Abstract
Fluorescence-guided surgery using 5-aminolevulinic acid (5-ALA) is now a widely-used modality for glioblastoma (GBM) treatment. However, intratumoral heterogeneity of fluorescence intensity may reflect different onco-metabolic programs. Here, we investigated the metabolic mechanism underlying the heterogeneity of 5-ALA fluorescence in GBM. Using an in-house developed fluorescence quantification system for tumor tissues, we collected 3 types of GBM tissues on the basis of their fluorescence intensity, which was characterized as strong, weak, and none. Expression profiling by RNA-sequencing revealed 77 genes with a proportional relationship and 509 genes with an inverse relationship between gene expression and fluorescence intensity. Functional analysis and in vitro experiments confirmed glutaminase 2 (GLS2) as a key gene associated with the fluorescence heterogeneity. Subsequent metabolite profiling discovered that insufficient NADPH due to GLS2 underexpression was responsible for the delayed metabolism of 5-ALA and accumulation of protoporphyrin IX (PpIX) in the high fluorescence area. The expression level of GLS2 and related NADPH production capacity is associated with the regional heterogeneity of 5-ALA fluorescence in GBM.
Introduction
Glioblastoma (GBM) is one of the most devastating cancers of the brain; the current best practice of multimodal treatment results in an overall survival rate of 27.2% in 2 years1. However, in the modern era of GBM management, it is now generally accepted that the extent of surgical resection significantly affects patient prognosis2. Diverse efforts for the improvement of the resection rate in GBM have recently been attempted that integrate the newest technologies, including fluorescence-guided surgery (FGS), intraoperative neurophysiological monitoring, intraoperative magnetic resonance imaging, and neuronavigation/ultrasound-assisted surgery3. Among them, FGS using 5-aminoleveulinic acid (5-ALA) has rapidly become a widely accepted practice for treatment of high-grade gliomas, owing to its ability to improve the extent of resection while preserving the patient’s functional status4–6. 5-ALA is a metabolic targeting agent and the natural precursor of the fluorescent protoporphyrin IX (PpIX) in the heme biosynthesis pathway7. The temporary accumulation of PpIX by overloaded exogenous 5-ALA emits enhanced red fluorescence (wavelength 600–720 nm) selectively in cancer cells under a blue light source (wavelength: 375–440 nm), thus enabling the identification of cancer boundaries otherwise hardly distinguishable with a white light microscope during GBM surgery8. The sensitivity and specificity of 5-ALA fluorescence have been reported to be as high as 85% and 75% in GBM9.
Despite the widespread clinical use of 5-ALA FGS in GBM and the extensive studies exploring its mechanism of selective fluorescence, the conclusive explanation has not yet been reported. Only sporadic reports on alterations in heme biosynthetic enzymes and porphyrin transporters have been proposed for the mechanism of the preferential accumulation of PpIX in cancer cells, but the evidences were inconsistent among the tumor types10. Among the suggested heme biosynthetic enzymes with changes in the expression or activity in cancer tissues, only ferrochelatase (FECH) has been found to be downregulated in GBM tissues compared with the normal brain11. However, there is no evidence of mutation in the FECH gene itself in GBM, thus suggesting that another indirect regulatory mechanism is responsible for the decreased FECH activity that is yet to be discovered. Among the altered porphyrin transporters in cancer cells, ATP-binding cassette sub-family B member 6 (ABCB6) shows higher expression in glioma tissue than in normal brain tissue and is correlated with 5-ALA/PpIX fluorescence12. However, the non-specific location of ABCB6 identified in cell organelles suggests that the porphyrin transport can be bidirectional, thus resulting in its weak contribution to intracellular PpIX accumulation13–15. No direct evidence of alterations for other heme biosynthetic enzymes or porphyrin transporters in human glioma tissues has been reported until now.
Considering the nature of the temporary accumulation of PpIX after 5-ALA administration in cancer cells, focusing on metabolic reprogramming in cancer may be a more reasonable way to investigate the mechanism of selective fluorescence. Studies on the mechanism of 5-ALA fluorescence in gliomas from the perspective of metabolomics are scarce. Reprogrammed metabolism is one of the hallmarks of cancer, including GBM16. Altered carbon metabolism involving enhanced aerobic glycolysis and a decreased tricarboxylic acid (TCA) cycle is a common feature of proliferating cancer cells17. We have previously shown that mutation of isocitrate dehydrogenase 1 (IDH1), one of the TCA cycle isozymes commonly mutated in lower grade gliomas, is related to the enhancement of PpIX accumulation and fluorescence by exogenous 5-ALA in malignant glioma cells18. Further evidence of crosstalk between the TCA cycle and the heme biosynthesis pathway in cancer has been demonstrated19. However, the onco-metabolomic relationships between the TCA cycle and the heme biosynthesis pathway in the absence of mutations in TCA cycle enzymes have not been elucidated. A novel hypothesis for the metabolic mechanism is needed to explain the selective 5-ALA-induced fluorescence in GBM tissue, because most GBM tissues express wild-type TCA enzymes20.
The issue of intratumoral heterogeneity has been proposed and proven in terms of genomics, proteomics and histology in many cancers21. With the recent advances in metabolomics, intratumoral metabolic heterogeneity has also emerged in the spotlight22,23. Regional heterogeneity of metabolic reprogramming in solid tumors is thought to be linked to various microenvironments and specific pathways implicated by genomic alterations22,24. Glioblastoma is one of the representative models for intratumoral heterogeneity with regard to genomic alterations and microscopic features. The interesting characteristic about 5-ALA fluorescence in GBM is that there is also regional heterogeneity in the fluorescence intensity25. The degree of fluorescence intensity in GBM has been reported to be correlated with cell density25. However, this intratumoral fluorescence heterogeneity is more likely to be the direct result of intratumoral metabolic heterogeneity. Therefore, the genomic and metabolomic mechanisms of 5-ALA fluorescence in GBM are expected to be determined by investigating the cause of intratumoral fluorescence heterogeneity.
In this study, we have identified glutaminase 2 (GLS2) as a key gene in the regulation of 5-ALA metabolism by transcriptome profiling by using multi-regional GBM samples with different fluorescence intensities. Additionally, in vitro experiments validated the relationship between GLS2 and 5-ALA fluorescence/PpIX accumulation in GBM cell lines, thus demonstrating that insufficient NADPH production, a major determinant of the heterogenous 5-ALA/PpIX fluorescence in correlation with GLS2 underexpression is a major determinant of the heterogeneous 5-ALA/PpIX fluorescence. On the basis of the observed findings, we proposed a hypothesis for the metabolic mechanism of 5-ALA fluorescence in GBM tissue.
