migration throughout the embryo in response to environmental cues. Migratory neural crest cells express Sox10, a critical regulator of neural crest migration and differentiation. Expression of Sox10 is controlled by multiple enhancers in chick, mice, and fish [45,58–61]. The chick cranial specific enhancer, 10E2, is regulated by neural crest specifiers Myb, Ets1, Sox9 [45]. In the trunk, Sox5 and Sox8 directly interact with the trunk specific enhancer, 10E1 [59]. Enhancer analysis in the mouse revealed seven enhancer regions that were directly bound by Pax3/7, Tfap2a, FoxD3, TCF/Lef, Sox10, as well as other Sox proteins [60,61]. Furthermore, an intronic enhancer in fish was uncovered, with direct binding sites of FoxD3, TCF/Lef, and Sox proteins [58].
Recently, tissue-specific transcriptomes have identified numerous transcription factors upregulated in the chick migratory cranial neural crest [62]. Furthermore, perturbation analysis revealed putative regulatory linkages between the upstream premigratory module and downstream transcription factors, including Lmo4, RxrG, Ltk [62], though direct connections and other linkages still need to be uncovered.
To maintain migratory ability yet remain plastic enough to respond to differential cues, the migration regulatory state must be stable. How differential migratory paths change regulatory states while both temporally and spatially responding to different cues remains unknown. As discussed below, distinct environmental signals activate downstream differentiation gene batteries to give rise to diverse array of derivatives.
Differentiation GRN circuits
Differentiation gene batteries interpret and differentially respond to a complex set of environmental signals. The gene batteries operate under a positive feed forward circuitry where initial neural crest regulators function together with locally activated differentiation effector genes. Below, we outline the most characterized terminal differentiation gene modules, which include five representative neural crest derivatives: melanocytes, chondrocytes, Schwann cells, the enteric nervous system, and the sympathetic nervous system (Figure 2).
Melanocytes
Melanocytes, which produce pigmentation of the skin and hair follicles, arise from neural crest cells at all axial levels [63]. Transcriptionally, Sox10 acts synergistically with Pax3/7 to bind and activate a proximal promoter of the critical pigmentation transcription factor Mitf [63–65]. Sox10 also directly activates Mef2c in mice, while Mef2c autoregulates itself [66]. In melanocytes, Wnt signaling induces expression of Mitf [67,68]. Melanocyte function depends on Mitf expression which, along with Sox10, directly controls the production of the melanin synthesis enzymes Dopachrome tautomerase (Dct), tyrosinase (Tyr), and Premelanosome protein (Pmel) [69,70]. Tfap2a is a pleiotropic transcription factor that has important roles in early neural crest specification as well as many differentiation gene batteries. Recently, Tfap2a will often co-occupy active regulatory elements of melanin synthesis genes with Mitf, including super-enhancers of melanin synthesis genes Dct and Pmel [40].
On the epigenetic level, HDAC1 is important for repressing Foxd3 and other neural crest fates as well as Sox10 to modulate its activity, allowing for melanocyte differentiation [65]. Taken together, these studies are building a detailed understanding of melanocytic differentiation at the gene regulatory level. These data will have an important role in understanding how melanocyte differentiation may be disrupted in diseases such as melanoma (Box 3).
Box 3. Using GRNs as tools to understand disease.
With the current state of the neural crest GRN, we can begin to extract information from and utilize it as a tool to understand neural crest derived diseases, neurocristopathies. Neurocristopathies are diseases, craniofacial malformations, and tumors of the neural crest. They include but are not limited to: melanoma, DiGeorge syndrome (craniofacial and heart defects), Treacher-Collins syndrome (craniofacial defects), Hirschsprung disease (aganglionic megacolon), pheochromocytoma, Schwannomas, and neuroblastoma. Neurocristopathies arise at various stages of neural crest development. Many mutations have been uncovered in numerous human disease. It is likely that underlying these diseases are mutations that cause a mis-wiring of GRN architecture. For instance, a nonpolyalanine repeat expansion mutation in PHOX2B alters a wildtype repression of a Sox10 enhancer to a transactivation of Sox10 leading to an imbalanced differentiation into glial versus neuronal lineages. This GRN mis-wiring event results in a higher incidence of Hirschsprung and neuroblastoma [88].
