Abstract
The advent of RNA interference (RNAi) technology has profoundly impacted molecular biology research and medicine but has also advanced the field of skin care. Both effector molecules of RNAi, short-interfering RNA molecules and microRNAs (miRNAs), have been explored for their relative impact and utility for treating a variety of skin conditions. These post-transcriptional RNA regulatory molecules down-modulate protein expression through targeting of the 3′ untranslated regions of messenger RNAs, leading to their degradation or repression through sequestration. As researchers hunt for genetic linkages to skin diseases, miRNA regulators have emerged as key players in the biology of keratinocytes, fibroblasts, melanocytes, and other cells of the skin. Herein, we attempt to coalesce the current efforts to combat various skin disorders and diseases through the development of miRNA-based technologies.
Key Points
| The relative contributions of various microRNAs to the pathogenesis of an increasing number of skin conditions have been elucidated as a result of expanded microRNA profiling efforts. |
| Mimics and antagomiRs of microRNAs that have been implicated in the pathogenesis of certain skin diseases have been tested in vitro and in vivo as therapeutic agents. |
| Both ultra-flexible liposomes and skin-penetrating peptides complexed with microRNA-based therapeutics have demonstrated utility as topical delivery platforms capable of transporting microRNAs across the skin barrier. |
Introduction
Skin Disorders and Diseases
The skin represents the primary barrier protecting the body from a horde of environmental insults. Among its many functions, skin protects against pathogen invasion and excessive water loss, provides sensation, facilitates the production of vitamin D, provides insulation, and regulates body temperature [1–4]. The skin can be grossly divided into two layers: the outer epidermis and the inner dermis. For the purposes of this review, we focus mostly on the epidermal layer, which is composed primarily (approximately 95%) of keratinocytes but also melanocytes, inflammatory cells, Langerhans cells, and Merkel cells [5].
Multiple skin disorders and diseases exist, and even a brief overview of all known skin conditions would be a herculean task. For the purposes of this review, we selected several conditions primarily on the basis of the degree with which microRNA (miRNA) involvement has been elucidated. As mentioned, additional emphasis has been given to disorders and diseases relevant to the epidermal layer of the skin, including psoriasis, acne, dermatitis, pigmentation disorders, skin aging, and ultraviolet (UV) light-induced skin damage.
RNA Interference and MicroRNAs (miRNAs)
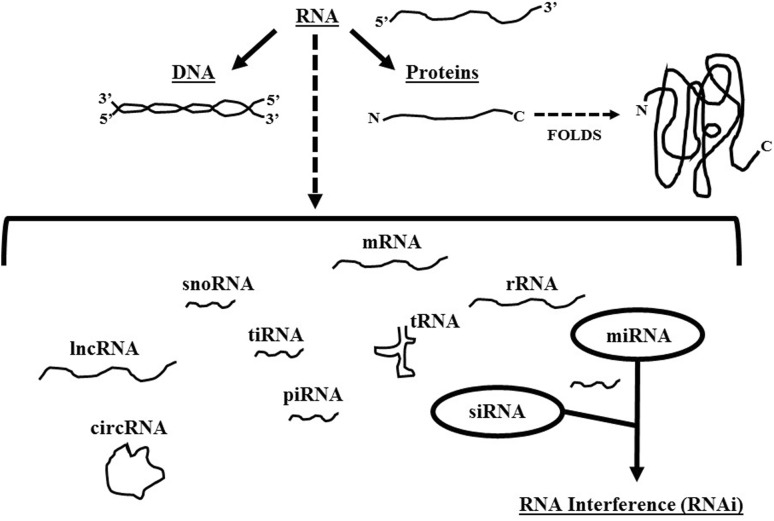
Within the past two decades, considerable advances have been made in our understanding of the multi-faceted role RNA plays in organismal biology as well as in the molecular pathogenesis of multiple diseases. Indeed, there has been an increase in the number of diversified forms of RNA that have been identified. Beyond the triad of messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA) involved in the process of gene expression, scientists have also discovered long non-coding RNA (lncRNA), circular non-coding RNA (circRNA), small nucleolar RNA (snoRNA), piwi-interacting RNA (piRNA), and tRNA-derived stress-induced RNA (tiRNA), to name a few (Fig. 1). Of interest to this review article are two forms of small non-coding RNAs (ncRNAs): short-interfering RNA (siRNA) and miRNA (or miR), with emphasis given to miRNAs.
Fig. 1.
Diverse roles for RNA. Schematic of the diversified roles for various RNA molecules, including but not limited to messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA), small nucleolar RNA (snoRNA), piwi-interacting RNA (piRNA), tRNA-derived stress-induced RNA (tiRNA), microRNA (miRNA), short interfering RNA (siRNA), long non-coding RNA (lncRNA), and circular non-coding RNA (circRNA). Emphasis is given to miRNAs and siRNAs that are the primary focus of this review article
The first report describing the technology platform termed RNA interference (or RNAi) in the context of Caenorhabditis elegans, the common earthworm, was published in 1998 by Andrew Fire and Craig Mello, both of whom received the Nobel Prize in Physiology and Medicine for their discovery [6–8]. Their seminal work described a pathway found in many eukaryotes known as RNAi, where small ncRNA molecules, of approximately 20 nucleotides (nts) in length, can knockdown expression of a target gene through partial or full complementary binding to an mRNA transcript, which culminates in the degradation of the mRNA or its translational repression.
The effector molecules of the RNAi pathway can be found in two forms: siRNAs and miRNAs. There are significant differences between how they are generated and how they function [9]. siRNAs are frequently derived from exogenous sources of double-stranded RNA (dsRNA) such as dsRNA virus genomes, though they can also be generated from genomic sources endogenously. In contrast, the precursors to miRNAs are expressed from the host genome in a regulated manner akin to protein-coding genes. Additionally, some miRNAs are delivered to the cell cytoplasm via viral infection such as in the case of Epstein Barr Virus. Moreover, siRNAs typically target a single gene, whereas miRNAs have the potential to regulate multiple gene targets [10]. Mature forms of both sets of small ncRNAs are produced via a cytoplasmic enzyme known as Dicer, which cleaves long dsRNA molecules into small approximately 20-nt long double-stranded fragments [11, 12]. These small double-stranded ncRNA fragments then separate into what is termed the passenger strand and the guide strand. The passenger strand is ultimately degraded, whereas the guide strand is loaded into the RNA-induced silencing complex (RISC), which contains a catalytic protein called Argonaute (AGO) that is responsible for the degradation of RNA transcripts that pair with the guide strand. siRNAs exhibit full complementarity to their target mRNA transcripts. In contrast, miRNAs only require partial complementarity for the cognate mRNA molecules that they bind through a region called the seed sequence [13]. This sequence flexibility allows for a certain promiscuity with respect to miRNA gene targets [10]. Like their siRNA counterparts, miRNAs can down-regulate gene expression by fating their bound mRNA transcripts for degradation [14]. However, unlike siRNAs, miRNAs can also affect a form of non-degradative translational repression. Also dissimilar from siRNAs are the multiple upstream biogenesis pathways for miRNAs [15].
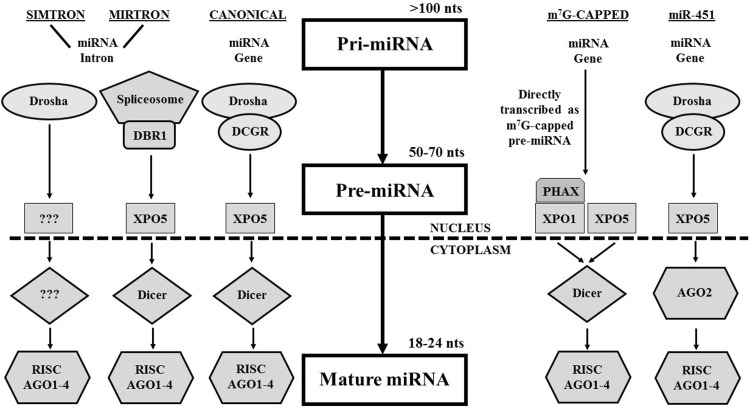
As mentioned, several cellular pathways exist for the biogenesis of miRNAs (Fig. 2), and such built-in redundancy highlights the evolutionary significance of this mode of post-transcriptional regulation of gene expression [15]. miRNAs typically start as a primary miRNA transcript (or pri-miRNA), approximately 1000 nts in length, encoded within intergenic as well as within intronic and exonic regions of the host genome [16]. The pri-miRNA is subsequently processed via different mechanisms into a shorter RNA molecule with a hairpin that is termed a pre-miRNA. The pre-miRNA is then exported from the nucleus to the cytoplasm via an exportin (XPO) molecule (most commonly XPO5) [17], where the hairpin pre-miRNA is cleaved into a double-stranded miRNA duplex by Dicer. These series of steps have been well-characterized with respect to what has been described as the canonical miRNA biogenesis pathway, where the expressed pri-miRNA is cleaved into the pre-miRNA hairpin via an enzymatic microprocessor complex involving two key proteins: drosha and digeorge critical region 8 (DGCR8) (Fig. 2) [18–21]. Several alternative miRNA biogenesis pathways exist, including but not limited to the mirtron pathway, simtron pathway, methyl7G-capped pathway, and miR-451 pathway [22–25]. The mirtron pathway bypasses the pri-miRNA to pre-miRNA cleavage induced by the Drosha/DGCR8 microprocessor complex, where intronic pri-miRNAs are spliced through the spliceosome complex into pre-miRNAs that are exported to the cytoplasm [26]. Although they are mirtron-like, simtrons distinguish themselves by how they do not require splicing for their formation; nor do they require many of the key components of most miRNA biogenesis pathways, including DGCR8, XPO5, Dicer, and AGO2 [22]. The methyl7G-capped pathway is distinct for several features, including a unique precursor RNA (pre-miRNA) molecule that is capped by 7-methylguanosine, that is subsequently exported from the nucleus by the PHAX-XPO1 or XPO5 transport pathways, and whose resulting guide strand is heavily favored for the 3p mature miRNA [25]. Finally, the miR-451 pathway has been characterized with respect to the titular singular miRNA species: miR-451, which follows nearly all of the same steps as the canonical pathway but is not processed into a mature miRNA in the cytoplasm by Dicer, but rather through the enzymatic activity of AGO2 [23, 24].
Fig. 2.