Results
Regional heterogeneity of 5-ALA fluorescence in GBM tissue
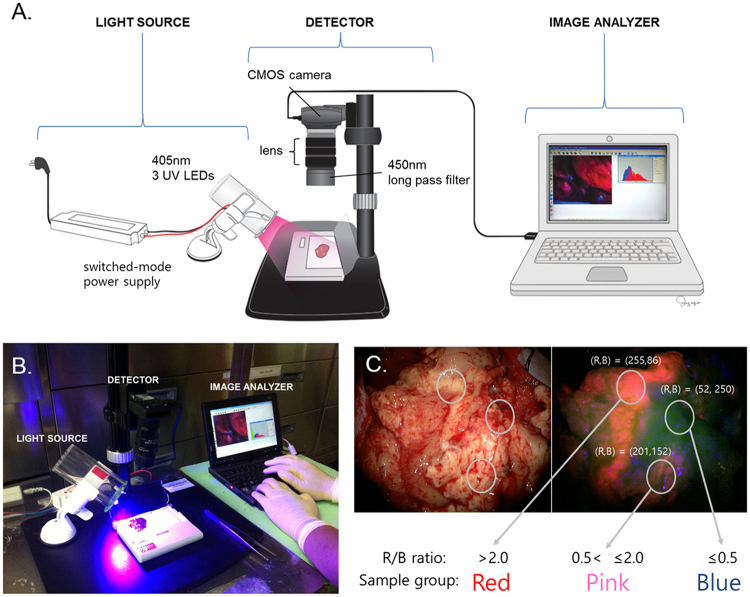
We devised an in-house system of fluorescence quantification in tumor tissues (Fig. 1). The fluorescence intensity was measured as the red/blue (R/B) ratio in captured images immediately after resection of the tumor during 5-ALA FGS. Using this system, we collected 3 classes of GBM tissues in each of 5 patients according to the fluorescence intensity as follows: strong (red, R/B ratio >2.0), weak (pink, 0.5 < R/B ratio ≤2.0), and none (blue, R/B ratio ≤0.5). All 5 patients showed clear intratumoral fluorescence heterogeneity in their GBM tissues (Supplementary Figure 1 and Supplementary Table 1). Individually separated samples were snap-frozen in liquid nitrogen before nucleic acid extraction, and parts of the tissues were sent for histological diagnosis. Each red, pink, and blue area showed different histological grades, IV, III, and II, respectively, in all patients (Supplementary Figure 2).
Figure 1.
Collection of glioblastoma samples according to the regional heterogeneity of fluorescence intensity. (A) Schematic illustration of the fluorescence measurement and quantification system for surgically obtained samples (all copyright of this artwork by Mi-Jin Jung of Biomedical Illustration & Design Company transferred to Nature Publishing Group). (B) Real situation of sample management immediately after tumor resection. (C) The samples were classified and acquired according to the quantified fluorescence intensity. The fluorescence intensities were measured according to the red/blue (R/B) ratios in the captured images and then were separated into the following 3 classes: strong (red, R/B ratio >2.0), weak (pink, 0.5 < R/B ratio ≤2.0), and none (blue, R/B ratio ≤0.5).
Key matched genomics for the regional heterogeneity of fluorescence
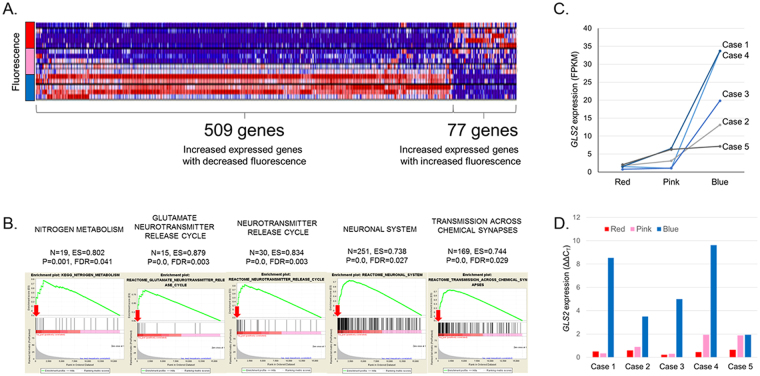
After the extraction of RNA from samples, RNA-sequencing of total RNA was performed to profile the gene expression levels in each sample. To identify the key genes related to fluorescence intensity, we first sorted out the genes that showed a linear trend of expression levels among the red, pink, and blue sample groups. A total of 3,344 genes were identified to have either an increasing or decreasing trend of expression in the red, pink, and blue groups (FDR < 0.01). Next, the genes were filtered further on the basis of the condition of a clear linear trend among the 3 groups by selecting the differentially expressed genes with fold changes greater than two between groups. The final result elicited 77 genes with positive correlation, and 509 genes with negative correlation between the expression level and fluorescence intensity (Fig. 2A and Supplementary Dataset).
Figure 2.
Identification of key genes associated with 5-aminolevulinic acid (5-ALA) fluorescence intensity in glioblastoma. (A) Results of RNA-sequencing of classified samples, which were analyzed according to differential expression along with the fluorescence intensity (fold change >2, p < 0.05, FDR < 0.01). Seventy-seven genes with positive correlation and 509 genes with negative correlation between the expression level and fluorescence intensity were identified. (B) Pathway analysis using gene set enrichment analysis (GSEA) resulted in 30 significantly enriched gene sets of (p < 0.01, FDR < 0.25). Among them, GLS2 (red arrows) was repeatedly found with a high enrichment score in the selected gene sets of metabolism and the neuronal system. (C) From the results of the RNA-seq FPKM values, GLS2 was highly expressed in areas of no fluorescence intensity, and the expression was decreased in areas of positive fluorescence in all samples. (D) The expression level of GLS2 in all samples, as measured by quantitative real-time PCR, confirmed the result of RNA-seq.
To identify the most plausible key genes for fluorescence heterogeneity, the list of candidate genes was classified according to predefined functional modules by using gene set enrichment analysis (GSEA) after being ranked on the basis of a score calculated from the adjusted p-value. Among the large numbers of gene sets from the GSEA that were enriched positively and negatively, 10 gene sets from the Kyoto Encyclopedia of Genes and Genomes (KEGG)pathway (http://www.genome.jp/kegg/) and 20 gene sets from the REACTOME pathway (http://reactome.org/) were displayed significantly with p-value < 0.01 and FDR < 0.25 (Supplementary Table 2). Because 5-ALA fluorescence is the result of the temporary intracellular accumulation of PpIX in cancer cells, we focused on gene sets related to cell metabolism as well as the neuronal system. Interestingly, GLS2 was repeatedly found with a high enrichment score in the selected gene sets (Fig. 2B). The expression of GLS2 was inversely correlated with the fluorescence intensity in fragments per kilobase of transcript per million mapped reads (FPKM) values (Fig. 2C). Additionally, we confirmed the expression level of GLS2 by quantitative real-time PCR (Fig. 2D). Therefore, it is plausible that GLS2 is a key candidate gene for heterogenous 5-ALA fluorescence intensity in GBM.
