SUMMARY
The three-dimensional arrangement of the human genome comprises a complex network of structural and regulatory chromatin loops important for coordinating changes in transcription during human development. To better understand the mechanisms underlying context-specific 3D chromatin structure and transcription during cellular differentiation, we generated comprehensive in situ Hi-C maps of DNA loops in human monocytes and differentiated macrophages. We demonstrate that dynamic looping events are regulatory rather than structural in nature and uncover widespread coordination of dynamic enhancer activity at preformed and acquired DNA loops. Enhancer-bound loop formation and enhancer-activation of preformed loops together form multi-loop activation hubs at key macrophage genes. Activation hubs connect 3.4 enhancers per promoter and exhibit a strong enrichment for Activator Protein 1 (AP-1) binding events, suggesting multi-loop activation hubs involving cell-type specific transcription factors may represent an important class of regulatory chromatin structures for the spatiotemporal control of transcription.
Keywords: CTCF, AP-1, Hi-C, DNA looping, cellular differentiation
INTRODUCTION
Less than 2% of the human genome codes for functional proteins (Alexander et al., 2010). Scattered throughout the rest of the genome are regulatory regions that can exert control over genes hundreds of thousands of base pairs away through the formation of DNA loops. Loop-based transcriptional regulation plays a part in many biological contexts but is critically important for human development and cellular differentiation (Krijger and de Laat, 2016). Alteration of DNA loops has been implicated in a variety of developmental abnormalities and human diseases (Hnisz et al., 2016; Montavon et al., 2011; Narendra et al., 2016).
Typically acting in cis at distances no larger than 2 Mb, DNA loops are often confined within structures known as topologically associating domains (TADs) (Dixon et al., 2012; Nora et al., 2012) (Dekker et al., 2013). TADs themselves are partitioned into two or more nuclear compartments of similar transcriptional activity (Imakaev et al., 2012; Lieberman-Aiden et al., 2009). At a local level, loops are controlled by proteins including CCCTC-binding factor (CTCF) cohesin, Nipped-B-like protein (NIPBL), MAU2 chromatid cohesion factor homolog (MAU2), and Wings apart-like protein homolog (WAPL) (Busslinger et al., 2017; Haarhuis et al., 2017; Heidari et al., 2014; Nora et al., 2017; Rao et al., 2014; Sofueva et al., 2013). Cohesin is first loaded onto DNA by NIPBL and MAU2 (Ciosk et al., 2000). Extrusion of DNA through the cohesion complex leads to enlargement of loops. Properly oriented CTCF binding can stop or pause the extrusion process. Ultimately cohesin is removed from chromatin by WAPL (Busslinger et al., 2017; Gandhi et al., 2006; Haarhuis et al., 2017). In addition to these cell type invariant mechanisms, several recent studies have demonstrated a role for tissue-specific transcription factors in loop formation (Krivega et al., 2014; Lee et al., 2017; Song et al., 2007). While our knowledge regarding the general characteristics and mechanisms of loops is improving, much less is known regarding the scope, mechanisms, and functional significance of dynamic looping events during biological processes such as cellular differentiation.
Rapidly evolving DNA sequencing-based technologies have provided increasingly comprehensive views of DNA looping in human cells and are improving our ability to address these salient questions. Hi-C is a genome-wide approach to detect contact frequencies between all mappable regions of the human genome. Application of this approach has revealed extensive chromatin reorganization, both within stable chromatin domains and in higher order compartment localization, during ES cell differentiation and across human tissues and cell-types (Dixon et al., 2015; Schmitt et al., 2016). However, the sequencing depth required to achieve high resolution maps of contact frequency as well as the high number of nonspecific ligation events have made the identification of loops using Hi-C problematic.
To circumvent these issues, multiple approaches have been developed that target small fractions of the genome, thus reducing the sequencing burden (Dostie et al., 2006; Fullwood et al., 2009; Mumbach et al., 2016). Application of one such approach, targeted chromosome conformation capture approach (5C), which interrogates small contiguous genomic stretches, identified examples of short-range non-CTCF loops that change in differentiating mouse embryonic stem cells at specific regulatory genes (Phillips-Cremins et al., 2013), and which to varying degrees are dynamically restored during somatic cell reprogramming (Beagan et al., 2016). Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET), which maps interactions between genomic regions bound by specific proteins, provides further examples of dynamic enhancer and promoter interactions between ES cells and differentiated B lymphocytes (Kieffer-Kwon et al., 2013). While useful for asking specific questions, such targeted approaches are unsuitable for identifying genome-wide intra-chromosomal interactions that are likely important for cellular differentiation. By interrogating only small, interspersed regions of the genome, approaches such as ChIA-PET, capture Hi-C, and HiChIP may not be able to distinguish differences in local chromatin compaction from true DNA loops (Rao et al., 2014).
The introduction of a modified Hi-C protocol, in situ Hi-C, in combination with the continually decreasing cost of genomic sequencing, has allowed for the unbiased genome-wide detection of DNA loops in human cells. Nuclear proximity ligation increases the efficiency of in situ Hi-C over existing protocols by reducing random ligation events, enabling higher resolution and detailed mapping of DNA loops at currently achievable sequencing depths. Importantly, the comprehensive nature of in situ Hi-C allows comparison of interaction frequencies to local backgrounds, something not possible with targeted methods like ChIA-PET, capture Hi-C, and HiChIP, producing quantifiable improvements in accuracy of loop detection (Rao et al., 2014). While multi-cell comparison of high resolution in situ Hi-C maps has identified thousands of DNA loops that are preserved across diverse cell types (Rao et al., 2014), examples of cell-type specific looping events also support a role for dynamic genome architecture regulating specific genes. However, this method has not been broadly applied to study dynamic looping in the context of human development and cellular differentiation.
Macrophages are phagocytic cells of the innate immune system that represent one of the body’s first defenses against invading pathogens. Macrophage-mediated inflammation has been identified as a key driver of multiple human disorders and diseases including atherosclerosis, diabetes, and cancer (Chawla et al., 2011; Moore et al., 2013; Noy and Pollard, 2014; Ostuni et al., 2015). The differentiation of monocytic precursors into mature macrophages is well characterized and key transcriptional regulators of this process have been identified including SPI1, MAFB, and AP-1 (Consortium et al., 2009; Kelly et al., 2000; Rosa et al., 2007; Valledor et al., 1998). However, the role of DNA looping in this process remains largely unexplored. Treatment of the monocytic leukemia cell line, THP-1, with phorbol myristate acetate (PMA), is a widely used model for studying monocyte-macrophage differentiation and provides an ideal system for studying the regulatory dynamics controlled via long-range interactions (Daigneault et al., 2010). First, THP-1 cells grow as a largely homogenous cell population with a near-diploid genetic background lacking major cytogenetic rearrangements typical of most established cell lines (Odero et al., 2000). Second, they exhibit high functional similarity to in vivo monocytes including the ability to differentiate into extremely pure populations of macrophages with over 95% of cells transitioning to macrophages following PMA treatment (Kouno et al., 2013; Lund et al., 2016). And finally, because these cells renew indefinitely, unlimited experiments can be performed on these same cells eliminating variability introduced by genetic differences.
Here, we apply in situ Hi-C in combination with other genomic methodologies to profile global changes in DNA looping events during the differentiation of human monocytes into macrophages. We show that the transcriptional dynamics of differentiating THP-1 cells are accompanied by changes in long-range DNA loops at key regulatory genes known to be important for macrophage development and function. Intersection with histone modification profiles reveals that transcriptional regulation is accompanied by both gained and pre-formed chromatin loops that acquire enhancer activity during differentiation. These gained and enhancer-activated loops form multiple-loop regulatory communities that connect single promoters to multiple distal enhancers. Strong enrichment of AP-1 binding sites is observed at the promoter-distal ends of these gained and activated loops. AP-1 activation hubs are similar in nature to specific, previously characterized multi-loop structures, such as the developmentally regulated beta globin locus, suggesting that activation hubs may represent a common mechanism for controlling the spatiotemporal expression of distinct regulatory genes important for other developmental contexts.