Multiple miRNA biogenesis pathways. Five different pathways by which mature microRNA molecules are generated, including canonical pathway, mirtron pathway, simtron pathway, 5′ methyl-guanosine pathway, and miR-451 pathway. AGO Argonaute proteins 1–4, DGCR8 DiGeorge critical region 8, RISC RNA-induced silencing complex, XPO5 exportin 5
As with their siRNA counterparts, artificial miRNAs (sometimes denoted as a miRNAs) have also been artificially designed to be complementary to specific recognition sequences within mRNAs of cellular factors [27, 28]. Another strategy is to deploy so-called anti-miRNA inhibitors or antagomiRs, which are chemically synthesized miRNA passenger strands that have been altered to bind the cognate cellular active strand irreversibly, producing an miRNA duplex that can no longer participate in RISC complex repression of a target gene, causing a gain-of-function effect [29, 30]. Alternatively, chemically synthesized mimics of naturally occurring miRNAs have also been introduced in cell culture and certain in vivo systems to modulate gene expression, often resulting in a loss-of-function effect (also referred to as miRNA replacement therapy) [30–32]. This approach essentially represents a second-generation RNAi-based therapy. We will delve into the expanding applications of miRNAs to characterize, ameliorate, and potentially eradicate several skin conditions.
miRNAs: Skin Therapy Applications
In recent years, the number of diseases with which miRNAs have been implicated has expanded dramatically, and as such it is unsurprising that various skin disorders and diseases have also found miRNA involvement. As of the writing of this review article, more than 2000 mature human miRNAs have been identified and curated in the most current version of the miRNA online database called miRbase (version 21) [33–35]. The bulk of miRNA discoveries with respect to skin conditions has centered on various skin cancers such as melanomas. One such important miRNA species is miR-203a, which has been ascribed a tumor-suppressor role in combating melanoma [36]. Another miRNA of interest to skin care is miR-29, whose post-transcriptional regulatory function dovetails with that of the tumor-suppressor protein p53 in the promotion of skin aging [37, 38]. As the list of miRNAs participating in the maintenance and dysregulation of skin health continues to grow, research into the translation of these discoveries into skin care applications will also expand. Indeed, currently, no fewer than 21 RNAi-based products are enrolled in clinical trials [39]; perhaps most notable is the inhibitory molecule of miR-122 being applied to treat hepatitis C virus (HCV) infection [40, 41]. miRNA profiling efforts have successfully identified the most abundant miRNA species within skin, which includes miR-152, miR-143, miR-126, miR-21, miR-27a, miR-214, miR-16, miR-203, miR-125b, miR-34a, miR-205, miR-27b, miR-30b, miR-125a, miR-191, miR-200a, miR-200b, miR-200c, miR-141, miR-429, miR-199a, miR-199b, miR-19b, miR-20, miR-17-5p, and miR-93 [42]. We highlight research findings in which the expression patterns of many of these miRNAs are altered as a result of different skin conditions.
Differential miRNA Profiles
A variety of biological samples have been examined for differential regulation of miRNA populations in response to certain disease conditions, including cancer, bacterial infections, and viral infections [43, 44]. Several technologies such as quantitative polymerase chain reaction (qPCR), miRNA arrays, and RNA-seq next-generation sequencing (NGS) have been developed to evaluate the presence and relative levels of miRNAs in cells, tissues, and other biological samples. Multiple studies have also shown that miRNAs fluctuate in response to various disorders and diseases of the skin. These distinct sets of miRNAs that are detectably up- and downregulated have been explored for their role in disease pathogenesis and their utility as biomarkers for diagnostic assays. The cumulative data regarding differentially expressed miRNAs in response to skin conditions have allowed for miRNA profiles to be developed, and these miRNAs represent potential therapeutic targets for these afflictions of the skin (Table 1).
Table 1.
Signature microRNA profiles of different skin conditions
| Skin condition | UV-A (6 h) [45] | UV-B (6 h) [45] | UV-A (7 d) [47] | Atopic dermatitis [48] | Psoriasis [48] | Vitiligo [58] | Vitiligo + T [58] |
|---|---|---|---|---|---|---|---|
| Cell type | Keratinocytes | Fibroblasts | Skin tissue | Skin tissue | PBMCs | ||
| miRNAs | (−) 10a | (−) 20b | (−) 1246 | (−) 122a | (−) 125b | (−) 3940-5p | (−) 224-3p |
| (−) 18b | (−) 23c | (−) 146a | (−) 133a-b | (−) 99b | (+) 224-3p | (−) 2682-3p | |
| (−) 98 | (−) 29c | (−) 3613-3p | (−) 326 | (−) 122a | (+) 2682-3p | (−) 4712-3p | |
| (−) 99b | (−) 30c | (−) 218 | (−) 215 | (−) 197 | (+) 4712-3p | (+) 3940-5p | |
| (−) 127-3p | (−) 96 | (−) 146b-5p | (−) 483 | (−) 100 | |||
| (−) 130b | (−) 98 | (−) 4281 | (−) 519a | (−) 381 | |||
| (−) 210 | (−) 181c | (−) 181c | (−) 335 | (−) 5186 | |||
| (−) 212 | (−) 218 | (+) 365 | (−) 133b | (−) 524 | |||
| (−) 323-3p | (−) 301a | (+) 30b | (−) 515-5p | (−) let-7e | |||
| (−) 330-3p | (−) 323-3p | (+) 30c | (+) let-7i | (−) 30c | |||
| (−) 376a | (−) 330-3p | (+) 148a | (+) 29a | (−) 365 | |||
| (−) 487b | (−) 335 | (+) 199a-3p | (+) 146a | (−) 133b | |||
| (−) 494 | (−) 376a | (+) 222 | (−) 10a | ||||
| (−) 598 | (−) 411 | (+) 24 | (−) 133a-b | ||||
| (+) 23b | (−) 494 | (+) 193a | (−) 22 | ||||
| (+) 96 | (−) 503 | (+) 199a | (−) 326 | ||||
| (+) 132 | (−) 532-5p | (+) 27a | (−) 215 | ||||
| (+) 191 | (−) 598 | (+) 21 | (+) 146b | ||||
| (+) 196b | (−) 600 | (+) 20a | (+) 20a | ||||
| (+) 224 | (+) let-7c | (+) 17-5p | (+) 146a | ||||
| (+) 340 | (+) 139-5p | (+) 106b | (+) 31 | ||||
| (+) 376c | (+) 191 | (+) 200a | |||||
| (+) 452 | (+) 339-3p | (+) 17-5p | |||||
| (+) 484 | (+) 361-5p | (+) 30e-5p | |||||
| (+) 501-5p | (+) 362-5p | (+) 141 | |||||
| (+) 574-3p | (+) 376c | (+) 203 | |||||
| (+) 886-5p | (+) 455-3p | (+) 142-3p | |||||
| (+) 501-5p | (+) 21 | ||||||
| (+) 106a | |||||||
The table shows the unique altered miRNA levels in response to specific skin conditions, including ultraviolet A and B damage, atopic dermatitis, psoriasis, and vitiligo. Indicated are the cell source and the human miRNAs that are up (+) or down (−) regulated. For UV-treated cells, the duration of exposure is indicated
miRNA microRNA, PBMCs peripheral blood mononuclear cells, UV ultraviolet, +T treatment with thymosin-α-1
In the case of skin damage from UV radiation, distinct miRNA profiles have been identified for UV-A and UV-B exposure. As shown in Table 1, a study that exposed primary keratinocytes to either UV-A or UV-B light for 6 h and examined for changes in miRNA expression levels revealed 27 and 28 miRNAs differentially regulated for UV-A and UV-B light, respectively [45]. Ten miRNAs were shared between the two groups. A separate study (not shown in Table 1) exposed keratinocytes to UV-B for different durations (4 and 24 h) and saw four unique miRNA profiles develop: miRNAs that increased at 4 h but decreased at 24 h, miRNAs that decreased at 4 h but increased at 24 h, miRNAs that consistently increased at 4 and 24 h, and miRNAs that continued to decrease from 4 to 24 h [46]. The first group comprised the following miRNAs: miR-326, miR-423-5p, miR-193b, and miR-542-5p. The second group consisted of miR-26a, let-7c, let-7f, miR-26a-2, miR-543, and miR-487b. The third group saw consistent upregulation of miR-31, miR-24, miR-27b, let-7a, let-7b, let-7g, miR-200b, miR-125b, miR-27a, miR-23a, miR-98, miR-221, miR-186, miR-30a, miR-22, miR-96, miR-16, miR-18b, miR-34a, miR-93, miR-185, miR-197, miR-365, miR-23b, and miR-29a. Finally, the last group saw overall downregulation of miR-489, miR-138-1, miR-138-2, miR-23a, miR-296-5p, miR-376b, miR-493, miR-126, and miR-143. To add another layer to the story, a different profile was detected when fibroblasts were exposed to UV-A (Table 1), where upregulation of miR-365, miR-30b, miR-30c, miR-148a, and miR-199a-5p, as well as the downregulation of miR-1246, miR-146a, miR-3613-3p, miR-218, miR-146b-5p, miR-4281, and miR-181c, was detected [47].
Beyond UV irradiation, differential miRNA expression profiles have been detected for several skin diseases, including but not limited to psoriasis, atopic dermatitis, and vitiligo (Table 1). Since the symptomology of psoriasis and atopic dermatitis can overlap to a certain extent, the discovery that the two conditions present distinct changes in miRNA levels can be adapted to aid in the diagnosis of the skin disease [48]. Importantly, this first report of distinctive miRNA dysregulation between psoriasis and atopic dermatitis was reinforced by subsequent studies that produced lists of differentially regulated miRNAs that overlapped considerably with what was previously identified and were detected by methodologies [49–51]. As shown in Table 1, psoriasis presented 29 differentially regulated miRNAs, whereas atopic dermatitis only had 21. Of the miRNAs identified in the study, eight were shared between the two skin diseases. Among the dysregulated miRNAs detected in all four studies, miR-203 was consistently observed to be upregulated, from which it can be inferred that miR-203 is an important factor in the pathogenesis of psoriasis and could be a potential therapeutic target. Both interleukin-17 receptor (IL-17R) and IL-22R have been shown to be involved the pathogenesis of psoriasis, which is interesting in that miR-197 was found to be downregulated in the miRNA-profiling effort with psoriatic skin (Table 1) and miR-197 is reportedly antagonistic to IL-17R and IL-22R [48, 52, 53]. Additional studies have examined whether differentially regulated miRNAs contribute to the effectiveness of photochemotherapy and phototherapy in treating psoriatic skin. For example, examinations of dysregulated miRNAs in response to treatment with UV-A and 8-methoxypsoralen (so-called PUVA therapy) revealed that miR-4516 is upregulated, which contributes to decreased expression of UBE2N, STAT3, and CDK6 proteins, resulting in apoptosis [54, 55]. Similarly, two reports observed the dysregulation of miR-21, miR-125b, miR-146a, and miR-203 as a result of narrow-band UV-B phototherapy [56, 57]. Cumulatively, this demonstrates that not only do miRNAs become differentially regulated as a result skin diseases, but also in response to the treatments applied to combat these conditions.