GLS2 attenuates 5-ALA fluorescence in GBM cells
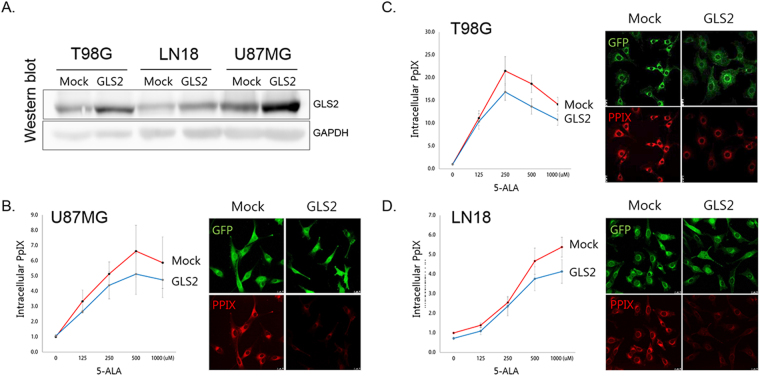
To verify the role of GLS2 in 5-ALA fluorescence in cancer cells, the lentiviral vector containing the GLS2 gene construct was established and transduced into GBM cell lines (T98G, U87MG, and LN18). In vitro measurements of intracellular PpIX after 5-ALA treatment showed a significant decrease (Fig. 3). Fluorescence was evident after 5-ALA treatment in GBM cell lines but was attenuated in cells with GLS2 transduction (Fig. 3). Therefore, it was confirmed that the underexpression of GLS2 is associated with increased 5-ALA fluorescence intensity.
Figure 3.
In vitro evidence of an association between GLS2 expression and 5-ALA fluorescence. (A) The overexpression of the GLS2 gene after transduction into three glioblastoma cell lines (T98G, LN18, and U87MG) was confirmed at the protein level. Western blot images are cropped for the GLS2/GAPDH blots and the full-length of gel image is included in Supplementary Figure 3. (B–D) All cell lines with GLS2 overexpression showed decreased PpIX accumulation and fluorescence intensity after 5-ALA treatment (p < 0.05, average value of 4 duplicate experiments). The concentration of PpIX after 5-ALA treatment was expressed as relative fluorescence units (RFUs) normalized against total cell protein levels. Confocal laser scanning microscope images were taken 2 hours after 5-ALA treatment. PpIX was visible as red fluorescence and was located mainly in the cytoplasm (green fluorescence protein (GFP)).
GLS2 induces NADPH production, and NADPH is consumed during 5-ALA metabolism
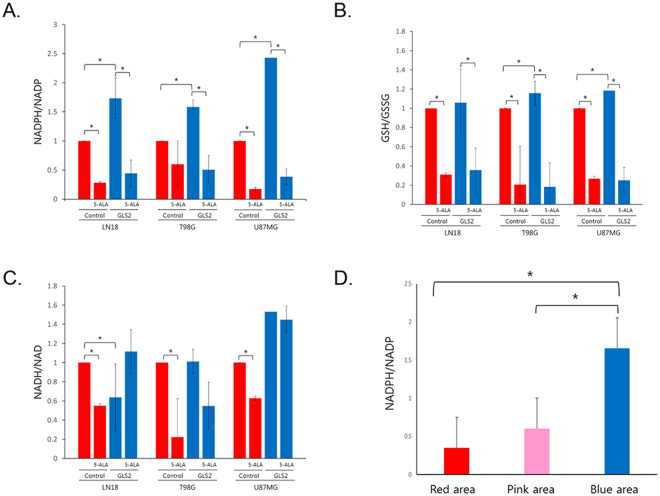
To screen for metabolic changes that might explain the difference in 5-ALA metabolism between cell lines with and without GLS2 expression, we profiled the metabolites from the lysates of GBM cell lines with or without GLS2 expression after 5-ALA exposure, by using liquid chromatography–mass spectrometry (LC-MS). Among the metabolites tested, NADPH production was significantly increased when GLS2 expression was enhanced, and NADPH was rapidly consumed after the introduction of 5-ALA (Fig. 4A). The ratio of reduced glutathione (GSH) to oxidized glutathione (GSSG) was also slightly increased with GLS2 expression, and it was decreased after 5-ALA treatment (Fig. 4B). The NADH/NAD ratio was decreased after 5-ALA treatment in cells with GLS2 underexpression, but changes in the NADH/NAD ratio were not observed in cells with GLS2 expression with or without 5-ALA treatment (Fig. 4C). There were no intracellular energy metabolism changes regarding GLS2 expression or 5-ALA treatment, as represented by AMP, ADP, and ATP levels (Supplementary Figure 4). Overall, NADPH/NADP and GSH/GSSG levels were significantly decreased after 5-ALA treatment, and GLS2 expression induced increased levels of NADPH/NADP, thus possibly increasing the capacity for 5-ALA metabolism.
Figure 4.
(A–C) Metabolite screening by using liquid chromatography-mass spectrometry (LC-MS) on glioblastoma cell lines with or without GLS2 expression after 5-aminolevulinic acid (5-ALA) treatment. Each sample was tested in triplicate. NADPH/NADP levels were significantly increased with GLS2 expression, but they decreased rapidly after 5-ALA treatment in all cell lines (A). GSH/GSSG levels were also decreased after 5-ALA treatment, but they increased with GLS2 expression in 2 cell lines with a relatively small amount of change (B). NADH/NAD levels were decreased after 5-ALA treatment only in cells without GLS2 expression (C). (D) Direct measurement of NADPH/NADP levels in glioblastoma tissues showing different fluorescence intensities (n = 30) confirmed increased NADPH/NADP levels in the no-tumor area without fluorescence.
To confirm the regional differences in the NADPH levels in GBM, the NADPH/NADP levels were measured in 10 paired GBM tissues located in the red, pink, and blue fluorescence areas. The NADPH/NADP level was significantly increased in the area without 5-ALA fluorescence compared with that with positive fluorescence (Fig. 4D).
