Abstract
Background
Bioactive activities of adzuki bean have been widely reported, however, the phytochemical information of adzuki bean is incomplete. The aim of this study was to characterize and quantify flavonoids and saponins in adzuki bean. High performance liquid chromatography with diode array detection and electro spray ionization-tandem multi-stage mass spectrometry (HPLC–DAD–ESI–MSn) were applied to do qualitative and quantitative analyses.
Results
A total of 15 compounds from adzuki bean were identified by HPLC–DAD–ESI–MSn. Among 15 compounds identified, four flavonoids (catechin, vitexin-4″-O-glucoside, quercetin-3-O-glucoside, and quercetin-3-O-rutinoside) and six saponins (azukisaponin I, II, III, IV, V, and VI) in adzuki bean were further quantified by external calibration method using HPLC–MS with the program of time segment and extract ion chromatogram (EIC) analysis.
Conclusions
Current qualitative and quantitative method based on HPLC and MS technique provides a scientific basis for in vitro and in vivo pharmacological study in the future.
Graphical abstract.
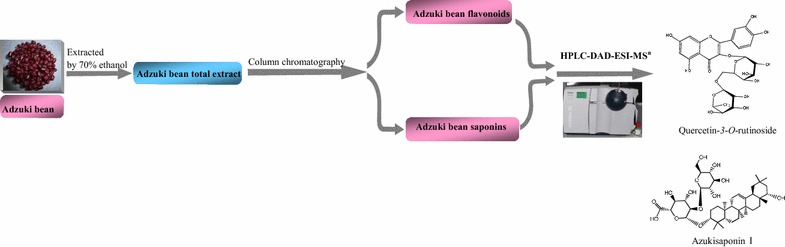
Isolation and characterization of flavonoids and saponins from adzuki bean.
Keywords: Adzuki bean, Flavonoids, Saponins, ESI–MSn, HPLC
Introduction
Adzuki bean is mainly produced and consumed in China and several other countries in East Asia. It has been used as a diuretic, antidote, and remedy for dropsy and beriberi in traditional Chinese medicine and also used as food for thousands of years. Extensive bioactivities of adzuki bean, such as anti-tumor [1, 2], anti-diabetes, [3, 4], antioxidant [5–8], and hepatoprotective effect [9] have been reported. These bioactivities are contributed by chemical constituents in beans, mainly including flavonoids and saponins. The previous articles showed that adzuki bean contained flavonoids such as (+) epicatechin, (+) catechin, quercetin, vitexin or their derivatives; [6, 10, 11] and saponins, such as azukisaponin I, II, III, IV, V, and VI [12] and AZ I [13], II, III, and IV [14]. The information, especially saponin information on adzuki bean, is incomplete, the name and structure of “AZ” and azukisaponin are confused. Moreover, there are limited articles in recent years to systematically and comprehensively investigate flavonoids and saponins of adzuki bean. Therefore, the present study aimed to establish a method to separate individual flavonoids and saponins from adzuki bean, characterize their chemical structures by HPLC–DAD–ESI–MSn, and further quantify them by HPLC–MS.
Materials and methods
Materials
Adzuki beans (Vigna angularis L.) were purchased from local market in Changchun, Jilin Province, and identified by Prof. Jinming Mu of Faculty of Agronomy in Jilin Agricultural University.
Chemicals and reagents
Chromatographic grade acetonitrile and methanol were purchased from Merck (Darmstadt, USA). (+) Catechin, (+) epicatechin, quercetin-3-O-rutinoside, quercetin-3-O-glucoside and vitexin-4″-O-glucoside were purchased from Sigma (St. Louis, MO, USA). Saponin standards of azukisaponin I, II, III, IV, V, and VI were isolated in our lab. Other chemicals, such as ethanol, methanol, and acetone were of analytical grade. Macro porous resins AB-8 were supplied by Nankai University. Polyamide resin was purchased from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China).
Sample preparation of flavonoids and saponins from adzuki bean
The flavonoids and saponins of adzuki bean were prepared according to the previous articles [15–18]. Extraction and isolation scheme of total extract, flavonoids and saponins from adzuki bean was shown in Fig. 1. Briefly, adzuki bean was ground, 14 kg of the powder was then extracted with 140 L of 70% ethanol for three times. The combined extracts were filtrated and concentrated to remove ethanol. The remaining aqueous solution was extracted with 14 L of petroleum ether at room temperature for three times. The aqueous phase was then extracted with 14 L of n-butanol at room temperature three times. The n-butanol layer was evaporated under vacuum to obtain 158.6 g of extract which was defined as adzuki bean total extract (ABTE). Flavonoids of adzuki bean were collected from the 45% ethanol elution fraction from AB-8 resin column after eluting with water. The collected crude flavonoids of adzuki bean were subjected to the second column with polyamide according to the literature [19–21], and the flavonoids were further obtained in 40% ethanol fraction from polyamide column after eluting with 10% ethanol. Finally, the enriched adzuki bean flavonoids (ABF) were obtained from the supernatant after precipitating with methanol–acetone. The enriched saponin of adzuki bean was collected in the 80% ethanol fraction from AB-8 resin column after eluting with 45% ethanol. With precipitation method, adzuki bean saponins (ABS) were further purified using precipitation method by adding methanol–acetone.
Fig. 1.
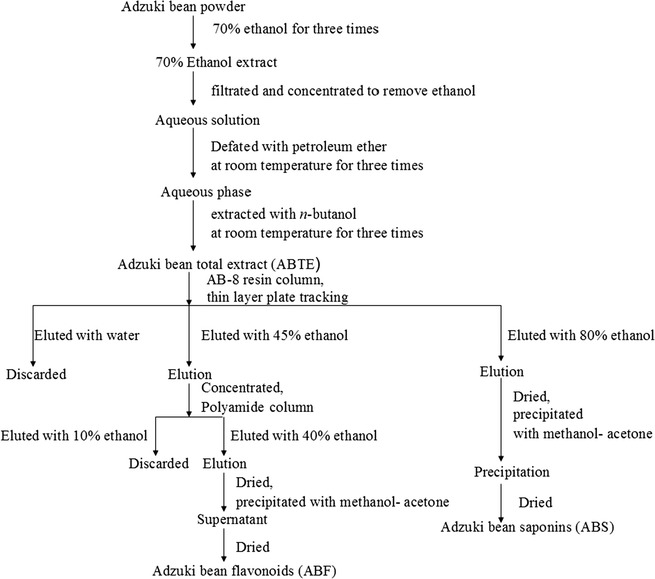
Extraction and isolation scheme of total extract, flavonoids and saponins from adzuki bean
High performance liquid chromatography analysis
The chemical constituents of adzuki bean total extract (ABTE), adzuki bean flavonoids (ABF) and adzuki bean saponins (ABS) were identified by liquid chromatography-ion trap mass spectrometry. HPLC analysis was performed on an Agilent 1100 series HPLC system equipped with degasser, binary pump, diode array detector and auto-sampler (San Francisco, USA). The separation was performed on a Phenomenex C8 column (150 × 2.0 mm, 5 μm). Gradient elution was performed using water containing 10 mM ammonium acetate (A) and acetonitrile (B). Initial conditions were 10% B for 10 min, changed to 15% B at 30 min and 25% B at 45 min, and then 35% B at 55 min, 45% B at 60 min and 55% B at 70 min. Flow rate was set at 0.2 mL/min, and UV absorption was measured at wavelength of 205 nm and 262 nm for saponins and flavonoids, respectively. The sample injection volume was 10 μL.