It will be critical to ascertain the underlying genetic circuitry of developing neural crest lineages at the onset of neurocristopathies. The role of individual genes necessary for onset of different neurocristopathies is an important first step for developing treatments; however, these genes regulate one another and interact within a network of many genes. Understanding the whole circuit can provide a new perspective on treatment options as well as disease ontogeny. Any aberrations within the regulatory network can confer downstream effects on other potential disease targets. Furthermore, having the potential to re-program/re-wire nonfunctioning gene networks using gene-editing techniques such as CRISPR/Cas9 could deliver a new avenue for ascertaining and treating with novel therapeutic targets.
Chondrocytes
Cranial neural crest-derived chondrocytes give rise to different elements of the craniofacial skeleton by forming the scaffold for facial bone. Sox9 in concert with Sox5/6 provide direct inputs into collagen gene, Col2a1 and cartilage differentiation marker, Agc1 [71,72]. A role for TGFβ pathway, through Smad3, in chondrogenic differentiation recruits histone acetyltransferase creb-binding protein (CBP)/p300 to allow for Sox9 activation of regulatory genes [73,74].
Schwann cells
Schwann cells are neuronal support cells that produce an insulating myelin sheath that surrounds axons of the peripheral nervous system and promote neuronal conduction by propagating electrical signals through often long peripheral nerves. Regulatory interactions important for Schwann cell development involve transcription factors that activate genes encoding myelin proteins and lipids as well as a repressor, Zeb2, which maintains a differentiated state [75,76].
The SoxE transcription factor, Sox10, activates POU domain transcription factor, Oct6, by directly binding to a Schwann cell specific enhancer (SCE) that is 10kb downstream of the Oct6 coding sequence [77–80]. Sox10 bound Oct6 recruits Brg1-containing BAF remodeling complexes and histone deacetylases HDAC1 and HDAC2 for proper enhancer activity [81,82]. The direct interactions of Sox10 and Oct6 to activate Krox20 also require BAF and HDAC1/2 for local remodeling and changes to the chromatin structure. Upon activation, this forms a coherent feed-forward circuit stabilizing the regulatory state for terminal Schwann cell differentiation. The combination of Krox20 and Sox10 controls terminal differentiation by co-occupying myelin gene regulatory regions for myelin basic protein (MBP), myelin protein zero (MPZ), proteolipid protein (PLP), and gap junction proteins Connexin32 and Connexin47 [76,83]. Furthermore, dysregulation of this gene battery leads to the formation of schwannomas, which are malignant tumors that form from abnormally differentiated Schwann cells (Box 3).
Enteric nervous system
The enteric nervous system (ENS) is comprised of a complex network of neurons and glia organized into ganglionic plexuses. The ENS regulates transport of food, digestion and absorption of nutrients, as well as other functions of the gastrointestinal tract [84]. Enteric neurons and glia arise from the vagal neural crest cells which migrate to the foregut mesenchyme. The receptor tyrosine kinase Ret signaling is responsible for the migration of enteric precursors to populate the entire gut. Transcription factor Sox10 is expressed in the developing ENS and interacts with Pax3 to synergistically activate a Ret enhancer [84,85]. Ret expression is also regulated by paired homeobox transcription factor, Phox2b[86]. Glial-derived neurotrophic factor (GDNF) activates Ret and co-receptor GDRα1 in the migrating enteric neural crest cells to activate a cascade of downstream pathways [87].
Within the ENS circuit, there is mutual repression of Sox10 and Phox2b. Whereas cells with upregulated Sox10 assume a glial fate, Phox2b+ cells form neuronal lineages. This reciprocal repression subcircuit within the ENS circuit is responsible for irreversibly tipping cell fate in one direction and ensuring a balance of glial and neuronal fates along the gut [88].
