Abstract
BACKGROUND: Oncolytic adenoviruses show promise in targeting gliomas because they do not replicate in normal brain cells. However, clinical responses occur only in a subset of patients. One explanation could be the heterogenic expression level of virus receptors. Another contributing factor could be variable activity of tumor antiviral defenses in different glioma subtypes. METHODS: We established a collection of primary low-passage cell lines from different glioma subtypes (3 glioblastomas, 3 oligoastrocytomas, and 2 oligodendrogliomas) and assessed them for receptor expression and sensitivity to human adenovirus (HAd) serotypes 3, 5, and 11p. To gauge the impact of antiviral defenses, we also compared the infectivity of the oncolytic adenoviruses in interferon (IFN)-pretreated cells with IFN-sensitive Semliki Forest virus (SFV). RESULTS: Immunostaining revealed generally low expression of HAd5 receptor CAR in both primary tumors and derived cell lines. HAd11p receptor CD46 levels were maintained at moderate levels in both primary tumor samples and derived cell lines. HAd3 receptor DSG-2 was reduced in the cell lines compared to the tumors. Yet, at equal multiplicities of infection, the oncolytic potency of HAd5 in vitro in tumor-derived cells was comparable to HAd11p, whereas HAd3 lysed fewer cells than either of the other two HAd serotypes in 72 hours. IFN blocked replication of SFV, while HAds were rather unaffected. CONCLUSIONS: Adenovirus receptor levels on glioma-derived cell lines did not correlate with infection efficacy and may not be a relevant indicator of clinical oncolytic potency. Adenovirus receptor analysis should be preferentially performed on biopsies obtained perioperatively.
Introduction
Diffusely infiltrating gliomas consist of WHO grade II to III astrocytic tumors; grade II to III oligodendrogliomas; and the most common form, grade IV glioblastomas [1], which feature a median life expectancy of 15 months with combined chemo- and radiotherapy [2]. Advanced gliomas typically feature inactivating mutations in tumor suppressors RB1, p53, and p16 and activating mutations in tyrosine kinases, such as epidermal growth factor receptor (EGFR) [3]. Mutations in isocitrate dehydrogenase (IDH) 1 and 2, and O-methylguanine-DNA-methyltransferase (MGMT) promoter methylation guide treatment options and prognosis of particularly low-grade gliomas [1], [4].
Oncolytic viruses offer an appealing option to conventional treatments because they can be selected to rely on oncogenic alterations for replication, averting toxicity to normal cells [5]. Human adenovirus serotype 5 (HAd5) has shown excellent safety in glioma patients, but tumor infection and regression only occur in a subset of patients [6], [7]. HAd5 uses the cellular tight junction protein coxsackie-adenovirus receptor (CAR) for entry, whose expression in tumors may generally be low and could account for some of the observed patient response heterogeneity [8], [9]. To improve the therapeutic efficacy of HAd5, its tropism has been increased by switching its receptor-binding domain, the so-called fiber knob, to that of other human adenovirus serotypes, such as HAd3 and HAd11p, which utilize more ubiquitous and cancer-overexpressed receptors for infection, desmoglein-2 (DSG-2) and cluster of differentiation 46 (CD46), respectively [10], [11], [12]. However, the expression patterns of also these receptors in tumors may be heterogenic, and as we and others have shown, the replication of some oncolytic viruses is also regulated intracellularly, particularly by components of the interferon (IFN)-inducible antiviral defense [13], [14], [15]. To complicate matters, most analyses of virus tropism and oncolytic efficacy to date have been made in established cell lines, which are prone to significant transcriptional and phenotypic deviation from the parental tumors they were derived from [16], [17], [18].
To consolidate the tumor grade-and-passage–dependent, virus entry-mediated, and IFN-regulated determinants of glioma permissiveness to oncolytic viruses, we established a collection of primary tumor cell lines from both low- and high-grade gliomas and compared the expression levels of CAR, DSG-2, and CD46 in the cell lines to the parental tumor tissue. We sought to correlate the different human adenovirus receptor levels to infection efficacy with the respective viruses, HAd5, HAd3, and HAd11p. To account for receptor-independent barriers to virus replication, we assessed whether IFN responsiveness of the primary tumor cell lines correlated with infection efficacy of these adenoviruses. Our results show that, within the glioma subgroups, individual tumors displayed highly variable levels of HAd receptors and that did not correlate with infection efficacy with any of the three HAd serotypes. In our limited sample set, there was also no association between virus serotype and glioma subtype. All tested glioma cells mounted effective antiviral defense, as observed by the complete inhibition of an IFN-sensitive reporter alphavirus [Semliki Forest virus (SFV)-VA7]. However, type I IFN-mediated antiviral signaling did not hinder replication of the tested adenoviruses. Our results support in general the continued development of oncolytic adenoviruses for glioma targeting and call for further investigation into the interconnection of virus tropism and therapeutic efficacy.
Materials and Methods
Patients and Tumor Sample Collection
Samples were obtained from 17 adult glioma patients (range 27-76 years) operated on the NeuroCenter of Kuopio University Hospital during May 2012 to March 2014 (Table 1). For clarity, the cases are indicated as follows: AO, oligoastrocytoma; AOD, anaplastic oligodendroglioma; GB, glioblastoma; and OD, oligodendroglioma. The locations of gliomas are illustrated on representative magnetic resonance images (Supplementary Figure S1). Original diagnoses were confirmed by a neuropathologist (T.R.). Procedures of this study, which were in accordance with the ethical standards of the Helsinki Declaration, were approved by Research Ethics Committee of the Hospital District of Northern Savo. The written consent was obtained from all patients. Samples collected perioperatively were placed in ice-cold Opti-Mem Reduced Serum medium (31985, Gibco) when aimed for cell culture. Samples for histology were placed in 4% paraformaldehyde (in 0.1 M phosphate buffer, pH 7.4) overnight at +4°C.
Table 1.
Characteristics of Glioma Patients
| Code-Subtype | Sex | Age | WHO Grade | Location of Tumor | Survival (Months) |
|---|---|---|---|---|---|
| Glioblastomas | |||||
| 003-GB | M | 67 | IV | Frontal | 49 |
| 022-GB | M | 75 | IV | Temporal | 11 |
| 027-GB | M | 67 | IV | Frontal | 35 |
| 038-GB | F | 68 | IV | Occipital | 23 |
| Oligodendrogliomas | |||||
| 004-OD | M | 44 | II | Frontal | A (57) |
| 008-OD§ | F | 40 | II-III | Insula | A (56) |
| 020-OD§ | M | 69 | II-III | Frontal | A (48) |
| 014-AOD* | M | 39 | III-IV | Temporal | 9 |
| Oligoastrocytomas | |||||
| 036-OA | M | 70 | II | Temporal | 7 |
| 015-OA§ | M | 35 | II-III | Temporal | A (51) |
| 025-OA§ | F | 31 | II-III | Frontal | A (44) |
| 039-OA | M | 51 | II-III | Frontal | A (37) |
| 002-OA§ | M | 42 | III | Parietal | 45 |
| 016-OA§ | F | 27 | III | Temporal + insula | A (51) |
| 026-OA†,§ | F | 61 | III | Frontal | 23 |
| 034-OA§ | M | 39 | III | Frontal | A (39) |
| Ependymomas | |||||
| 035-E‡ | M | 76 | II | 4th ventricle | 4 |
A, alive (in parenthesis months since operation); E, ependymoma; F, female; M, male.
Oligodendroglioma 4 and 12 year earlier.
Anaplastic oligodendroglioma 4 years earlier.
Ependymoma 8 years earlier.