RESULTS
High resolution maps of DNA structure in human monocytes and macrophages
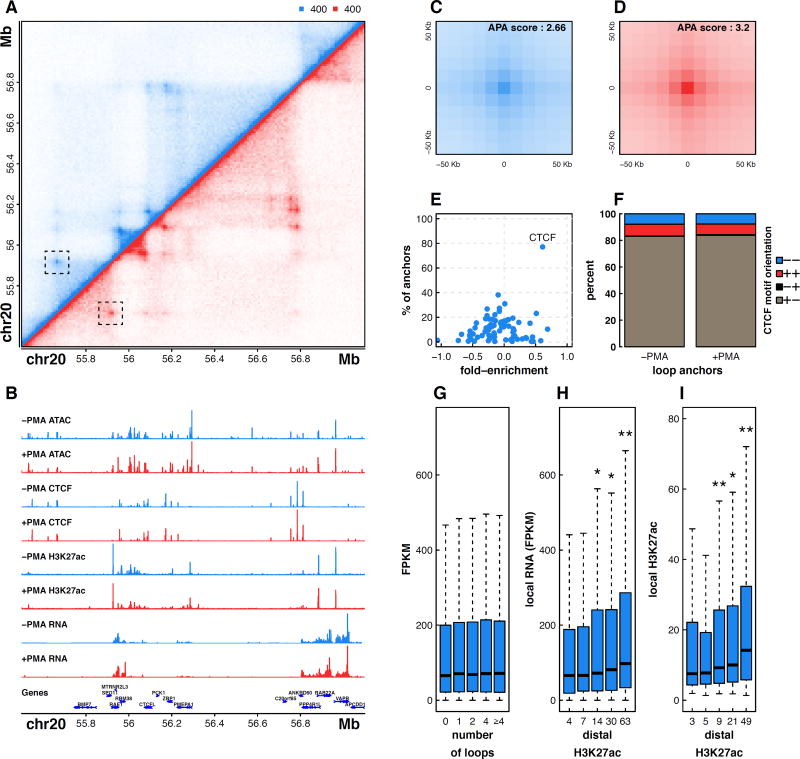
We generated high-resolution genome-wide chromatin interaction maps of THP-1 cells before and after exposure to PMA using in situ Hi-C. Sequencing to a depth of greater than 5 billion reads per sample, we generated contact maps with a bin resolution of 10 Kb (Fig. 1A). In addition, we mapped changes in transcript abundance (RNA-seq), chromatin accessibility (ATAC-seq), CTCF occupancy, and histone H3K27 acetylation (H3K27ac) levels (ChIP-seq), a histone modification associated with active enhancers (Fig. 1B) (Creyghton et al., 2010). Using an approach developed by Rao et al, we identified 16067 and 16335 loops in untreated and treated THP-1 cells respectively (Tables S1,S2)(Rao et al., 2014). Loop detection by Juicer yielded very similar results (Fig. S1) (Durand et al., 2016). The accuracy of the resulting loop calls was supported by high scoring Aggregate Peak Analysis (APA) plots (Fig. 1C–D)(Rao et al., 2014). Motif analysis at loop anchors revealed a strong enrichment for CTCF binding and bias for inward oriented CTCF motifs, consistent with previous reports (Fig. 1E–F)(Rao et al., 2014). However, we also identify clear examples of CTCF-independent looping (Fig. S2), raising critical questions regarding the mechanisms driving the formation and maintenance of DNA loops in human cells and highlighting the value of genome-wide interaction mapping.
Figure 1. Multi-omic mapping of chromatin architecture in untreated and +PMA treated THP-1 cells.
(A) Hi-C contact matrix depicting normalized contact frequencies for untreated THP-1 cells (blue, top left) and PMA treated THP-1 cells (red, bottom right). One loop is highlighted. (B) ATAC-seq, ChIP-seq, and RNA-seq signal tracks. APA plots showing aggregated signal across all loops in both untreated (C) and PMA treated THP-1 cells (D). (E) Motif enrichment in loop anchors of untreated THP-1 cells. (F) Distribution of CTCF motif orientations at loop anchors. (G) RNA FPKM values of genes binned by the number of loops that connect to their promoter. (H) RNA FPKM values of genes binned by the histone H3K27ac signal at the promoter-distal end of a loop. (I) H3K27ac signal binned by the histone H3K27ac signal at the other end of a loop. Significant differences (p < 0.05, Wilcox Rank Sum Test) vs subset to the immediate left or all subsets to the left are indicated by one or two asterisks respectively. See also Figures S1 and S2.
To explore the regulatory nature of DNA looping in monocytes and macrophages we next intersected our loop calls with H3K27ac signal and transcript abundance. Comparison of relative transcript level and the number of long-range interactions that a promoter is connected to reveals no significant relationship between gene activity and number of DNA loops (Fig. 1G). However, we find that both transcription and local H3K27ac are higher for regions connected to distal DNA elements with elevated H3K27ac, implying active enhancer-gene interactions (Fig. 1H–I). Overall, this suggests that the activity and chromatin context at distal regulatory element connections, rather than the mere presence or number of DNA loops, is a major determinant of gene expression and perhaps an important element controlling dynamic transcription of regulatory genes during cellular differentiation.
Macrophage-specific transcription is associated with both dynamic and pre-formed DNA loops
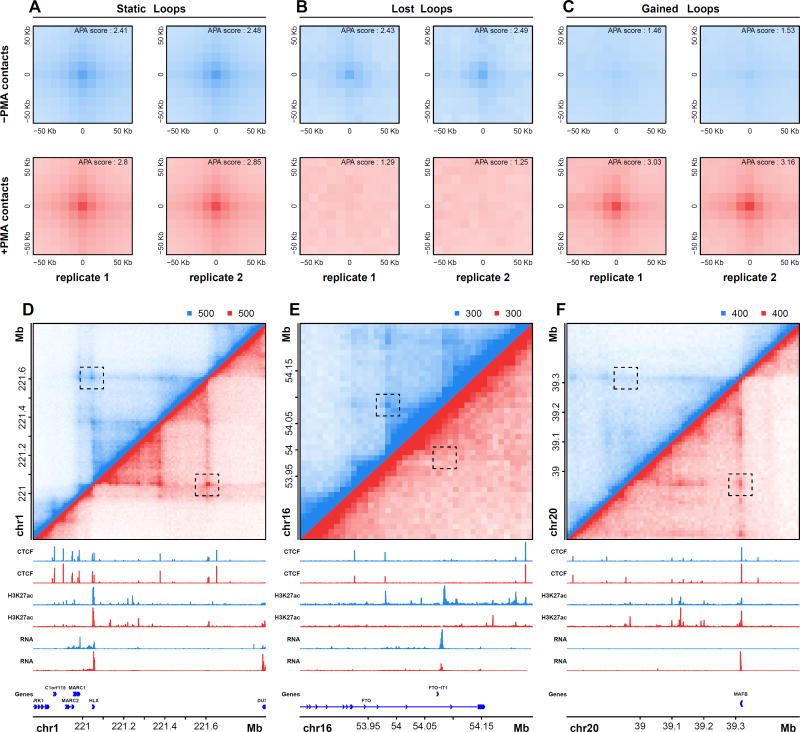
We next leveraged our Hi-C data to determine whether differentiation of THP-1 cells is accompanied by dynamic looping events. Using a method that explicitly tests for changes relative to local background, we identified 217 differential looping events following PMA treatment (DESeq2, p < 0.001; STAR Methods, Table S3). APA analysis illustrates both the reproducibility of dynamic interactions across biological replicates and the significant difference in contact frequencies before and after differentiation, suggesting our stringent approach identifies loops that are entirely lost or gained during macrophage development (Fig. 2A–C). Examples of loops that were static, lost, or gained during differentiation are shown in Figure 2D–F and Figure S3A–C. Loops gained during differentiation are more abundant (184 vs 33) and overlap more genes (88 vs 8) than loops lost during differentiation, suggesting that loop formation as opposed to loop disruption may play a broader role in macrophage development.
Figure 2. Detection and visualization of differential looping events during differentiation.
APA plots for loops that are static (A), lost (B), or are gained during PMA-induced differentiation of THP-1 cells (C). Individual plots for each biological replicate and condition are shown. Hi-C contract matrices, ChIP-seq signal tracks, RNA-seq signal tracks, and genes are shown for examples of static (D), lost (E), or gained (F) loops. See also Figure S3.
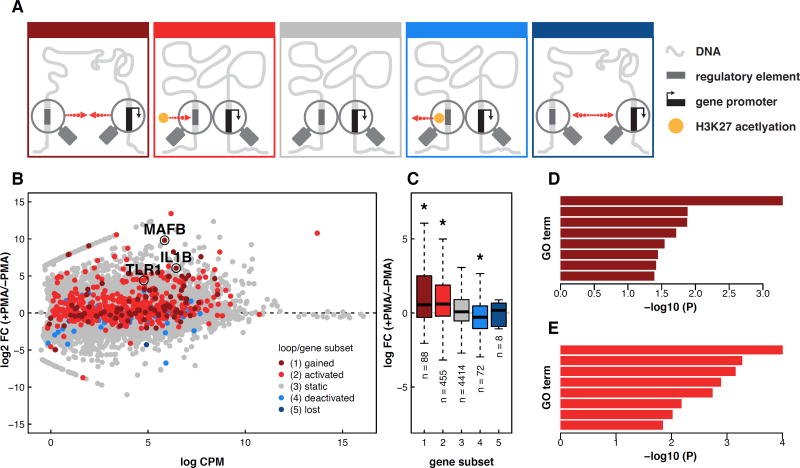
Visual inspection of the data indicated that both static and dynamic loops link gene promoters to distal regulatory elements marked by changes in H3K27ac, suggesting that preformed loops may also play a role in transcriptional regulation during differentiation. To investigate this further, we categorically divided our loops into five subsets; (1) loops that disappeared during differentiation (‘lost’), (2) loops that did not change significantly during differentiation but contained an anchor region that harbored a decreasing promoter-distal H3K27ac peak (‘deactivated’), (3) loops that did not change and that did not overlap any dynamic promoter-distal H3K27ac peaks (‘static’), (4) loops that did not change during differentiation but contained an anchor region that harbored an increasing promoter-distal H3K27ac peak (‘activated’), and (5) loops that are acquired during differentiation (‘gained’) (Fig. 3A). We next considered how genes whose promoters overlapped anchors of each of these loop sets changed during differentiation (Fig 3B–C). For deactivated and activated enhancer-loop sets, only genes at the distal end of the dynamic enhancer were considered. Only eight genes were found at lost loops and they showed no significant difference in expression when compared to static loops. However, deactivated loops connected regions with decreased H3K27ac to 72 genes whose expression significantly decreased during differentiation. In contrast, activated and gained loops overlapped 455 and 88 genes, respectively, that increased in expression during differentiation. These five loop subsets also correlated with transcription-start-site (TSS) usage as measured by the FANTOM consortium (Fig S3D) (Consortium et al., 2009). Overall, these results support two modes of distal gene regulation during THP-1 differentiation. In the first mode, new loop formation connects distal regulatory elements to the promoters of key regulatory genes that are dynamically expressed during differentiation. In the second mode, changes in enhancer activity may control the expression of target genes through stable chromatin loops that do not change during differentiation.