Also important within the ENS circuitry are bHLH transcription factors Achaete-scute (Ascl1) and Hand2 [89,90]. Ascl1 deletion in mice results in downregulation of calbindin, tyrosine hydroxylase, and vasoactive intestinal peptide (VIP), all of which have an important role in development of neuronal subtypes [91]. Ascl1 also has been shown to downregulate Sox10, which is important for ensuring production of neurons [92]. Mutations in Hand2 cause downregulation of neuronal differentiation leading to a loss of of nNOS, calretinin, and VIP neuronal subtypes in mice [89,93,94]. Recently, an elegant paper has characterized the neuronal and glial profile of the mammalian gut [95]. Another recent study performed a transcriptome analysis of sorted enteric neurons in zebrafish to uncover novel genes involved in ENS development [96]. Understanding all of the neuronal subtypes and the transcriptional and regulatory landscapes within the gut will further delineate neural gene regulatory circuits underlying the complexity of the enteric nervous system.
Sympathetic nervous system
The sympathetic nervous system is the branch of the autonomic nervous system responsible for the flight-or-fight response and needed to maintain organ homeostasis, heart rate, and many other functions in response to an external challenge. Sympathetic neurons arise primarily from trunk neural crest. In response to BMP signaling at the dorsal aorta, a cascade of transcription factors is activated within a differentiation gene battery [97]. Smad4-dependent and –independent BMP signaling activates Ascl1 and Phox2b. Through a positive feedback mechanism, Phox2b is required for maintenance of Ascl1 expression [97,98]. Downstream of the initial activation of Phox2b and Ascl1 is the activation of Phox2a, Hand2, and zinc finger proteins Gata2 and Gata3, and Insm1 [97,99,100].
Sympathetic neurons are primarily adrenergic, producing norepinephrine via enzymes tyrosine hydroxylase (TH) and dopamine beta hydroxylase (DBH), which are directly bound by transcription factors Tfap2a and Phox2a to activate their transcription [99,101]. The sympathetic nervous system differentiation circuit is laced with positive feedback loops that result in expression of norepinephrine and neurogenesis regulators. Understanding the genetic program controlling normal development of the sympathoadrenal lineage may also inform upon the aberrant regulatory network that leads to onset of neuroblastoma, thus helping identify novel therapeutic targets.
Concluding Remarks
This overview of the gene regulatory network underlying neural crest development highlights recent advances, from initial induction to differentiation into a subset of derivatives. This combination of studies in numerous vertebrate models allows us to infer and update a GRN as viewed in a Biotapestry model (Figure 2) [102]. As more linkages are validated, regulatory interactions within this robust GRN will become increasingly resolved. This neural crest GRN provides a useful tool to understand developmental processes, disease, cancer (Box 3), and evolution (Box 4) at a systems-level.
Box 4. Using GRNs to understand morphological novelty.
Morphological novelties arise due to changes in the regulatory program. The neural crest is a cell type that is unique to vertebrates, raising the question of whether evolution of neural crest was preceded by co-opted regulatory programs from other tissues into the neural crest lineage. Questions such as these can be answered by GRN comparisons to interrogate which nodes underlie trait diversity. For example, comparisons between jawed and jawless like the sea lamprey may lead to identification of traits as well as GRN states that were present in early vertebrates. Here, we focus on regulatory architecture comparison between vertebrates. For a discussion on invertebrate chordates, see Green et al 2015 [104].
Jaws are unique to jawed vertebrates, arising by modification of the anterior pharyngeal arches. Other features unique to jawed vertebrates include ondontoblasts that produce dentine, sympathetic neurons, and vagal-derived enteric neural crest [104,105]. While there are astonishing similarities between the early specification neural crest GRNs of jawed and jawless vertebrates [106], tracing regulatory differences between jawed and jawless vertebrates may have lead to formation of new cells types, thus correlating with the divergence of these cell fates.