Hypothesis for the metabolic mechanism of 5-ALA fluorescence in GBM tissue
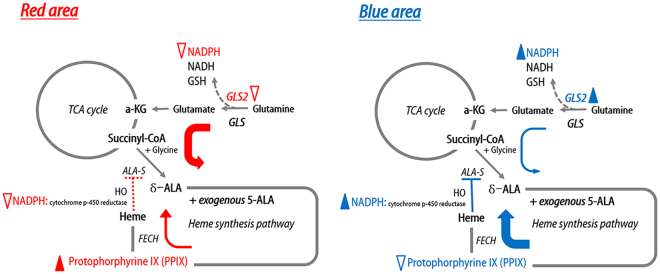
On the basis of the evidence obtained, we deduced that GLS2 expression and the intracellular NADPH level are key players in the 5-ALA fluorescence in GBM tissue. There is crosstalk between the TCA cycle and heme synthesis pathway through succinyl coenzyme A (Succinyl-CoA) and glycine to form 5-ALA by ALA synthase19,26–30. The rate-limiting step of the heme synthesis pathway is ALA synthase, which is negatively regulated by heme [which is usually degraded by heme oxygenase (HO-1)], via a feedback mechanism26. In this step, HO degrades heme in concert with NADPH cytochrome P450 reductase, and it has been shown that HO activity is increased by NADPH or NADH31. Therefore, if GLS2 were underexpressed, NADPH production would be decreased, and insufficient NADPH would result in decreased HO activity, thereby saturating FECH because of heme synthesis pathway over-activation. Thus, exogenous 5-ALA cannot be metabolized rapidly, and PpIX accumulates under such conditions (Fig. 5). However, if GLS2 expression were normal, there would be sufficient NADPH production to facilitate HO activity in the normal negative feedback mechanism of 5-ALA synthesis. Thus, sufficient FECH activity allows for exogenous 5-ALA to be metabolized without delay (Fig. 5). Therefore, the regional heterogeneity of GLS2 expression in GBM tissue is responsible for the 5-ALA fluorescence heterogeneity.
Figure 5.
Schematic illustration of the hypothesis for the regional heterogeneity of 5-aminolevulinic acid (5-ALA) fluorescence intensity in glioblastoma. At the area of strong fluorescence intensity (red area), decreased expression of glutaminase 2 (GLS2) results in insufficient production of NADPH, thus hindering the negative feedback for ALA-synthase (ALA-S). Under such conditions, ferrochelatase (FECH) is saturated and hence metabolizes a large amount of exogenous 5-ALA, thus resulting in the temporary accumulation of protoporphyrin IX (PpIX). At the area of no fluorescence intensity (blue area), GLS2 expression is normal, and sufficient NADPH is produced to support the normal feedback mechanism for ALA-synthase. In this case, adequate FECH activity metabolizes exogenous 5-ALA without delay. Filled arrowhead; sufficient amount or activity of molecule or enzyme, Open arrowhead; insufficient amount or activity of molecule or enzyme, Thick color arrow; smooth metabolic process. Thin color arrow; delayed metabolic process. Dashed red line; abnormal feedback mechanism. Solid blue line; normal feedback mechanism., Dashed arrow; abbreviated metabolic process.
Discussion
Glutamine is an essential fuel source for cancer cell metabolism, which is required for cancer growth and proliferation32. Glutamine is converted by glutaminases to ammonium ion and glutamate in the mitochondria, and the glutamate further catabolized to alpha-ketoglutarate (α-KG), which enters the TCA cycle. There are two glutaminase types in mammals, as Kreb had originally described, which are tissue specific33. Of the two glutaminase isoforms, GLS (kidney-type) is more broadly expressed in normal tissue except for the liver, whereas GLS2 (liver-type) is exclusively expressed in the liver, brain, and pancreas34. Interestingly, evidence has shown that GLS and GLS2 differ in their regulation and activity. GLS is inhibited by its product glutamate, which is not the case with GLS2; however, GLS2, but not GLS, is activated by its product ammonia in vitro 35. In terms of the oncogenic process, GLS expression is regulated by the oncogene c-myc 36, whereas GLS2 is induced by the tumor suppressor p5332,33. Additionally, GLS is known to have oncogenic properties36, whereas GLS2 acts as a tumor suppressor33. Silencing GLS induces apoptosis in glioma cells, whereas GLS2 overexpression suppresses malignant properties37. Moreover, the switching to GLS upregulation in combination with GLS2 repression in hepatoma cells has been found to be accompanied by malignant transformation in an experimental setting38. Cells with elevated GLS2 expression show increased glutamate concentrations, increased mitochondrial respiration, higher GSH and NADH levels, and decreased reactive oxygen species (ROS) levels32,33. Additionally, the knock-down of GLS2 increases the intracellular ROS levels after radiation by decreasing the production of antioxidant GSH, NADH and NADPH34. However, knowledge concerning the role of GLS2 in cancer is relatively limited compared with that of GLS.
There is substantial evidence that glutamine metabolism is crucial in gliomas39. The lack of GLS2 expression has been reported in human malignant glioma samples35. Additionally, low GLS2 expression is a specific property of tumor cells of glial origin39. Transfection of T98G human GBM cells with GLS2 decreases cell proliferation, survival, and migration40. Moreover, transfection with GLS2 alters the expression pattern of many genes relevant to malignancy40. The opposite effect of GLS and GLS2 on cell growth has also been demonstrated in GBM cells39. However, glutamine metabolism is heterogeneous in GBM. There is evidence that glutamine synthetase (GS) activity is variable within the GBM microenvironment in that some cells exhibit high GS expression and glutamine self-sufficiency, whereas others exhibit low GS levels and dependence on exogenous glutamine41. Another report has shown striking differences in the glutamate level between GBM and adjacent normal brain tissues, thus supporting regional metabolic heterogeneity of GBMs42. The results of the present study are in line with the previously mentioned observations regarding GBM, that are the importance of glutamine metabolism involving GLS2, ii) the tumor suppressor effect of GLS2, and iii) the regional metabolic heterogeneity in tumor tissues. Considering these findings together, we conclude that regional metabolic heterogeneity in GBM results in low GLS2 expression in the malignant area with low NADPH and GSH levels (high ROS) and high GLS2 expression in the lower grade area with high NADPH and GSH levels (low ROS).
In the present study, we propose that NADPH is a metabolic mediator of 5-ALA fluorescence intensity. Both NADPH and GSH were significantly depleted after 5-ALA treatment. However, the NADPH level was increased more than that of GSH after GLS2 overexpression. Additionally, in vitro evidence, as well as human tissue samples of GBM, showed that 5-ALA fluorescence was inversely correlated with GLS2 expression and NADPH level. There is growing evidence in cancer metabolomics highlighting the role of NADPH in oncogenic signaling. The increased demand of NADPH to participate in reductive biosynthesis and redox homeostasis is essential in cancer cells in hypoxic microenvironments24. Additionally, there are various mechanisms by which glutamine affects NADPH metabolism and redox control36. Canonical NADPH production pathways include a complex source such as the pentose phosphate pathway, serine-glycine metabolism, malic enzyme, IDH, and glutamate dehydrogenase (Supplementary Figure 5A)43. These metabolic pathways are frequently altered in cancers. One good example is mutant IDH1 glioma (Supplementary Figure 5B). It has been reported that IDH activity is the main supplier of NADPH in the human brain and glioma, as compared with the rodent brain, and it depends primarily on the pentose phosphate pathway44. NADPH production is decreased by more than 40% in gliomas with the IDH1 mutation38,45. Our previous study has also shown decreased NADPH production (and increased consumption) in IDH1-mutated malignant gliomas and its implications in 5-ALA fluorescence enhancement18. The lack of NADPH inhibits the reduction of GSSG to GSH, which is required for ROS clearance46. However, most GBMs express wild-type IDH1, thus suggesting the existence of other major metabolic reprogramming mechanisms influencing NADPH production (Supplementary Figure 5C). To our knowledge, no data on the direct measurement of NADPH in human GBM tissue have been reported. However, on the basis of our results and evidence from other cancers and in vitro experiments, GBM tissue is in a harsh microenvironment state of high ROS with an insufficient level of ROS scavengers such as GSH and NADPH. A recent study has shown compensatory upregulation of IDH1 to meet the NADPH demand in GBMs45. The effects of glutamine or glutamate on NADPH production can be conveyed directly by glutamate dehydrogenase or indirectly by glutamine-derived malate46,47. Therefore, an aberrant glutamine pathway originating from GLS2 underexpression is likely to contribute to the NADPH deficit in GBM.