Figure 3. Expression and function of genes correlate with dynamic loop type and distal chromatin state.
(A) Schematic depictions of five loop classes. Red arrows indicate direction of change during PMA-induced differentiation of THP-1 cells. ‘Ac’ refers to a change in H3K27ac as detected by ChIP-seq. (B) Counts per million vs log fold change for all transcripts measured by RNA-seq. Genes are colored according to the class of loop found at their promoter. (C) RNA FPKM values for each gene subset. Selected GO terms enriched in genes sets at gained (D) and activated (E) loop anchors.
Gene ontology (GO) enrichment analysis further revealed that genes overlapping gained or activated loop anchors are enriched for biological processes related to macrophage function (Fisher’s Exact Test, p < .05; Fig. 3D–E, Tables S4–5). Specific examples of loop-associated genes important for macrophage development and function include (1) Interleukin 1 beta (IL1β), a canonical proinflammatory cytokine whose activation initiates widespread signaling and transcriptional changes in macrophages and neighboring cells, (2) V-maf musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB), an AP-1 family transcription factor and essential regulator of differentiation and self-renewal during haematopoiesis, and (3) Toll-Like Receptor 1 (TLR1), a cell surface membrane receptor critical for foreign pathogen recognition and stimulation of innate immune response. Overall, the enrichment of macrophage-related genes at gained and activated loop anchors suggests that cell-type specific transcription may be controlled through both re-wiring of enhancer-promoter interactions and through modulating enhancer activities at the promoter-distal end of pre-formed loops.
It is important to note that gene transcription is a complex process which is regulated by a myriad of factors including 3D chromatin structure, DNA methylation, TF binding, and modification of chromatin-associated proteins. While 2070 genes are upregulated during differentiation, gained and activated loops contact the promoters of only 527 genes. Moreover, despite a clear correlation with increased transcription, only 156 of these genes are significantly upregulated during differentiation. While our stringent methods for differential loop calling likely underestimate the number of dynamic loops, these results highlight the fact that chromatin structure is just one of many factors involved in transcriptional control.
Loops gained during differentiation are regulatory rather than structural in nature
One of the proposed functions of DNA loops is to bring distal regulatory elements such as enhancers into close physical proximity of their target genes to drive increased expression. However, many loops do not overlap gene promoters and are thought to play a structural role, such as forming insulated neighborhoods which facilitate stochastic interactions between enhancers and promoters (Ong and Corces, 2014)(Dowen et al., 2014). To explore whether loops formed during differentiation are mediating direct enhancer-promoter interactions or whether they are more structural in nature, we integrated our loops with H3K27ac signal which serves as a proxy for enhancer activity.
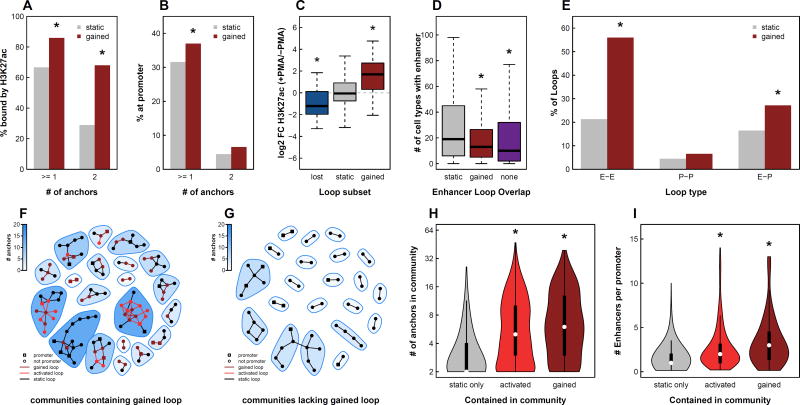
Gained loops were enriched for both H3K27ac and gene promoters compared to static loops (Fisher’s Exact Test, p < 0.01; Fig 4A–B). Strikingly, 90% gained loops connected two ‘regulatory elements’ (i.e. an enhancer to another enhancer, a promoter to another promoter, or an enhancer to a promoter) compared to only 42% of static loops. There was an especially strong enrichment for loops with H3K27ac peaks at both ends (Fisher’s Exact Test, p = 1.4 × 10−27; Fig 4A). Greater than 60% of gained loops contained H3K27ac sites at both anchors, a 2.4-fold increase compared to static loops (Fig 4A). In agreement with these results, fold changes of H3K27ac signal at loop anchors were positively correlated with changes in DNA looping (Fig 4C). Moreover, enhancers at dynamic loop anchors showed increased cell-type specificity compared to those at static loop anchors, in agreement with previous studies (Fig 4D) (Rao et al., 2014; Roadmap Epigenomics et al., 2015; Sanyal et al., 2012; Smith et al., 2016). These findings suggest that changes in chromatin structure during PMA-induced differentiation of THP-1 cells primarily facilitate direct connections between enhancers and promoters and likely play a role regulating cell-type specific gene transcription as opposed to forming structural elements such as insulated neighborhoods.
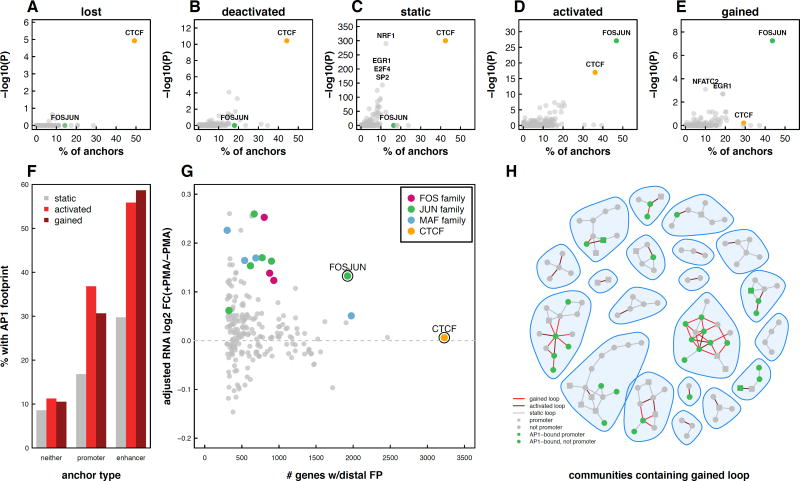
Figure 4. Gained and activated loops form multi-loop multi-enhancer activation hubs.
(A) The percent of static or gained loops with H3K27ac peaks at the anchors. Asterisks indicate p < 10−16 based on Fisher’s Exact Test. (B) The percent of static or gained loops with gene promoters at the anchors. Asterisks indicate p < 10−4 based on Fisher’s Exact Test. (C) Fold changes of H3K27ac peaks at lost, static, and gained loop anchors. Asterisks indicate p < 10−3 based on Wilcox Rank Sum Test. (D) Enhancers that overlapped with static loop anchors, gained loop anchors, or no loop anchors were intersected with enhancers from 98 cell types assayed by the Roadmap Epigenomics Mapping Consortium. The number of cell types containing each enhancer is depicted as a box plot. Asterisk indicates p < 10−4 based on Wilcox Rank Sum Test. (E) The percent of static or gained loops that connect an enhancer to an enhancer, a promoter to a promoter, or an enhancer to a promoter. Asterisks indicate p < 10−3 based on Fisher’s Exact Test. 20 randomly chosen interaction communities either containing (F) or lacking (G) a gained loop. Circles indicate loop anchors and lines indicate DNA loops. (H) Violin plots depicting the number of anchors per community for communities lacking gained/activated loops (static), communities containing activated loops, and communities containing gained loops. (I) Violin plots depicting the ratio of enhancers to promoters for communities lacking gained/activated loops (static), communities containing activated loops, and communities containing gained loops. See also Figure S4.
Formation of multi-loop activation hubs during macrophage development
We next explored the combinations of regulatory elements that are brought together by loops formed during differentiation (Fig 4E). Surprisingly, despite enrichment for enhancer-promoter interactions (1.7 fold compared to static loops), direct enhancer-promoter interactions accounted for only 27% of gained loops (Fig 4E). The majority (56%) of gained loops connected an enhancer to another enhancer, a 2.6-fold enrichment compared to static loops. While the functional significance of enhancer-promoter loops is well established, the role of enhancer-enhancer interactions is less obvious.