Conserved in the early specification network are the importance of transcription factors Pax7, Msx1, and Zic1 in the sea lamprey [106]. Similar to the jawed vertebrate GRN (Figure 2), knockdown of these neural plate border transcription factors results in a downregulation of foxd3 and soxE1 [106]. A key difference in the early gene networks of lamprey is a lack of cranial specific transcription factors, Ets1 and Twist. Could a lack of these transcription factors in early regulatory modules cause distinct traits to be missing in lamprey? Or could a heterochronic shift in gene expression lead to novel regulatory interactions and circuitry in jawed vertebrates? Further investigation of the jawless vertebrate GRN is sure to uncover such discrepancies.
Throughout evolution, differentiation gene batteries arose to give rise to novel cell types. It is likely that differentiation gene batteries were “plugged in” to the upstream GRN hierarchy by gain of function cis-regulatory changes in differentiation genes, which led to their control by upstream neural crest specifiers. Regulatory analyses such as ATAC-seq will help to uncover differences between lamprey and jawed vertebrates in regulatory DNA at these time points.
Trends.
The neural crest GRN provides a unique look at regulatory hierarchies underlying multipotency and such developmental events as specification, delamination, migration, and differentiation into various derivatives.
Interrogating direct connections is now possible on a larger scale with a variety of new technological advances.
Rewiring of GRN architecture has been shown to reprogram trunk neural crest into a cranial identity, which delivers a powerful route for novel therapeutic approaches.
Gene regulatory networks provide a foundation from which to understand onset of neurocristopathies.
Regulatory changes within the neural crest GRN lead to hypotheses concerning the evolution of morphological novelties such as jaws.
Outstanding Questions Box.
What are the axial level gene regulatory network differences between cranial, vagal, trunk, and sacral neural crest cells, and are those differences responsible for the suite of derivatives each axial level is fated to become?
Does neural crest induction occur via a gain of developmental potential at the neural plate border or by maintenance of early embryonic stem cell properties?
How does the regulatory state of the migratory neural crest transition to initiate differential downstream gene batteries?
How can bioinformatics tools and advanced technologies such as RNA-seq and ATAC-seq be exploited to uncover the entirety of the neural crest GRN complete with direct linkages?
At what hierarchical level do gene regulatory network circuits change to give rise to morphological novelties such as jaws, sympathetic ganglia, and the vagal-derived enteric nervous system?
Acknowledgments
Work in the laboratory of M.E.B. is supported by NIH R01DE02415, R01NS08690, and HD037105. M.L.M. is funded by the Helen Hay Whitney Foundation.
Glossary
- Apomorphy
the neural crest is an apomorphy in that it is a derived trait that is unique to vertebrates and present in their last common ancestor
- Enhancer
short, regulatory regions of DNA that is bound by transcription factors to control the activation and transcription of a particular gene
- Epithelial-Mesenchymal-Transition
a series of complex cell state changes (ie. structural remodeling, de-adhesion, delamination) that transforms an epithelial cell to a mesenchymal cell
- Multipotent
a cell that can develop into many cell types
- Neurocristopathy
syndromes that involve the dysregulation of one or more neural crest cell populations
- Regulatory state
the combination of transcription factors and signaling molecules expressed in a given cell type at a specific time
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 2.Hall BK. The neural crest as a fourth germ layer and vertebrates as quadroblastic not triploblastic. Evol Dev. 2000;2:3–5. doi: 10.1046/j.1525-142x.2000.00032.x. [DOI] [PubMed] [Google Scholar]
- 3.Hall BK. Evolutionary Biology. Springer; Boston, MA: 1998. Germ Layers and the Germ-Layer Theory Revisited; pp. 121–186. [Google Scholar]
- 4.Le Douarin N. A biological cell labeling technique and its use in experimental embryology. Dev Biol. 1973;30:217–222. doi: 10.1016/0012-1606(73)90061-4. [DOI] [PubMed] [Google Scholar]