Electro spray ionization-tandem multi-stage mass spectrometry (ESI–MSn) analysis
ESI–MS analysis was carried out on an Esquire 4000 ion trap mass spectrometer (Bruker–Daltonics, Bremen, Germany) with an electrospray ionization (ESI) interface. The instrument was operated at an ionization voltage of +4000 V and source temperature of 300 °C. Nitrogen was used as nebulizer gas at 30 psi and drying gas at a flow rate of 9 L/min. Collision energy was optimized for each compound. Three time segments were used in mass spectrometric acquisition in order to optimize the instrumental parameters for each compound to increase the peak intensity. The full scan of ions ranging from m/z 100 to m/z 1200 in the negative ion mode was used. Retention times and MS chromatograms of all flavonoids and saponins were confirmed by authentic standards, respectively. The HPLC chromatograms and total ion chromatograms (TIC) were obtained using the above method.
Results and discussion
Optimization of sample preparation
Flavonoids distribute in nature widely, especially in a large number of biologically active natural products. Most flavonoids exhibit two major maximum UV absorption wavelengths, namely the range of 240–285 nm and the range of 300–400 nm. While most saponins exhibit no ultraviolet absorption. The current results revealed the presence of flavonoids mainly in 45% ethanol fractions and the presence of saponins mainly in 80% ethanol fractions by AB-8 resin column. It was reported that AB-8 resin was good at separating chemical constituent according to the polarity [22]. After that, polyamide column was employed to further purify the flavonoids. 40% Ethanol eluent from polyamide column was obtained, and the fraction was rich in flavonoids. Precipitation with methanol–acetone was applied to further separate flavonoids and saponins from adzuki bean. Flavonoids existed in the supernatant fraction, while saponins presented in the precipitate fraction. Finally, adzuki bean total extract (ABTE), adzuki bean flavonoids (ABF) and adzuki bean saponins (ABS) (Fig. 1) were obtained and utilized for HPLC–DAD–ESI–MSn analysis.
Optimization of HPLC–DAD–ESI–MSn conditions
A binary mobile phase of water/acetic acid (98:2, v/v) (solvent A) and water/acetonitrile/acetic acid (78:20:2, v/v/v) (solvent B) with gradient program to separate flavonoids such as quercetin derivatives, cinnamic acid derivatives and kaempferol derivatives were applied previously [10]. A mixture of solvent A (HPLC water containing 0.05% TFA) and solvent B (acetonitrile: methanol: TFA = 30:10:0.05) was also used to separate phenolics including flavonoids and their derivatives in another report [23]. In current article, several aqueous mobile phases, consisting of methanol, water or acetonitrile and water (with or without adjusting pH value), or different buffers (such as ammonium acetate, ammonium formate and formic acid), with altered flow rates, and different gradient compositions, were used to optimize HPLC chromatographic conditions. The results showed that the mobile phase of water containing 10 mM ammonium acetate combined with mobile phase B containing acetonitrile were feasible for HPLC–MS system. The flow rate was set at 0.2 mL/min, the gradient eluting conditions were 10% B for 10 min, changed to 15% B at 30 min, 25% B at 45 min, 35% B at 55 min, 45% B at 60 min and 55% B at 70 min. Such conditions exhibited good separation for both flavonoids and saponins (Fig. 2). Previously, Alltima C18 column [23], and Phenomenex Luna C18 column [10] were respectively used to separate flavonoids such as catechin, vitexin, and quercetin. In current article, different chromatographic columns such as C18 column, C8 column, purchased from different companies (such as Agilent, Waters, Phenomenex, Shimadzu) were attempted, and finally the Phenomenex C8 column (150 × 2.0 mm, 5 μm) were selected. As regards to the selection of the wavelength, 205 nm was employed to detect oleanene-glucuronides in commercial edible beans [24], while 230 nm [25], 280 nm [23], and 320 nm [10] were used to monitor phenolics including flavonoids and their derivatives. Therefore, the current working wavelength at 205 nm for detecting saponins and the wavelength of 262 nm for detecting flavonoids were set according to the preliminary experiments. The HPLC chromatograms of adzuki bean total extract (205 and 262 nm) were presented in Fig. 2a. The peaks of HPLC chromatogram detected at 262 nm disappeared after 50 min, while the peaks of HPLC chromatogram detected under 205 nm showed up after 50 min. Figure 2b showed the chromatogram (262 nm) of adzuki bean flavonoids. Figure 2c exhibited the chromatogram (205 nm) of adzuki bean saponins.
Fig. 2.
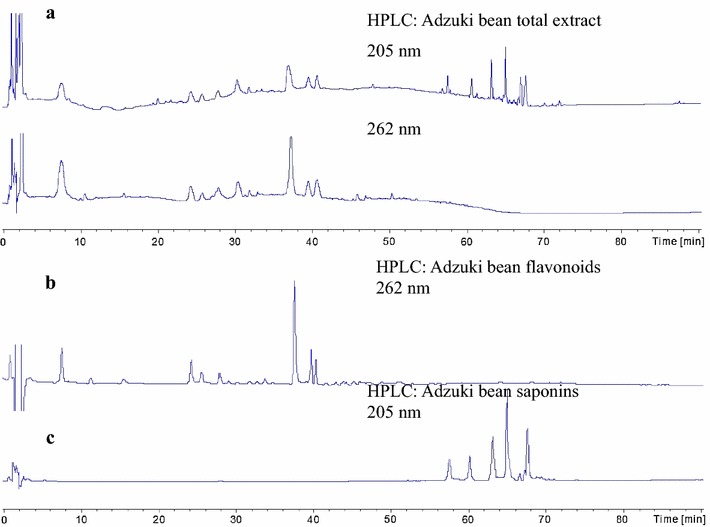
HPLC–DAD chromatograms of adzuki bean extracts. a HPLC–DAD chromatogram of adzuki bean total extract at 205 and 262 nm, respectively; b HPLC–DAD chromatogram of adzuki bean flavonoids at 262 nm; c HPLC–DAD chromatogram of adzuki bean saponins at 205 nm
Subsequently, electro spray ionization (ESI) conditions for detecting flavonoids and saponins were optimized. A direct infusion experiment was firstly employed in negative ion detection mode. Under the optimized MS conditions, the m/z 289 precursor ion was identified as catechin. The m/z 609 precursor ion, which produced the m/z 463, m/z 301 daughter ions, was identified as quercetin-3-O-rutinoside. The m/z 463 precursor ion, which produced the m/z 301 daughter ion, was identified as quercetin-3-O-glucoside. The m/z 431 precursor ion was identified as vitexin. The m/z 593 precursor ion, which produced the m/z 431 daughter ion, was identified as vitexin 4″-O-glucoside. In order to detect the chemical constituents effectively and simultaneously, the program of time segments with different MS conditions were used in this article. Figure 3 showed HPLC–ESI–MS total ion chromatograms of adzuki bean samples. The authors focused on 15 major peaks as marked in Fig. 3a for further structural analysis. Similar to the saponins standards, the m/z 779, m/z 795, m/z 809, m/z 971, m/z 941, and m/z 1133 precursor ions were for azukisaponin I, II, III, IV, V, and VI, respectively. The detailed MSn information of azukisaponins was discussed in the following identification analysis.
Fig. 3.