A separate study examined the differential miRNA expression profile in circulating peripheral blood mononuclear cells (PBMCs) in response to vitiligo, since vitiligo is an auto-immune disorder [58]. Only four miRNAs were found to be dysregulated in PBMCs, where miR-3940-5p was downregulated and miR-224-3p, miR-2682-3p, and miR-4712-3p were upregulated. When the thymosin-α-1 (T) immune modulator was introduced, the miRNA dysregulation was reversed, with miR-3940-5p upregulated and miR-224-3p, miR-2682-3p, and miR-4712-3p downregulated.
Certainly, with time, the number of unique miRNA profiles for different skin disorders and diseases will expand, particularly as the technology improves. The power of these findings comes from the ability to further diagnose and understand these skin conditions as well as in the identification of potential targets for therapeutic approaches, which is detailed in the following section.
Therapeutic Application of miRNAs
As more miRNAs have been identified as key players in the molecular pathogenesis of cancer and infectious diseases, so too has the number of miRNAs implicated in the manifestation of various skin disorders and diseases. With the delineation of the role of each of these culprit miRNAs, scientists have theorized ways to treat these skin conditions through the application of miRNA mimics (miRNA replacement therapy) and miRNA inhibitors (antagomiR therapy) [29–32, 59]. We discuss some examples of naturally occurring miRNAs that have been investigated for their potential utility in combating a series of skin conditions selected for this review, including aberrant pigmentation, skin aging, UV damage to skin, acne, psoriasis, and acute dermatitis (Table 2).
Table 2.
Skin therapeutic applications of microRNAs
| Skin therapeutic application | microRNA | Target(s) | References |
|---|---|---|---|
| Psoriasis | miR-203 | SOCS3, p63 | Wang et al. [145] Huang et al. [103] Bracke et al. [102] Joyce et al. [49] Sonkoly et al. [146] Bostjancic et al. [147] |
| miR-146a | IL-17, IRAK1, TRAF6 | Srivastava et al. [99] Meisgen et al. [148] Sonkoly et al. [146] Bostjancic et al. [147] |
|
| miR-217 | GRHL2 | Zhu et al. [101] | |
| miR-99a miR-99b miR-100 |
IGF-1R | Joyce et al. [49] Lerman et al. [50] Sonkoly et al. [48] |
|
| miR-125b | FGFR2 | Xu et al. [68] | |
| miR-181b | TLR4 | Feng et al. [98] | |
| miR-31 | STK40 | Wang et al. [145] Xu et al. [100] |
|
| miR-197 | IL-17R, IL-22R | Sonkoly et al. [48] Lerman et al. [53] Elharrar et al. [52] |
|
| Atopic dermatitis | miR-146a | CCL5, IRAK1, TRAF6 | Urgard et al. [109] Rebane et al. [106] Sonkoly et al. [146] |
| miR-155 | CTLA-4 | Sonkoly et al. [108] | |
| UV-B damage | miR-141 | PTEN | Li et al. [86] |
| Moisturizing | miR-434-5p | HYAL | Chen et al. [142] |
| Pigmentation | miR-434-5p | TYR | Wu et al. [67] |
| miR-145 | SOX9, TYR, TRP1, MITF | Dynoodt et al. [65] | |
| miR-25 | MITF | Zhu et al. [69] | |
| miR-125b | TYR, DCT | Kim et al. [149] | |
| miR-155 | SOCS1, IFITM1, TRP1 | Sahmatova et al. [72] | |
| miR-211 | TGF-β receptor | Dai et al. [64] | |
| miR-330-5p | TYR | Rambow et al. [66] | |
| miR-137 | MITF | Bemis et al. [70] | |
| miR-182 | MITF, FOXO3 | Segura et al. [71] | |
| Acne | miR-143 | TLR2 | Xia et al. [95] |
| miR-105 | TLR2 | Benakanakere et al. [94] | |
| Skin aging | miR-29 | p53 | Li et al. [73] |
| miR-155 | c-Jun | Song et al. [84] |
CCL5 chemokine ligand 5, CTLA-4 cytotoxic T-lymphocyte-associated antigen 4, DCT dopachrome tautomerase, FGFR2 fibroblast growth factor receptor 2, FOXO3 Forkhead box protein O3, GRLH2 grainy head like transcription factor 2, HYAL hyaluronidase, IFITM1 interferon induced transmembrane protein 1, IGF-1R insulin-like growth factor-1 receptor, IL interleukin, IRAK1 interleukin-1 receptor associated kinase 1, miRNA microRNA, MITF microphthalmia-associated transcription factor, PTEN phosphatase and tensin homolog, SOCS suppressor of cytokine signaling, SOX9 Sry-related high mobility group protein 9, STK40 serine/threonine kinase 40, TGF transforming growth factor, TLR toll-like receptor, TRAF6 tumor necrosis factor receptor associated factor 6, TRP1 TYR-related protein 1, TYR tyrosinase, UV ultraviolet
Skin Pigmentation
Multiple skin pigmentation disorders would potentially benefit from miRNA-based therapeutic applications, including vitiligo, albinism, aging spots (such as solar lentigo), freckles, and melasma. Many of these skin disorders involve a dysregulation at one or more steps in the melanin synthesis pathway. This multi-step pathway sees the amino acid tyrosine enzymatically converted into dihydroxyphenylalanine then dopaquinone by tyrosinase (TYR), which is followed by oxidation of dopaquinone into dopachrome. Dihydroxyindole or dihydroxyindole-2-carboxylic acid are formed from dopachrome and finally convert to eumelanin. TYR and its related proteins (such as TYR-related protein 1[TRP1]) are further regulated by the microphthalmia-associated transcription factor (MITF) [60–63]. As shown in Table 2, multiple miRNAs have been implicated in this signaling pathway. Both TYR and MITF have been explored as therapeutic targets for de-pigmentation [64–67]. A 2008 report cited miR-434-5p as an upstream regulator of the expression of not only TYR but also hyaluronidase (HYAL) [67]. This suggested that miR-434-5p could potentially be utilized to induce de-pigmentation while simultaneously increasing the moisture content of skin.
Another miRNA that could potentially be applied to treating aberrant skin pigmentation is miR-211, which was shown to target transforming growth factor (TGF)-β receptor in an MITF-dependent fashion [64]. Others include miR-25, miR-125b, miR-137, and miR-182, all of which target MITF; miR-330-5p, which targets TYR; and miR-145, which is a key regulator of many genes in the pigmentation process, such as TYR, TRP1, and MITF [65, 66, 68–71]. In cell culture, it was shown that miR-155 contributed to the pathogenesis of vitiligo by downregulating multiple genes associated with melanogenesis, including suppressor of cytokine signaling 1 (SOCS1) and TRP1, and that increased levels of miR-155 contributed to development of the skin disease [72]. In this particular instance, an antagomiR of miR-155 could potentially assist in the suppression of vitiligo symptoms by decreasing the effective levels of endogenous miR-155 [72]. One can certainly conclude from this small sampling of specific miRNAs that multiple miRNA candidates have the potential to be translated into skin care products for the treatment of aberrant pigmentation.
Skin Aging
Skin aging occurs via either intrinsic or extrinsic means [39]. The intrinsic source of skin aging is the genetics of the individual that change with chronological age. Extrinsic sources of aging are all of the environmental insults that impact the skin over time, including but certainly not limited to UV irradiation (photo-aging), particulate matter accumulation (PM-25), and other environmental factors. Much of aging and skin-aging research has revolved around the cellular progression into senescence, where the cell is no longer proliferating. Indeed, it has been shown that the onset of age-related diseases can be delayed through the removal of senescent cells. Evidence is accumulating to show that specific miRNAs play key roles in the development or reversal of cellular senescence and could conceivably be adapted as therapies for skin aging.
Considerable evidence supports a significant role for miR-29 in the aging of skin [73]; it is also a noted regulator of collagen production in the skin [74, 75]. The downstream effects of miR-29 regulation appear to be centered on tumor-suppressor protein p53 [37, 38, 76, 77]. These findings have been the basis of an anti-skin-aging treatment that incorporates an antagomiR of miR-29 [38]. Downregulation of miR-29a-3p was shown to substantially decrease senescence in skin fibroblasts [78].
Other miRNAs that have reported involvement in the aging process and may have therapeutic anti-skin-aging applications include miR-17-5p, miR-21, miR-23a-3p, miR-30a-5p, miR-34a, miR-130b, miR-138, miR-155, miR-181a, and miR-181b. The term longevimiR has been ascribed to the cell proliferation-promoting miR-17-5p, which is downregulated in senescent cells [79]. Although miR-21 has been described as having significant oncogenic potential, one report defined a role for it in the development of cellular senescence in endothelial cells with potential applicability to other cell and tissue types [80]. The hyaluronan synthase 2 (HAS2) gene is targeted by miR-23a-3p in human skin samples and fibroblasts, which has implications for the relative moisture content of the skin [81]. Senescent skin fibroblasts had upregulated miR-23a-3p, whereas non-senescent cells had much reduced levels of this miRNA. In the case of miR-34a, senescent fibroblasts showed its upregulation, which targeted histone deacetylase sirtuin-1 (SIRT1) that resulted in a downstream increase in p53-driven senescence [82, 83]. Similarly, another report found that downregulation of miR-29a-3p, miR-30a-5p, and miR-34a led to reduced senescence in skin fibroblasts [78]. With respect to photo-aging of the skin, miR-155 was observed to be downregulated in dermal fibroblasts that were exposed to UV-A, whereas c-Jun expression increased. It was revealed that miR-155 directly targets c-Jun and that miR-155 potentially counter acts the proliferative potential of UV-A-induced activation of c-Jun [84]. Finally, primary keratinocytes entering replicative senescence showed increased levels of miR-138, miR-181a, and miR-181b, all three of which directly targeted SIRT1 [85]. The same report also showed that miR-130b was upregulated in senescent primary keratinocytes, which targeted the p63 protein [85].
UV Damage
As described in the miRNA profile section of this review, several dysregulated miRNAs have been reported to be up- or downregulated in response to UV-A- and UV-B-induced skin damage. These same miRNAs have also been explored for their potential to repair UV-damaged skin. Among these miRNAs is miR-141, which has several documented targets, among them the anti-proliferative phosphatase, phosphatase and tens in homolog (PTEN) (Table 2) [86].