- 5.Simoes-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couly G, et al. Determination of the identity of the derivatives of the cephalic neural crest: incompatibility between Hox gene expression and lower jaw development. Development. 1998;125:3445–3459. doi: 10.1242/dev.125.17.3445. [DOI] [PubMed] [Google Scholar]
- 8.Creazzo Tony L, et al. ROLE OF CARDIAC NEURAL CREST CELLS IN CARDIOVASCULAR DEVELOPMENT. 2003;60:267–286. doi: 10.1146/annurev.physiol.60.1.267. http://dx.doi.org/10.1146/annurev.physiol.60.1.267. [DOI] [PubMed] [Google Scholar]
- 9.Le Douarin N. Development Of The Peripheral Nervous System From The Neural Crest. Annu Rev Cell Dev Biol. 1988;4:375–404. doi: 10.1146/annurev.cb.04.110188.002111. [DOI] [PubMed] [Google Scholar]
- 10.Espinosa-Medina I, et al. The sacral autonomic outflow is sympathetic. Science. 2016;354:893–897. doi: 10.1126/science.aah5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. Development. 1975;34:125–154. [PubMed] [Google Scholar]
- 12.Le Lievre CS, et al. Restrictions of developmental capabilities in neural crest cell derivatives as tested by in vivo transplantation experiments. Dev Biol. 1980;77:362–378. doi: 10.1016/0012-1606(80)90481-9. [DOI] [PubMed] [Google Scholar]
- 13.Simoes-Costa M, Bronner ME. Reprogramming of avian neural crest axial identity and cell fate. Science. 2016;352:1570–1573. doi: 10.1126/science.aaf2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groves AK, LaBonne C. Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Dev Biol. 2014;389:2–12. doi: 10.1016/j.ydbio.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roellig D, et al. Dynamic transcriptional signature and cell fate analysis reveals plasticity of individual neural plate border cells. Elife. 2017;6:e21620. doi: 10.7554/eLife.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuhlmiller TJ, García-Castro MI. FGF/MAPK signaling is required in the gastrula epiblast for avian neural crest induction. Development. 2012;139:289–300. doi: 10.1242/dev.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Castro MI, et al. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- 18.Leung AW, et al. WNT/β-catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development. 2016;143:398–410. doi: 10.1242/dev.130849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schille C, et al. Ror2 signaling is required for local upregulation of GDF6 and activation of BMP signaling at the neural plate border. Development. 2016;143:3182–3194. doi: 10.1242/dev.135426. [DOI] [PubMed] [Google Scholar]
- 20.Basch ML, et al. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- 21.Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Developmental Dynamics. 2009;238:716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichane M, et al. Hairy2 functions through both DNA-binding and non DNA-binding mechanisms at the neural plate border in Xenopus. Dev Biol. 2008;322:368–380. doi: 10.1016/j.ydbio.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 23.Hu N, et al. DNA methyltransferase3A as a molecular switch mediating theneural tube-to-neural crest fate transition. Genes Dev. 2012;26:2380–2385. doi: 10.1101/gad.198747.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, et al. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development. 2009;136:3267–3278. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garnett AT, et al. BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development. 2012;139:4220–4231. doi: 10.1242/dev.081497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buitrago-Delgado E, et al. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science. 2015;348:1332–1335. doi: 10.1126/science.aaa3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato T, et al. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- 28.Simoes-Costa M, et al. Axud1 Integrates Wnt Signaling and Transcriptional Inputs to Drive Neural Crest Formation. Dev Cell. 2015;34:544–554. doi: 10.1016/j.devcel.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simões-Costa MS, et al. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genet. 2012;8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallin J, et al. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J Biol Chem. 2001;276:30350–30358. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- 31.Barembaum M, Bronner ME. Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Dev Biol. 2013;382:567–575. doi: 10.1016/j.ydbio.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milet C, et al. Pax3 and Zic1 drive induction and differentiation of multipotent, migratory, and functional neural crest in Xenopus embryos. Proc Natl Acad Sci USA. 2013;110:5528–5533. doi: 10.1073/pnas.1219124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plouhinec JL, et al. Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev Biol. 2014;386:461–472. doi: 10.1016/j.ydbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strobl-Mazzulla PH, et al. Epigenetic landscape and miRNA involvement during neural crest development. Developmental Dynamics. 2012;241:1849–1856. doi: 10.1002/dvdy.23868. [DOI] [PubMed] [Google Scholar]