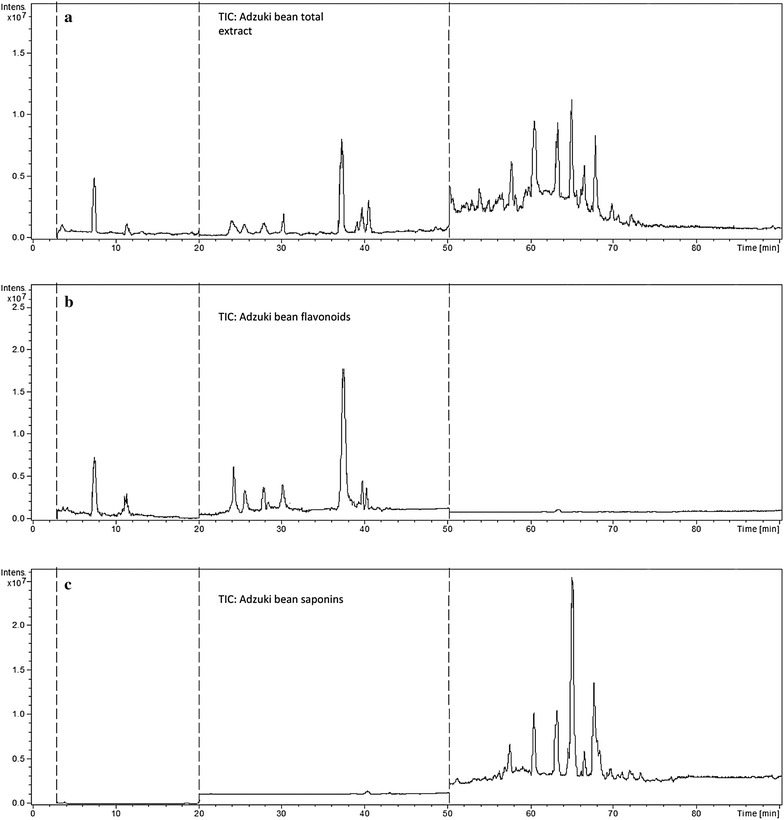
HPLC–ESI–MS total ion chromatograms of adzuki bean extracts. a HPLC–ESI–MS total ion chromatogram of adzuki bean total extract; b HPLC–ESI–MS total ion chromatogram of adzuki bean flavonoids; c HPLC–ESI–MS total ion chromatogram of adzuki bean saponins
Analysis of flavonoids in adzuki bean by HPLC–ESI–MSn
A total of 15 major peaks in HPLC–ESI–MS total ion chromatograms of adzuki bean total extract were marked in Fig. 3a. The information of the retention times, m/z for the [M−H]− ions and the collision induced dissociation (CID) fragments of peaks were listed in Table 1. Peaks 1–9 were identified as flavonoids by HPLC–DAD–ESI–MSn by comparing the retention times and ESI–MSn spectra with those of authentic standards, and the chemical structures of these flavonoids were shown in Fig. 4.
Table 1.
HPLC–ESI–MSn data of identified flavonoids and saponins in adzuki bean
| Peak no. | Retention time (min) | –MS [M−H]− (m/z) | Daughter ion of MS2 (m/z) | Daughter ion of MS3 (m/z) | Identity |
|---|---|---|---|---|---|
| 1 | 7.5 | 451 | 289 | (+) Catechin-7-O-β-d-glucopyranoside | |
| 2 | 11.3 | 289 | (+) Catechin | ||
| 3 | 24.1 | 613 | 451 | 289 | (+) Epicatechin-7-O-β-d-glucopyranoside-glucoside |
| 4 | 25.7 | 451 | 289 | (+) Epicatechin-7-O-β-d-glucopyranoside | |
| 5 | 27.9 | 625 | 493 463 |
463, 301 301 |
Quercetin-3-glucoside–glucoside |
| 6 | 30.1 | 739 | 593 431 |
431, 413 | Vitexin-4″-O-glucoside-rhamnose |
| 7 | 37.5 | 609 | 463 | 301 | Quercetin-3-O-rutinoside |
| 8 | 39.6 | 463 | 301 | Quercetin-3-O-glucoside | |
| 9 | 40.5 | 593 | 431 | 413 | Vitexin-4″-O-glucoside |
| 10 | 57.7 | 971 | 809 647 629 485 |
647, 485 485 |
Azukisaponin IV |
| 11 | 60.4 | 1133 | 809 629 471 |
629, 471 | Azukisaponin VI |
| 12 | 63.2 | 941 | 795 633 457 |
633, 615, 457 615, 457 |
Azukisaponin V |
| 13 | 64.9 | 795 | 633 457 |
615, 457 | Azukisaponin II |
| 14 | 66.4 | 779 | 617 599 441 |
599, 441 | Azukisaponin I |
| 15 | 67.7 | 809 | 647 471 |
471 | Azukisaponin III |
Fig. 4.
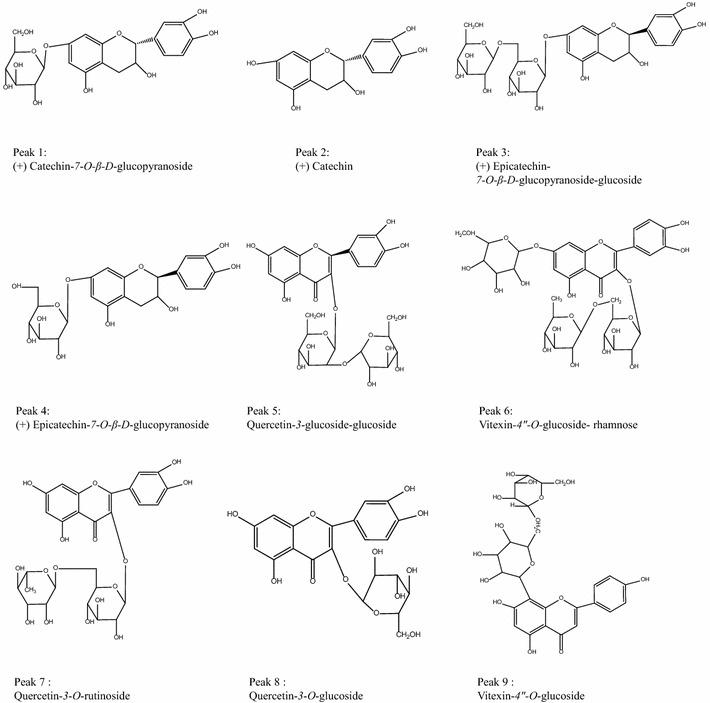
Chemical structures of flavonoids in adzuki bean identified by HPLC–ESI–MSn
For peak 1, the retention time was 7.5 min, and the m/z 451 precursor ion was presented in Fig. 5A-1. As shown in Fig. 5A-2, the m/z 289 daughter ion was the main fragment ion of the parent ion at m/z 451. Moreover, the m/z 289 ion was the precursor ion of (+) catechin. Therefore, peak 1 was identified as (+) catechin-7-O-β-d-glucopyranoside according to the above information and the previous article [26]. The retention time of peak 2 was 11.3 min, and peak 2 was identified to be (+) catechin comparing the retention time and the ESI (−)-MS spectra with the authentic standard (+) catechin. The m/z 289 precursor ion for peak 2 was presented in Fig. 5B. Peak 3 was speculated to be (+) epicatechin-7-O-β-d-glucopyranoside-glucoside with the retention time 24.1 min and m/z 613 (Fig. 5C-1) as the precursor ion [M−H]−. As shown in Fig. 5C-2, the m/z 451 and m/z 289 daughter ions were the main fragment ions of the parent ion at m/z 613.
Fig. 5.