Several compounds have been tested for protective effects against UV-B-induced skin damage in skin keratinocytes (specifically the HaCaT keratinocyte cell line). For example, arctiin reportedly provides photo-protection to HaCaT cells from UV-B light by stimulating an alteration in the expression of certain miRNAs [87–89]. These reports saw upregulation of miR-125a-3p, miR-205-3p, miR-21-3p, and miR-29b-1-5p and downregulation of 62 distinct miRNAs. Troxerutin is another tested material that also effected a change in miRNA expression patterns: miR-125a-3p, miR-205-3p, miR-21-3p, miR-34b-5p, and miR-181a-2-3p were upregulated and 63 miRNAs were downregulated [90, 91]. Notably, both compounds offering photo-protective effects induced changes in the abundance of similar arrays of miRNA species.
Acne
Skin acne afflicts more than 600 million people globally and frequently ranks as the eighth most common disease among humans. Acne (or acne vulgaris) presents symptoms over a long duration that include greasy skin, pimples, whiteheads, blackheads, and occasional scarring of the skin [92, 93]. These symptoms present when skin oil and dead skin material clog hair follicles. Scientists estimate that more than 80% of all cases of acne vulgaris have a genetic route cause, often implicating tumor necrosis factor (TNF)-α and IL-1-α. TNF-α was shown to be post-transcriptionally regulated by miR-105 in keratinocyte cell culture [94], which modulated inflammatory signaling through TLR2 and TLR4 (Table 2). Another potential causative factor is the presence of an anaerobic bacterial species on skin called Propionibacterium acnes, though its specific role in acne development has not been fully elucidated. Intriguingly, it appears that a combination of both genetic factors and P. acnes can contribute to the manifestation of acne vulgaris, with a recent report citing the involvement of miR-143 in P. acnes-induced acne manifestation [95]. In the study, Staphylococcus epidermidis activated TLR2, which stimulated expression of miR-143 that led to a biofeedback loop, where miR-143 targeted the TLR2 mRNA for downregulation (Table 2). The miR-143-induced TLR2 downregulation subsequently abrogated the P. acnes-stimulated skin inflammation. As mentioned, miR-105 was demonstrated to also target TLR2 in keratinocytes, presenting yet another miRNA that could conceivably be translated into an acne therapeutic [94].
Psoriasis
Psoriasis represents a severe auto-immune disease of the skin that is often characterized by abnormal patches of red, itchy, scaly skin. More than 90% of all cases of psoriasis can be classified as plaque psoriasis (or psoriasis vulgaris), where patients present with red patches topped with white scales. Typically, this condition is treated with topical formulas containing either or both corticosteroids and vitamin D combined with various moisturizers and emollients. Evidence points to psoriasis being a genetic disease that is precipitated by an environmental insult. As an auto-immune disease, the genetic components have been linked to multiple immune system-related genes, including but not limited to IL-23R, IL-12ß, and the locus of the major histocompatibility complex (MHC) [96, 97]. As post-transcriptional gene regulators, miRNAs have also been explored as factors in this genetic skin condition. Several miRNAs have been reportedly linked to psoriasis, including miR-203a, miR-146a, miR-217, miR-99a, miR-181b, miR-31, and miR-125b (Table 2) [50, 68, 98–103].
The most highly expressed miRNA in psoriatic skin keratinocytes is miR-31, which directly targets serine/threonine kinase 40 (STK40), noted for antagonizing pro-inflammatory nuclear factor (NF)-κB signaling, thus demonstrating the inflammation-inducing potential of miR-31 [100]. This is further reinforced by the finding that TGF-β1 stimulates upregulation of miR-31 [100]. In contrast, many of the aforementioned miRNAs (Table 2) are downregulated in psoriatic skin as they promote cellular differentiation and are antagonistic to the aberrant proliferation and inflammation observed with psoriasis. For example, miR-99a, miR-125b, and miR-217 target pro-proliferative factors insulin-like growth factor-1 receptor (IGF-1R), fibroblast growth factor receptor 2 (FGFR2), and grainy head-like transcription factor 2 (GRLH2), respectively [50, 68, 101]. Therefore, hypothetically, miRNA-replacement therapy using mimics of miR-99a, miR-125b, and miR-217 could conceivably be used to treat psoriasis. Another investigation showed that the application of miR-146a mimics could ameliorate the inflammatory symptoms of psoriasis through targeting of IL-17 [99]. Similarly, miR-181b demonstrated anti-inflammatory activity through the targeting of toll-like receptor 4 (TLR4) [98]. With each new miRNA discovered to be a player in the molecular pathogenesis of psoriasis, a new target is added to the arsenal of potential miRNA-based treatments for the disease. Furthermore, as an example of how RNAi can potently affect the pathogenesis of psoriasis, a group tested the involvement of a cellular protein, insulin-like growth factor-binding protein 7 (IGFBP7), which had been observed at decreased levels in psoriatic skin, by transfecting keratinocytes with an siRNA targeted to the IGFBP7 mRNA [104]. Transfection with the IGFBP7 siRNA resulted in the onset of psoriasis-like effects in the keratinocyte culture. As such, it is conceivable that an antagomiR to a naturally occurring miRNA targeting the IGFBP7 mRNA would increase endogenous levels of IGFBP7 and potentially diminish the psoriatic pathology.
Atopic Dermatitis
Atopic dermatitis (or atopic eczema) is a skin disorder similar to but distinct from psoriasis and other forms of dermatitis and is characterized by itchy, red, swollen, and cracked skin. This skin disorder is likely to affect nearly 20% of the population during their life and, like psoriasis, there is believed to be a genetic element to the condition. At least two miRNAs have been identified as playing a role in the molecular pathogenesis of this disorder: miR-146a and miR-155 (Table 2) [105–108]. Two different groups have found miR-155 to be overexpressed in patients with atopic dermatitis, where it targeted cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4). In the case of miR-146a, it was reported that the NF-κB pro-inflammatory signaling pathway was disrupted by miR-146a targeting chemokine ligand 5 (CCL5). It was then concluded that both miRNAs had the potential to be developed into therapeutics to combat atopic dermatitis, and efforts are already underway for an miR-146a-based application [109].
Topical miRNA Delivery Platforms
A variety of methodologies are currently being developed for the deployment of miRNA-based therapeutics for multiple skin diseases and disorders. These different approaches could be distilled down to two broad categories: topical and systematic delivery. Topical delivery can be further divided into physical and biochemical delivery systems. To thoroughly review all of the aforementioned strategies would be an enormous task; therefore, discussion in this review is limited to advances in biochemical vehicles for topical delivery of miRNA-based therapeutics. Topical-based approaches are attractive because the treatment is applied directly to the affected tissue while avoiding the dilution and off-target effects often associated with systemic administration of therapeutic molecules. Furthermore, topical applications can be performed by the patient without professional assistance, which also lends to the technology being utilized by both pharmaceutical and cosmetic industries. However, topical-based delivery systems are not without caveats, which include but are not limited to the need for consistent application for treatment effectiveness and the difficulty in administering a sustained or delayed therapeutic release. Despite some drawbacks, topical treatments incorporating siRNAs or miRNAs represent a new paradigm in combating skin conditions, where in lieu of bio-active compounds being utilized to repair skin tissue, the cells of the skin are able to fix themselves by being provided the genetic instructions to do so.
Liposomes
Conventional liposomes are frequently employed for the deployment of miRNAs and other RNAi molecules in both in vitro and in vivo settings, but these lipid-based carrier vehicles are usually introduced by injection, not by topical application. The concept of topical delivery of an RNAi-based or other nucleic acid-based therapeutic is attractive for multiple reasons: (1) the target site of the disease state is directly accessible, (2) the release of the treatment is easily controlled, (3) treatment efficacy and/or side effects resulting from treatment are easily observed, (4) the treatment is not diluted as it can be with systemic introduction, and (5) off-target effects are minimized. Despite these advantages, topically applied RNAi-based therapeutics have had difficulty penetrating the stratum corneum barrier of the skin [110, 111].
To address this problem, certain groups have explored the preparation of ultra-deformable liposomes (UDLs) or elastic liposomes that can effectively squeeze through the pores of the skin [112]. Collectively, these research efforts to produce UDLs have led to the development to two broad categories of specialized liposomes with augmented skin penetration capability: ethosomes and transfersomes. As the name would suggest, ethosomes represent liposomes with ethanol included in their composition as a penetration enhancer. Alternatively, transfersomes utilize an edge activator, often a single-chain surfactant such as sodium cholate (SC) or Tween 80 to allow for greater deformability. One effort to marry UDL technology to the topical delivery of RNAi-based therapeutics resulted in the production of surfactant-ethanol-cholesterol-osomes (SECosomes) for the delivery of siRNAs across the skin to epidermal cells (Table 3) [113, 114]. The SECosomes comprised a cationic lipid, 2,3-dioleoyloxy-propyl-trimethylammonium chloride (DOTAP); a helper lipid, cholesterol (Chol); a single-chain surfactant (edge activator), SC; and ethanol (penetration enhancer) [115]. This particular formulation represents a combined approach, essentially transethosomes, which have shown enhanced topical delivery efficiency of other molecules [116]. While this lipid formulation improved the transfection efficiency of siRNAs in the study, its skin penetration was limited to the uppermost stratum corneum. Later, the same group attempted to optimize their topical RNAi delivery system by altering the chemistry of the elastic liposomes by eliminating the SC and adding another helper lipid, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE). The most successful alteration from this study produced a liposome with DOTAP, DOPE, Chol, and ethanol (Table 3) [117]. Indeed, this lipid substitution improved the delivery efficiency of multiple RNAi-based therapeutics, including siRNAs, pre-miRNAs, and antagomiRs, and improved the degree of skin penetration without reaching the dermal layer. The promise of this approach is reinforced by the augmented skin penetration being observed both in vitro and with intact human skin.
Table 3.