IDHR132 mutation.
Supplementary Figure S1.
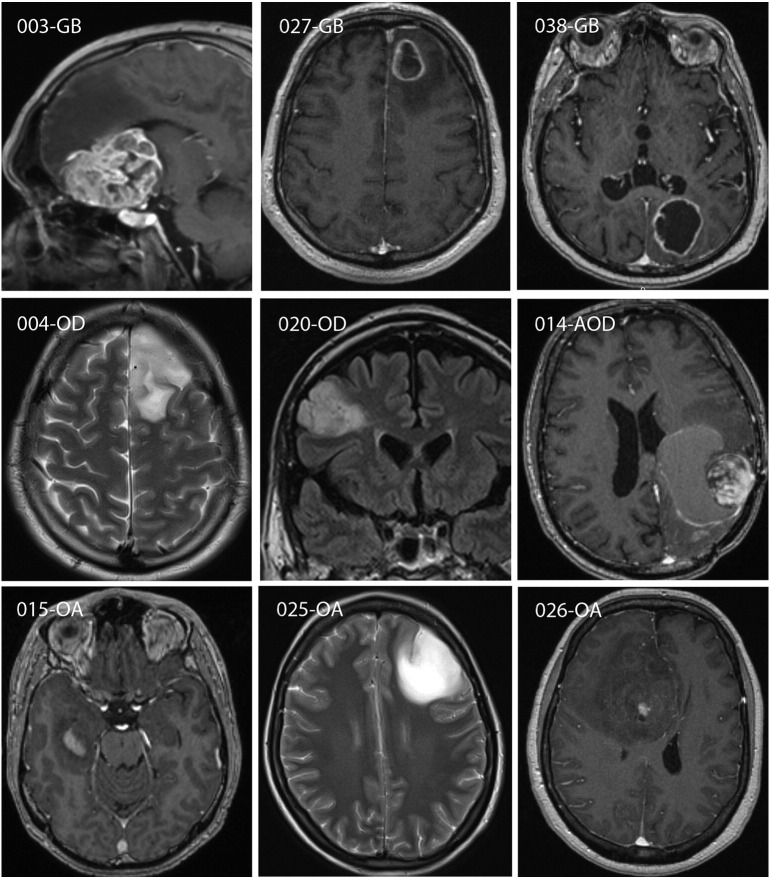
Magnetic resonance imaging characteristics of gliomas. Postcontrast T1W and T2W/FLAIR images were obtained preoperatively. Contrast enhancement was present for 003-GB, 027-GB, 038-GB, 014-AOD, 015-OA, and 026-OA but not for 004-OD, 020-OD, and 025-OA. Postcontrast T1W images are shown for enhancing and T2W/FLAIR images for nonenhancing gliomas.
Culturing of Cells
Obtained tumor was cut into smaller pieces with scalpel and McIlwain Tissue chopper (thickness 500 μm). Pieces were passed through a 21G needle followed by trituration and centrifugation. A portion of samples was treated enzymatically for 45 minutes at 37°C with Accutase (A6964, Sigma Aldrich) or 5 mg/ml collagenase (C0130, Sigma Aldrich) and 400 U/ml DNase I (D4527, Sigma Aldrich). Red blood cells were lysed with ammonium chloride (07800, Stem Cell Technologies). After two sequential centrifugations and washings, cells were suspended into culturing medium, and live cells were quantified by trypan blue exclusion. Cells were seeded in uncoated flasks (25 cm2, Greiner CellStar) and cultivated under serum-free and serum-containing conditions at 37°C in 5% CO2. Serum-free medium consisted of Dulbecco's modified Eagle's medium/F12 medium (21331, Gibco) containing 1% B27 supplement (12587 or 17504, Gibco), 1% N2 supplement (17502, Gibco), 1% Glutamax-I (Gibco 35050), 10 mM HEPES (H0887, Sigma), 20 ng/ml EGF (AF100-15, Peprotech), 10 ng/ml FGF (100-18B, Peprotech), 100 U/ml penicillin–0.1 mg/ml streptomycin (P0781, Sigma), and 1.25 μg/ml fungizone (15290, Gibco). Serum-containing medium included Dulbecco's modified Eagle's medium/F12 medium containing 1% Glutamax-I, 100 U/ml penicillin–0.1 mg/ml streptomycin, 1.25 μg/ml fungizone, and 10% heat-inactivated fetal bovine serum (Bioclear). Cells were fed twice a week by replacing with 50% of fresh medium, with first feeding performed 1 day after plating. Serum-free conditions were switched to adherent cultures after day 4. Both in serum-free and in serum-containing cultures, plates coated with ECM gel (1:20 dilution) (E6909, Sigma) gave the most reliable growth. For cell culture splitting, cells were detached with Accutase and split 1:2 up to 1:4. Procedures for additional cell lines and preparation of rat primary cortical cells are described in Supplementary Methods.
Immunohistochemistry of Paraffin-Embedded Sections and Immunocytochemistry of Cultivated Cells
Paraformaldehyde-fixed samples were embedded in paraffin, cut into 7-μm sections, and stained with hematoxylin/eosin (H&E). For immunohistochemistry, antigen retrieval was done in citrate buffer (pH 6.0, 20 minutes), and endogenous peroxidase was blocked with 0.3% H2O2–methanol (20 minutes). For NeuN and CAR stainings, 0.1% Triton X-100 was included after peroxidase blocking. Sections were blocked with 1% BSA in PBS (A9647, Sigma Aldrich) and 1% normal goat/horse serum and stained using Vectastain ABC kits (PK-4001 or PK-6102, Vector Laboratories). Primary antibodies for cell type characterization were against astrocytic-like cells [polyclonal rabbit anti–glial fibrillary acidic protein (GFAP), Z0334, DAKO, 1:10,000], oligodendrocyte-like cells [monoclonal mouse anti–2′, 3′-cyclic nucleotide 3′-phosphodiesterase (CNPase), MAB326R, Millipore, 1:500], neurons (monoclonal mouse anti-NeuN, Millipore MAB377, 1:10,000), and proliferating cells (monoclonal mouse anti-human Ki-67 antigen, M7240, DAKO, 1:2000). Primary antibodies for virus receptor studies were against coxsackie-adenovirus receptor (monoclonal mouse anti-CAR, 05-644, Millipore, 1:50), desmoglein-2 (rabbit polyclonal anti-DSG-2, H-145, sc-20,115, Santa Cruz Biotechnology Inc., 1:300), and cluster of differentiation 46 (polyclonal rabbit anti-CD46, H-294, sc-9098, Santa Cruz Biotechnology Inc., 1:100). Color reaction was performed with 3,3′-diaminobenzidine (D4293, Sigma Aldrich) for all other primary antibodies except for DSG-2, which was detected by ImmPACT NovaRED staining (SK-4805, Vector Laboratories). Sections were counterstained by Delafield's hematoxylin and analyzed under Olympus AX70 microscope using 10× and 40× objectives. Quantification of antigen expression in tumors was done as follows: distribution (no expression = 0; <25% expression = 1; 25%-50% expression = 2; >50% expression =3) × intensity (weak/moderate = ½; strong = 1). Results were transformed into scoring (0.5-1 = +; 1.5-2 = ++; 3 = +++).