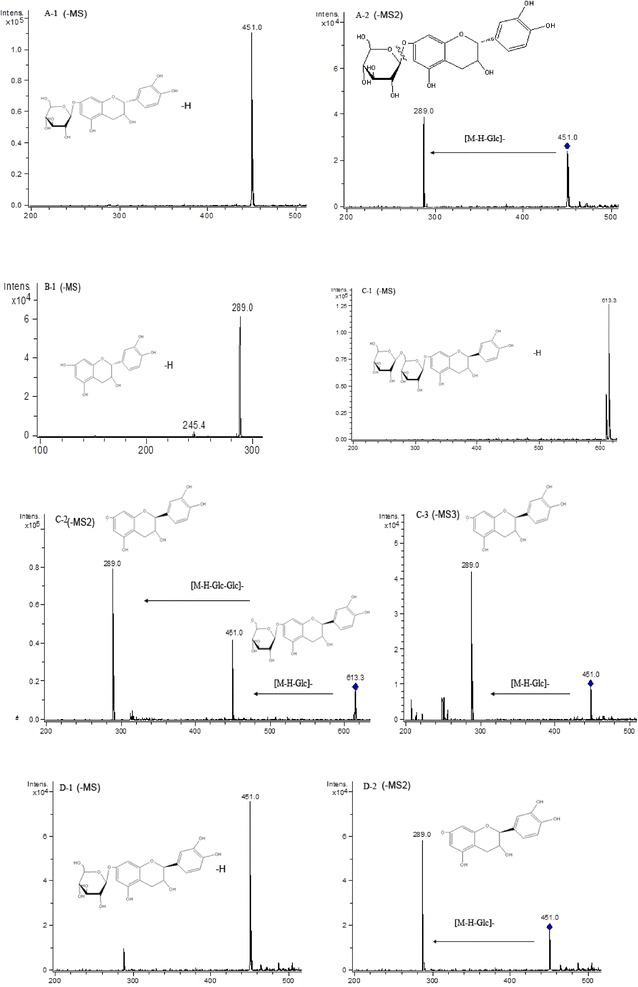
ESI (−) MS, MS2, and MS3 spectra of identified flavonoids in adzuki bean. Peak 1 (A): A-1 MS spectrum of peak 1 ([M−H]−), A-2 MS2 spectrum of the ion at m/z 451. Peak 2 (B): B-1 MS spectrum of peak 2 ([M−H]−). Peak 3 (C): C-1 MS spectrum of peak 3 ([M−H]−), C-2 MS2 spectrum of the ion at m/z 613, C-3 MS3 spectrum of the ion at m/z 451. Peak 4 (D): D-1 MS spectrum of peak 4 ([M−H]−), D-2 MS2 spectrum of the ion at m/z 451
The peak 4 gave the retention time 25.7 min, which was different from the retention time of (+) catechin-7-O-β-d-glucopyranoside, and its parent ion [M−H]− was at m/z 451 (Fig. 5D-1). As shown in Fig. 5D-2, the m/z 289 daughter ions were the main fragment ions of the parent ion at m/z 451. Comparing the HPLC–MS results of the standards of catechin to epicatechin, peak 4 was speculated to be epicatechin with one glucoside. According to the previous article [26], the peak 4 was speculated to be (+) epicatechin-7-O-β-d-glucopyranoside. For peak 5, the precursor ion m/z 625 with the retention time 27.9 min was observed in Fig. 6E-1. The CID of peak 5 produced main fragment ions at m/z 493, and m/z 463 in the ESI–MS2 and MS3 spectra (Fig. 6E-2, E-3). Hence, the peak 5 was speculated to be quercetin-3-O-glucoside-glucoside. The precursor ion of peak 6 was at m/z 739 (Fig. 6E-1), and one of the daughter ions was at m/z 593 of peak 6 (Fig. 6E-2), which was also the parent ion of peak 9 (Fig. 7I-1). Peak 6 lost an ion m/z 146, this suggested that the compound of peak 6 contained a rhamnose as compared to peak 9. In the ESI–MS3 spectrum (Fig. 6F-3), the fragmentation ion at m/z 431 produced from MS2 of the fragment ion at m/z 593. Peak 6 was speculated to be vitexin 4″-O-glucoside-rhamnose. Peaks 7 and 8 were respectively identified to be quercetin-3-O-rutinoside and quercetin-3-O-glucoside according to the retention times (37.5 min and 39.6 min, respectively) and MS information of their standards (Figs. 6, 7G, H).
Fig. 6.
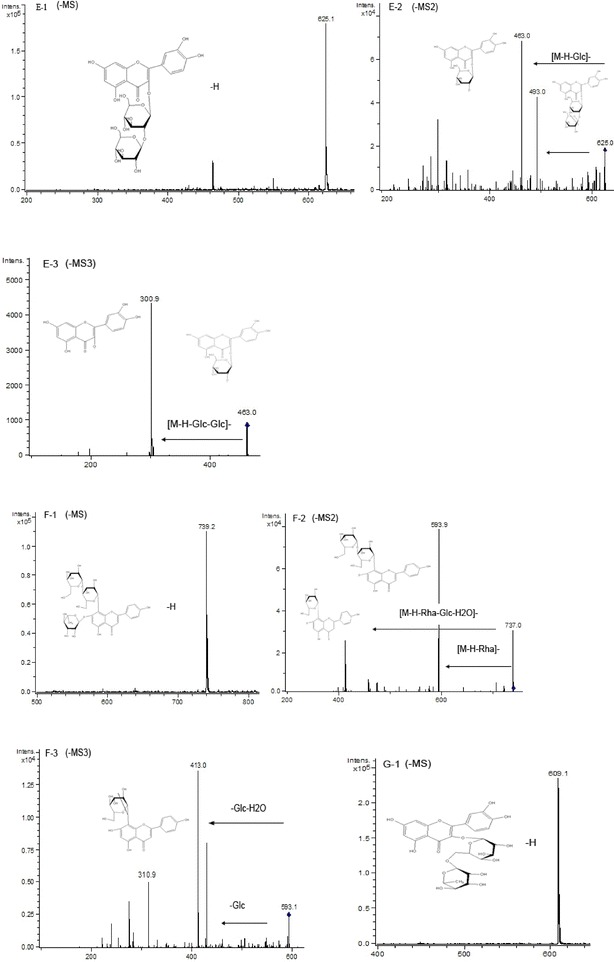
ESI (−) MS, MS2, and MS3 spectra of identified flavonoids in adzuki bean. Peak 5 (E): E-1 MS spectrum of peak 5 ([M−H]−), E-2 MS2 spectrum of the ion at m/z 625, E-3 MS3 spectrum of the ion at m/z 463. Peak 6 (F): F-1 MS spectrum of peak 6 ([M−H]−), F-2 MS2 spectrum of the ion at m/z 739, F-3 MS3 spectrum of the ion at m/z 593. Peak 7 (G): G-1 MS spectrum of peak 7 ([M−H]−)
Fig. 7.