One possible explanation for the large proportion of enhancer-enhancer loops is the presence of interaction hubs involving a single promoter and multiple enhancers that all interact with each other. A fully connected hub with one promoter and N enhancers would contain N enhancer-promoter interactions and (N)!/2(N−2)! enhancer-enhancer interactions. For all values greater than N = 3, there would be more enhancer-enhancer loops than enhancer-promoter loops. To determine if our loops were forming such hubs we built interaction networks and detected communities of interacting anchor regions using a fast greedy modularity optimization algorithm (Clauset et al., 2004). We then classified these communities into two subsets: those containing a gained loop and those without. Twenty randomly chosen communities representing each subset are shown in Figure 4F–G. Inspection of these subsets revealed stark differences. First, communities involving gained loops contained significantly more loop anchors (mean = 8.3 vs 3.6) compared to communities lacking gained loops (Wilcoxon Rank Sum Test, p = 3.7 * 10−32; Fig. 4H). This finding holds true even when accounting for the higher likelihood of a gained loop falling within a larger community (permutation test, p < 0.001; Fig. S4). Second, the average ratio of enhancers to promoters per community was significantly higher (mean = 3.4 vs 1.6) in communities containing a PMA-specific loop (Wilcoxon Rank Sum Test, p = 7.4 * 10−15; Fig. 4I). Since communities involving gained loops had, on average, more than 3 enhancers it is consistent to observe more enhancer-enhancer than enhancer-promoter interactions in this subset.
These findings demonstrate that loops gained and activated during differentiation form hubs linking multiple distal enhancers to gene promoters. Such interactive hubs have been previously reported including one that forms during erythroid differentiation bringing multiple distal DNase hypersensitive sites into close proximity with either fetal or adult globin genes (Palstra et al., 2003; Tolhuis et al., 2002) (Kim and Dean, 2012; Krivega and Dean, 2016). However, to the best of our knowledge this is the first report that such hubs are the primary location of newly formed and activated loops during cellular differentiation. Whether this phenomenon is specific to macrophage development or a more broadly applicable aspect of cellular differentiation remains to be understood.
Dynamic re-wiring of chromatin architecture links distal AP-1 binding sites to key macrophage regulatory genes
To better characterize the cell-stage specific loops observed in our high resolution in situ Hi-C experiments, we mapped transposase-accessible DNA genome-wide by ATAC-seq in THP-1 cells (Buenrostro et al., 2015). Sequence and motif analysis of the exact transposase insertion sites can be used to predict which protein(s) may be present using an approach called TF footprinting (Fig. S5A). Footprinting of ATAC-seq libraries from untreated and PMA-treated THP-1 cells suggests increased binding at JUN, FOS, and other AP-1-related target sequences, and decreased binding at sites targeted by interferon regulatory factors IRF8 and IRF9 (Fig. S5B). In agreement with these results, we observed upregulation of genes encoding AP-1 family transcription factors, particularly for members of the MAF, JUN, and FOS AP-1 subfamilies, and significant downregulation of IRF8 (Fig. S5C). The upregulation of AP-1 transcription factor levels and corresponding increase in TF occupancy at AP-1 target sequences is consistent with the broad role of AP-1 in controlling cellular differentiation and proliferation (Eferl and Wagner, 2003; Shaulian and Karin, 2002).
We next leveraged TF footprinting analysis to specifically identify the transcription factors present at anchor regions of each class of loops (Fig. 5A–E). As expected, CTCF binding is strongly enriched at static chromatin loops, consistent with both motif enrichment and ChIP-seq experiments in THP-1 cells (Fig. 5C). Similar levels of CTCF are observed at lost and deactivated loop anchors. In contrast, gained and activated loops exhibited a striking enrichment for AP-1 target sequences (>40% of loop anchors, Fig. 5D–E). Interestingly, CTCF footprints are far less enriched at gained and activated interaction sites. ChIP-seq data reveals that CTCF is bound at these loop anchors but that CTCF binding is significantly weaker compared to static loop anchors. The functional significance of this difference is unclear but it further underscores the characteristic differences between static and gained/activated loops (Fig. S5D–F).
Figure 5. AP-1 enriched at enhancers containing loop anchors in both gained and activated loops.
Scatter plots depicting the percent of lost (A), deactivated (B), static (C), activated (D), and gained (E) anchors that overlap TF footprints and the −log 10 p-value of enrichment (Fisher’s Exact Test) for each TF. (F) Percent of loop anchors that overlap an AP-1 footprint as a function of loop subset and promoter or enhancer overlap (G) RNA fold-change of genes connected via a loop to distal TF footprints. FOS, JUN, and MAF points represent individual FOS-, JUN-, and MAF-related TF footprints. Log2 fold-changes were median normalized to account for the fact that genes at loop anchors exhibited a shift towards upregulation during differentiation. (H) 20 randomly chosen interaction communities containing a gained loop are shown. Loop anchors containing AP-1 footprints are indicated in green. See also Figure S5.
AP-1 binding was particularly enriched at gained and activated loop anchors with active enhancers (Fig. 5F), suggesting that AP-1 may be positively regulating gene transcription via binding at distal regulatory elements. To explore this possibility, we determined the relationship between distal TF binding and gene expression (Fig. 5G). Indeed, distal binding of AP-1 proteins, as measured by TF footprinting, correlated with increased expression of connected genes (Fig. 5G). While increased expression of genes that looped to distal FOSJUN motifs was particularly prominent, other FOS, JUN, and MAF family protein footprints showed a similar effect.
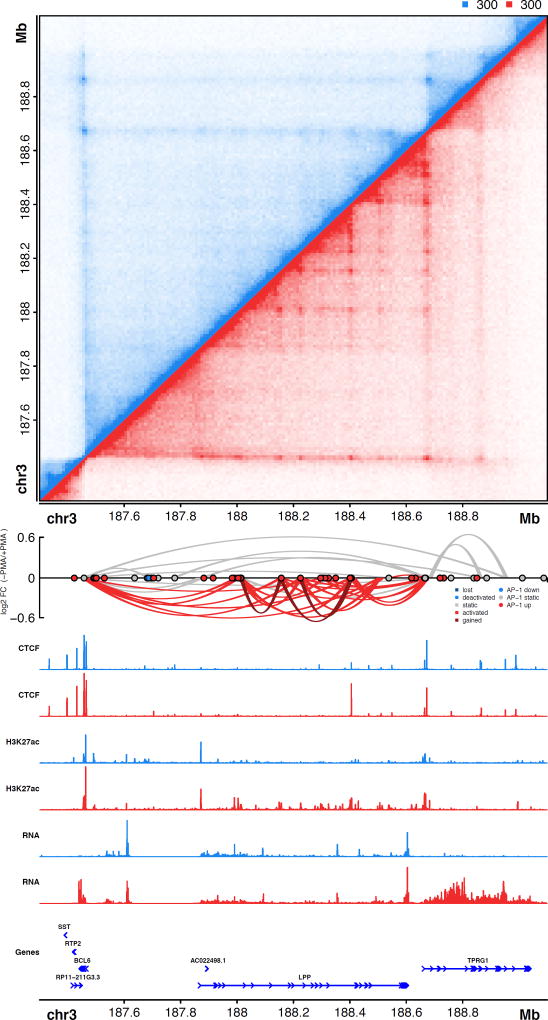
Taken together, the results presented here reveal multiple characteristics of promoter-distal gene regulation during macrophage development. Gained and pre-formed loops form multi-loop activation hubs marked by active enhancers and AP-1 binding sites. These hubs harbor more than 3 distal regulatory elements per promoter and are associated with increases in gene transcription. A clear example of a newly formed activation hub that exhibits all of these characteristics is observed at the TPRG1/BCL6 locus on chromosome 3 (Fig. 6). A complex network of AP-1 bound loci, located primarily within introns of the LPP gene, connect distal enhancers to the promoter regions of TPRG1 and BCL6. Intriguingly, LPP, whose promoter is not involved in any gained or activated loops, exhibits only a moderate change in gene expression (1.4-fold increase). In contrast, TPRG1 and BCL6, which are both associated with gained and activated loops, increase in expression by more than 25 fold. Further examples of AP-1 bound activation hubs are observed at key macrophage regulatory genes including MAFB and IL1β (Fig. S6A–C). The gain of AP-1 enhancer-promoter interactions are in certain cases marked as well by CTCF binding, suggesting AP-1 binding may be directed towards gene promoters for gene activation by CTCF (Fig. S6A). However, in other cases dynamic AP-1 looping-interaction sites are not marked by CTCF binding (Fig. S6C), or marked by non-dynamic CTCF binding (Fig. S6B), suggesting the chromatin interactome at these genes might be directed either through AP-1-mediated interactions or through additional factors.
Figure 6. AP-1 bound activation hub formed during PMA induced differentiation of THP-1 cells on chromosome 3.
(Top) Hi-C contact matrix depicting normalized contact frequencies in untreated THP-1 cells (blue, top left) and PMA treated THP-1 cells (red, bottom right). (Middle) Depiction of loops, loop fold changes (y-axis), loop subset (color of loops), and differential AP-1 footprints (circles). (Bottom) ChIP-seq signal tracks, RNA-seq signal tracks, and gene structures. See also Figure S6.
DISCUSSION
AP-1, a heterodimeric transcription factor comprising various combinations of FOS, JUN, MAF, ATF, and CREB family proteins, has been known to play a pivotal role in leukocyte development for decades (Liebermann et al., 1998; Valledor et al., 1998). However, its participation in gene regulation via DNA looping during macrophage development has not been previously described. Nevertheless, locus- and gene-specific examples of AP-1 bound DNA loops have been reported (Chavanas et al., 2008; Qiao et al., 2015), supporting a role for AP-1 family proteins in three-dimensional regulation of target genes and the broader, genome-wide participation of AP-1 characterized in the present study. Given its role across diverse cellular differentiation pathways (Eferl and Wagner, 2003; Shaulian and Karin, 2002), we speculate that the composition of the AP-1 transcription factor complex may contribute to re-wiring of chromatin interactions in a cell-type and tissue-specific manner. However, given the extraordinary number of potential transcription factor combinations that may co-bind at AP-1 consensus motifs (Mechta-Grigoriou et al., 2001), which cannot be determined directly by footprinting methods, future studies aimed at comprehensively mapping this combinatorial landscape would shed significant insight into the precise proteins underlying AP-1 related looping events.