For immunocytochemistry, cells were fixed in 4% paraformaldehyde (10 minutes); washed with 1× PBS (3 × 5 minutes); and treated for NeuN, CNPase, CD46, and DSG-2 staining with 50 mM NH4Cl–1× PBS (10 minutes). Cells were permeabilized with 0.1% Triton X-100–1% BSA–10% normal goat serum (S2000-500, Biowest)–1× PBS (1 hour) and incubated with primary antibodies overnight at +4°C (1:500 for GFAP and CNPase, 1:750 for NeuN, 1:100 for CAR and CD46, 1:300 for DSG-2). Detection was with Alexa Fluor 594–conjugated secondary antibodies (A 11032 or A 11012, Molecular Probes, 1 hour). Nuclei were stained with 5 μM DAPI (D1306, Molecular Probes). Cells were observed and photographed by Zeiss Axio Observer Z1 microscope via 20× or 40× objectives.
Western Blotting
Confluent cells were lysed in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Igepal (MP Biomedicals), 0.5% deoxycholate (Sigma Aldrich), and 0.1% SDS with Complete Mini protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche) and stored at −80°C until analyzed. A431 control cell lysate (ab7909) was purchased from Abcam. Aliquots were run on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis gels and blotted to Hybond-ECL membranes (GE Healthcare). Proteins were detected with primary antibodies against CAR (1:500, H-300, Santa Cruz Biotechnology Inc.), CD46 (1:500), DSG-2 (1:500), S100 (1:500, ab868, Abcam), p53 (1:500, ab131442, Abcam), EGFR (1:200, ab2430, Abcam), isocitrate dehydrogenase (wild-type IDH1, 1:600, ab94571, Abcam), IDH1 R132H (the most common IDH1 mutant, 1:500, DIA-H09, Dianova, USA), MGMT (1:1000, ab108630, Abcam), and β-actin (1:2500, C4, Santa Cruz Biotechnology). Blots were developed with anti-rabbit-Cy5 and anti-mouse-Cy3 secondary antibodies of Amersham ECL Plex Western blotting system (GE Healthcare).
Viruses and Infections In Vitro
Cells were infected with the replication-competent Ad of serotype 3 recombinant rAd3ΔE3GFP [19], referred here as HAd3; the conditionally replicating adenovirus serotype 5 construct Ad5-CRAD [20], referred to as HAd5; or the replication competent serotype 11p virus RCAd11pE1GFP [21], referred to as HAd11p. These viruses were amplified in A549 human lung carcinoma cells and functional titers obtained by TCID50 assay.
Patient glioma-derived cells (12,000 or 15,000/well) and commercial cell lines (A549, 293, A172, U87-MG; 25,000/well) were plated on 48-well plates and monitored using Zeiss Axio Observer Z1 microscope with 10× objective. For study of virus IFN sensitivity, cells were pretreated for 4 hours with human IFN β (500 U/ml or 2000 U/ml; PPT-262, Biosite). SFV VA7-EGFP produced in BHK-21 cells was used as a reference for an IFN-sensitive virus [22], [23].
Cell Viability
Cell viability was assayed with MTT Cell Proliferation Kit I (Roche) reading absorbances by VICTOR plate reader (PerkinElmer-Wallac, Turku, Finland). Results are expressed as means of absorbance difference (A595nm − A650nm).
Statistical Analyses
Cell viability data were analyzed with unpaired t test or one-way analysis of variance (followed by Bonferroni test) (Origin and GraphPad software).
Results and Discussion
Establishment of Primary Human Glioma Cell Lines
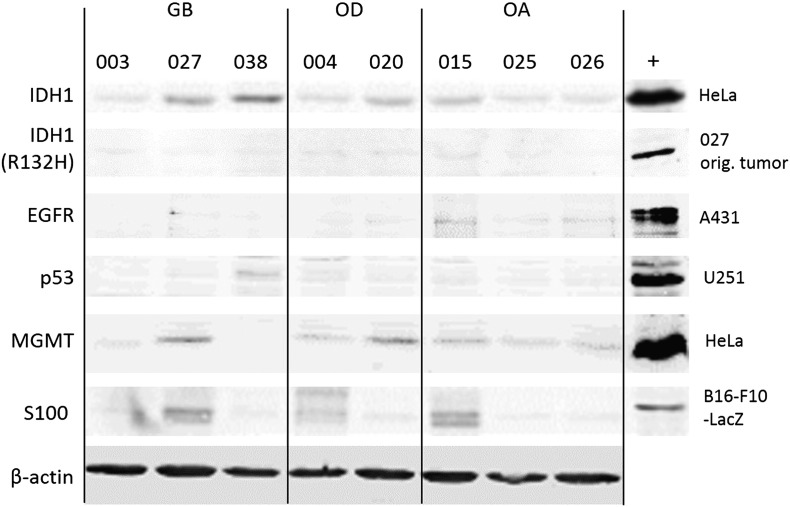
Primary surgical samples were obtained from 17 patients featuring diffusely infiltrating gliomas from grade II to IV (Table 1, Supplementary Figure S1). Superior cell viability was achieved when cells from the perioperatively collected glioma samples were extracted purely mechanically omitting proteolytic enzymes (not shown). We were able to establish cell lines from 14 of the 17 patient samples in serum containing medium and in two thirds of cases in serum-free conditions using serum-free neurosphere and monolayer techniques [24], [25]. Because of favorable growth properties, we chose to continue work with eight cell lines representing each of the glioma subtypes (three glioblastomas, two oligodendrogliomas, three oligoastrocytomas). Although cells grown in serum-free conditions may more faithfully retain glioma phenotypic features than cells grown in serum-containing medium [26], [27], they grow much more slowly and were prone to detach. Therefore, to ensure sufficient cell numbers for functional experiments, cell lines kept in serum-containing medium were used, with infections performed on cells in passages 6 to 14. To provide basal biological information about the eight chosen cell lines, we assessed by Western blotting the expression of common glioma hallmarks, such as select genes commonly deleted, mutated, amplified, or otherwise indicative of glioma growth, including TP53, IDH1, MGMT, and EGFR. Overall, none of these markers were prominently expressed in the established cell lines (Supplementary Figure S2).
Supplementary Figure S2.
Western blot analysis of IDH1, IDH1 (R132H), EGFR, p53, MGMT, and S100 in the established glioma cell lines. “+” indicates positive control cells. IDH1 represents wild type, and IDH1 (R132H) is the most common mutant of IDH1.