There are controversies in the clinical evidence of the connection between 5-ALA fluorescence and the abnormal status of the TCA cycle in malignant gliomas18,48. However, it is accepted that reprogrammed cancer cell metabolism induces the saturation of FECH and PpIX accumulation by activating heme biosynthesis, owing to TCA cycle metabolite accumulation49. We suggest another mechanism of aberrant NADPH production and consumption for this crosstalk between the TCA cycle and the heme biosynthesis pathway. As mentioned above, high demand and an insufficient supply of NADPH is the hallmark of the core GBM area, which showed increased 5-ALA fluorescence. Additionally, those areas are under high oxidative stress with increased ROS. Human HO-1 activity has an absolute stoichiometric requirement for NADPH50. HO-1 is the rate-limiting enzyme catalyzing the oxidative degradation of cellular heme to liberate free iron. Therefore, the lack of NADPH results in the significant disturbance of the HO-1 activity, which in turn activates the heme biosynthesis pathway, owing to the absence of a feedback mechanism by heme. The result of an overactivated heme biosynthesis pathway decreases the capacity for 5-ALA metabolism. Indeed, a large amount of exogenous 5-ALA administration in cancer cells in this state results in the temporary delay of metabolism and PpIX accumulation, owing to the saturation of FECH. This hypothesis is well correlated with the clinical phenomenon of 5-ALA FGS in which fluorescence exists temporarily in cancer cells between 3 and 9 hours after 5-ALA administration and is washed out completely within a day. The rapid consumption of NADPH and GSH after 5-ALA treatment, as shown in our experiments, also supports their small capacity for metabolism.
In conclusion, although the FGS of malignant glioma using 5-ALA allows for the identification of cancer cells otherwise indistinguishable from normal tissue, there is 5-ALA fluorescence heterogeneity in GBM tissues, which reflects spatial metabolic heterogeneity. This study demonstrated an inverse correlation between the 5-ALA fluorescence intensity and GLS2 expression in intratumoral tissues of GBM. Moreover, we revealed the metabolic aspects of the mechanism, confirming NADPH as a key molecule in the process. These discoveries improve knowledge about the mechanism of metabolic heterogeneity in GBMs in relation to clinically observable phenomena. NADPH may be developed as a metabolic surrogate marker for the degree of malignancy, given a background of intratumoral heterogeneity. It is expected to contribute to non-invasive molecular diagnostic method development and therapeutic target identification.
Materials and Methods
Patient samples
We used prospectively collected samples of 5 histologically verified GBM patients who had undergone FGS for this study. For FGS, 20 mg/kg of 5-ALA (Gliolan®; Medac, Wedel, Germany) was administered orally 3–4 hours before anesthesia induction. The main procedure was carried out 5–7 hours after 5-ALA intake. Surgical resection was performed using a Leica M720 OH5 microscope (Leica, Wetzlar, Germany) equipped with an FL400 fluorescence module or a Zeiss Pentero equipped with a fluorescent 400-nm UV light and filters (Zeiss, Oberkochen, Germany). Immediately after the tumor resection, the samples were analyzed for the fluorescence intensity and were classified into 3 groups by using an in-house-devised fluorescence intensity quantification system (see below). The fluorescence intensity was measured on the basis of the red/blue (R/B) ratio in captured images and was divided into the strong (red, R/B ratio >2.0), weak (pink, 0.5 < R/B ratio ≤2.0), and none (blue, R/B ratio ≤0.5) groups. The samples were partly snap-frozen in liquid nitrogen as soon as possible and were stored at −80 °C; a portion of each sample was sent to the pathology department for histological diagnosis.
Another set of samples from 10 GBM patients were collected in the same manner for subsequent NADPH/NADP measurement. This study was performed under the approval of the Institutional Review Board of Seoul National University Hospital, and all experiments were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all patients for the usage of samples.
Fluorescence intensity quantification system
The custom-made fluorescence intensity quantification system was composed of a fluorescence excitation source and a fluorescence image detection system (Fig. 1). Three UV LEDs (405 nm, P8D240, LMH Korea, Hwasung-si, Gyeonggi-do, Korea) were used as a fluorescence excitation source, and SMPS (switched mode power supply) was used as a stable power supplier. The fluorescence image detection system consisted of an optical lens (MNL35M23, Thorlabs, Newton, NJ, USA), an optical long pass filter (450 nm, FEL0450, Thorlabs, Newton, NJ, USA), and a CMOS camera (DCC1645C, Thorlabs, Newton, NJ, USA). The captured images from the fluorescence image detection system were processed to extract fluorescence intensity.
RNA-sequencing
Before the extraction of RNA, tissues were ground in liquid nitrogen. Total RNA from ground tissues was isolated with an RNeasy Lipid Tissue Mini Kit (Cat No.74804, Qiagen, Hilden, Germany) and was subjected to DNase I treatment (Cat No. 79254, Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The RNA integrity was assessed with a Bioanalyzer system (Agilent, New Haven, CT, USA), and tumor RNAs with an RNA integrity number (RIN) ≥6.5 were subjected to RNA-seq (Supplementary Table 3). RNA-seq libraries were generated using a TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). cDNA libraries were sequenced on the HiSeq. 2000 platform (Illumina, San Diego, CA, USA) to obtain approximately 100 million paired-end reads. After removal of poor-quality raw reads containing the adaptor sequence, more than 10% of unknown bases or low-quality bases, the remaining reads were aligned by STAR51 to the human reference genome with the GENCODE reference gene annotation version 19. The sequencing reads were counted for genes by using the HTSeq-count52. Expression profiles were measured with the unit of FPKM.
Differentially expressed gene and pathway analysis
After filtering genes with low-sequencing depth across experiments whose mean read counts were less than 20, we statistically evaluated the linear trend of expression levels among the three sample groups to identify differentially expressed genes (FDR < 0.01). Specifically, the generalized linear model that included a patient and sample group as covariates was fitted and statistically tested for the linear trend of gene expression over the ordered factors of sample groups using edgeR53. In addition, we required the differentially expressed genes to show fold changes greater than two between groups, to identify genes with a clear linear trend. GSEA was performed for gene set level analysis with the likelihood ratio statistic as an input score54.