The upregulation of macrophage-related genes through both pre-existing DNA loops and through dynamic long-range interactions agrees with previous gene-specific examples of loop-dependent gene regulation within distinct developmental contexts. At the beta globin locus, one of the best studied examples of long-range gene regulation (Kim and Dean, 2012), novel loop formation between locus control elements during blood cell development are required and sufficient for appropriate gene activation (Deng et al., 2012; Deng et al., 2014). In contrast, stimulation of IMR90 cells with TNFα activates enhancers at the promoter-distal anchors of pre-existing loops but does not induce large scale changes to 3D chromatin architecture (Jin et al., 2013). The identification of both static and dynamic loop-based mechanisms in various biological contexts suggests that both phenomena represent important paradigms for dynamic gene regulation. Our data reveals that these two types of regulatory looping mechanisms co-occur at specific loci forming multi-loop activation hubs at key macrophage regulatory genes. The hubs often connect multiple distal enhancers to a single gene promoter and are associated with strong upregulation of gene transcription.
AP-1 bound activation hubs are reminiscent of dynamic chromatin structures found at the beta-globin locus in which multiple distal sites loop to the active beta globin genes during specific stages in erythroid cell development (Tolhuis et al., 2002). Analogous to AP-1 interactions targeting macrophage-specific genes, the beta-globin locus is organized into an “Active Chromatin Hub” that requires regulatory transcription factors such as ELKF, GATA1, LDB1, and FOG1 (Drissen et al., 2004; Song et al., 2007; Vakoc et al., 2005). Indeed, we believe that the multi-loop communities identified herein may have far-reaching implications for how chromosome organization instructs transcription in other cellular contexts and throughout human development.
Finally, because Hi-C and other chromatin profiling assays query DNA loops and DNA-protein interactions across cell populations, it is impossible to determine from these data sets whether all loops in a hub exist at the same time or if we are observing multiple subpopulations of cells with exclusive subsets of DNA looping events. Single cell Hi-C, while useful for comparison against aggregate cell populations, also struggle to discriminate between these two possibilities, as only a single anchor-to-anchor ligation event is generated per allele by chromosome conformation capture. The prevalence of enhancer-enhancer contacts leads us to speculate that these loops often do form in the same cells. By doing so, activation hubs could increase the local concentrations of enhancers and distally bound transcription factors at gene promoters contributing to increased transcription. Further development of computational methods and experimental methods to identify multi-loop communities, such as Concatemer Ligation Assay (“COLA”), should help address some of these pressing questions in chromatin structure and gene regulation (Darrow et al., 2016).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
For information regarding resources and reagents please contact the lead contact, Michael Snyder (mpsnyder@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human THP-1 cells were obtained from ATCC and passaged in growth medium containing RPMI-1640 (Corning), 10% fetal bovine serum, and 1% penicillin streptomycin. THP-1 differentiation was carried out at a final concentration of 100 nM PMA (Sigma-Aldrich P1585-1MG) for 72 hours, followed by trypsinization and isolation of adherent THP-1 derived macrophages with TrypLE (ThermoFisher).
METHOD DETAILS
in situ Hi-C protocol
In situ Hi-C was performed exactly as described by Rao et al (Rao et al., 2014). Cells were crosslinked in 1% v/v formaldehyde for ten minutes with stirring and quenched by adding 2.5M glycine to a final concentration of 0.2M for 5 minutes with rocking. Cells were pelleted by spinning at 300 G for 5 minutes at 4 degrees C. Cells were washed with cold PBS and spun again prior to freezing in liquid nitrogen. Cells were lysed with 10mM Tris-HCl pH8.0, 10mM NaCl, 0.2% Igepal CA630 and protease inhibitors (Sigma, P8340) for 15 minutes on ice. Cells were pelleted and washed once more using the same buffer. Pellets were resuspended in 50ul of 0.5% SDS and incubates for 5–10 minutes at 62’C. Next reactions were quenched with 145ul of water and 25ul of 10% Triton X-100 (Sigma, 93443) at 37’C for 15 minutes. Chromatin was digested overnight with 25ul of 10× NEBuffer2 and 10U of MboI at 37’C with rotation. Reactions were incubated at 62’C for 20 minutes to inactivate MboI and then cooled to room temperature. Fragment overhangs were repaired by adding 37.5 ul of 0.4mM biotin-14-dATP, 1.5 ul of 10mM dCTP, 1.5 ul of 10mM dGTP, 1.5 ul of 10mM dTTP, and 8 ul of 5U/ul DNA Polymerase I, Lar (Klenow) Fragment and incubating at 37’C for 1 hour. Ligation was performed by adding 667 ul of water, 120 ul of 10× NEB T4 DNA ligase buffer, 100 ul of 10% Triton X-100, 12 ul of 10 mg/ml BSA, and 1 ul of 2000 U/ul T4 DNA Ligase and incubating at room temperature for 4 hours. Samples were pelleted at 2500 G and resuspended in 432 ul water, 18 ul 20 mg/ml proteinase K, 50 ul 10% SDS, 46 ul 5M NaCl and incubated for 30 minutes at 55’C. The temperature was raised to 68’C and incubated overnight. Samples were cooled to room temperature. 874 ul of pure ethanol and 55 ul of 3M sodium acetate, pH 5.2 were added to each tube which were subsequently incubated for 15 minutes at −80’C. Tubes were spun at max speed at 2’C for 15 minutes and washed twice with 70% ethanol. The resulting pellet was resuspended in 130 ul of 10mM Tric-HCl, pH8 and incubated at 37’C for 15 minutes. DNA was sheared using an LE220 Covaris Focused-ultrasonicator to a fragment size of 300–500 bp. Sheared DNA was size selected using AMPure XP beads. 110 ul of beads were added to each reaction and incubated for 5 minutes. Using a magnetic stand supernatant was removed and added to a fresh tube. 30ul of fresh AMPure XP beads were added and incubated for 5 minutes. Beads were separated on a magnet and washed two times with 700 ul of 70% ethanol without mixing. Beads were left to dry and then sample was eluted using 300 ul of 10 mM Tris-HCl, pH 8. 150 of 10 mg/ml Dynabeads MyOne Streptavidin T1 beads were washed resuspended in 300 ul of 10 mM Tris HCl, pH 7.5. This solution was added to the samples and incubated for 15 minutes at room temperature. Beads were washed twice with 600ul Tween Washing Buffer (TWB; 250 ul Tris-HCl, pH 7.5, 50 ul 0.5 M EDTA, 10 ml 5M NaCl, 25 ul Tween 20, and 39.675 ml water) at 55’C for 2 minutes with shaking.
in situ Hi-C library preparation
Sheared ends were repaired by adding 88 ul 1× NEB T4 DNA ligase buffer with 1mM ATP, 2 ul of 25 mM dNTP mix, 5 ul of 10U/ul NEB T4 PNK, 4ul of 3U/ul NEB T4 DNA polymerase I, 1ul of 5U/ul NEB DNA polymerase I, Large (Klenow) Fragment and incubating at room temperature for 30 minutes. Beads were washed two more times with TWB for 2 minutes at 55’C with shaking. Beads were washed once with 100 ul 1× NEBuffer 2 and resuspended in 90 ul of 1× NEBuffer 2, 5 ul of 10 mM dATP, 5ul of 5U/ul NEB Klenow exo minus, and incubated at 37’C for 30 minutes. Beads were washed two more times with TWB for 2 minutes at 55’C with shaking. Beads were washed once in 50 ul 1× Quick Ligation reaction buffer and resuspended in 50 ul 1× Quick Ligation reaction buffer. 2 ul of NEB DNA Quick ligase and 3 ul of an illumina indexed adapter were added and the solution was incubated for 15 minutes at room temperature. Beads were reclaimed using the magnet and washed two more times with TWB for 2 minutes at 55’C with shaking. Beads were washed once in 100 ul 10 mM Tris-HCl, pH8 and resuspended in 50 ul 10 mM Tris-HCl, pH8. Hi-C libraries were amplified for 7–12 cycles in 5 ul PCR primer cocktail, 20 ul of Enhanced PCR mix, and 25 ul of DNA on beads. The PCR settings included 3 minutes of 95’C followed by 7–12 cycles of 20 seconds at 98’C, 15 seconds at 60’C, and 30 seconds at 72’C. Samples were then held at 72’C for 5 minutes before lowering to 4’C until samples were collected. Amplified samples were brought to 250 ul with 10 mM Tris-HCl, pH 8. Samples were separated on a magnet and supernatant was transferred to a new tube. 175 ul of AMPure XP beads were added to each sample and incubated for 5 minutes. Beads were separated on a magnet and washed once with 700 ul of 70% ethanol. Supernatant was discarded. 100 ul of 10 mM Tris-HCl and 70 ul of fresh AMPure XP beads were added and the solution was incubated for 5 minutes at room temperature. Beads were separated with a magnet and washed twice with 700 ul 70% ethanol. Beads were left to dry until cracking started to be observed and eluted in 25 ul of Tris HCl, pH 8.0. The resulting libraries were quantified by Qubit and Bioanalyzer prior to sequencing.