Cell Type and Virus Receptor Expression in Patient Tumors and Derived Cell Lines
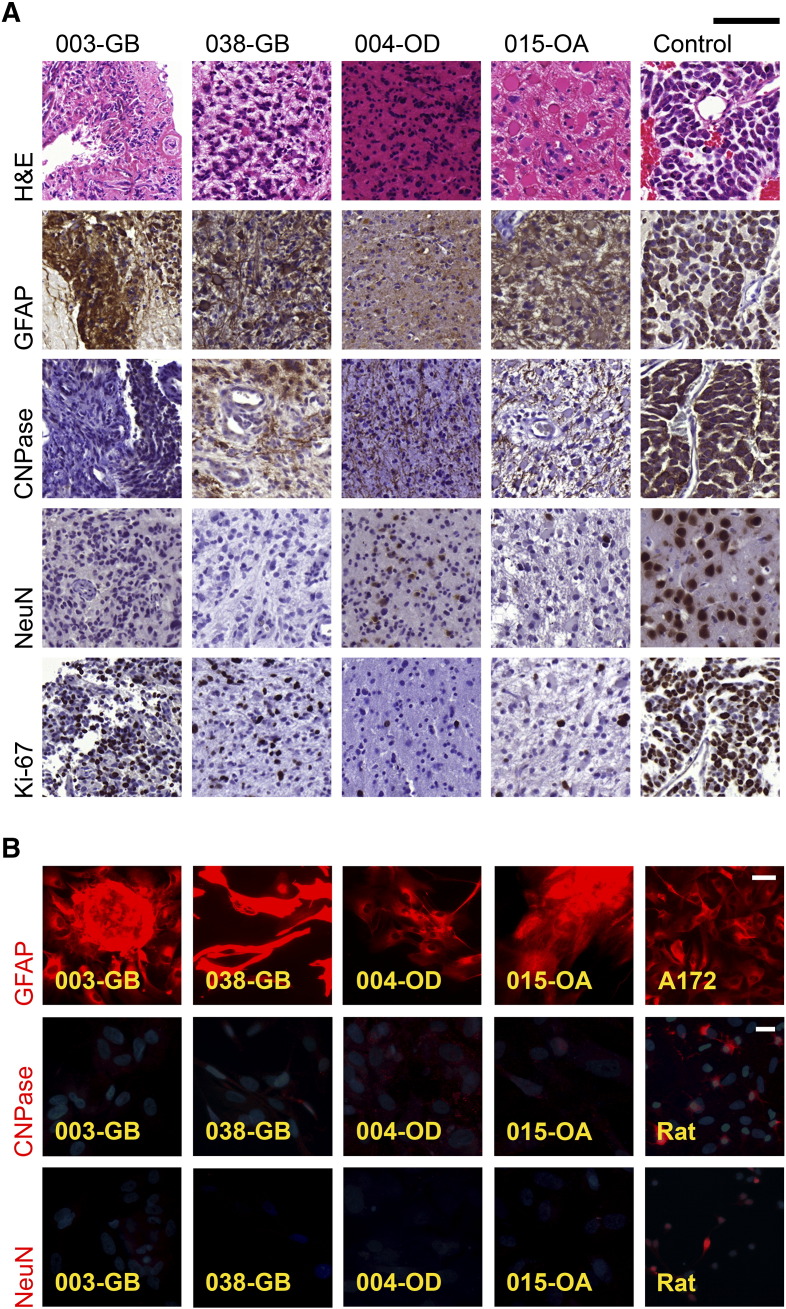
We next compared expression of cell-specific differentiation markers and virus receptors in the primary glioma tissue samples and the derived cell lines by immunohisto-/fluorescence labeling (summarized in Table 2). Astrocytic marker GFAP was strongly expressed in both the parental tumors and the derived cell lines in all cases (Supplementary Figure S3), in agreement with other studies and as expected for gliomas [17], [18], [27], [28], [29]. The oligodendrocytic marker CNPase was found in sporadic cells in tumor samples but was only detected in a punctate pattern in cultured cells originating from one oligoastrocytoma case, 025-OA, and from both the oligodendroglioma cases, 004-OD and 020-OD. As expected, the neuronal marker NeuN was found only sporadically in the original tumors and was absent from all the derived cell lines, in line with the ontogeny of gliomas [30], [31].
Table 2.
Expression Analysis of Parental Tumors and Glioma-Derived Cell Lines
| Cell Type Markers |
Virus Receptors |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NeuN |
CNPase |
GFAP |
CAR |
CD46 |
DSG-2 |
|||||||
| Code | Parental Tumor | Cell Line | Parental Tumor | Cell Line | Parental Tumor | Cell Line | Parental Tumor | Cell Line | Parental Tumor | Cell Line | Parental Tumor | Cell Line |
| 003-GB | − | − | + | − | +++ | ++ | ++ | − | + | + | + | − |
| 027-GB | + | − | − | − | ++ | ++ | + | − | +++ | +++ | ++ | + |
| 038-GB | − | − | ++ | − | ++ | +++ | + | +/− | +++ | ++ | +++ | +/− |
| 004-OD | + | − | ++ | + | + | ++ | + | − | + | +++ | + | + |
| 020-OD | + | − | + | + | + | ++ | + | − | ++ | +++ | +++ | +/− |
| 015-OA | + | − | + | − | + | ++ | + | − | +++ | +++ | +++ | + |
| 025-OA | + | − | ++ | + | + | ++ | + | − | + | ++ | +++ | − |
| 026-OA | − | − | + | − | +++ | ++ | + | − | + | ++ | +++ | − |
− = no expression; +/− = negligible expression; + = weak expression; ++ = moderate expression; +++ = strong expression.
Supplementary Figure S3.
Glial (GFAP and CNPase), neuronal (NeuN), and proliferating (Ki-67) cells in glioma tissues and cultured glioma cells. (A) Diversity of tumors was evident from paraffin sections of patient gliomas in which hematoxylin (blue color) was used as counterstain. Some tumors, e.g., oligodendroglioma 004-OD, were morphologically compact. H&E staining shows gemistocytes with large eosinophilic cytoplasm and peripheral nuclei in 015-OA parental tumor. Notable are, for example, the highly proliferating, Ki-67–positive (brown color) cells in glioblastomas and anaplastic oligodendroglioma. 014-OAD was used as control in all cases except in NeuN staining in which neurons from rat brain are shown. Scale bar 100 μm. (B) Fluorescence images of immunocytochemically (red color) stained cultured cells. GFAP images do not contain nuclei stain. In the other images, nuclei are stained with DAPI (blue color). Cultured cells derived from patient gliomas possessed astrocytic marker GFAP and, to a lesser extent, oligodendrocytic marker CNPase. A172 cells and primary cultures from rat cortex were used as positive controls. Scale bar 50 μm and 20 μm in GFAP and other images, respectively.
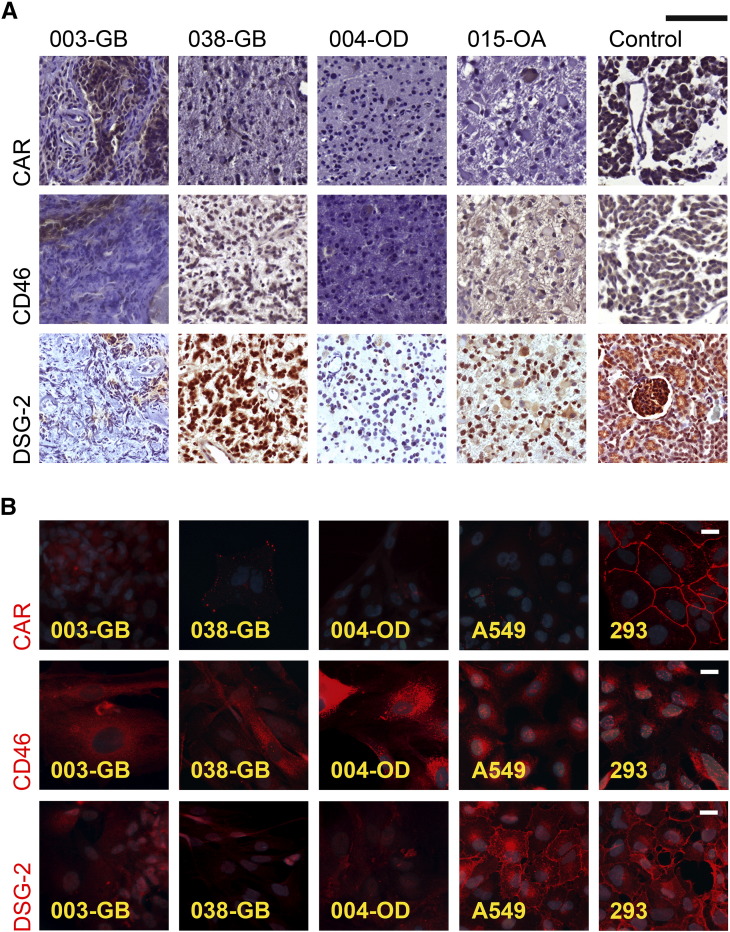
Efficacy of adenovirus infection of cultured cells has been shown to depend on cognate virus receptor expression levels [8], [9], [32]. Correspondingly, it is possible that the unfavorable results in recent phase II and III clinical trials treating glioblastoma patients with human adenovirus serotype 5–derived constructs [6], [7] may partly derive from insufficient expression of CAR on glioma cells [32]. In our samples, CAR was expressed at low level in sporadic cells in the primary tumors and was almost absent in the derived cell lines (Table 2, Figure 1). Although CAR has been detected in several commercial and low-passage primary glioma cell lines in earlier studies [9], [32], [33], [34], [35], [36], [37], the general expression level also in these studies has been low. Moreover, as CAR has inhibitory properties for cancer invasion, its expression is generally reduced in advanced gliomas compared to lower-grade tumors [9], [38], [39].