- 35.Aybar MJ, et al. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130:483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- 36.Schorle H, et al. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 37.de Crozé N, et al. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci USA. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knight RD, et al. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755–5768. doi: 10.1242/dev.00575. [DOI] [PubMed] [Google Scholar]
- 39.Luo T, et al. Induction of neural crest in Xenopus by transcription factor AP2α. PNAS. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seberg HE, et al. TFAP2 paralogs regulate melanocyte differentiation in parallel with MITF. PLoS Genet. 2017;13:e1006636. doi: 10.1371/journal.pgen.1006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holzschuh J, et al. Noradrenergic neurons in the zebrafish hindbrain are induced by retinoic acid and require tfap2a for expression of the neurotransmitter phenotype. Development. 2003;130:5741–5754. doi: 10.1242/dev.00816. [DOI] [PubMed] [Google Scholar]
- 42.Bellmeyer A, et al. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 43.Betancur P, et al. Expression and function of transcription factor cMyb during cranial neural crest development. Mech Dev. 2014;132:38–43. doi: 10.1016/j.mod.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell DR, et al. Prdm1a directly activates foxd3 and tfap2a during zebrafish neural crest specification. Development. 2013;140:3445–3455. doi: 10.1242/dev.096164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betancur P, et al. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc Natl Acad Sci USA. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vadasz S, et al. Pax7 is regulated by cMyb during early neural crest development through a novel enhancer. Development. 2013;140:3691–3702. doi: 10.1242/dev.088328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cano A, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 48.Ferronha T, et al. LMO4 is an essential cofactor in the Snail2-mediated epithelial-to-mesenchymal transition of neuroblastoma and neural crest cells. J Neurosci. 2013;33:2773–2783. doi: 10.1523/JNEUROSCI.4511-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strobl-Mazzulla PH, Bronner ME. A PHD12-Snail2 repressive complex epigenetically mediates neural crest epithelial-to-mesenchymal transition. J Cell Biol. 2012;198:999–1010. doi: 10.1083/jcb.201203098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tien CL, et al. Snail2/Slug cooperates with Polycomb repressive complex 2 (PRC2) to regulate neural crest development. Development. 2015;142:722–731. doi: 10.1242/dev.111997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers CD, et al. Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. J Cell Biol. 2013;203:835–847. doi: 10.1083/jcb.201305050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheung M, et al. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Barriga EH, et al. The hypoxia factor Hif-1α controls neural crest chemotaxis and epithelial to mesenchymal transition. J Cell Biol. 2013;201:759–776. doi: 10.1083/jcb.201212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 55.Liu JAJ, et al. Phosphorylation of Sox9 is required for neural crest delamination and is regulated downstream of BMP and canonical Wnt signaling. Proc Natl Acad Sci USA. 2013;110:2882–2887. doi: 10.1073/pnas.1211747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.ten Berge D, et al. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabadán MA, et al. Delamination of neural crest cells requires transient and reversible Wnt inhibition mediated by Dact1/2. Development. 2016;143:2194–2205. doi: 10.1242/dev.134981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dutton JR, et al. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev Biol. 2008;8:105. doi: 10.1186/1471-213X-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murko C, Bronner ME. Tissue specific regulation of the chick Sox10E1 enhancer by different Sox family members. Dev Biol. 2017;422:47–57. doi: 10.1016/j.ydbio.