Cell culture
Human glioma cell lines T98G, U87MG and LN18 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, Gran Island, NY, USA), 100 units/ml penicillin and 100 μg/ml streptomycin sulfate (Welgene, Daegu, Korea).
Gene constructs
Human GLS2 (GenBank Accession No. NM_013267) cDNA obtained from Origene (Rockville, MD, USA) was cloned into the GenTarget’s expression lentivector. The vector contains a GFP-puromycin dual marker under a Rous sarcoma virus (RSV) promoter, and the cloned GLS2 was expressed under the enhanced constitutive elongation fator-1 alpha (EF1a) promoter (Supplementary Figure 6).
Transient transfection and lentiviral transduction
Expression lentiviral particles were produced in 293 T cells in DMEM medium according to the manufacturer’s protocol. The viruses were collected and filtered through 0.45-μm filters. Virus titers were measured via GFP-positive cells after transduction into TH1080 cells; the concentrations of the titers were 1.25 × 107 IFU/mL for Mock and 1.15 × 107 IFU/mL for GLS2.
The T98G, U87MG and LN18 cell lines were transduced with the corresponding lentiviruses for 24 hours in the presence of 4 μg/mL polybrene. The transduced cells were selected using puromycin (Cat. No. A7793, Sigma-Aldrich, St Louis, MO, USA), and the transduction efficacy was observed by fluorescence microscopy using green filters (Leica, Wetzlar, Germany). The transduction efficiency of each cell line expressing GFP was over 90%, and then the cells were analyzed using a FACS Calibur cell sorter (BD Bioscience, San Jose, CA, USA) equipped with a 530-nm filter (bandwidth, 15 nm), a 585-nm filter (bandwidth, 21 nm), and Cell-Quest software (BD Bioscience, San Jose, CA).
Quantitative real-time PCR (qPCR)
For real-time PCR, total RNA from the transduced cells was extracted using a PureLink® RNA Mini Kit (Cat No. 12183018 A. Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was performed using 1 μg of total RNA and Maxime RT premix (Cat. No. 25082, iNtRON, Seongnam, Korea). The mRNA level of GLS2 was determined using TaqMan Gene Expression Assays (Assay ID: Hs00998733_m1 for GLS2 and Hs02786624_g1 for GAPDH) and TaqMan Universal PCR MasterMix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. The relative expression was calculated using the ΔΔCT method and was normalized to GAPDH expression.
Western blot analysis
Western blot analysis was performed as described previously55. In brief, cells were lysed in PRO-PREP™ solution (Cat. No. 17081, iNtRON, Seongnam, Korea) supplemented with protease inhibitors (Roche Applied Science, Indianapolis, IN, USA). Whole cell lysates precleared by centrifugation were used for immunoblotting. Thirty micrograms of each total protein sample was loaded per lane, separated by SDS-PAGE, transferred to a nitrocellulose filter and subjected to immunoblotting with appropriate antibodies. The anti-GAPDH antibody was from Cell Signaling Technology (Cat. No. 2118, Beverly, MA, USA), the anti-glutaminase 2 antibody was from Abcam (Cat. No. ab113509, Cambridge, MA, USA), and HRP-conjugated IgGs were from Jackson ImmunoResearch Laboratories (Cat. No. 111-035-003, West Grove, PA, USA). The bands were detected using a PicoEPD Western Reagent Kit (ELPis Biotech, Daejeon, Korea), and immunoblots were visualized using a ChemiDoc XRS System (Bio-Rad, Redmond, WA, USA).
In vitro 5-ALA treatment
We purchased 5-ALA from Sigma-Aldrich (Cat. No. A7793, St. Louis, MO, USA) and dissolved it in phosphate-buffered saline (PBS, pH 7.4) to make a 500-mM stock solution. Cells were seeded into 24-well plates and then were incubated in 500 μl of medium overnight. After 5-ALA treatment with the indicated concentration for 2 hours, cells were rinsed twice with PBS and then incubated in complete medium until analysis.
Quantification of PpIX fluorescence
To quantify intracellular PpIX fluorescence, we followed a previously described method with modifications (32). Specifically, after 5-ALA treatment, cells were lysed with 60 μl of 0.2% (vol/vol) Triton X-100 on ice for 5 minutes and were centrifuged at 13,000 rpm for 5 minutes at 4 °C. Next, 5 μl of cell supernatant was transferred and subjected to the BCA Protein Assay, and the results were used to normalize the detected relative fluorescence units (RFUs) of PpIX. To detect PpIX fluorescence, 100 μl of methanolic perchloric acid, consisting of 5.6% (vol/vol) perchloric acid in 50% (vol/vol) methanol, was added to the sample and incubated at 37 °C in the dark for 15 minutes. The sample was centrifuged at 13,000 rpm for 5 minutes at 4 °C. Next, 100 μl of supernatant was transferred into 96-well black plates, and the fluorescence intensity was measured using an Infinite M200 PRO instrument (TECAN, Männedorf, Switzerland) and MagellanTM software at a fluorescence excitation wavelength of 400 ± 9 nm and an emission wavelength of 645 ± 20 nm.
To visualize PpIX fluorescence, cells were fixed with 4% paraformaldehyde in PBS at room temperature for 10 minutes, washed again with PBS twice, and mounted using Slow Fade Gold (Cat No. S36936, Invitrogen, Carlsbad, California, USA). The fluorescence images were acquired under 400-fold magnification using a Leica TCS SP8 X confocal laser scanning microscope (Leica Microsystems GmbH, Mannheim, Germany) equipped with a Leica Plan APO 0.4NA objective lens and a PMT detector. The data were analyzed using Leica LASX software (Leica Microsystems GmbH). PpIX fluorescence was excited at 405 nm, and images were collected in the red channel using a 595-nm long-pass emission filter. Confocal images of PpIX fluorescence were collected with a Leica TCS SP8 X confocal laser-scanning microscope (Leica Microsystems GmbH, Mannheim, Germany) at 1024 × 1024 pixels using a HC PL APO CS 40 × water immersion objective (1.10 NA; Leica). Excitation wavelength of 405 nm (UV diode) and peak emission wavelength of 595 nm were used to observe PPIX fluorescence.
Metabolite screening via LC-MS
Metabolite extraction was performed on LN18, T98G, U87 cells (3 × 106 cells). The harvested cells were quickly washed with cold PBS and were extracted directly with 500 μL of a cold methanol/ACN/water (5:3:2, v/v) mixture. The samples were centrifuged at 28,000 g for 20 min at 4 °C. The supernatant was collected and dried with a centrifugal vacuum evaporator (Vision, Seoul, Korea), and the pellets were collected for protein quantification with a BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA). The dried extracts were dissolved in 20 μL of a mixture of acetonitrile/water (1:1, v/v) and then were transferred into glass vials with micro-inserts for small-volume injections.