Figure 1.
Expression of adenovirus receptors (CAR, CD46, and DSG-2) in glioma tissues and cultured glioma cells. (A) Adenovirus receptor expression (brown color) was variable in paraffin sections of patient gliomas for which hematoxylin (blue color) was used as counterstain. Anaplastic oligodendroglioma 014-AOD was used as positive control in CAR and CD46 staining and rat kidney in DSG-2 staining. Among gliomas especially, 014-AOD had abundant expression of adenovirus receptors. Scale bar 100 μm. (B) Fluorescence images of immunocytochemically (red color) stained cultured cells. Nuclei were stained with DAPI (blue color). Especially CD46, receptor for serotype 11p, was well preserved among patient glioma-derived cells. A549 and 293 cells were used as positive controls. Scale bar 20 μm.
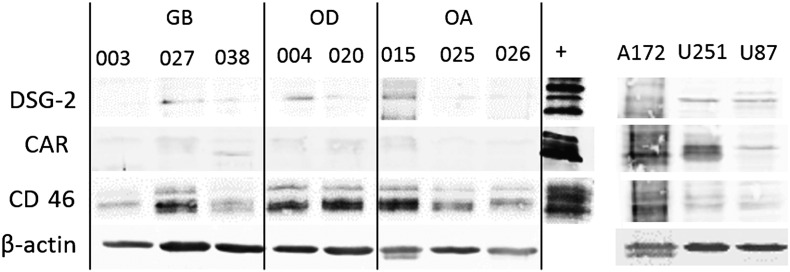
DSG-2 belongs to the cadherin family of transmembrane glycoproteins and serves as receptor for HAd3 entry, but little is known about DSG-2 expression in gliomas. In colon carcinoma xenografts, loss of DSG-2 led to reduced epithelial cell proliferation and tumor growth [40]. In the present study, DSG-2 subjectively appeared more abundant in the parental tumors than CAR, and its expression in the derived cell lines was more obvious than CAR, although still weak compared to the positive controls (Table 2, Figure 1). Adenovirus 11p receptor CD46 was maintained at moderate/strong levels in both primary tumors and derived cell lines (Table 2, Figure 1), consistent with previous investigations [32], [36], [37], [41]. The results by immunofluorescence staining in fixed cells were largely mirrored by Western blotting of corresponding cell lysates (Supplementary Figure S4).
Supplementary Figure S4.
Western blot analysis of adenovirus receptor expression in established and commercially available glioma cell lines (A172, U251, and U87-MG). “+” indicates positive control cells; A431 for DSG-2, 025 (original tumor lysate) for CAR, and U251 for CD46.
Glioma Permissiveness to Oncolytic Adenoviruses In Vitro Maybe Not Primary Receptor Dependent
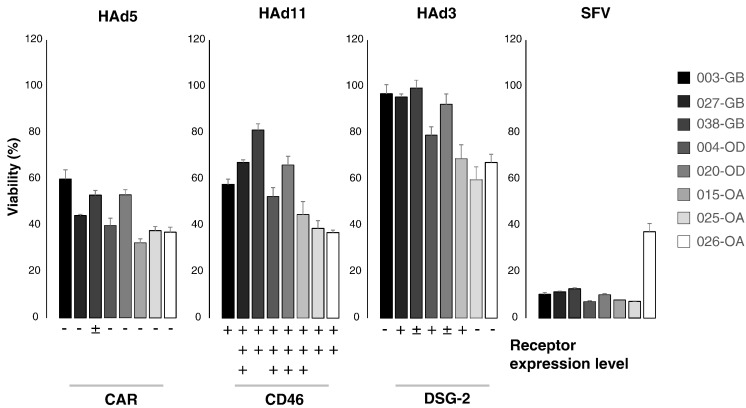
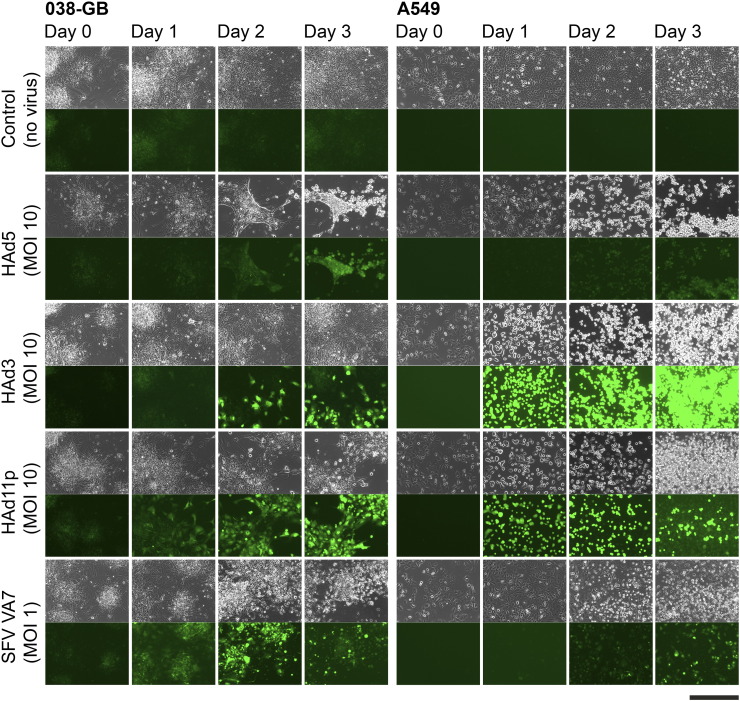
Given the small sample size in the present study, it was no surprise that HAd serotype receptor expression did not correlate with any particular glioma subclass. Next, to determine whether the glioma subclass and/or virus receptor levels would predict infection efficacy with the corresponding oncolytic adenoviruses, established cell lines were infected at multiplicity of infection (MOI) 10 and followed under fluorescence microscope for 72 hours, at the end of which oncolytic potency was determined by viability assay. SFV VA7 served as a broad-tropism oncolytic control, which also potentially can reveal the presence of basal antiviral defenses. Our results showed comparable tumor killing by HAd5 and HAd11p in all tested cell lines (Figure 2). Expression of GFP was more intense in the HAd11p-infected cells than in the HAd5-infected cells (Supplementary Figure S5), which is likely due to differences in the respective virus designs (in HAd5 GFP expression is transcriptionally linked to E1B), as the cytopathic effects indicative of virus replication were very similar between the two serotypes. In contrast, HAd3 reduced cell viability by at most 40% and then only in the lower-grade glioma cell lines (particularly from oligoastrocytomas), with the glioblastoma cell lines showing only minor loss of viability (Figure 2). Based on GFP expression, in 72 hours, HAd3 had only infected a fraction of primary glioma cells compared to the other two serotypes (Supplementary Figure S5). SFV infection progressed similarly in all samples, except in 026-OA, where infection appeared less efficient than in the other cell lines, constituting an interesting case for further study with regard to cell-intrinsic resistance to oncolytic SFV replication (Figure 2).