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Werner T, et al. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wahlbuhl M, et al. Transcription factor Sox10 orchestrates activity of a neural crest-specific enhancer in the vicinity of its gene. Nucleic Acids Res. 2012;40:88–101. doi: 10.1093/nar/gkr734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simoes-Costa M, et al. Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome Res. 2014;24:281–290. doi: 10.1101/gr.161182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mort RL, et al. The melanocyte lineage in development and disease. Development. 2015;142:620–632. doi: 10.1242/dev.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elworthy S, et al. Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development. 2003;130:2809–2818. doi: 10.1242/dev.00461. [DOI] [PubMed] [Google Scholar]
- 65.Greenhill ER, et al. An iterative genetic and dynamical modelling approach identifies novel features of the gene regulatory network underlying melanocyte development. PLoS Genet. 2011;7:e1002265. doi: 10.1371/journal.pgen.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Agarwal P, et al. The MADS box transcription factor MEF2C regulates melanocyte development and is a direct transcriptional target and partner of SOX10. Development. 2011;138:2555–2565. doi: 10.1242/dev.056804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin EJ, et al. Wnt and BMP signaling govern lineage segregation of melanocytes in the avian embryo. Dev Biol. 2001;233:22–37. doi: 10.1006/dbio.2001.0222. [DOI] [PubMed] [Google Scholar]
- 68.Takeda K, et al. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J Biol Chem. 2000;275:14013–14016. doi: 10.1074/jbc.c000113200. [DOI] [PubMed] [Google Scholar]
- 69.Potterf SB, et al. Analysis of SOX10 function in neural crest-derived melanocyte development: SOX10-dependent transcriptional control of dopachrome tautomerase. Dev Biol. 2001;237:245–257. doi: 10.1006/dbio.2001.0372. [DOI] [PubMed] [Google Scholar]
- 70.Ludwig A, et al. Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Letters. 2004;556:236–244. doi: 10.1016/s0014-5793(03)01446-7. [DOI] [PubMed] [Google Scholar]
- 71.Lefebvre V, et al. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bell DM, et al. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 73.Furumatsu T, et al. Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J Biol Chem. 2005;280:8343–8350. doi: 10.1074/jbc.M413913200. [DOI] [PubMed] [Google Scholar]
- 74.Tsuda M, et al. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem. 2003;278:27224–27229. doi: 10.1074/jbc.M303471200. [DOI] [PubMed] [Google Scholar]
- 75.Quintes S, et al. Zeb2 is essential for Schwann cell differentiation, myelination and nerve repair. Nat Neurosci. 2016;19:1050–1059. doi: 10.1038/nn.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stolt CC, Wegner M. Schwann cells and their transcriptional network: Evolution of key regulators of peripheral myelination. Brain Res. 2016;1641:101–110. doi: 10.1016/j.brainres.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 77.Schreiner S, et al. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007;134:3271–3281. doi: 10.1242/dev.003350. [DOI] [PubMed] [Google Scholar]
- 78.Ghazvini M, et al. A cell type-specific allele of the POU gene Oct-6 reveals Schwann cell autonomous function in nerve development and regeneration. EMBO J. 2002;21:4612–4620. doi: 10.1093/emboj/cdf475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jagalur NB, et al. Functional dissection of the Oct6 Schwann cell enhancer reveals an essential role for dimeric Sox10 binding. J Neurosci. 2011;31:8585–8594. doi: 10.1523/JNEUROSCI.0659-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mandemakers W, et al. A distal Schwann cell-specific enhancer mediates axonal regulation of the Oct-6 transcription factor during peripheral nerve development and regeneration. EMBO J. 2000;19:2992–3003. doi: 10.1093/emboj/19.12.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacob C, et al. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci. 2011;14:429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- 82.Weider M, et al. Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev Cell. 2012;23:193–201. doi: 10.1016/j.devcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 83.Schlierf B, et al. Expression of connexin47 in oligodendrocytes is regulated by the Sox10 transcription factor. J Mol Biol. 2006;361:11–21. doi: 10.1016/j.jmb.2006.05.072. [DOI] [PubMed] [Google Scholar]