RNA-seq
For each replicate, approximately 5 million treated or untreated THP-1 cells were collected and washed with 1× ice cold PBS. RNA was extracted using the Dynabeads mRNA Direct kit according to manufacturer directions. Sequencing libraries were prepared using the Epicenter ScriptSeq V2 kit according to manufacturer supplied protocol. The resulting libraries were quantified by Qubit, mixed in equal concentrations, and assayed by Bioanalyzer prior to sequencing using Illumina HiSeq 2500 technology.
ChIP-seq
For each biological replicate, approximately 10 million treated or untreated THP-1 cells were resuspended in growth media at 1 × 106 cells/mL. Fixation was performed for 10 minutes with rotation at room temperature in 1% formaldehyde; cross-linking was then quenched in 200 mM glycine with rotation for 5 minutes. Cross-linked cells were then pelleted and resuspended in 1× RIPA lysis buffer, followed by chromatin shearing via sonication (3 cycles using a Branson sonicator: 30 seconds on, 60 seconds off; 15 additional cycles on a Bioruptor sonicator: 30 seconds on, 30 seconds off). Individual ChIP experiments were performed on pre-cleared chromatin using antibody-coupled Dynabead protein G (ThermoFisher) magnetic beads. Anti-histone H3 (acetyl K27) antibody was obtained from Abcam (ab4729), CTCF antibody was obtained from Millipore (07-729). 3–5 ug of antibody per ChIP was coupled to 18 uL beads and rotated overnight with sheared chromatin at 4°C. Beads were then washed 5× in ChIP wash buffer (Santa Cruz), 1× in TE, and chromatin eluted in TE + 1% SDS. Cross-linking was reversed by incubation at 65°C overnight, followed by digestion of RNA (30 min RNase incubation at 37°C) and digestion of protein (30 min proteinase K incubation at 45°C). ChIP DNA was then purified on a minElute column (Qiagen), followed by DNA library preparation and size selection of 350–550 bp fragments via gel extraction (Qiagen).
ATAC-seq
For each biological replicate, approximately 50,000 treated or untreated THP-1 cells were collected and washed with 1× ice cold PBS. Cells were pelleted via centrifugation and resuspended in lysis buffer (10mM Tris-HCl, pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% IGEPAL CA-630). Cells were again pelleted by centrifugation and the supernatant discarded. Transposition was carried out for 30 minutes at 37°C using the Nextera DNA library prep kit (Illumina, cat#FC-121-1030). DNA was subsequently purified on a minElute column (Qiagen), and PCR amplified using the NEBNext high-fidelity master mix (NEB cat#M0541) with nextera PCR primers and barcodes. PCR amplification was monitored as described (Buenrostro et al. 2015), and gel purified to remove contaminating primer-dimer species.
QUANTIFICATION AND STATISTICAL ANALYSIS
in situ Hi-C loop calling and differential analysis
In situ Hi-C data sets were processed as described by Rao et al with minor differences to FDR calculations and final filtering parameters. MboI fragments of the human hg19 reference genome were determined using hicup_digester with the following command: “hicup_digester -g Human_hg19 -re1 ^GATC,MboI”. Hi-C fastq files were split into small files of 5 million reads each. Reads were aligned to the human reference genome hg19 using bwa mem version 0.7.12 with default parameters (Li and Durbin, 2010). Read pairs, in which both ends mapped uniquely were retained. Reads likely to mapping of a ligation junction were also retained as described by Rao et al (Rao et al., 2014). Each read was assigned to a single fragment using bedtools (Quinlan and Hall, 2010). Read pairs were filtered for unique combinations of chromosomes, start positions, and strand orientations to remove potential artifacts from PCR duplication. Such filtering was applied after merging sequencing duplicates but prior to merging data sets arising from different library preparations. Filtered reads from each pair of SAM files were combined into a single bed paired end file. Reads with mapping scores (MAPQ) below 30 were filtered out from subsequent analyses. We next built contact matrices for various resolutions including 5, 10 and 100Kb. For each resolution we binned all fragments according to their midpoint and then counted the read pairs that corresponded to each pair of fragments. Only intra chromosomal matrices were constructed. We constructed two type of contact matrices. For differential loop calling we built matrices for each biological replicate separately. For visualization we combined biological replicates into a single contact matrix for each sample (i.e. un-treated and PMA-treated THP-1 cells). Matrices were balanced according to a method proposed by Knight and Ruiz (Knight and Ruiz, 2013). Bins with less than 25 pixels of non-zero values were discarded from the normalization procedure. After balancing we calculated the expected normalized contacts for each distance for each chromosome separately. Noise associated with distances with few counts was mitigated by merging distances until more than 400 counts were achieved.
P values describing the observed contact frequencies given local background contact frequencies were determined for pixels at 10Kb resolution as described by Rao et al. For all pixels representing genomic bins separated by less than 2 million base pairs, various metrics were collected. For each pixel we defined several local neighborhoods as described by Rao et al; donut, horizontal, vertical, and lower right. Values of p=2 and w = 5 were used. The donut neighborhood is defined as pixels that are greater than p and less than or equal to w pixels away from the primary pixel in either the x or y directions. The other three neighborhoods are subsets of the donut neighborhood. The horizontal neighborhood is defined as pixels that are greater than p and less than or equal to w pixels away from the primary pixel in the x direction and greater less than p pixels away in the y direction. The horizontal neighborhood is defined as pixels that are greater than p and less than or equal to w pixels away from the primary pixel in the y direction and less than p pixels away in the x direction. And the lower right neighborhood is all pixels with × values greater than the primary pixel but less than or equal to w pixels away, pixels with y values less than the primary pixel and less than or equal to w pixels away, and pixels with coordinates that are more than p pixels away in either the x or y direction. For each neighborhood, summed normalized contact frequencies were determined. If summed values were less than 16, w was increased until either greater than 16 counts were reached or w was equal to 20. For each neighborhood, summed expected contact frequencies were determined as well. For each neighborhood the ratio of observed / expected counts was determined. This ratio was multiplied by the expected value of the primary pixel to determine the expected normalized contacts for each pixel analyzed. This value was converted to an expected raw contact count by multiplying by the corresponding normalization factors determined by our matrix balancing step. P values of differences between observed raw counts and expected raw count, as estimated from each of the four local neighborhoods, were was determined determined using the R programming language and the function ppois with lower.tail = FALSE. P values for all pixels tested on all chromosomes were corrected for multiple hypothesis testing using the Benjamini-Hochberg method.
Loops were determined by further clustering and filtering of significant pixels in a similar fashion as described by Rao et al. All pixels with corrected p values less than or equal to 0.05 were clustered using the DBSCAN algorithm with epsilon = 20 Kb. For each cluster the pixel with the highest normalized counts was retained and annotated with the number of pixels in its cluster. These pixels were filtered for the following parameters; fold-change of observed vs expected (donut) > 1.75, fold-change of observed vs expected (horizontal) > 1.5, fold-change of observed vs expected (vertical) > 1.5, fold-change of observed vs expected (lower right) > 1.5, the sum of the adjusted p-values for all four neighborhoods < 0.001, and the number of pixels in the cluster > 2. Finally, to avoid artifacts due to local neighborhoods that contained regions of repetitive nature, pixels within 50Kb of a genomic bin that was discarded by the matrix normalization step were removed.
Hi-C normalization for visualization
Visualizing differences between to Hi-C contact matrices can be complicated by different sequencing depths between data sets as well as differences in average interaction frequencies as a function of distance. To allow for accurate visual comparison of contact matrices between two samples all PMA-treated matrices were normalized such that the median normalized contact frequency for each genomic distance was identical between untreated and PMA-treated cells. For all distances plotted, the median contact frequency was determined for each data set. For each distance an offset was determined by dividing the median untreated value but the median PMA-treated value. All PMA-treated normalized contact frequencies were multiplied by this offset factor prior to plotting.
Aggregate peak analysis
In order to assess the quality of loop calls we generated aggregate peak analysis (APA) plots and scores. These analyses aggregate the signal of pixels of loops as well as the pixels surrounding them, the local background. For each loop in a given set of loops, the normalized observed contact frequencies were collected for the pixel representing the loop as well as for pixels within 10 bins in both the x and y directions. To normalize for loops at different distances, each pixel was divided by the expected normalized interaction frequency at that distance to give an observed over expected ratio. Median observed over expected ratios for each position in the matrix were calculated and plotted as a heatmap. APA scores were determined by dividing the center pixel value by the median value of the nine pixels in the lower right section of the APA plot.
Motif enrichment at loop anchors
To determine the frequency of transcription factor motif overlap with loop anchors, we downloaded TF motifs determined by Factorbook and intersected the motifs with our 10Kb loop anchors using the bedtools intersect function (Quinlan and Hall, 2010; Wang et al., 2013). To determine the expected frequency of overlap we shuffled the assignments of motif and genomic region 100 times and performed the same analysis. Unannotated motifs, those that started with “UAK”, were not included.