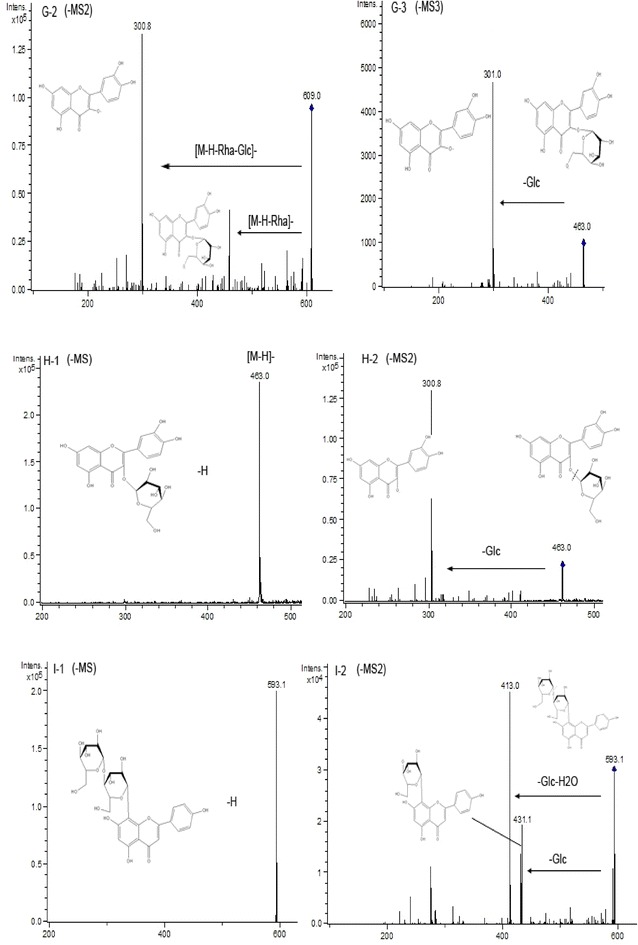
ESI (−) MS, MS2, and MS3 spectra of identified flavonoids in adzuki bean. Peak 7 (G): G-2 MS2 spectrum of the ion at m/z 609, G-3 MS3 spectrum of the ion at m/z 463. Peak 8 (H): H-1 MS spectrum of peak 8 ([M−H]−), H-2 MS2 spectrum of the ion at m/z 463. Peak 9 (I): I-1 MS spectrum of peak 9 ([M−H]−), I-2 MS2 spectrum of the ion at m/z 593
For peak 9, the de-protonated molecular ion [M−H]− was at m/z 593(Fig. 7I-1), which molecular weight can be 594. In the ESI–MS2 spectrum (Fig. 7I-2), the fragment ions at m/z 431 and m/z 413 were the daughter ions from the precursor ion m/z 593. Based on the above information and the retention time of the standard vitexin and vitexin-4″-O-glucoside, peak 9 was finally confirmed to be vitexin-4″-O-glucoside. Similar to peak 6, the daughter ions m/z 431 and m/z 413 were also observed in the ESI–MS2 spectrum (Fig. 6F-2).
Analysis of saponins in adzuki bean by HPLC–ESI–MSn
Peaks 10–16 (Fig. 3) were identified to be saponins of adzuki bean according to the retention times and MS information. Their structures were shown in Fig. 8. The following were the detailed analysis procedures. The molecular weight of peak 10 was confirmed to be 972 due to the existence of ion [M−H]− at m/z 971 (Fig. 9A-1), and four fragment ions of MS2 at m/z 971 (809 [M−H-Glc]−, m/z 674 [M−H-Glc-Glc]−, m/z 629 [M−H-Glc-Glc-H2O]−, m/z 485 [M−H-Glc-Glc-Glc]−) (Fig. 9A-2), and the existences of fragment ions of MS3 at m/z 809 and m/z 647 (Fig. 9A-3), respectively. It was in tune with the standard and the previous article [27]. Taken together, peak 10 was identified as azukisaponin IV (Fig. 12).
Fig. 8.
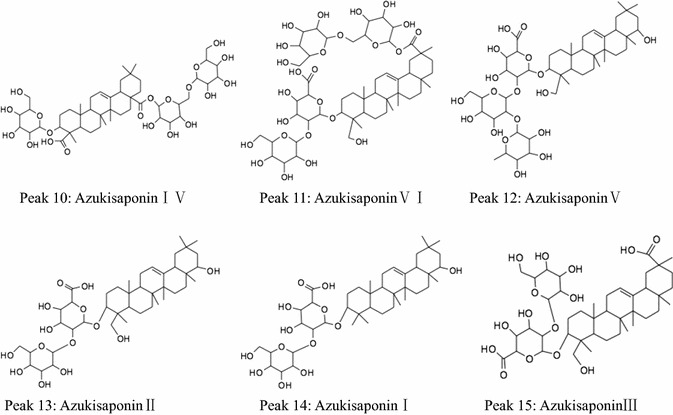
Chemical structures of saponins in adzuki bean identified by HPLC–ESI–MSn
Fig. 9.
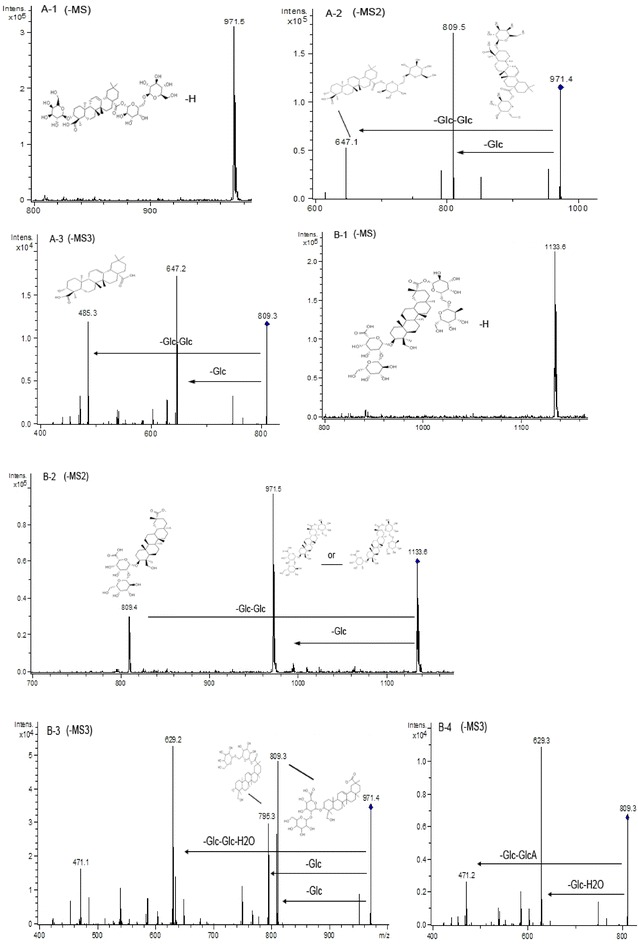
ESI (−) MS, MS2, and MS3 spectra of identified saponins in adzuki bean. Peak 10 (A): A-1 MS spectrum of peak 10 ([M−H]−), A-2 MS2 spectrum of the ion at m/z 971, A-3 MS3 spectrum of the ion at m/z 809. Peak 11 (B): B-1 MS spectrum of peak 11 ([M−H]−), B-2 MS2 spectrum of the ion at m/z 1133, B-3 MS3 spectrum of the ion at m/z 971; B-4 MS3 spectrum of the ion at m/z 809
Fig. 12.