Topical delivery methods for both short interfering RNAs and microRNAs
| Delivery platform | Properties | miRNA/siRNA | Target | Skin condition | References |
|---|---|---|---|---|---|
| Liposomes—“transfersomes” | Contain edge activator (sodium cholate) | siRNA | Myosin Va exon F (melanosome transport) | Pigmentation change | Geusens et al. [114] |
| Liposomes—“transethosomes” (SECosomes) | Contain edge activator (sodium cholate) and penetration enhancer (ethanol) | siRNA | Myosin Va exon F (melanosome transport) | Pigmentation change | Geusens et al. [115] |
| Liposomes—“transethosomes” (DDC642) | Contain edge activator (DOPE) and penetration enhancer (ethanol) | siRNA pre-miRNA antagomiR |
Defensin Beta 4 (siRNA and pre-miR-145) Myosin Va exon F (pre-miR-145) SOCS3 (antagomiR to miR-203) |
Psoriasis treatment | Desmet et al. [150] |
| Cell-penetrating peptides | TAT peptide | siRNA | RelA (NF-κB family member) | Atopic dermatitis treatment | Uchida et al. [151, 152] |
| SPACE peptide decorated ethosomes | siRNA | GAPDH | NA | Chen et al. [125] | |
| Self-delivering RNAi (Accell) | Chemically modified siRNAs allowing for greater stability and cellular uptake | siRNA | LUC2P-2 (luciferase reporter) | NA | Hegde et al. [141] |
The multiple delivery vehicles and methods being explored for the topical administration of short interfering RNA/microRNA therapeutics and their penetration through the barrier of the stratum corneum of the upper epidermis
GAPDH glyceraldehyde 3-phosphate dehydrogenase, miRNAs microRNAs, NA not applicable, NF nuclear factor, RNAi RNA interference, siRNAs short interfering RNAs, SOCS suppressor of cytokine signaling, SPACE skin penetrating and cell entering, TAT trans-activating transcription activator
Cell-Penetrating Peptides
Cell-penetrating peptides (CPPs) have also been explored in some instances for their ability to bind nucleic acids covalently or non-covalently and facilitate their entry through the skin [118]. Much of the evidence to date supports non-covalent binding of siRNAs and miRNAs to CPPs as the more robust approach with a greater biological response [119]. The three best studied examples are the trans-activating transcription activator (TAT) peptide, poly-arginine (poly-R), and the skin penetrating and cell entering (SPACE) peptide. The TAT from the human immunodeficiency virus (HIV) represents the first CPP to be discovered (Table 3) [120–123]. Three mechanisms have been proposed to account for the ability of CPPs to penetrate the cell plasma membrane: through a cell-mediated endocytosis pathway, direct physical penetration of the plasma membrane, or through a transition state that allows translocation of the peptide and its cargo.
Later studies of CPPs revealed a minimal peptide domain necessary for cell penetration, which frequently involved a series of positively charged amino acids, including arginine and lysine. Poly-R peptides conjugated covalently or non-covalently to a nucleotide cargo molecule such as siRNA have been studied in both in vitro and in vivo settings with mixed results [124]. Another example of a CPP is the SPACE peptide. The conjugation of the SPACE peptide to siRNAs targeted to specific proteins in skin cells has been tested for efficacy as a topical application multiple times (Table 3) [125, 126]. In one notable instance, an siRNA targeted to the MITF transcription factor, which is important in the molecular pathogenesis of melasma, was conjugated to the SPACE peptide and applied in vivo to the skin in a cream that effectively reduced the levels of TYR and TRP1, producing a marked decrease in melanin content in skin lesions [127]. The cumulative knowledge obtained from these research efforts with CPPs and RNAi-based therapeutics could be applied to the development of an optimized means of topically delivering RNAi-based therapeutics for skin conditions.
Chitosan
Chitosan protein has been investigated for its usage in a variety of bio-medical applications, particularly in the area of wound healing [128, 129]. Chitosan as a material brings a lot of advantages in that it is biocompatible, biodegradable, and antimicrobial and has been found to be non-toxic both in vitro and in vivo. Moreover, the US FDA has already approved its use for wound-healing applications. Its polycationic properties have led to chitosan being explored in many instances to deliver DNA, siRNA, and miRNAto various types of cells, though the transfection efficiency of chitosan reagents is reportedly low [130–132]. The utility of chitosan–miRNA complexes for RNAi therapy has been extensively studied in a variety of cell types [133–136]. Multiple investigations have attempted to improve the low transfection efficiency of the chitosan-based siRNA/miRNA complexes through the incorporation of additional components such as hyaluronic acid and CPPs [137, 138]. Therefore, modified chitosan might ultimately become a favored delivery vehicle for topical RNAi therapy.
Self-Delivering RNAi
Delivery of naked siRNAs and miRNAs to the skin does not result in efficient skin penetration and transfection of the desired skin cells. This is due to a combination of factors, including the negative charge of the siRNA and miRNA molecules, their relatively large size (approximately 13–14 kD), and the presence of nucleases on the skin. The chemical modification of the siRNAs and miRNAs themselves to be more stable and amenable to skin penetration and transfection has also been explored for topical RNAi delivery. One reported success is the Accell self-delivering RNAi system (produced by Dharmacon, Lafayette, CO, USA), which obviates the need for transfection reagents, viruses, and specialized instruments for siRNA delivery. Through a proprietary series of chemical modifications, the Accell siRNA molecules exhibit increased efficiency at reducing gene expression of targeted mRNA transcripts as well as increased stability, target specificity, and cell uptake. This system has already been employed to transfect keratinocytes in cell culture [139] as well as skin in vivo (Table 3) [140, 141]. Although proven effective, the cost and proprietary nature of this technology limit its deployment in the skin care industry. Potentially, more cost-effective measures will be available for the production of miRNA mimics as well as antagomiRs in the future, which would certainly revolutionize how these molecules could be utilized for topical application of RNAi therapeutics.
Concluding Remarks
The recent advances in the identification of miRNAs that contribute to various skin conditions open up a whole new arena of skin care therapeutics. From moisturizing, photo-aging, and whitening to acne, dermatitis, and psoriasis, the potential applications of miRNA-based and, more broadly, RNAi-based, skin care products seem boundless. Two key hurdles that need to be overcome for the translation of miRNA research to skin care products are ensuring that there are no off-target effects of the miRNAs employed and the development of an effective delivery system for the miRNA therapeutic. Both obstacles are certainly surmountable, and it is highly likely that miRNA-based skin care products will soon be commercially available.
With respect to miRNA-based therapeutics producing off-target effects, the further elucidation of the downstream targets of different naturally occurring miRNAs will ultimately provide enhanced predictive value. This particular issue is especially important when considering the regulatory aspects of bringing a new therapeutic to the market. Even siRNAs with their limited target promiscuity due to their exquisite sequence complementarity requirement have the potential to have side effects given how far upstream these effector molecules are of multiple signaling pathways. To that end, current miRNA-profiling efforts are necessary and indispensable if RNAi-based treatments are to be commercialized, allowing scientists and physicians to accurately predict whether more harm than good would result from the usage of a particular siRNA/miRNA mimic or antagomiR. In some instances, off-target effects may actually provide additional benefit as can be inferred from miR-434-5p for skin care. This miRNA post-transcriptionally targets TYR, which has utility in treating aberrant skin pigmentation but also down-modulates expression of HYAL, which can add a moisturizing benefit. Regardless, efforts have already been described to limit the side targets of miR-434-5p and allow for safe miRNA-induced reduction in TYR activity [67, 142]. Combining the knowledge gleaned from miRNA-profiling efforts and the artificial refinement and narrowing of the associated targets for a particular miRNA will greatly improve the safety of RNAi-based treatments.
Current experimental topical delivery platforms for RNAi-based therapeutics described herein have attempted to surmount the skin barrier either through novel lipid chemistries allowing for ultra-flexible liposomes or through the incorporation of skin-penetrating peptides into the liposome bilayer. Topical application of prospective RNAi-based treatments is an attractive mode of delivery for a variety of reasons articulated in this review, and the breakthrough vehicle will likely employ a combination of the strategies outlined. Liposomes represent a well-developed technology, and large-scale production is possible with relative ease. Similarly, peptide synthesis has been thoroughly optimized and is quite cost effective. Therefore, the production of peptide-modified ultra-flexible liposomes for the enhanced penetration of RNAi effector molecules into the skin should prove to be an economically attractive platform for both cosmetic and pharmaceutical industries.
A final consideration for the development and commercialization of RNAi-based treatments for skin conditions, and indeed for other types of disease, is that these effector molecules allow for the targeting of disease pathways that were previously undruggable by standard treatment approaches [143]. Particularly when contemplating therapeutic strategies for genetic diseases, especially those manifesting from a single nucleotide polymorphism, standard methodologies with conventional drugs fall short [144]. The validation and scale-up design of a cost-effective delivery system for siRNAs and miRNAs will be the critical factor in unleashing the power of the RNAi-based therapeutics: a new class of treatments that empower cells to repair themselves.
Acknowledgements
The authors of this manuscript wish to thank Mr. Isaiah Negron and Ms. Brianna Tokar for their objective review of this manuscript, fruitful discussions, and suggested revisions.
Compliance with ethical standards
Conflict of interest
The authors of this manuscript have no conflicts of interest.
Funding
No funding was received by either author toward the preparation of this manuscript.
Contributor Information
Paul Lawrence, Phone: (631) 638-1109, Email: Paul.Lawrence@biocogent.com.
Joseph Ceccoli, Phone: (631) 638-1300, Email: Joseph.Ceccoli@biocogent.com.