Differential Loop analysis
Detection of differential loops can be complicated by larger scale changes chromatin structure that do indeed change interaction frequency but do not change looping per se. Such large-scale structural changes include changes in chromatin compaction, changes in domain boundaries, and duplication of genomic regions. To specifically detect changes in looping we devised a method using DESeq2 that looked for changes in the enrichment of pixels representing DNA loops compared to local background interaction frequencies (Love et al., 2014). First, we collected information for all pixels that were identified as loops in either untreated or PMA-treated THP-1 cells. For each loop pixel we collected the following information from each sample and each biological replicate: raw interaction counts, expected interaction counts, and normalization factors. We collected that same information for local background pixels defined as pixels that were greater than 2 and less than 6 pixels away from the loop pixel in both x and y directions. Using this data we built a DESeq2 counts matrix and sample table as follows. Counts for all pixels in all samples in all biological replicates formed the columns of the count matrix and the rows represented different loops. A corresponding table described the relationship among the columns of the count matrix: each pixel belonging to a sample, a biological replicate, and either representing a loop ‘L’ or a background ‘B’ pixel. We analyzed this data for differential enrichment of the loop over background across condition, using the DESeq2 design formula “~ rep + sample + rep:sample + pixel_type + sample: pixel_type”. Raw counts required normalization to account for differential sequencing depth in each genomic bin and distance-dependent interaction frequencies, while sample-specific sequencing depth was controlled via terms in the design formula. A per-pixel normalization factor was calculated by multiplying the normalization factor for bin 1, the normalization factor for bin 2, and the expected interaction frequency. These normalization factors were centered around a value of 1 and added to the DESeq2 analysis. The DESeq2 differential pipeline was run with settings betaPrior=FALSE and dispersion fitType="local". The resulting loops were filtered for those with a p value of < 0.001 to produce our final set of differential loops.
Comparison to Juicer pipeline
Hi-C libraries were also analyzed using the Juicer pipeline. Data was processed for 5,929,803,301 Hi-C read pairs in untreated THP-1 cells, yielding 3,935,374,088 Hi-C contacts and 5,522,487,839 Hi-C read pairs in PMA-treated THP-1 cells, yielding 3,789,121,851 Hi-C contacts. Loops were annotated using HiCCUPS at 5kB and 10kB resolutions with default Juicer parameters. This yielded a list of 14,964 loops in untreated THP-1 cells and 22,615 loops in PMA-treated THP-1 cells. All the code used in the above steps is publicly available at (github.com/theaidenlab). Loops identified by Juicer were filtered for those that were greater or equal to 50Kb and less than or equal to 2Mb. In order to determine overlap between our loops and Juicer we had to consider not only the pixel that was picked to represent the loop but all enriched pixels within a cluster. Therefore, for Juicer loops we picked the centroid of the cluster +/− the radius of the cluster for each loop anchor. We then rank ordered our loop calls by increasing p values and determined the percent of our loops that were also found in the Juicer loops calls. We did this for 20 subsets of our loop calls ranging from the top 5% of our loop calls to 100% of our loop calls. These overlaps were performed for both data sets.
Loop community detection and analysis
Non-directed graphs were constructed from loops using the R package igraph. Communities were determined using the fastgreedy.community function with default parameters.
Comparison to FANTOM CAGE data
CAGE data generated by the FANTOM4 consortium describing TSS usage (level 3) was used. For each TSS and time point we extracted the median tpm value. For each time point we calculated the log 2 ratio compared to the 0 hour time point. The resulting values were centered around zero. Sets of TSS that overlapped each loop subset were determined and visualized as a boxplot.
Gene Ontology enrichment
GO enrichment was performed on genes whose promoter overlapped either a gained or activated loop. Promoters were defined as 2000 base pair regions upstream of a gene TSS. For activated loops, only genes at the distal end of an upregulated H3K27ac mark were considered. The background gene set used for these analyses was a list of all genes whose promoters overlapped a loop anchor in either treated or untreated cells. GO enrichment was performed for biological process gene ontologies using the goana function from the R package limma (Ritchie et al., 2015). GO terms were filtered for adjusted p value < 0.05. Selected enriched GO terms were plotted.
RNA-seq analysis and visualization
Paired end sequencing reads were trimmed using trim galore version 0.40 with command-line settings “trim_galore -q 20 --trim1 --paired” and subsequently aligned to gencode v19 transcripts using kallisto with default parameters. For analysis of differential genes, kallisto outputs were processed using tximport. Genes with less than 2 counts per million were filtered out. Statistical significance was determined using the glmTreat function in edgeR with a fold change cutoff of 2. For genome-track visualization purposes, RNA-seq reads were aligned to the hg19 reference genome using tophat. Duplicate reads were removed using from picard-tools version 1.92. The resulting reads were normalized for sequencing depth and mappability using the align2rawsignal pipeline with options “-of=bg -n=5 -l=1 -w=200 -mm=30” (https://align2rawsignal.googlecode.com).
ChIP-seq peak calling and visualization
Paired end sequencing reads were trimmed using trim galore version 0.40 with command-line settings “trim_galore -q 20 --trim1 --paired” and subsequently aligned to the hg19 reference genome using bowtie1 version 1.1.1 with settings “bowtie -q --phred33-quals -X 2000 -m 1 --fr -p 8 -S --chunkmbs 400”. Mapped reads were merged across technical and sequencing replicates, and duplicate reads removed using picard tools version 1.92. Peaks were identified for each sample and biological replicate using MACS2 version 2.1.0 with command line options “macs2 callpeak --bdg -t -g hs”. For analysis of differential chromatin accessibility, raw ATAC-seq reads were extracted for each condition over a merged set of ATAC-seq peaks and statistical significance determined using the glmTreat function in edgeR with a fold change cutoff of 2. For genome-track visualization purposes, ATAC-seq reads were normalized for sequencing depth and mappability using the align2rawsignal pipeline with options “-of=bg -n=5 -l=200 -w=200 -mm=30” (https://align2rawsignal.googlecode.com).
CTCF motif orientation analysis
Loops were filtered to retain those that overlapped a CTCF binding site, as determined by ChIP-seq, at both anchors. These loops were further filtered for those that overlapped a single CTCF motif as determined by Factorbook (Wang et al., 2013). We then calculated the percent of those remaining loops that contained each of the four possible pairwise combinations of motifs.
Enhancer definition
Precise classification of genomic enhancers requires extensive experimental validation. However, several features of chromatin can be used to infer enhancer activity with reasonable accuracy. For all of the analyses in this paper enhancers were defined as regions with an H3K27ac peak as determined by ChIP-seq. H3K27ac peaks that overlapped a gene promoter (i.e. the 2000 bp region upstream of a UCSC hg19 known gene transcription start site) were removed from this list.
ATAC-seq peak calling and visualization
Paired end sequencing reads were trimmed using trim galore version 0.40 with command-line settings “trim_galore -q 20 --trim1 --paired” and subsequently aligned to the hg19 reference genome using bowtie2 version 2.2.4 with settings “bowtie2 -t --sensitive”. Mapped reads were merged across technical and sequencing replicates, and duplicate reads removed using picard tools version 1.92. Peaks were identified for each sample and biological replicate using MACS2 version 2.1.0 with command line options “macs2 callpeak --bdg --nomodel -t -g hs”. For analysis of differential chromatin accessibility, raw ATAC-seq reads were extracted for each condition over a merged set of ATAC-seq peaks and statistical significance determined using the glmTreat function in edgeR with a fold change cutoff of 2. For genome-track visualization purposes, ATAC-seq reads were normalized for sequencing depth and mappability using the align2rawsignal pipeline with options “-of=bg -n=5 -l=200 -w=200 -mm=30” (https://align2rawsignal.googlecode.com).
Transcription Factor Footprinting Analyses
Transcription factor footprinting was broken into two steps: (A) identify bound TF motifs in THP-1 monocytes and THP-1 derived macrophages and (B) Determine differential footprinting scores before and after PMA treatment for each motif identified in THP-1 cells. Step (A): Putative TF footprints were identified using the Protein Interaction Quantification (PIQ) footprinting algorithm (Sherwood et al., 2014) against the JASPAR core vertebrate database of TF motifs (http://jaspar.genereg.net). First, motif matching was performed for 516 known TF target sequences against the hg19 reference genome using the PIQ package pwmmatch.exact.r script. Second, filtered ATAC-seq alignment reads were converted into binary RData files using the PIQ package pairedbam2rdata.r script. Third, TF footprint scores were determined for each motif match using the PIQ package pertf.bg.r and common.r scripts with default settings. Putative TF footprints were filtered at a positive predictive value (PPV) cutoff of 0.7 and for footprints that intersect ATAC-seq peaks identified in THP-1 cells. Altogether, 2,731,616 TF footprints were identified post-filtering with a median 3,693 binding sites per unique TF motif. Step (B): Analysis of dynamic TF binding was performed using the Wellington-bootstrap algorithm for differential footprinting (Piper et al., 2015) against all post-filtering TF footprints identified by PIQ. First, differential footprinting was applied using the pyDNase wellington-bootstrap.py script with the command-line option for ATAC-seq input “-A”. Second, differential footprint scores were determined using the pyDNase dnase_ddhs_scorer.py script with the command-line option for ATAC-seq input (“-A”). Differential footprint scores (DFP) were altogether adjusted by median normalization, followed by median differential footprint analysis for each independent factor. Wellington DFPs strongly correlate with changes in PIQ PPV values (p < 2.2e-16), and using a cutoff of +/− 2 standard deviations, we identify a total of 33,130 decreasing and 95,898 increasing TF binding events.