Analysis was carried out using an Agilent 1100 Series liquid chromatography system, and an auto-sampler set at 4 °C (Agilent, Santa Clara, California, USA). Chromatographic separation was performed by injection of 4 μl of the sample on a ZIC-pHILIC Polymeric Beads Peek column (150 × 2.1 mm, 5 μm, Merck kGaA, Darmstadt, Germany) at 35 °C and a 0.15 mL/min flow rate, using 10 mM ammonium carbonate (pH = 9.0) in distilled water as mobile phase A and acetonitrile (ACN) as mobile phase B. The linear gradient was as follows: 80% B at 0 minutes, 35% B at 10 minutes, 5% B at 12 minutes, 5% B at 25 minutes, 80% B at 25.1 minutes, and 80% B at 35 minutes.
During the LC-MS experiments, mass spectra were acquired in negative ion mode using an API 2000 Mass Spectrometer (AB/SCIEX, Framingham, MA, USA) equipped with an electrospray ionization (ESI) source. The parameters of the ESI source operation were as follows: −4.5 kV of ion spray voltage; curtain gas (nitrogen), ion source gas 1 (nitrogen), and ion source gas 2 (nitrogen) pressures at 15, 70, and 90, respectively; temperature of the heater (turbo) gas at 550 °C. For the detection, MRM was performed with the m/z values of the precursor and fragment ions as indicated in the Supplementary Table 4, which were established with standard compounds. MRM was performed and controlled with Analyst 1.6 Software. Chromatographic peaks were identified by comparing the retention times, m/z values of the parent and daughter ions with those in the HMDB (www.hmdb.ca), METLIN (https://metlin.scripps.edu) and Massbank (www.massbank.jp) databases and those of the standards.
NADPH and NADP measurement
The ratios of NADPH/NADP + in the GBM cell lines were calculated using the NADP/NADPH Quantification Kit (BioVision, Mountain View, CA, USA) according to the manufacturer’s instructions. In brief, cell pellets were lysed in extraction buffer with vigorous shaking in an automatic homogenizer (Automill™; Tokken, Chiba, Japan). After centrifugation, the supernatant was filtered through a 10-kDa spin column (Cat. No. ab113509, Abcam, Cambridge, MA, USA) to remove NADPH-consuming enzymes. To detect the total NADPH/NADP + (NADPt), 50 μl of the extracted supernatant was placed into 96-well plates. To detect NADPH only, heat was applied to the supernatant sample at 60°C for 30 min in a heating block (Throughout this process, all NADP + was decomposed, whereas the NADPH remained intact). Then, 50 μl of NADP-decomposed NADPH samples were added to 96-well plates. Next, 100 μl of the cycling mixture was also added to each well, and the plate was incubated for 5 min to convert NADP to NADPH. Ten microliters of NADPH developer was continuously added, and the plates were incubated for 1 hour. Finally, the plates were read at 450 nm by using a microplate reader (Bio-Rad, Hercules, CA, USA).
Statistical Analyses
All statistical analyses were performed using a free statistical software package, R (version 3.0.2; http://www.r-project.org/), and IBM SPSS Statistics software (version 21; SPSS, Chicago, IL, USA). The data were expressed as the means ± SEM. Student’s t-test was used for the comparison of mean values. Statistical significance was accepted at a level of p < 0.05 (*).
Electronic supplementary material
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2015-R1D1A1A09057171) in Korea, and the Seoul National University Hospital Research Fund (3020150350). We like to thank Min Jung Park, R.N. for the support in summarization of clinical data and financial management of experiments. We also like to thank Ms. Mi-Jin Jung of Biomedical Illustration & Design Company for her artwork used in this manuscript.
Author Contributions
S.K., and J.E.K., performed the in vitro experiments and wrote the paper. Y.H.K. wrote the paper. T.H. analyzed the genomic data, and wrote the paper. J.-Y.S., and J.-I.K. analyzed the genomic data. S.K.K. performed fluorescence analysis of samples. W.J.X. performed the metabolite analysis. H.C. and H.-C.K. invented the fluorescence measurement system. H.R.C., A.C., T.C., Y.S., and Y.-S.D. performed the in vitro experiments. J.W.K., and D.G.K. reviewed the paper. S.-H.P. performed the histological study. H.K., and S.H.C. contributed to the analyses and review the paper. S.P., and S.-H.L. supervised the study. C.-K.P. developed the concept of the study, supervised the work, and wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Sojin Kim, Ja Eun Kim, Yong Hwy Kim and Taeyoung Hwang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12557-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet Oncology. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Brown TJ, et al. Association of the Extent of Resection With Survival in Glioblastoma: A Systematic Review and Meta-analysis. JAMA oncology. 2016;2:1460–1469. doi: 10.1001/jamaoncol.2016.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stummer W. Commentary: Combining 5-Aminolevulinic Acid Fluorescence and Intraoperative Magnetic Resonance Imaging in Glioblastoma Surgery: A Histology-Based Evaluation. Neurosurgery. 2016;78:484–486. doi: 10.1227/NEU.0000000000001107. [DOI] [PubMed] [Google Scholar]
- 4.Tonn JC, Stummer W. Fluorescence-guided resection of malignant gliomas using 5-aminolevulinic acid: practical use, risks, and pitfalls. Clin Neurosurg. 2008;55:20–26. [PubMed] [Google Scholar]
- 5.Stummer W, et al. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. The Lancet Oncology. 2006;7:392–401. doi: 10.1016/S1470-2045(06)70665-9. [DOI] [PubMed] [Google Scholar]
- 6.Mansouri A, et al. The role of 5-aminolevulinic acid in enhancing surgery for high-grade glioma, its current boundaries, and future perspectives: A systematic review. Cancer. 2016;122:2469–2478. doi: 10.1002/cncr.30088. [DOI] [PubMed] [Google Scholar]
- 7.Stummer W, et al. In vitro and in vivo porphyrin accumulation by C6 glioma cells after exposure to 5-aminolevulinic acid. J Photochem Photobiol B. 1998;45:160–169. doi: 10.1016/S1011-1344(98)00176-6. [DOI] [PubMed] [Google Scholar]
- 8.Stummer, W. et al. Intraoperative detection of malignant gliomas by 5-aminolevulinic acid-induced porphyrin fluorescence. Neurosurgery 42, 518–525; discussion 525–516 (1998). [DOI] [PubMed]
- 9.Ferraro N, et al. The role of 5-aminolevulinic acid in brain tumor surgery: a systematic review. Neurosurgical review. 2016;39:545–555. doi: 10.1007/s10143-015-0695-2. [DOI] [PubMed] [Google Scholar]