References
- 1.Baroni A, Buommino E, De Gregorio V, Ruocco E, Ruocco V, Wolf R. Structure and function of the epidermis related to barrier properties. Clin Dermatol. 2012;30:257–262. doi: 10.1016/j.clindermatol.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Bikle DD, Pillai S. Vitamin D metabolite production and function in keratinocytes. Ann N Y Acad Sci. 1988;548:27–44. doi: 10.1111/j.1749-6632.1988.tb18790.x. [DOI] [PubMed] [Google Scholar]
- 3.Pillai S, Bikle DD, Elias PM. Vitamin D and epidermal differentiation: evidence for a role of endogenously produced vitamin D metabolites in keratinocyte differentiation. Skin Pharmacol. 1988;1:149–160. doi: 10.1159/000210769. [DOI] [PubMed] [Google Scholar]
- 4.Proksch E, Brandner JM, Jensen JM. The skin: an indispensable barrier. Exp Dermatol. 2008;17:1063–1072. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan FL, Bernstein IA. Molecular aspects of control in epidermal differentiation. Mol Cell Biochem. 1976;12:171–179. doi: 10.1007/BF01741715. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Montgomery MK, Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 1998;14:255–258. doi: 10.1016/S0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 8.Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 12.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll AP, Goodall GJ, Liu B. Understanding principles of miRNA target recognition and function through integrated biological and bioinformatics approaches. Wiley Interdiscip Rev RNA. 2014;5:361–379. doi: 10.1002/wrna.1217. [DOI] [PubMed] [Google Scholar]
- 14.Dalmay T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013;54:29–38. doi: 10.1042/bse0540029. [DOI] [PubMed] [Google Scholar]
- 15.Faller M, Guo F. MicroRNA biogenesis: there’s more than one way to skin a cat. Biochim Biophys Acta. 2008;1779:663–667. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38:257–268. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 20.Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can’t cut it without a partner. Curr Biol. 2005;15:R61–R64. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y, Han J, Yeom KH, Jin H, Kim VN. Drosha in primary microRNA processing. Cold Spring Harb Symp Quant Biol. 2006;71:51–57. doi: 10.1101/sqb.2006.71.041. [DOI] [PubMed] [Google Scholar]
- 22.Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Sestan N, Steitz JA. Mammalian 5’-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SL, Kim H, Ying SY. Intron-mediated RNA interference and microRNA (miRNA) Front Biosci. 2008;13:2216–2230. doi: 10.2741/2836. [DOI] [PubMed] [Google Scholar]
- 27.Du J, Guo X, Gao S, Luo J, Gong X, Hao C, Yang B, Lin T, Shao J, Cong G, Chang H. Induction of protection against foot-and-mouth disease virus in cell culture and transgenic suckling mice by miRNA targeting integrin alphav receptor. J Biotechnol. 2014;187:154–161. doi: 10.1016/j.jbiotec.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Gismondi MI, Ortiz XP, Curra AP, Asurmendi S, Taboga O. Artificial microRNAs as antiviral strategy to FMDV: structural implications of target selection. J Virol Methods. 2014;199:1–10. doi: 10.1016/j.jviromet.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Simonson B, Das S. MicroRNA therapeutics: the next magic bullet? Mini Rev Med Chem. 2015;15:467–474. doi: 10.2174/1389557515666150324123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, Bader AG. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 34.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr Protoc Bioinform. 2010;12:12–19. doi: 10.1002/0471250953.bi1209s29. [DOI] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Zhang ZW. Expression of miR-203 is decreased and associated with the prognosis of melanoma patients. Int J Clin Exp Pathol. 2015;8:13249–13254. [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Z, Klein JD, Mitch WE, Zhang L, Martinez I, Wang XH. MicroRNA-29 induces cellular senescence in aging muscle through multiple signaling pathways. Aging (Albany NY) 2014;6:160–175. doi: 10.18632/aging.100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ugalde AP, Ramsay AJ, de la Rosa J, Varela I, Marino G, Cadinanos J, Lu J, Freije JM, Lopez-Otin C. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 2011;30:2219–2232. doi: 10.1038/emboj.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang P, Chen J, Li T, Zhu YY. Use of small RNA as antiaging cosmeceuticals. J Cosmet Sci. 2013;64:455–468. [PubMed] [Google Scholar]
- 40.Janssen HL, Kauppinen S, Hodges MR. HCV infection and miravirsen. N Engl J Med. 2013;369:878. doi: 10.1056/NEJMc1307787. [DOI] [PubMed] [Google Scholar]
- 41.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 42.Banerjee J, Chan YC, Sen CK. MicroRNAs in skin and wound healing. Physiol Genom. 2011;43:543–556. doi: 10.1152/physiolgenomics.00157.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui L, Qi Y, Li H, Ge Y, Zhao K, Qi X, Guo X, Shi Z, Zhou M, Zhu B, et al. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. PLoS One. 2011;6:e27071. doi: 10.1371/journal.pone.0027071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duy J, Koehler JW, Honko AN, Schoepp RJ, Wauquier N, Gonzalez JP, Pitt ML, Mucker EM, Johnson JC, O’Hearn A, et al. Circulating microRNA profiles of Ebola virus infection. Sci Rep. 2016;6:24496. doi: 10.1038/srep24496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraemer A, Chen IP, Henning S, Faust A, Volkmer B, Atkinson MJ, Moertl S, Greinert R. UVA and UVB irradiation differentially regulate microRNA expression in human primary keratinocytes. PLoS One. 2013;8:e83392. doi: 10.1371/journal.pone.0083392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou BR, Xu Y, Permatasari F, Liu WL, Li W, Guo XF, Huang QH, Guo Z, Luo D. Characterization of the miRNA profile in UVB-irradiated normal human keratinocytes. Exp Dermatol. 2012;21:317–319. doi: 10.1111/j.1600-0625.2012.01465.x. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Zhou BR, Hua LJ, Guo Z, Luo D. Differential miRNA profile on photoaged primary human fibroblasts irradiated with ultraviolet A. Tumour Biol. 2013;34:3491–3500. doi: 10.1007/s13277-013-0927-4. [DOI] [PubMed] [Google Scholar]
- 48.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, Zhang W, Bowcock AM. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. 2011;20:4025–4040. doi: 10.1093/hmg/ddr331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lerman G, Avivi C, Mardoukh C, Barzilai A, Tessone A, Gradus B, Pavlotsky F, Barshack I, Polak-Charcon S, Orenstein A, et al. MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS One. 2011;6:e20916. doi: 10.1371/journal.pone.0020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zibert JR, Lovendorf MB, Litman T, Olsen J, Kaczkowski B, Skov L. MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci. 2010;58:177–185. doi: 10.1016/j.jdermsci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Elharrar E, Masalha M, Lerman G, Leibowitz-Amit R, Kassem R, Harats M, Sidi Y, Avni D. Positive-negative feedback loop between miR-197 and IL-17A signaling in human keratinocytes. Immun Res. 2016;12:1–8. [Google Scholar]
- 53.Lerman G, Sharon M, Leibowitz-Amit R, Sidi Y, Avni D. The crosstalk between IL-22 signaling and miR-197 in human keratinocytes. PLoS One. 2014;9:e107467. doi: 10.1371/journal.pone.0107467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chowdhari S, Saini N. hsa-miR-4516 mediated downregulation of STAT3/CDK6/UBE2N plays a role in PUVA induced apoptosis in keratinocytes. J Cell Physiol. 2014;229:1630–1638. doi: 10.1002/jcp.24608. [DOI] [PubMed] [Google Scholar]
- 55.Chowdhari S, Saini N. Gene expression profiling reveals the role of RIG1 like receptor signaling in p53 dependent apoptosis induced by PUVA in keratinocytes. Cell Signal. 2016;28:25–33. doi: 10.1016/j.cellsig.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 56.Ele-Refaei AM, El-Esawy FM. Effect of narrow-band ultraviolet B phototherapy and methotrexate on MicroRNA (146a) levels in blood of psoriatic patients. Dermatol Res Pract. 2015;2015:145769. doi: 10.1155/2015/145769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu X, Nylander E, Coates PJ, Nylander K. Effect of narrow-band ultraviolet B phototherapy on p63 and microRNA (miR-21 and miR-125b) expression in psoriatic epidermis. Acta Derm Venereol. 2011;91:392–397. doi: 10.2340/00015555-1086. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Wang K, Liang J, Yang H, Dang N, Yang X, Kong Y. Differential expression analysis of miRNA in peripheral blood mononuclear cells of patients with non-segmental vitiligo. J Dermatol. 2015;42:193–197. doi: 10.1111/1346-8138.12725. [DOI] [PubMed] [Google Scholar]
- 59.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, Kawamata M, Kelnar K, Bader AG, Brown D, Ochiya T. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 2010;18:181–187. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schallreuter KU. A review of recent advances on the regulation of pigmentation in the human epidermis. Cell Mol Biol (Noisy-le-grand) 1999;45:943–949. [PubMed] [Google Scholar]
- 61.Schallreuter KU, Kothari S, Chavan B, Spencer JD. Regulation of melanogenesis–controversies and new concepts. Exp Dermatol. 2008;17:395–404. doi: 10.1111/j.1600-0625.2007.00675.x. [DOI] [PubMed] [Google Scholar]
- 62.Schallreuter KU, Rokos H, Chavan B, Gillbro JM, Cemeli E, Zothner C, Anderson D, Wood JM. Quinones are reduced by 6-tetrahydrobiopterin in human keratinocytes, melanocytes, and melanoma cells. Free Radic Biol Med. 2008;44:538–546. doi: 10.1016/j.freeradbiomed.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 63.Schallreuter KU, Wood JM. The importance of l-phenylalanine transport and its autocrine turnover to l-tyrosine for melanogenesis in human epidermal melanocytes. Biochem Biophys Res Commun. 1999;262:423–428. doi: 10.1006/bbrc.1999.1241. [DOI] [PubMed] [Google Scholar]
- 64.Dai X, Rao C, Li H, Chen Y, Fan L, Geng H, Li S, Qu J, Hou L. Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF-beta receptor 2. Pigment Cell Melanoma Res. 2015;28:217–222. doi: 10.1111/pcmr.12334. [DOI] [PubMed] [Google Scholar]
- 65.Dynoodt P, Mestdagh P, Van Peer G, Vandesompele J, Goossens K, Peelman LJ, Geusens B, Speeckaert RM, Lambert JL, Van Gele MJ. Identification of miR-145 as a key regulator of the pigmentary process. J Invest Dermatol. 2013;133:201–209. doi: 10.1038/jid.2012.266. [DOI] [PubMed] [Google Scholar]