Figure 2.
Relationship between viability and receptor expression level in glioma cell lines. Viability is determined 3 days after infection with adenovirus (HAd) or SFV and is given as percentage of control (100%; average ± S.E.M., n = 3 parallel wells/group) at the same time point. Cells were derived from glioblastoma (003-GB, 027-GB, and 038-GB), oligodendroglioma (004-OD and 020-OD), and oligoastrocytoma (015-OA, 025-OA, and 026-OA) patients. MOI was 10 for adenoviruses (HAd5, HAd3, and HAd11p) and 1 for SFV.
Supplementary Figure S5.
Human cells infected with adenoviruses (HAd5, HAd3, and HAd11p) and SFV VA7. Phase contrast (upper) and fluorescence (lower) microscopy images were captured before infection and up to 3 days after infection. 038-GB cells were derived from glioblastoma patient, while A549 human lung carcinoma cells served as controls. Scale bar 500 μm.
Interestingly, while CAR was virtually absent in the established glioma cell lines, infection and oncolytic potency of HAd5 were at least as pronounced as for HAd11p, whose receptor CD46 was clearly expressed in all studied cell lines (Figure 1, Figure 2). Overall, in our limited sample set, the level of these adenovirus receptors did not correlate with oncolytic potency in vitro (which serves as an indicator for infection efficiency). However, because numerous previous studies have demonstrated a functional role for the virus receptors in adenovirus oncolytic potency [8], [9], [35], [42], [43], it is preferable to measure virus receptor levels perioperatively rather than from derived cell lines due to potential loss of expression during passaging. Moreover, the therapeutic efficacy with oncolytic viruses in general is not solely determined by infection capacity but also by immunogenicity of virus-induced cell death [44], which complicates predictions of clinical outcome based on infection data in vitro.
While relative comparisons between these adenovirus serotypes in our study is difficult due to fundamental differences in virus design, the HAd3 fiber knob attached onto an HAd5-based backbone, termed HAd5/3, did display greater infection capacity than the parental HAd5 virus [45], arguing for the possibility to optimize oncolytic adenoviruses for glioma targeting further by combining the strongest human adenovirus backbone with the best receptor-binding domain. Serotype 3 also has DSG-2-dependent tight-junction-opening properties which have not been described for other serotypes, making the virus still an appealing contender for glioma targeting [46].
Gliomas Capable of Mounting Interferon-Dependent Antiviral Responses But These Not Hindering Human Adenovirus Oncolysis
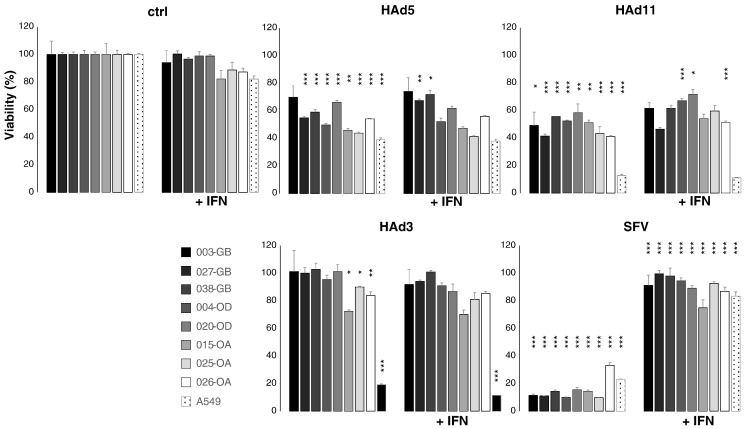
We previously demonstrated that some human commercial glioma cell lines as well as murine gliomas can mount both intrinsic and IFN-mediated resistance against oncolytic SFV [23]. To test whether primary human glioma cell lines can mount IFN-triggered antiviral defenses and whether such resistance could limit the replication of the three different adenovirus serotypes used in the present study, we pretreated for 4 hours our eight primary cell lines, two commercial glioma cell lines (A172 and U87), and IFN-responsive A549 cells with recombinant human IFN beta (2000 U/ml) and infected them with the three human adenovirus serotypes. SFV was used as an indicator for productive antiviral defense. The experiments revealed that only SFV replication was blocked by the IFN pretreatment, demonstrating that the primary glioma cell lines are capable of responding to IFN (Figure 3). While it has been shown that serotype 5 oncolytic adenovirus replicates more slowly in several types of epithelial cancer cells featuring constitutive expression of select IFN-stimulated genes, such as MxA [13], [14], it is possible that such resistance differs functionally from the response to brief IFN pretreatment. Alternatively, as SFV (control virus) was inhibited, we conclude that oncolytic adenoviruses can replicate even in IFN-exposed glioma cells, and thus lack of clinical efficacy may result from yet undiscovered factors.
Figure 3.
Effect of exogenous interferon on infection with adenovirus (HAd) and SFV. Viability of human cells is determined 3 days after infection and is given as percentage of control (ctrl, 100%; average ± S.E.M., n = at least 3 parallel wells/group) at the same time point. Cells were derived from glioblastoma (003-GB, 027-GB, and 038-GB), oligodendroglioma (004-OD and 020-OD), and oligoastrocytoma (015-OA, 025-OA, and 026-OA) patients. A549 human lung carcinoma cells were used as controls. MOI was 10 for adenoviruses (HAd5, HAd3, and HAd11p) and 1 for SFV. IFN-β (2000 U/ml) was applied 4 hours before cells were challenged with viruses. *P < .05, **P < .01, ***P < .001 (unpaired t test, comparisons: control versus virus infected or virus infected versus virus infected + IFN).
Conclusions
We describe here establishment of new glioma cell lines derived not only from glioblastomas but also from lower-grade gliomas including oligoastrocytomas and oligodendrogliomas. Along with clinical data and original tumors, their immunophenotypic characterization is presented. We show that, compared to human adenoviruses (serotypes 3, 5, and 11p), the VA7-EGFP alphavirus vector based on SFV is superior in infection and killing of low-passage human glioma cells. Human adenoviruses might, however, be more suitable for therapeutic use, as they seem capable of replicating despite IFN response, although recently also more IFN-I resistant versions of SFV have been described by us [47]. To sum up, clinical efficacy may be influenced by factors not fully reflected under culture conditions.
The following are the supplementary data related to this article.
Supplementary material
Funding
This study was supported by Academy of Finland (grant 137958); Finnish Cancer Foundations; Cancer Society of North Savo; Emil Aaltonen Foundation; Medicinska Understödsföreningen Liv och Hälsa; State Funding for University Hospitals; and the University strategic funding for the Cancer Center of Eastern Finland.
Conflict of Interest
Ya-Fang Mei states that adenovirus serotype 11p vector is under European patent process (application EP20100822318). Other authors declare no conflicts of interest.
Acknowledgements
We thank Katariina Helin, Marita Parviainen, and Minna Rautiainen (Kuopio University Hospital, Finland) for coordinating issues related to patient samples.
Contributor Information
Minna Niittykoski, Email: mijoni@gmail.com.
Mikael von und zu Fraunberg, Email: mikael.fraunberg@kuh.fi.
Miika Martikainen, Email: miika.martikainen@igp.uu.se.
Tuomas Rauramaa, Email: tuomas.rauramaa@uef.fi.
Arto Immonen, Email: arto.immonen@kuh.fi.
Susanna Koponen, Email: susanna.koponen@kuh.fi.