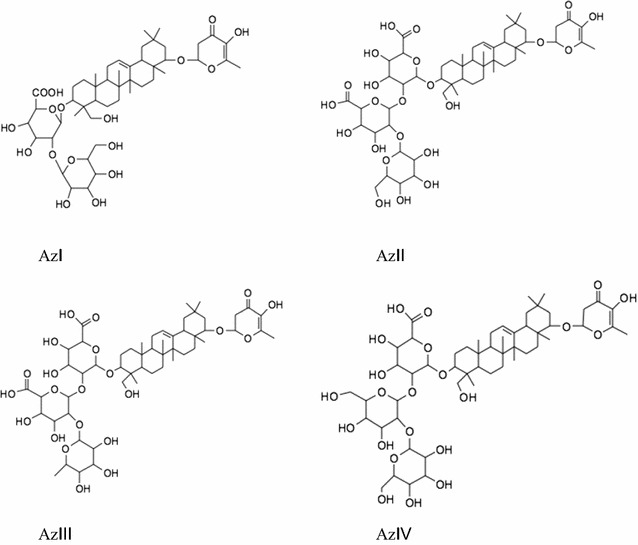
Chemical structures of Az saponins from adzuki bean
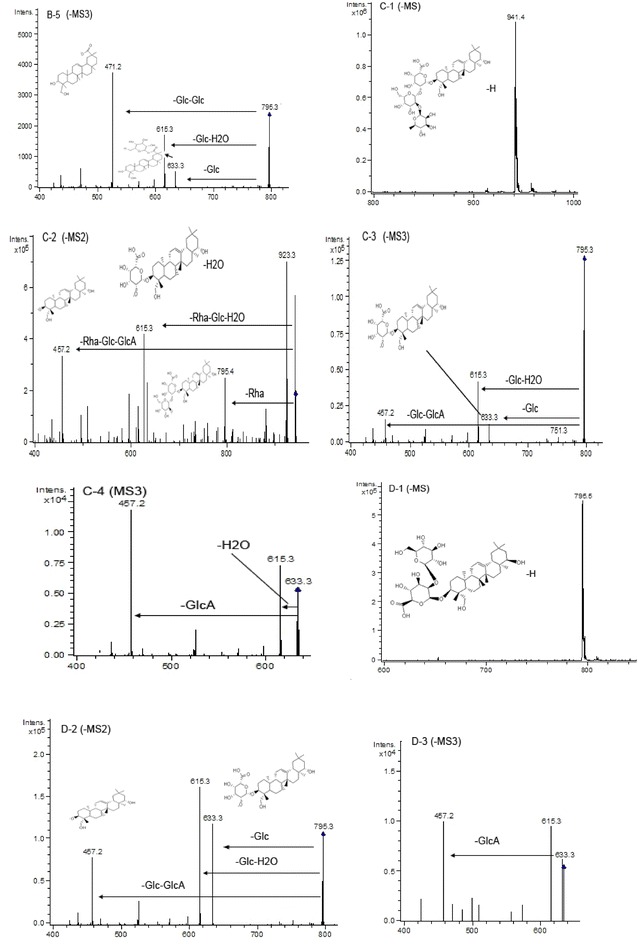
The CID of peak 11 with the [M−H]− ion at m/z 1133 (Fig. 9B-1) resulted in fragments at m/z 971, m/z 809, m/z 795, and m/z 471 (Fig. 9B-2). MS3 spectrum at m/z 971 ([M−H-Glc]−), m/z 809 ([M−H-Glc-Glc]−) and m/z 795 ([M−H-Glc-Glc-Glc-H2O]−) were presented in Fig. 9B-3, B-4 and Fig. 10B-5, respectively. It was the same as the previous article [12]. So peak 11 was identified as azukisaponin VI.
Fig. 10.
ESI (−) MS, MS2, and MS3 spectra of identified saponins in adzuki bean. Peak 11 (B): B-5 MS3 spectrum of the ion at m/z 795. Peak 12 (C): C-1 MS spectrum of peak 3 ([M−H]−), C-2 MS2 spectrum of the ion at m/z 941, C-3 MS3 spectrum of the ion at m/z 795; C-4 MS3 spectrum of the ion at m/z 633. Peak 13 (D): D-1 MS spectrum of peak 13 ([M−H]−), D-2 MS2 spectrum of the ion at m/z 795, D-3 MS3 spectrum of the ion at m/z 633
The retention time of peak 12 was 63.2 min and the molecular ion was at m/z 941 (Fig. 10C-1). CID of the molecular ion of peak 12 produced three predominant fragments at m/z 795 ([M−H-Rha]−), m/z 633 ([M−H-Rha-Glc]−), and m/z 457 ([M−H-Rha-Glc]−) (Fig. 10C-2). Its MS3 spectrum at m/z 795 exhibited the fragment ions at m/z 633 and m/z 457, in which the ion m/z 795 lost a glucosyl, and a glucosyl with a glucuronic residue (Fig. 10C-3). MS3 spectrum at m/z 633 also produced the fragment ion m/z 457, which lost a glucuronic residue from the m/z 633 (Fig. 10C-4). It was identified to be azukisaponin V, which was consistent with the previous articles [24, 28, 29].
The retention time of peak 13 was 64.9 min, and the precursor ion was at m/z 795 (Fig. 10D-1), which suggested its molecular weight was 796. Its MS2 spectrum at m/z 795 and MS3 spectrum at m/z 633 were shown in Fig. 10D-2 and D-3, respectively. The identification of azukisaponin II of peak 13 was based on the above information and the similarity of MS2 with those reported by Kinjo et al. [24, 30].
Mass spectrometric analysis of peak 14 with the retention time at 66.4 min indicated that the molecular ion [M−H]− present at m/z 779 (Fig. 11E-1). The fragment ions of m/z 617, m/z 599, and m/z 441 produced from the negative ESI-MS2 spectrum at m/z 779 ([M−H]−) in Fig. 11E-2. Its MS3 spectrum at m/z 617 further confirmed the result (Fig. 11E-3). Therefore, peak 14 was identified as azukisaponin I based on the above results and the previous article [12].
Fig. 11.
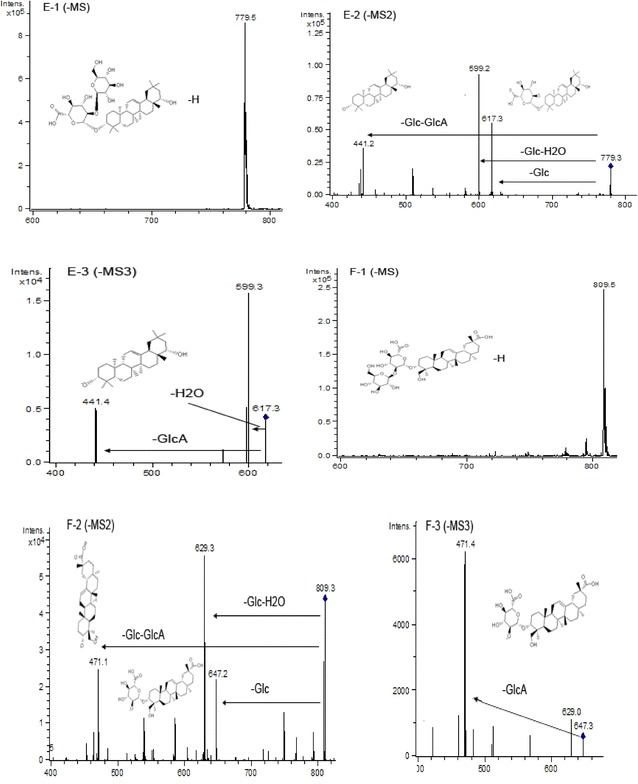
ESI (−) MS, MS2, and MS3 spectra of identified saponins in adzuki bean. Peak 14 (E): E-1 MS spectrum of peak 14 ([M−H]−), E-2 MS2 spectrum of the ion at m/z 779, E-3 MS3 spectrum of the ion at m/z 617. Peak 15 (F): F-1 MS spectrum of peak 15 ([M−H]−), F-2 MS2 spectrum of the ion at m/z 809, F-3 MS3 spectrum of the ion at m/z 647
The molecular ion of peak 15 was at m/z 809 ([M−H]−) (Fig. 11F-1), and the molecular weight of peak 15 was 810. The fragment ion of m/z 647 indicated the loss of a glucose residue and m/z 471 indicated the losses of a glucose residue and a glucuronic residue. The detailed results were found in the MS2 spectrum at m/z 809 (Fig. 11F-2). In its MS3 spectrum, the main daughter ion at m/z 471 ([M−H-Glc-GlcA]−) was found from the fragment ion at m/z 647 (Fig. 11F-3). Moreover, it was consistent with the results reported by Kitagawa et al. [12]. Finally, peak 15 was identified as azukisaponin III.