Footprint enrichment at loop anchors
To determine enrichment of TF at loop anchors we first intersected all TF footprints with loop anchors using the bedtools intersect function. For each TF we determined the number of footprints of that TF that overlapped an anchor, number of footprints of that TF that did not overlap an anchor, the number of footprints of other TFs that overlapped an anchor, and the number of footprints of other TFs that did not overlap an anchor. Using these values, we built a contingency table and performed fishers exact test in R. The resulting p values were corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure.
DATA AND SOFTWARE AVAILABILITY
Data availability
The sequencing data from this study is publicly available through the Gene Expression Omnibus (GEO) series number GSE96800 (ChIP-seq, ATAC-seq, RNA-seq) and the Sequencing Read Archive (SRA) accession number PRJNA385337 (in situ Hi-C). Processed Hi-C data is also available through Juicebox (http://www.aidenlab.org/juicebox/).
Software and data resources
Transcription factor footprinting scripts were obtained from https://bitbucket.org/thashim/piq-single (PIQ R scripts pwmmatch.exact.r, pairedbam2rdata.r, pertf.bg.r and common.r) and http://pythonhosted.org/pyDNase/ (Wellington-bootstrap python scripts wellington-bootstrap.py and dnase_ddhs_scorer.py).
Supplementary Material
Table S1. DNA Loops in untreated THP-1 cells, Related to STAR methods.
Table S2. DNA Loops in PMA-treated THP-1 cells, Related to STAR methods.
Table S3. Differential Loops, Related to STAR methods.
Table S4. Gene Ontology enrichments at gained loop anchors, Related to Figure 3.
Table S5. Gene Ontology enrichments at activated loop anchors, Related to Figure 3.
Highlights.
Loop dynamics shown by deeply sequenced in situ Hi-C during macrophage differentiation
Multi-loop interaction communities identified surrounding key macrophage genes.
Enhancers form multi-loop activation hubs through both static and newly acquired loops
Activation hubs are enriched for AP-1 bound long-range enhancer interactions
Acknowledgments
DHP is supported by National Institutes of Health (NIH), National Human Genome Research Institute (NHGRI) grant R00HG008662 and the Damon Runyon Cancer Research Foundation (DRG-2122-12). KVB is supported by NIH, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant F32DK107112. GTH is supported by NIH grant NIH T32HG000044. MIL is supported by NIH, National Cancer Institute (NCI) grant CA142538-07. DVS was supported by an NIH T32 fellowship (HG000044) and a Genentech Graduate Fellowship. This research was supported by NIH Centers of Excellence in Genomic Science (CEGS) grant 5P50HG00773502. ELA is supported by an NIH New Innovator Award (1DP2OD008540), an NSF Physics Frontiers Center Award (PHY-1427654, Center for Theoretical Biological Physics), the NHGRI Center for Excellence for Genomic Sciences (HG006193), the Welch Foundation (Q-1866), an NVIDIA Research Center Award, an IBM University Challenge Award, a Google Research Award, a Cancer Prevention Research Institute of Texas Scholar Award (R1304), a McNair Medical Institute Scholar Award, an NIH 4D Nucleome Grant U01HL130010, an NIH Encyclopedia of DNA Elements Mapping Center Award UM1HG009375, and the President’s Early Career Award in Science and Engineering. Illumina sequencing services were performed by the Stanford Center for Genomics and Personalized Medicine. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health. We thank Craig D Wenger and Neva C Durand for helpful advice, guidance, and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The sequencing data from this study is publicly available through the Gene Expression Omnibus (GSE96800) and the Sequencing Read Archive (PRJNA385337). Processed Hi-C data is also available through Juicebox (http://www.aidenlab.org/juicebox/).
AUTHOR CONTRIBUTIONS
Conceptualization, DHP, KVB; Methodology, DHP, KVB, DVS; Software, DHP, KVB, IA, MSS, MIL, ELA; Formal Analysis, DHP, KVB; Funding Acquisition, DHP, MPS; Visualization, DHP, KVB; Writing, DHP, KVB; Validation, GTH, MCB.
References
- Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11:559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- Beagan JA, Gilgenast TG, Kim J, Plona Z, Norton HK, Hu G, Hsu SC, Shields EJ, Lyu X, Apostolou E, et al. Local Genome Topology Can Exhibit an Incompletely Rewired 3D-Folding State during Somatic Cell Reprogramming. Cell Stem Cell. 2016;18:611–624. doi: 10.1016/j.stem.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21–29. doi: 10.1002/0471142727.mb2129s109. 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature. 2017;544:503–507. doi: 10.1038/nature22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas S, Adoue V, Mechin MC, Ying S, Dong S, Duplan H, Charveron M, Takahara H, Serre G, Simon M. Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylarginine deiminase 3 gene in differentiated keratinocyte. PLoS One. 2008;3:e3408. doi: 10.1371/journal.pone.0003408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Molecular cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- Consortium, F. Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One. 2010;5:e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, Sun Z, Huang SC, Sanborn AL, Machol I, Shamim M, et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A. 2016;113:E4504–4512. doi: 10.1073/pnas.1609643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, Aiden EL. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Current biology : CB. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yanez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 2017;169:693–707. e614. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, Zhang MQ, Snyder MP. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19:1987–1997. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Dean A. Chromatin loop formation in the beta-globin locus and its role in globin gene transcription. Mol Cells. 2012;34:1–5. doi: 10.1007/s10059-012-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight PA, Ruiz D. A fast algorithm for matrix balancing. Ima Journal of Numerical Analysis. 2013;33:1029–1047. [Google Scholar]
- Kouno T, de Hoon M, Mar JC, Tomaru Y, Kawano M, Carninci P, Suzuki H, Hayashizaki Y, Shin JW. Temporal dynamics and transcriptional control using single-cell gene expression analysis. Genome Biol. 2013;14:R118. doi: 10.1186/gb-2013-14-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PH, de Laat W. Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol. 2016;17:771–782. doi: 10.1038/nrm.2016.138. [DOI] [PubMed] [Google Scholar]
- Krivega I, Dale RK, Dean A. Role of LDB1 in the transition from chromatin looping to transcription activation. Genes & development. 2014;28:1278–1290. doi: 10.1101/gad.239749.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivega I, Dean A. Chromatin looping as a target for altering erythroid gene expression. Ann N Y Acad Sci. 2016;1368:31–39. doi: 10.1111/nyas.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Krivega I, Dale RK, Dean A. The LDB1 Complex Co-opts CTCF for Erythroid Lineage-Specific Long-Range Enhancer Interactions. Cell reports. 2017;19:2490–2502. doi: 10.1016/j.celrep.2017.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann DA, Gregory B, Hoffman B. AP-1 (Fos/Jun) transcription factors in hematopoietic differentiation and apoptosis. Int J Oncol. 1998;12:685–700. doi: 10.3892/ijo.12.3.685. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund ME, To J, O'Brien BA, Donnelly S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J Immunol Methods. 2016;430:64–70. doi: 10.1016/j.jim.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13:709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Bulajic M, Dekker J, Mazzoni EO, Reinberg D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016;30:2657–2662. doi: 10.1101/gad.288324.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944. e922. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odero MD, Zeleznik-Le NJ, Chinwalla V, Rowley JD. Cytogenetic and molecular analysis of the acute monocytic leukemia cell line THP-1 with an MLL-AF9 translocation. Genes Chromosomes Cancer. 2000;29:333–338. [PubMed] [Google Scholar]
- Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36:229–239. doi: 10.1016/j.it.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Assi SA, Cauchy P, Ladroue C, Cockerill PN, Bonifer C, Ott S. Wellington-bootstrap: differential DNase-seq footprinting identifies cell-type determining transcription factors. BMC Genomics. 2015;16:1000. doi: 10.1186/s12864-015-2081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Shiue CN, Zhu J, Zhuang T, Jonsson P, Wright AP, Zhao C, Dahlman-Wright K. AP-1-mediated chromatin looping regulates ZEB2 transcription: new insights into TNFalpha-induced epithelial-mesenchymal transition in triple-negative breast cancer. Oncotarget. 2015;6:7804–7814. doi: 10.18632/oncotarget.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics, C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, Masella B, Guarini A, Fatica A, Peschle C, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A. 2007;104:19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, Li Y, Lin S, Lin Y, Barr CL, et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O'Donnell CW, Lewis S, Barkal AA, van Hoff JP, Karun V, Jaakkola T, Gifford DK. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD Boundaries Constrain Cell-Type-Specific Looping Interactions between Promoters and Distal Elements around the CFTR Locus. Am J Hum Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Valledor AF, Borras FE, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol. 1998;63:405–417. doi: 10.1002/jlb.63.4.405. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, Moore J, Pierce BG, Dong X, Virgil D, et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013;41:D171–176. doi: 10.1093/nar/gks1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. DNA Loops in untreated THP-1 cells, Related to STAR methods.
Table S2. DNA Loops in PMA-treated THP-1 cells, Related to STAR methods.
Table S3. Differential Loops, Related to STAR methods.
Table S4. Gene Ontology enrichments at gained loop anchors, Related to Figure 3.
Table S5. Gene Ontology enrichments at activated loop anchors, Related to Figure 3.
Data Availability Statement
The sequencing data from this study is publicly available through the Gene Expression Omnibus (GEO) series number GSE96800 (ChIP-seq, ATAC-seq, RNA-seq) and the Sequencing Read Archive (SRA) accession number PRJNA385337 (in situ Hi-C). Processed Hi-C data is also available through Juicebox (http://www.aidenlab.org/juicebox/).