- 84.Nagy N, Goldstein AM. Enteric nervous system development: A crest cell’s journey from neural tube to colon. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lang D, et al. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pattyn A, et al. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 87.Asai N, et al. Targeted mutation of serine 697 in the Ret tyrosine kinase causes migration defect of enteric neural crest cells. Development. 2006;133:4507–4516. doi: 10.1242/dev.02616. [DOI] [PubMed] [Google Scholar]
- 88.Nagashimada M, et al. Autonomic neurocristopathy-associated mutations in PHOX2B dysregulate Sox10 expression. J Clin Invest. 2012;122:3145–3158. doi: 10.1172/JCI63401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.D’Autréaux F, et al. Hand2 is necessary for terminal differentiation of enteric neurons from crest-derived precursors but not for their migration into the gut or for formation of glia. Development. 2007;134:2237–2249. doi: 10.1242/dev.003814. [DOI] [PubMed] [Google Scholar]
- 90.Lei J, Howard MJ. Targeted deletion of Hand2 in enteric neural precursor cells affects its functions in neurogenesis, neurotransmitter specification and gangliogenesis, causing functional aganglionosis. Development. 2011;138:4789–4800. doi: 10.1242/dev.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Memic F, et al. Ascl1 Is Required for the Development of Specific Neuronal Subtypes in the Enteric Nervous System. J Neurosci. 2016;36:4339–4350. doi: 10.1523/JNEUROSCI.0202-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim J, et al. SOX10 Maintains Multipotency and Inhibits Neuronal Differentiation of Neural Crest Stem Cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 93.D’Autréaux F, et al. Expression level of Hand2 affects specification of enteric neurons and gastrointestinal function in mice. Gastroenterology. 2011;141:576–587. e1–6. doi: 10.1053/j.gastro.2011.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hendershot TJ, et al. Expression of Hand2 is sufficient for neurogenesis and cell type-specific gene expression in the enteric nervous system. Dev Dyn. 2007;236:93–105. doi: 10.1002/dvdy.20989. [DOI] [PubMed] [Google Scholar]
- 95.Lasrado R, et al. Lineage-dependent spatial and functional organization of the mammalian enteric nervous system. Science. 2017;356:722–726. doi: 10.1126/science.aam7511. [DOI] [PubMed] [Google Scholar]
- 96.Roy-Carson S, et al. Defining the transcriptomic landscape of the developing enteric nervous system and its cellular environment. BMC Genomics. 2017;18:290. doi: 10.1186/s12864-017-3653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huber K. The sympathoadrenal cell lineage: specification, diversification, and new perspectives. Dev Biol. 2006;298:335–343. doi: 10.1016/j.ydbio.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 98.Morikawa Y, et al. BMP signaling regulates sympathetic nervous system development through Smad4-dependent and -independent pathways. Development. 2009;136:3575–3584. doi: 10.1242/dev.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morrison MA, et al. Studying the peripheral sympathetic nervous system and neuroblastoma in zebrafish. Methods Cell Biol. 2016;134:97–138. doi: 10.1016/bs.mcb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 100.Howard MJ, et al. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 2000;127:4073–4081. doi: 10.1242/dev.127.18.4073. [DOI] [PubMed] [Google Scholar]
- 101.Kim HS, et al. Regulation of the tyrosine hydroxylase and dopamine beta-hydroxylase genes by the transcription factor AP-2. J Neurochem. 2001;76:280–294. doi: 10.1046/j.1471-4159.2001.00044.x. [DOI] [PubMed] [Google Scholar]
- 102.Longabaugh WJR, et al. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 103.Trinh LA, et al. Biotagging of Specific Cell Populations in Zebrafish Reveals Gene Regulatory Logic Encoded in the Nuclear Transcriptome. Cell Rep. 2017;19:425–440. doi: 10.1016/j.celrep.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Green SA, et al. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Green SA, et al. Ancient evolutionary origin of vertebrate enteric neurons from trunk-derived neural crest. Nature. 2017;544:88–91. doi: 10.1038/nature21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sauka-Spengler T, et al. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]