In the previous articles, other saponins were found in adzuki bean, namely Az I with the molecular weight of 922 [13], Az II with the molecular weight of 1098, Az III with the molecular weight of 1082, and Az IV with the molecular weight of 1084 [14]. The main differences of these saponins with the saponins of the present article were at the C-21 in Fig. 12. In the present article, no Az saponins were detected in adzuki bean samples. The reason may be their limited contents in adzuki bean or the procedure of preparation for adzuki bean samples.
Quantification of flavonoids and saponins in adzuki bean
The program of time segments of MS analysis was employed to enhance sensitivity for flavonoids and saponins analysis. Among 15 compounds identified, four flavonoids (catechin, vitexin-4″-O-glucoside, quercetin-3-O-glucoside, and quercetin-3-O-rutinoside) and six saponins (azukisaponin I, II, III, IV, V, and VI) in adzuki bean samples were further quantified by external calibration using HPLC–MS methods with the program of “time segments” and extract ion chromatogram (EIC) analysis.
For making a standard curve of flavonoids, seven standard working solutions with 1, 2, 5, 20, 50 and 80, and 100 ng/mL were made by diluting from high concentration stock solutions, and analyzed by HPLC–DAD–MSn using the above conditions, sequentially. Seven levels of saponins standard working solutions with 2, 5, 10, 40, and 60, 80, and 100 ng/mL were used to construct the standard curves. The curve of peak area (Y) versus flavonoid standard concentration (X) was plotted. The linear regression equation were Y = 42,769 X + 3 × 106, (catechin, R2 = 0.994), Y = 2 × 106 X + 5 × 106, (quercetin-3-O-rutinoside, R2 = 0.9976), Y = 2 × 106 X + 2 × 107, (quercetin 3-glucoside, R2 = 0.9943), Y = 3 × 106 X + 7 × 107, (vitexin-4″-O-glucoside, R2 = 0.9969), Y = 2 × 106 X + 4 × 107, (azukisaponin IV, R2 = 0.9916), Y = 3 × 106 X + 2 × 107, (azukisaponin VI, R2 = 0.9906), Y = 2 × 106 X + 3 × 107, (azukisaponin V, R2 = 0.9985), Y = 4 × 106 X + 2 × 107, (azukisaponin II, R2 = 0.9923), Y = 1 × 106 X + 4 × 107, (azukisaponin I, R2 = 0.9911), and Y = 1 × 106 X − 1 × 107, (azukisaponin III, R2 = 0.9909), respectively.
The limit of detection (LOD) was calculated with Signal/Noise ratio better than 3, and the limit of quantification (LOQ) was calculated with Signal/Noise ratio better than 10. In the present article, the range for LOD of flavonoids standards was from 0.30 to 0.81 ng/mL, while the range for LOQ of saponins was from 0.7 to 1.42 ng/mL. The results showed that the contents of catechin (49.4 mg/g), quercetin-3-O-rutinoside (404.7 mg/g), quercetin-3-O-glucoside (90.1 mg/g) and vitexin-4″-O-glucoside (74.6 mg/g) in adzuki bean flavonoids extract were much higher than that of the adzuki bean total extract (12.4, 225.9, 21.4, and 36.7 mg/g, respectively). Meanwhile, the contents of azukisaponin IV (11.4 mg/g), azukisaponin VI (206.3 mg/g), azukisaponin V (283.2 mg/g), azukisaponin II (389.7 mg/g), azukisaponin I (5.4 mg/g), and azukisaponin III (27.6 mg/g) in adzuki bean saponins extract were much higher than that of adzuki bean total extract (6.6, 20.0, 165.9, 186.9, 8.9, and 79.0 mg/g, respectively) (Table 2).
Table 2.
Flavonoids and saponins contents in extracts from Adzuki Bean
| Peak no. | Compounds | Contents (mg/g ABTE) | Contents (mg/g ABF) | Contents (mg/g ABS) |
|---|---|---|---|---|
| 2 | Catechin | 12.37 | 49.39 | ND |
| 7 | Quercetin-3-O-rutinoside | 225.99 | 404.73 | ND |
| 8 | Quercetin-3-O-glucoside | 21.37 | 90.08 | ND |
| 9 | Vitexin-4″-O-glucoside | 36.66 | 74.60 | ND |
| 10 | Azukisaponin IV | 6.63 | ND | 11.40 |
| 11 | Azukisaponin VI | 20.04 | ND | 206.35 |
| 12 | Azukisaponin V | 165.99 | ND | 283.21 |
| 13 | Azukisaponin II | 186.99 | ND | 389.73 |
| 14 | Azukisaponin I | 8.90 | ND | 5.42 |
| 15 | Azukisaponin III | 79.03 | ND | 27.58 |
ABTE adzuki bean total extract, ABF adzuki bean flavonoids, ABS adzuki bean saponins, ND not detected
Conclusions
Flavonoids and saponins of adzuki bean have been produced by column chromatography and solvent precipitation. The present study has established a powerful method using HPLC–DAD–ESI–MSn in electro spray negative mode to separate and characterize nine flavonoids and six saponins in adzuki bean rapidly. A simple and sensitive method has been established for quantification of flavonoids and saponins in adzuki bean samples. Current preparation and analysis of flavonoids and saponins from adzuki bean could promote pharmacological experiments and attain much more reasonable experimental results.
Authors’ contributions
RL conducted lab work, data processing, statistical analysis and manuscript drafting. ZC conducted parts of lab work. ZC and BX made experimental design, conducted quality control for lab work, and took charge in manuscript revision and paper submission. All authors read and approved the final manuscript.
Acknowledgements
We thank Jinming Mu of Faculty of Agronomy in Jilin Agricultural University for identifying adzuki bean.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Funding
This project is jointly supported UIC internal grant (Project Codes: R201627 and R201714) from Beijing Normal University-Hong Kong Baptist University United International College, China.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rui Liu, Email: rayfayliuruifei@163.com.
Zongwei Cai, Email: zwcai@hkbu.edu.hk.
Baojun Xu, Phone: +86 756 3620636, Email: baojunxu@uic.edu.hk.