- 66.Rambow F, Bechadergue A, Saintigny G, Morizot F, Mahe C, Larue L. miR-330-5p targets tyrosinase and induces depigmentation. J Invest Dermatol. 2014;134:2846–2849. doi: 10.1038/jid.2014.231. [DOI] [PubMed] [Google Scholar]
- 67.Wu D, Chen JS, Chang DC, Lin SL. Mir-434-5p mediates skin whitening and lightening. Clin Cosmet Investig Dermatol. 2008;1:19–35. doi: 10.2147/ccid.s4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu N, Brodin P, Wei T, Meisgen F, Eidsmo L, Nagy N, Kemeny L, Stahle M, Sonkoly E, Pivarcsi A. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J Invest Dermatol. 2011;131:1521–1529. doi: 10.1038/jid.2011.55. [DOI] [PubMed] [Google Scholar]
- 69.Zhu Z, He J, Jia X, Jiang J, Bai R, Yu X, Lv L, Fan R, He X, Geng J, et al. MicroRNA-25 functions in regulation of pigmentation by targeting the transcription factor MITF in Alpaca (Lama pacos) skin melanocytes. Domest Anim Endocrinol. 2010;38:200–209. doi: 10.1016/j.domaniend.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 70.Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 71.Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sahmatova L, Tankov S, Prans E, Aab A, Hermann H, Reemann P, Pihlap M, Karelson M, Abram K, Kisand K, et al. MicroRNA-155 is dysregulated in the skin of patients with vitiligo and inhibits melanogenesis-associated genes in melanocytes and keratinocytes. Acta Derm Venereol. 2016;96:742–747. doi: 10.2340/00015555-2394. [DOI] [PubMed] [Google Scholar]
- 73.Li T, Yan X, Jiang M, Xiang L. The comparison of microRNA profile of the dermis between the young and elderly. J Dermatol Sci. 2016;82:75–83. doi: 10.1016/j.jdermsci.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 74.Cheng J, Wang Y, Wang D, Wu Y. Identification of collagen 1 as a post-transcriptional target of miR-29b in skin fibroblasts: therapeutic implication for scar reduction. Am J Med Sci. 2013;346:98–103. doi: 10.1097/MAJ.0b013e318267680d. [DOI] [PubMed] [Google Scholar]
- 75.Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE, Michel BA, Distler JH, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheumatol. 2010;62:1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 76.Boominathan L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 2010;29:613–639. doi: 10.1007/s10555-010-9257-9. [DOI] [PubMed] [Google Scholar]
- 77.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 78.Yan X, Serre C, Bergeron L, Mur L, Busuttil V, Botto JM, Domloge N. Modulation of a specific pattern of microRNAs, including miR-29a, miR-30a, and miR-34a, in cultured human skin fibroblasts, in response to the application of a biofunctional ingredient that protects against cellular senescence in vitro. J Cosmet Dermatol Sci Appl. 2015;5:332–342. [Google Scholar]
- 79.Dellago H, Bobbili MR, Grillari J. MicroRNA-17-5p: at the crossroads of cancer and aging-a mini-review. Gerontology. 2017;63:20–28. doi: 10.1159/000447773. [DOI] [PubMed] [Google Scholar]
- 80.Dellago H, Preschitz-Kammerhofer B, Terlecki-Zaniewicz L, Schreiner C, Fortschegger K, Chang MW, Hackl M, Monteforte R, Kuhnel H, Schosserer M, et al. High levels of oncomiR-21 contribute to the senescence-induced growth arrest in normal human cells and its knock-down increases the replicative lifespan. Aging Cell. 2013;12:446–458. doi: 10.1111/acel.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rock K, Tigges J, Sass S, Schutze A, Florea AM, Fender AC, Theis FJ, Krutmann J, Boege F, Fritsche E, et al. miR-23a-3p causes cellular senescence by targeting hyaluronan synthase 2: possible implication for skin aging. J Invest Dermatol. 2015;135:369–377. doi: 10.1038/jid.2014.422. [DOI] [PubMed] [Google Scholar]
- 82.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song J, Liu P, Yang Z, Li L, Su H, Lu N, Peng Z. MiR-155 negatively regulates c-Jun expression at the post-transcriptional level in human dermal fibroblasts in vitro: implications in UVA irradiation-induced photoaging. Cell Physiol Biochem. 2012;29:331–340. doi: 10.1159/000338488. [DOI] [PubMed] [Google Scholar]
- 85.di Val Cervo PR, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, Saintigny G, Dellambra E, Odorisio T, Mahe C, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109:1133–1138. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li W, Di W, Hua L, Zhou B, Guo Z, Luo D. UVB suppresses PTEN expression by upregulating miR-141 in HaCaT cells. J Biomed Res. 2011;25:135–140. doi: 10.1016/S1674-8301(11)60017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bae S, Lim KM, Cha HJ, An IS, Lee JP, Lee KS, Lee GT, Lee KK, Jung HJ, Ahn KJ, An S. Arctiin blocks hydrogen peroxide-induced senescence and cell death though microRNA expression changes in human dermal papilla cells. Biol Res. 2014;47:50. doi: 10.1186/0717-6287-47-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cha HJ, Lee GT, Lee KS, Lee KK, Hong JT, Lee NK, Kim SY, Lee BM, An IS, Hahn HJ, et al. Photoprotective effect of arctiin against ultraviolet B-induced damage in HaCaT keratinocytes is mediated by microRNA expression changes. Mol Med Rep. 2014;10:1363–1370. doi: 10.3892/mmr.2014.2326. [DOI] [PubMed] [Google Scholar]
- 89.Lee GT, Cha HJ, Lee KS, Lee KK, Hong JT, Ahn KJ, An IS, An S, Bae S. Arctiin induces an UVB protective effect in human dermal fibroblast cells through microRNA expression changes. Int J Mol Med. 2014;33:640–648. doi: 10.3892/ijmm.2014.1616. [DOI] [PubMed] [Google Scholar]
- 90.Cha HJ, Lee KS, Lee GT, Lee KK, Hong JT, Lee SN, Jang HH, Lee JH, Park IC, Kim YR, et al. Altered miRNA expression profiles are involved in the protective effects of troxerutin against ultraviolet B radiation in normal human dermal fibroblasts. Int J Mol Med. 2014;33:957–963. doi: 10.3892/ijmm.2014.1647. [DOI] [PubMed] [Google Scholar]
- 91.Lee KS, Cha HJ, Lee GT, Lee KK, Hong JT, Ahn KJ, An IS, An S, Bae S. Troxerutin induces protective effects against ultraviolet B radiation through the alteration of microRNA expression in human HaCaT keratinocyte cells. Int J Mol Med. 2014;33:934–942. doi: 10.3892/ijmm.2014.1641. [DOI] [PubMed] [Google Scholar]
- 92.Bhambri S, Del Rosso JQ, Bhambri A. Pathogenesis of acne vulgaris: recent advances. J Drugs Dermatol. 2009;8:615–618. [PubMed] [Google Scholar]
- 93.Eichenfield LF, Del Rosso JQ, Mancini AJ, Cook-Bolden F, Stein GL, Desai S, Weiss J, Pariser D, Zeichner J, Bhatia N, Kircik L. Evolving perspectives on the etiology and pathogenesis of acne vulgaris. J Drugs Dermatol. 2015;14:263–272. [PubMed] [Google Scholar]
- 94.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, Knudsen TB, Kinane DF. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem. 2009;284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xia X, Li Z, Liu K, Wu Y, Jiang D, Lai Y. Staphylococcal LTA-induced miR-143 inhibits Propionibacterium acnes-mediated inflammatory response in skin. J Invest Dermatol. 2016;136:621–630. doi: 10.1016/j.jid.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 96.Filer C, Ho P, Smith RL, Griffiths C, Young HS, Worthington J, Bruce IN, Barton A. Investigation of association of the IL12B and IL23R genes with psoriatic arthritis. Arthritis Rheumatol. 2008;58:3705–3709. doi: 10.1002/art.24128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith RL, Warren RB, Eyre S, Ho P, Ke X, Young HS, Griffiths CE, Worthington J. Polymorphisms in the IL-12beta and IL-23R genes are associated with psoriasis of early onset in a UK cohort. J Invest Dermatol. 2008;128:1325–1327. doi: 10.1038/sj.jid.5701140. [DOI] [PubMed] [Google Scholar]
- 98.Feng C, Bai M, Yu NZ, Wang XJ, Liu Z. MicroRNA-181b negatively regulates the proliferation of human epidermal keratinocytes in psoriasis through targeting TLR4. J Cell Mol Med. 2017;21:278–285. doi: 10.1111/jcmm.12963. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Srivastava A, Nikamo P, Lohcharoenkal W, Li D, Meisgen F, Landen NX, Stahle M, Pivarcsi A, Sonkoly E. MicroRNA-146a suppresses IL-17-mediated skin inflammation and is genetically associated with psoriasis. J Allergy Clin Immunol. 2017;139:550–561. doi: 10.1016/j.jaci.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 100.Xu N, Meisgen F, Butler LM, Han G, Wang XJ, Soderberg-Naucler C, Stahle M, Pivarcsi A, Sonkoly E. MicroRNA-31 is overexpressed in psoriasis and modulates inflammatory cytokine and chemokine production in keratinocytes via targeting serine/threonine kinase 40. J Immunol. 2013;190:678–688. doi: 10.4049/jimmunol.1202695. [DOI] [PubMed] [Google Scholar]
- 101.Zhu H, Hou L, Liu J, Li Z. MiR-217 is down-regulated in psoriasis and promotes keratinocyte differentiation via targeting GRHL2. Biochem Biophys Res Commun. 2016;471:169–176. doi: 10.1016/j.bbrc.2016.01.157. [DOI] [PubMed] [Google Scholar]
- 102.Bracke S, Desmet E, Guerrero-Aspizua S, Tjabringa SG, Schalkwijk J, Van Gele M, Carretero M, Lambert J. Identifying targets for topical RNAi therapeutics in psoriasis: assessment of a new in vitro psoriasis model. Arch Dermatol Res. 2013;305:501–512. doi: 10.1007/s00403-013-1379-9. [DOI] [PubMed] [Google Scholar]
- 103.Huang RY, Li L, Wang MJ, Chen XM, Huang QC, Lu CJ. An exploration of the role of MicroRNAs in psoriasis: a systematic review of the literature. Medicine (Baltimore) 2015;94:e2030. doi: 10.1097/MD.0000000000002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nousbeck J, Ishida-Yamamoto A, Bidder M, Fuchs D, Eckl K, Hennies HC, Sagiv N, Gat A, Gini M, Filip I, et al. IGFBP7 as a potential therapeutic target in psoriasis. J Invest Dermatol. 2011;131:1767–1770. doi: 10.1038/jid.2011.108. [DOI] [PubMed] [Google Scholar]
- 105.Rebane A. microRNA and allergy. Adv Exp Med Biol. 2015;888:331–352. doi: 10.1007/978-3-319-22671-2_17. [DOI] [PubMed] [Google Scholar]
- 106.Rebane A, Runnel T, Aab A, Maslovskaja J, Ruckert B, Zimmermann M, Plaas M, Karner J, Treis A, Pihlap M, et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin Immunol. 2014;134(836–847):e811. doi: 10.1016/j.jaci.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 107.Ma L, Xue HB, Wang F, Shu CM, Zhang JH. MicroRNA-155 may be involved in the pathogenesis of atopic dermatitis by modulating the differentiation and function of T helper type 17 (Th17) cells. Clin Exp Immunol. 2015;181:142–149. doi: 10.1111/cei.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, Xu N, Meisgen F, Wei T, Bradley M, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126:581–589. doi: 10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 109.Urgard E, Lorents A, Klaas M, Padari K, Viil J, Runnel T, Langel K, Kingo K, Tkaczyk E, Langel U, et al. Pre-administration of PepFect6-microRNA-146a nanocomplexes inhibits inflammatory responses in keratinocytes and in a mouse model of irritant contact dermatitis. J Control Release. 2016;235:195–204. doi: 10.1016/j.jconrel.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 110.Geusens B, Sanders N, Prow T, Van Gele M, Lambert J. Cutaneous short-interfering RNA therapy. Expert Opin Drug Deliv. 2009;6:1333–1349. doi: 10.1517/17425240903304032. [DOI] [PubMed] [Google Scholar]