Ville Leinonen, Email: ville.leinonen@kuh.fi.
Markus Vähä-Koskela, Email: markus.vaha-koskela@helsinki.fi.
Qiwei Zhang, Email: Zhang.Qiwei@yahoo.com.
Florian Kühnel, Email: kuehnel.florian@mh-hannover.de.
Ya-Fang Mei, Email: ya-fang.mei@umu.se.
Seppo Ylä-Herttuala, Email: seppo.ylaherttuala@uef.fi.
Juha E. Jääskeläinen, Email: juha.e.jaaskelainen@kuh.fi.
Ari Hinkkanen, Email: ari.hinkkanen@utu.fi.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Walker C, Baborie A, Crooks D, Wilkins S, Jenkinson MD. Biology, genetics and imaging of glial cell tumours. Br J Radiol. 2011;84:S90–S106. doi: 10.1259/bjr/23430927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weller M, Pfister SM, Wick W, Hegi ME, Reifenberger G, Stupp R. Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol. 2013;14:e370–379. doi: 10.1016/S1470-2045(13)70168-2. [DOI] [PubMed] [Google Scholar]
- 5.Chiocca EA, Abbed KM, Tatter S, Louis DN, Hochberg FH, Barker F, Kracher J, Grossman SA, Fisher JD, Carson K. A phase I open-label, dose-escalation, multi-institutional trial of injection with an E1B-attenuated adenovirus, ONYX-015, into the peritumoral region of recurrent malignant gliomas, in the adjuvant setting. Mol Ther. 2004;10:958–966. doi: 10.1016/j.ymthe.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 6.Immonen A, Vapalahti M, Tyynelä K, Hurskainen H, Sandmair A, Vanninen R, Langford G, Murray N, Ylä-Herttuala S. AdvHSV-tk gene therapy with intravenous ganciclovir improves survival in human malignant glioma: a randomised, controlled study. Mol Ther. 2004;10:967–972. doi: 10.1016/j.ymthe.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann JK, Chiocca EA. Glioma virus therapies between bench and bedside. Neuro Oncol. 2014;16:334–351. doi: 10.1093/neuonc/not310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asaoka K, Tada M, Sawamura Y, Ikeda J, Abe H. Dependence of efficient adenoviral gene delivery in malignant glioma cells on the expression levels of the coxsackievirus and adenovirus receptor. J Neurosurg. 2000;92:1002–1008. doi: 10.3171/jns.2000.92.6.1002. [DOI] [PubMed] [Google Scholar]
- 9.Fuxe J, Liu L, Malin S, Philipson L, Collins VP, Pettersson RF. Expression of the coxsackie and adenovirus receptor in human astrocytic tumors and xenografts. Int J Cancer. 2003;103:723–729. doi: 10.1002/ijc.10891. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami Y, Li H, Lam JT, Krasnykh V, Curiel DT, Blackwell JL. Substitution of the adenovirus serotype 5 knob with a serotype 3 knob enhances multiple steps in virus replication. Cancer Res. 2003;63:1262–1269. [PubMed] [Google Scholar]
- 11.Kuhn I, Harden P, Bauzon M, Chartier C, Nye J, Thorne S, Reid T, Ni S, Lieber A, Fisher K. Directed evolution generates a novel oncolytic virus for the treatment of colon cancer. PLoS One. 2008;3:e2409. doi: 10.1371/journal.pone.0002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauss R, Sova P, Liu Y, Li ZY, Tuve S, Pritchard D, Brinkkoetter P, Möller T, Wildner O, Pesonen S. Epithelial phenotype confers resistance of ovarian cancer cells to oncolytic adenoviruses. Cancer Res. 2009;69:5115–5125. doi: 10.1158/0008-5472.CAN-09-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monsurró V, Beghelli S, Wang R, Barbi S, Coin S, Di Pasquale G, Bersani S, Castellucci M, Sorio C, Eleuteri S. Anti-viral state segregates two molecular phenotypes of pancreatic adenocarcinoma: potential relevance for adenoviral gene therapy. J Transl Med. 2010;8:10. doi: 10.1186/1479-5876-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liikanen I, Monsurrò V, Ahtiainen L, Raki M, Hakkarainen T, Diaconu I, Escutenaire S, Hemminki O, Dias JD, Cerullo V. Induction of interferon pathways mediates in vivo resistance to oncolytic adenovirus. Mol Ther. 2011;19:1858–1866. doi: 10.1038/mt.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vähä-Koskela M, Tähtinen S, Grönberg-Vähä-Koskela S, Taipale K, Saha D, Merisalo-Soikkeli M, Ahonen M, Rouvinen-Lagerström N, Hirvinen M, Veckman V. Overcoming tumor resistance by heterologous adeno-poxvirus combination therapy. Mol Ther Oncolytics. 2015;1:14006. doi: 10.1038/mto.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torsvik A, Stieber D, Enger PØ, Golebiewska A, Molven A, Svendsen A, Westermark B, Niclou SP, Olsen TK, Chekenya Enger M. U-251 revisited: genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med. 2014;3:812–824. doi: 10.1002/cam4.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomès D, de Néchaud B, Maunoury R, Moura Neto V, Brigaudeau C, Labrousse F, Dupouey P. Glial fibrillary acidic protein expression in a new human glioma cell line in culture before and after xenogenic transplantation into nude mice. Acta Neuropathol. 1997;94:376–384. doi: 10.1007/s004010050722. [DOI] [PubMed] [Google Scholar]
- 18.Quentmeier H, Osborn M, Reinhardt J, Zaborski M, Drexler HG. Immunocytochemical analysis of cell lines derived from solid tumors. J Histochem Cytochem. 2001;49:1369–1378. doi: 10.1177/002215540104901105. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Su X, Seto D, Zheng BJ, Tian X, Sheng H, Li H, Wang Y, Zhou R. Construction and characterization of a replication-competent human adenovirus type 3-based vector as a live-vaccine candidate and a viral delivery vector. Vaccine. 2009;27:1145–1153. doi: 10.1016/j.vaccine.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 20.Wirth T, Zender L, Schulte B, Mundt B, Plentz R, Rudolph KL, Manns M, Kubicka S, Kühnel F. A telomerase-dependent conditionally replicating adenovirus for selective treatment of cancer. Cancer Res. 2003;63:3181–3188. [PubMed] [Google Scholar]
- 21.Mei Y-F, Wu H, Hutenby K, Silver J. Complete replication-competent adenovirus 11p vectors with E1 or E3 insertions show improved heat stability. Virology. 2016;497:198–210. doi: 10.1016/j.virol.2016.07.026. [DOI] [PubMed] [Google Scholar]
- 22.Vähä-Koskela MJ, Tuittila MT, Nygårdas PT, Nyman JK, Ehrengruber MU, Renggli M, Hinkkanen AE. A novel neurotropic expression vector based on the avirulent A7(74) strain of Semliki Forest virus. J Neurovirol. 2003;9:1–15. doi: 10.1080/13550280390173382. [DOI] [PubMed] [Google Scholar]