References
- 1.Itoh T, Itoh Y, Mizutani M, Fujishiro K, Furuichi Y, Komiya T, Hibasami H. A hot water extract of adzuki (Vigna angularis) induces apoptosis in cultured human stomach cancer cells. J Jpn Soc Food Sci. 2002;49:39–344. doi: 10.3136/nskkk.49.339. [DOI] [Google Scholar]
- 2.Itoh T, Itoh Y, Hibasami H, Katsuzaki H, Imai K, Furuichi Y, Komiya T. Isolation of a substance inducing apoptosis of cultured human gastric cancer cells from a hot-water extract of adzuki bean (Vigna angularis) J Jpn Soc Nutr Food Sci. 2005;58:281–287. doi: 10.4327/jsnfs.58.281. [DOI] [Google Scholar]
- 3.Itoh T, Kita N, Kurokawa Y, Kobayashi M, Horio F, Furuichi Y. Suppressive effect of a hot water extract of adzuki beans (Vigna angularis) on hyperglycemia after sucrose loading in mice and diabetic rats. Biosci Biotechnol Biochem. 2004;68:421–2426. doi: 10.1271/bbb.68.2421. [DOI] [PubMed] [Google Scholar]
- 4.Itoh T, Furuichi Y. Lowering serum cholesterol level by feeding a 40% ethanol-eluted fraction from HP-20 resin treated with hot water extract of adzuki beans (Vigna angularis) to rats fed a high-fat cholesterol diet. Nutrition. 2009;25:318–321. doi: 10.1016/j.nut.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Beecher GR, Holden JM, Haytowitz DB, Gebhardt SE, Prior RL. Lipophilic and hydrophilic antioxidant capacities of common foods in the United States. J Agric Food Chem. 2004;52:4026–4037. doi: 10.1021/jf049696w. [DOI] [PubMed] [Google Scholar]
- 6.Yao Y, Cheng X, Wang L, Wang S, Ren GA. Determination of potential α-glucosidase inhibitors from adzuki beans (Vigna angularis) Int J Mol Sci. 2011;12:6445–6451. doi: 10.3390/ijms12106445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sreerama YN, Takahashi Y, Yamaki K. Phenolic antioxidants in some Vigna species of legumes and their distinct inhibitory effects on α-glucosidase and pancreatic lipase activities. J Food Sci. 2012;77:927–933. doi: 10.1111/j.1750-3841.2012.02848.x. [DOI] [PubMed] [Google Scholar]
- 8.Xu BJ, Chang SKC. Comparative study on anti-proliferation properties and cellular antioxidant activities of commonly consumed food legumes against nine human cancer cell lines. Food Chem. 2012;134:1287–1296. doi: 10.1016/j.foodchem.2012.02.212. [DOI] [PubMed] [Google Scholar]
- 9.Han KH, Fukushima M, Ohba K, Shimada K, Sekikawa M, Chiji H, Lee CH, Nakano M. Hepatoprotective effects of the water extract from adzuki bean hulls on acetaminophen-induced damage in rat liver. J Nutr Sci Vitaminol. 2004;50:380–383. doi: 10.3177/jnsv.50.380. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Meckling KA, Marcone MF, Kakuda Y, Tsao R. Synergistic, additive, and antagonistic effects of food mixtures on total antioxidant capacities. J Agric Food Chem. 2011;59:960–968. doi: 10.1021/jf1040977. [DOI] [PubMed] [Google Scholar]
- 11.Yao Y, Cheng X, Wang S, Wang L, Ren G. Influence of altitudinal variation on the antioxidant and anti-diabetic potential of adzuki bean (Vigna angularis) Int J Mol Sci. 2012;63:117–124. doi: 10.3109/09637486.2011.604629. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa I, Wang HK, Saito M, Yoshikawa M. Saponin and sapogenol. XXXII. Chemical constituents of the seeds of Vigna angularis (Willd.) Ohwi et Ohashi. (2). Azukisaponins I, II, III, and IV. Chem Pharm Bull. 1883;31:674–682. doi: 10.1248/cpb.31.674. [DOI] [Google Scholar]
- 13.Iida T, Yoshiki Y, Kahara T, Okubo K, Ohrui H. A saponin conjugated with 2,3-dihydro-2,5-dihydroxy-6-methyl-4H-pyran-4-one from Vigna angularis. Phytochemistry. 1997;45:1507–1509. doi: 10.1016/S0031-9422(97)00169-6. [DOI] [Google Scholar]
- 14.Iida T, Yoshiki Y, Okubo K, Ohrui H, Kinjo J, Nohar T. Triterpenoid saponins from Vigna angularis. Phytochemistry. 1999;51:1055–1058. doi: 10.1016/S0031-9422(99)00148-X. [DOI] [Google Scholar]
- 15.Jia G, Lu X. Enrichment and purification of madecassoside and asiaticoside from Centellaasiatica extracts with macro porous resins. J Chromatogr A. 2008;1193:36–141. doi: 10.1016/j.chroma.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Itoh T, Kobayashi M, Horio F, Furuichi Y. Hypoglycemic effect of hot-water extract of adzuki bean (Vigna angularis) in spontaneously diabetic KK-Ay mice. Nutrition. 2009;25:34–141. doi: 10.1016/j.nut.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Xie LL, Ren L, Lai XS, Cao JH, Mo QY, Chen WW. Study on extraction and purification process of total ginsenosides from Radix Ginseng. J Chin Med Mater. 2009;32:1602–1605. [PubMed] [Google Scholar]
- 18.Wang ZP, Gao Y, Li WM, Liu J. Study on purification of total flavonoids and saponins of Astragalus with macro porous resin. J Chin Med Mater. 2010;33:1163–1166. [PubMed] [Google Scholar]
- 19.Wu XR, Liu ZG, Yan RL, Sun WF. Separation and purification of total flavonoids in Smilax glabra by polyamide resins. J Chin Med Mater. 2009;32:1606–1609. [PubMed] [Google Scholar]
- 20.Wei Q, Dai Y, Wu Y, Zhang WJ. Study on enriching total flavonoids from Folium chrysanthemiwith polyamide and macro porous resin. J Chin Med Mater. 2011;34:1285–1288. [PubMed] [Google Scholar]
- 21.Ke X, Chen L, Song H, Pang J. Study on total flavonoids purified from Folium Gynuraedivaricatae by macro-porous resin combined with ZTC. China J Chin Mat Med. 2012;37:1219–1223. [PubMed] [Google Scholar]
- 22.Liu R, Zhang JZ, Liu WC, Kimura Y, Zheng YN. Anti-obesity effects of protopanaxdiol types of ginsenosides isolated from the leaves of American ginseng (Panaxquinquefolius L.) in mice fed with a high-fat diet. Fitoterapia. 2010;81:1079–1087. doi: 10.1016/j.fitote.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Yao Y, Cheng X, Wang L, Wang S, Ren G. Biological potential of sixteen legumes in China. Int J Mol Sci. 2011;12:7048–7058. doi: 10.3390/ijms12107048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinjo J, Hatakeyama M, Udayama M, Tsutanaga Y, Yamashita M, Nohara T, Yoshiki Y, Okubo K. HPLC profile analysis of oleanene-glucuronides in several edible beans. Biosci Biotechnol Biochem. 1998;62:429–433. doi: 10.1271/bbb.62.429. [DOI] [PubMed] [Google Scholar]
- 25.Muhetaer K, Wang HT, Tu PF, Jiang Y. RP-HPLC determination of catechin-7-O-β-d-glucopyranoside in Vignae semen. Chin Pharm J. 2011;46:778–780. [Google Scholar]
- 26.Itoh T, Hori Y, Atsumi T, Toriizuka K, Nakamura M, Maeyama T, Ando M, Tsukamasa Y, Ida Y, Furuichi Y. Hot water extract of adzuki (Vigna angularis) suppresses antigen-stimulated degranulation in rat basophilic leukemia RBL-2H3 cells and passive cutaneous anaphylaxis reaction in mice. Phytother Res. 2011;26:1003–1011. doi: 10.1002/ptr.3660. [DOI] [PubMed] [Google Scholar]
- 27.Yoshiteru O, Takatomi O, Hiroshi H. Validity of the Oriental medicines. Part 66. Structures of dianosides G, H and I, triterpenoid saponins of Dianthus superbus var. longicalycinus herbs. Planta Med. 1984;50:254–258. doi: 10.1055/s-2007-969692. [DOI] [PubMed] [Google Scholar]
- 28.Kang S, Lee YS, Lee EB. Isolation of azukisaponin V possessing leucocyte migration inhibitory activity from Melilotus officinalis. SaengyakHakhoechi. 1987;18:89–93. [Google Scholar]
- 29.Avunduk S, Mitaine-Offer AC, Alankuş-Calişkan O, Miyamoto T, Senol SG, Lacaille-Dubois MA. Triterpene glycosides from the roots of Astragalusflavescens. J Nat Prod. 2008;71:141–145. doi: 10.1021/np0703500. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto S, Kofuji S, Kuroyanagi M, Ueno A, Sekita S. Saponins from Trifolium repens. Phytochemistry. 1992;31:1773–1777. doi: 10.1016/0031-9422(92)83145-O. [DOI] [PubMed] [Google Scholar]