- 111.Geusens B, Strobbe T, Bracke S, Dynoodt P, Sanders N, Van Gele M, Lambert J. Lipid-mediated gene delivery to the skin. Eur J Pharm Sci. 2011;43:199–211. doi: 10.1016/j.ejps.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 112.Kumar A, Pathak K, Bali V. Ultra-adaptable nanovesicular systems: a carrier for systemic delivery of therapeutic agents. Drug Discov Today. 2012;17:1233–1241. doi: 10.1016/j.drudis.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 113.Bracke S, Carretero M, Guerrero-Aspizua S, Desmet E, Illera N, Navarro M, Lambert J, Del Rio M. Targeted silencing of DEFB4 in a bioengineered skin-humanized mouse model for psoriasis: development of siRNA SECosome-based novel therapies. Exp Dermatol. 2014;23:199–201. doi: 10.1111/exd.12321. [DOI] [PubMed] [Google Scholar]
- 114.Geusens B, Lambert J, De Smedt SC, Buyens K, Sanders NN, Van Gele M. Ultradeformable cationic liposomes for delivery of small interfering RNA (siRNA) into human primary melanocytes. J Control Release. 2009;133:214–220. doi: 10.1016/j.jconrel.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 115.Geusens B, van Gele M, Braat S, De Smedt SC, Stuart MC, Prow T, Sanchez W, Roberts MS, Sanders N, Lambert J. Flexible nanosomes (SECosomes) enable efficient siRNA delivery in cultured primary skin cells and in the viable epidermis of ex vivo human skin. Adv Funct Mater. 2010;20:4077–4090. doi: 10.1002/adfm.201000484. [DOI] [Google Scholar]
- 116.Ascenso A, Raposo S, Batista C, Cardoso P, Mendes T, Praca FG, Bentley MV, Simoes S. Development, characterization, and skin delivery studies of related ultradeformable vesicles: transfersomes, ethosomes, and transethosomes. Int J Nanomed. 2015;10:5837–5851. doi: 10.2147/IJN.S86186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Desmet E, Bracke S, Forier K, Taevernier L, Stuart MC, De Spiegeleer B, Raemdonck K, Van Gele M, Lambert J. Characterization data on the topical carrier DDC642. Data Brief. 2016;7:1204–1210. doi: 10.1016/j.dib.2016.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Menegatti S, Zakrewsky M, Kumar S, De Oliveira JS, Muraski JA, Mitragotri S. De novo design of skin-penetrating peptides for enhanced transdermal delivery of peptide drugs. Adv Healthc Mater. 2016;5:602–609. doi: 10.1002/adhm.201500634. [DOI] [PubMed] [Google Scholar]
- 119.Boisguerin P, Deshayes S, Gait MJ, O’Donovan L, Godfrey C, Betts CA, Wood MJ, Lebleu B. Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv Drug Deliv Rev. 2015;87:52–67. doi: 10.1016/j.addr.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Crombez L, Aldrian-Herrada G, Konate K, Nguyen QN, McMaster GK, Brasseur R, Heitz F, Divita G. A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev. 2007;59:134–140. doi: 10.1016/j.addr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 122.Morris CB, Thanawastien A, Sullivan DE, Clements JD. Identification of a peptide capable of inducing an HIV-1 Tat-specific CTL response. Vaccine. 2001;20:12–15. doi: 10.1016/S0264-410X(01)00271-7. [DOI] [PubMed] [Google Scholar]
- 123.Wagstaff KM, Jans DA. Protein transduction: cell penetrating peptides and their therapeutic applications. Curr Med Chem. 2006;13:1371–1387. doi: 10.2174/092986706776872871. [DOI] [PubMed] [Google Scholar]
- 124.Kim WJ, Christensen LV, Jo S, Yockman JW, Jeong JH, Kim YH, Kim SW. Cholesteryl oligoarginine delivering vascular endothelial growth factor siRNA effectively inhibits tumor growth in colon adenocarcinoma. Mol Ther. 2006;14:343–350. doi: 10.1016/j.ymthe.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 125.Chen M, Zakrewsky M, Gupta V, Anselmo AC, Slee DH, Muraski JA, Mitragotri S. Topical delivery of siRNA into skin using SPACE-peptide carriers. J Control Release. 2014;179:33–41. doi: 10.1016/j.jconrel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hsu T, Mitragotri S. Delivery of siRNA and other macromolecules into skin and cells using a peptide enhancer. Proc Natl Acad Sci U S A. 2011;108:15816–15821. doi: 10.1073/pnas.1016152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yi X, Zhao G, Zhang H, Guan D, Meng R, Zhang Y, Yang Q, Jia H, Dou K, Liu C, et al. MITF-siRNA formulation is a safe and effective therapy for human melasma. Mol Ther. 2011;19:362–371. doi: 10.1038/mt.2010.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dreifke MB, Jayasuriya AA, Jayasuriya AC. Current wound healing procedures and potential care. Mater Sci Eng C Mater Biol Appl. 2015;48:651–662. doi: 10.1016/j.msec.2014.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Patrulea V, Ostafe V, Borchard G, Jordan O. Chitosan as a starting material for wound healing applications. Eur J Pharm Biopharm. 2015;97:417–426. doi: 10.1016/j.ejpb.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 130.Borchard G. Chitosans for gene delivery. Adv Drug Deliv Rev. 2001;52:145–150. doi: 10.1016/S0169-409X(01)00198-3. [DOI] [PubMed] [Google Scholar]
- 131.Rudzinski WE, Aminabhavi TM. Chitosan as a carrier for targeted delivery of small interfering RNA. Int J Pharm. 2010;399:1–11. doi: 10.1016/j.ijpharm.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 132.Singha K, Namgung R, Kim WJ. Polymers in small-interfering RNA delivery. Nucleic Acid Ther. 2011;21:133–147. doi: 10.1089/nat.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cosco D, Cilurzo F, Maiuolo J, Federico C, Di Martino MT, Cristiano MC, Tassone P, Fresta M, Paolino D. Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer treatment of multiple myeloma. Sci Rep. 2015;5:17579. doi: 10.1038/srep17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaban K, Salva E, Akbuga J. In vitro dose studies on chitosan nanoplexes for microRNA delivery in breast cancer cells. Nucleic Acid Ther. 2017;27:45–55. doi: 10.1089/nat.2016.0633. [DOI] [PubMed] [Google Scholar]
- 135.Louw AM, Kolar MK, Novikova LN, Kingham PJ, Wiberg M, Kjems J, Novikov LN. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomedicine. 2016;12:643–653. doi: 10.1016/j.nano.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 136.Santos-Carballal B, Aaldering LJ, Ritzefeld M, Pereira S, Sewald N, Moerschbacher BM, Gotte M, Goycoolea FM. Physicochemical and biological characterization of chitosan-microRNA nanocomplexes for gene delivery to MCF-7 breast cancer cells. Sci Rep. 2015;5:13567. doi: 10.1038/srep13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, Xiao X, Yang Y, Sheng W, Wu Y, Zeng Y. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 138.Layek B, Lipp L, Singh J. Cell penetrating peptide conjugated chitosan for enhanced delivery of nucleic acid. Int J Mol Sci. 2015;16:28912–28930. doi: 10.3390/ijms161226142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kovalenko A, Kim JC, Kang TB, Rajput A, Bogdanov K, Dittrich-Breiholz O, Kracht M, Brenner O, Wallach D. Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. J Exp Med. 2009;206:2161–2177. doi: 10.1084/jem.20090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hickerson RP, Flores MA, Leake D, Lara MF, Contag CH, Leachman SA, Kaspar RL. Use of self-delivery siRNAs to inhibit gene expression in an organotypic pachyonychia congenita model. J Invest Dermatol. 2011;131:1037–1044. doi: 10.1038/jid.2010.426. [DOI] [PubMed] [Google Scholar]
- 141.Hegde V, Hickerson RP, Nainamalai S, Campbell PA, Smith FJ, McLean WH, Pedrioli DM. In vivo gene silencing following non-invasive siRNA delivery into the skin using a novel topical formulation. J Control Release. 2014;196:355–362. doi: 10.1016/j.jconrel.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen JS, Wu DT. Application of intronic microRNA agents in cosmetics. Methods Mol Biol. 2013;936:325–341. doi: 10.1007/978-1-62703-083-0_25. [DOI] [PubMed] [Google Scholar]
- 143.Wu SY, Lopez-Berestein G, Calin GA, Sood AK. RNAi therapies: drugging the undruggable. Sci Transl Med. 2014;6:240ps247. doi: 10.1126/scitranslmed.3008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Seyhan AA. RNAi: a potential new class of therapeutic for human genetic disease. Hum Genet. 2011;130:583–605. doi: 10.1007/s00439-011-0995-8. [DOI] [PubMed] [Google Scholar]
- 145.Wang MJ, Xu YY, Huang RY, Chen XM, Chen HM, Han L, Yan YH, Lu CJ. Role of an imbalanced miRNAs axis in pathogenesis of psoriasis: novel perspectives based on review of the literature. Oncotarget. 2017;8:5498–5507. doi: 10.18632/oncotarget.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs: novel regulators in skin inflammation. Clin Exp Dermatol. 2008;33:312–315. doi: 10.1111/j.1365-2230.2008.02804.x. [DOI] [PubMed] [Google Scholar]
- 147.Bostjancic E, Glavac D. Importance of microRNAs in skin morphogenesis and diseases. Acta Dermatovenerol Alp Pannonica Adriat. 2008;17:95–102. [PubMed] [Google Scholar]
- 148.Meisgen F, Landen NX, Wang A, Rethi B, Bouez C, Zuccolo M, Gueniche A, Stahle M, Sonkoly E, Breton L, Pivarcsi A. MiR-146a negatively regulates TLR2-induced inflammatory responses in keratinocytes. J Invest Dermatol. 2014;134:1931–1940. doi: 10.1038/jid.2014.89. [DOI] [PubMed] [Google Scholar]
- 149.Kim KH, Bin BH, Kim J, Dong SE, Park PJ, Choi H, Kim BJ, Yu SJ, Kang H, Kang HH, et al. Novel inhibitory function of miR-125b in melanogenesis. Pigment Cell Melanoma Res. 2014;27:140–144. doi: 10.1111/pcmr.12179. [DOI] [PubMed] [Google Scholar]
- 150.Desmet E, Bracke S, Forier K, Taevernier L, Stuart MC, De Spiegeleer B, Raemdonck K, Van Gele M, Lambert J. An elastic liposomal formulation for RNAi-based topical treatment of skin disorders: proof-of-concept in the treatment of psoriasis. Int J Pharm. 2016;500:268–274. doi: 10.1016/j.ijpharm.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 151.Uchida T, Kanazawa T, Kawai M, Takashima Y, Okada H. Therapeutic effects on atopic dermatitis by anti-RelA short interfering RNA combined with functional peptides Tat and AT1002. J Pharmacol Exp Ther. 2011;338:443–450. doi: 10.1124/jpet.111.180042. [DOI] [PubMed] [Google Scholar]
- 152.Uchida T, Kanazawa T, Takashima Y, Okada H. Development of an efficient transdermal delivery system of small interfering RNA using functional peptides, Tat and AT-1002. Chem Pharm Bull (Tokyo) 2011;59:196–201. doi: 10.1248/cpb.59.196. [DOI] [PubMed] [Google Scholar]