- 23.Ruotsalainen J, Martikainen M, Niittykoski M, Huhtala T, Aaltonen T, Heikkilä J, Bell J, Vähä-Koskela M, Hinkkanen A. Interferon-β sensitivity of tumor cells correlates with poor response to VA7 virotherapy in mouse glioma models. Mol Ther. 2012;20:1529–1539. doi: 10.1038/mt.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fael Al-Mayhani TM, Ball SLR, Zhao JW, Fawcett J, Ichimura K, Collins PV, Watts C. An efficient method for derivation and propagation of glioblastoma cell lines that concerves the molecular profile of their original tumors. J Neurosci Methods. 2009;176:192–199. doi: 10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 27.Christensen K, Aaberg-Jessen C, Andersen C, Goplen D, Bjerkvig R, Kristensen BW. Immunohistochemical expression of stem cell, endothelial cell, and chemosensitivity markers in primary glioma spheroids cultured in serum-containing and serum-free medium. Neurosurgery. 2010;66:933–947. doi: 10.1227/01.NEU.0000368393.45935.46. [DOI] [PubMed] [Google Scholar]
- 28.Matyja E, Taraszewska A, Zabek M. Phenotypic characteristics of GFAP-immunopositive oligodendroglial tumors. Part I: immunohistochemical study. Folia Neuropathol. 2001;39:19–26. [PubMed] [Google Scholar]
- 29.Zhang L, Yamane T, Satoh E, Amagasaki K, Kawataki T, Asahara T, Furuya K, Nukui H, Naganuma H. Establishment and partial characterization of five malignant glioma cell lines. Neuropathology. 2005;25:136–143. doi: 10.1111/j.1440-1789.2005.00601.x. [DOI] [PubMed] [Google Scholar]
- 30.Koperek O, Gelpi E, Birner P, Haberler C, Budka H, Hainfellner JA. Value and limits of immunohistochemistry in differential diagnosis of clear cell primary brain tumors. Acta Neuropathol. 2004;108:24–30. doi: 10.1007/s00401-004-0856-9. [DOI] [PubMed] [Google Scholar]
- 31.Preusser M, Laggner U, Haberler C, Heinzl H, Budka H, Hainfellner JA. Comparative analysis of NeuN immunoreactivity in primary brain tumors: conclusions for rational use in diagnostic histopathology. Histopathology. 2006;48:438–444. doi: 10.1111/j.1365-2559.2006.02359.x. [DOI] [PubMed] [Google Scholar]
- 32.Brouwer E, Havenga MJ, Ophorst O, de Leeuw B, Gijsbers L, Gillissen G, Hoeben RC, ter Horst M, Nanda D, Dirven C. Human adenovirus type 35 vector for gene therapy of brain cancer: improved transduction and bypass of pre-existing antivector immunity in cancer patients. Cancer Gene Ther. 2007;14:211–219. doi: 10.1038/sj.cgt.7701010. [DOI] [PubMed] [Google Scholar]
- 33.Lamfers ML, Grill J, Dirven CM, Van Beusechem VW, Geoerger B, Van Den Berg J, Alemany R, Fueyo J, Curiel DT, Vassal G. Potential of the conditionally replicative adenovirus Ad5-Delta24RGD in the treatment of malignant gliomas and its enhanced effect with radiotherapy. Cancer Res. 2002;62:5736–5742. [PubMed] [Google Scholar]
- 34.Fueyo J, Alemany R, Gomez-Manzano C, Fuller GN, Khan A, Conrad CA, Liu TJ, Jiang H, Lemoine MG, Suzuki K. Preclinical characterization of the antiglioma activity of a tropism-enhanced adenovirus targeted to the retinoblastoma pathway. J Natl Cancer Inst. 2003;95:652–660. doi: 10.1093/jnci/95.9.652. [DOI] [PubMed] [Google Scholar]
- 35.Skog J, Edlund K, Widegren B, Salford LG, Wadell G, Mei Y-F. Efficient internalization into low-passage glioma cell lines using adenoviruses other than type 5: an approach for improvement of gene delivery to brain tumors. J Gen Virol. 2004;85:2627–2638. doi: 10.1099/vir.0.80084-0. [DOI] [PubMed] [Google Scholar]
- 36.Ulasov IV, Tyler MA, Zheng S, Han Y, Lesniak MS. CD46 represents a target for adenoviral gene therapy of malignant glioma. Hum Gene Ther. 2006;17:556–564. doi: 10.1089/hum.2006.17.556. [DOI] [PubMed] [Google Scholar]
- 37.Paul CPL, Everts M, Glasgow JN, Dent P, Fisher PB, Ulasov IV, Lesniak MS, Stoff-Khalili MA, Roth JC, Preuss MA. Characterization of infectivity of knob-modified adenoviral vectors in glioma. Cancer Biol Ther. 2008;7:786–793. doi: 10.4161/cbt.7.5.5421. [DOI] [PubMed] [Google Scholar]
- 38.Douglas JT, Kim M, Sumerel LA, Carey DE, Curiel DT. Efficient oncolysis by a replicating adenovirus (Ad) in vivo is critically dependent on tumor expression of primary ad receptors. Cancer Res. 2001;61:813–817. [PubMed] [Google Scholar]
- 39.Huang KC, Altinoz M, Wosik K, Larochelle N, Koty Z, Zhu L, Holland PC, Nalbantoglu J. Impact of the coxsackie and adenovirus receptor (CAR) on glioma cell growth and invasion: requirement for the C-terminal domain. Int J Cancer. 2005;113:738–745. doi: 10.1002/ijc.20623. [DOI] [PubMed] [Google Scholar]
- 40.Kamekura R, Kolegraff KN, Nava P, Hilgarth RS, Feng M, Parkos CA, Nusrat A. Loss of the desmosomal cadherin desmoglein-2 suppresses colon cancer cell proliferation through EGFR signaling. Oncogene. 2014;33:4531–4536. doi: 10.1038/onc.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wohlfahrt ME, Beard BC, Lieber A, Kiem H-P. A capsid-modified, conditionally replicating oncolytic adenovirus vector expressing TRAIL leads to enhanced cancer cell killing in human glioblastoma models. Cancer Res. 2007;67:8783–8790. doi: 10.1158/0008-5472.CAN-07-0357. [DOI] [PubMed] [Google Scholar]
- 42.Miller CR, Buchsbaum DJ, Reynolds PN, Douglas JT, Gillespie GY, Mayo MS, Raben D, Curiel DT. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res. 1998;58:5738–5748. [PubMed] [Google Scholar]
- 43.Candolfi M, Curtin JF, Xiong W-D, Kroeger KM, Liu C, Rentsendorj A, Agadjanian H, Medina-Kauwe L, Palmer D, Ng P. Effective high-capacity gutless adenoviral vectors mediate transgene expression in human glioma cells. Mol Ther. 2006;14:371–381. doi: 10.1016/j.ymthe.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartlett DL, Liu Z, Sathaiah M, Guo Z, He Y, Guo ZS. Oncolytic viruses as therapeutic cancer vaccines. Mol Cancer. 2013;12:103. doi: 10.1186/1476-4598-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulasov IV, Shah N, Kaverina NV, Lee H, Lin B, Lieber A, Kadagidze ZA, Yoon J-G, Schroeder B, Hothi P. Tamoxifen improves cytopathic effect of oncolytic adenovirus in primary glioblastoma cells mediated through autophagy. Oncotarget. 2015;6:3977–3987. doi: 10.18632/oncotarget.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Excoffon KJDA, Bowers JR, Sharma P. Alternative splicing of viral receptors: A review of the diverse morphologies and physiologies of adenoviral receptors. Recent Res Dev Virol. 2014;9:1–24. [PMC free article] [PubMed] [Google Scholar]
- 47.Martikainen M, Niittykoski M, von und zu Fraunberg M, Immonen A, Koponen S, van Geenen M, Vähä-Koskela M, Ylösmäki E, Jääskeläinen JE, Saksela K. MicroRNA-attenuated clone of virulent Semliki Forest virus overcomes antiviral type I interferon in resistant mouse CT-2A glioma. J Virol. 2015;89:10637–10647. doi: 10.1128/JVI.01868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material