- 10.Yang X, Palasuberniam P, Kraus D, Chen B. Aminolevulinic Acid-Based Tumor Detection and Therapy: Molecular Mechanisms and Strategies for Enhancement. International journal of molecular sciences. 2015;16:25865–25880. doi: 10.3390/ijms161025865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng L, et al. Silencing of ferrochelatase enhances 5-aminolevulinic acid-based fluorescence and photodynamic therapy efficacy. Br J Cancer. 2011;104:798–807. doi: 10.1038/bjc.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao SG, et al. Increased expression of ABCB6 enhances protoporphyrin IX accumulation and photodynamic effect in human glioma. Annals of surgical oncology. 2013;20:4379–4388. doi: 10.1245/s10434-011-2201-6. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto K, et al. Effects of plasma membrane ABCB6 on 5-aminolevulinic acid (ALA)-induced porphyrin accumulation in vitro: tumor cell response to hypoxia. Photodiagnosis and photodynamic therapy. 2015;12:45–51. doi: 10.1016/j.pdpdt.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Tsuchida M, Emi Y, Kida Y, Sakaguchi M. Human ABC transporter isoform B6 (ABCB6) localizes primarily in the Golgi apparatus. Biochemical and biophysical research communications. 2008;369:369–375. doi: 10.1016/j.bbrc.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 15.Paterson JK, et al. Human ABCB6 localizes to both the outer mitochondrial membrane and the plasma membrane. Biochemistry. 2007;46:9443–9452. doi: 10.1021/bi700015m. [DOI] [PubMed] [Google Scholar]
- 16.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell metabolism. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 18.Kim JE, et al. Mechanism for enhanced 5-aminolevulinic acid fluorescence in isocitrate dehydrogenase 1 mutant malignant gliomas. Oncotarget. 2015;6:20266–20277. doi: 10.18632/oncotarget.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frezza C, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477:225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 20.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch DR. Tumor Heterogeneity–A ‘Contemporary Concept’ Founded on Historical Insights and Predictions. Cancer research. 2016;76:4–6. doi: 10.1158/0008-5472.CAN-15-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta D, Pratx G. Imaging metabolic heterogeneity in cancer. Molecular cancer. 2016;15:4. doi: 10.1186/s12943-015-0481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nature cell biology. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stummer, W. et al. 5-Aminolevulinic acid-derived tumor fluorescence: the diagnostic accuracy of visible fluorescence qualities as corroborated by spectrometry and histology and postoperative imaging. Neurosurgery74, 310–319, discussion 319–320 (2014). [DOI] [PMC free article] [PubMed]
- 26.Abraham NG, Kappas A. Pharmacological and clinical aspects of heme oxygenase. Pharmacological reviews. 2008;60:79–127. doi: 10.1124/pr.107.07104. [DOI] [PubMed] [Google Scholar]
- 27.Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochimica et biophysica acta. 2006;1763:723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda H, Casas A, Batlle A. Aminolevulinic acid: from its unique biological function to its star role in photodynamic therapy. The international journal of biochemistry & cell biology. 2005;37:272–276. doi: 10.1016/j.biocel.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 29.Ishizuka M, et al. Novel development of 5-aminolevurinic acid (ALA) in cancer diagnoses and therapy. International immunopharmacology. 2011;11:358–365. doi: 10.1016/j.intimp.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 30.Colditz MJ, Leyen K, Jeffree RL. Aminolevulinic acid (ALA)-protoporphyrin IX fluorescence guided tumour resection. Part 2: theoretical, biochemical and practical aspects. Journal of clinical neuroscience: official journal of the Neurosurgical Society of Australasia. 2012;19:1611–1616. doi: 10.1016/j.jocn.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Abraham NG, Lin JH, Dunn MW, Schwartzman ML. Presence of heme oxygenase and NADPH cytochrome P-450 (c) reductase in human corneal epithelium. Investigative ophthalmology & visual science. 1987;28:1464–1472. [PubMed] [Google Scholar]
- 32.Suzuki S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu W, et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang L, et al. Knock-down of glutaminase 2 expression decreases glutathione, NADH, and sensitizes cervical cancer to ionizing radiation. Biochimica et biophysica acta. 2013;1833:2996–3005. doi: 10.1016/j.bbamcr.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Szeliga M, Sidoryk M, Matyja E, Kowalczyk P, Albrecht J. Lack of expression of the liver-type glutaminase (LGA) mRNA in human malignant gliomas. Neuroscience letters. 2005;374:171–173. doi: 10.1016/j.neulet.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 36.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szeliga M, Bogacinska-Karas M, Kuzmicz K, Rola R, Albrecht J. Downregulation of GLS2 in glioblastoma cells is related to DNA hypermethylation but not to the p53 status. Molecular carcinogenesis. 2016;55:1309–1316. doi: 10.1002/mc.22372. [DOI] [PubMed] [Google Scholar]
- 38.Bleeker FE, et al. The prognostic IDH1(R132) mutation is associated with reduced NADP+ -dependent IDH activity in glioblastoma. Acta neuropathologica. 2010;119:487–494. doi: 10.1007/s00401-010-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szeliga M, Albrecht J. Opposing roles of glutaminase isoforms in determining glioblastoma cell phenotype. Neurochemistry international. 2015;88:6–9. doi: 10.1016/j.neuint.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Szeliga M, et al. Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia. 2009;57:1014–1023. doi: 10.1002/glia.20825. [DOI] [PubMed] [Google Scholar]
- 41.Krall AS, Christofk HR. Rethinking glutamine addiction. Nature cell biology. 2015;17:1515–1517. doi: 10.1038/ncb3278. [DOI] [PubMed] [Google Scholar]
- 42.Marcus HJ, Carpenter KL, Price SJ, Hutchinson PJ. In vivo assessment of high-grade glioma biochemistry using microdialysis: a study of energy-related molecules, growth factors and cytokines. Journal of neuro-oncology. 2010;97:11–23. doi: 10.1007/s11060-009-9990-5. [DOI] [PubMed] [Google Scholar]
- 43.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Atai NA, et al. Differential activity of NADPH-producing dehydrogenases renders rodents unsuitable models to study IDH1R132 mutation effects in human glioblastoma. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2011;59:489–503. doi: 10.1369/0022155411400606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahl DR, et al. Glioblastoma Therapy Can Be Augmented by Targeting IDH1-Mediated NADPH Biosynthesis. Cancer research. 2017;77:960–970. doi: 10.1158/0008-5472.CAN-16-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nature reviews Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+ -dependent malic enzyme. The Journal of biological chemistry. 1984;259:6215–6221. [PubMed] [Google Scholar]
- 48.Jaber, M. et al. The Value of 5-Aminolevulinic Acid in Low-grade Gliomas and High-grade Gliomas Lacking Glioblastoma Imaging Features: An Analysis Based on Fluorescence, Magnetic Resonance Imaging, 18F-Fluoroethyl Tyrosine Positron Emission Tomography, and Tumor Molecular Factors. Neurosurgery78, 401–411; discussion 411 (2016). [DOI] [PMC free article] [PubMed]
- 49.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenhunen R, Marver HS, Schmid R. The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proceedings of the National Academy of Sciences of the United States of America. 1968;61:748–755. doi: 10.1073/pnas.61.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi F. 5-ALA Sonodynamic Therapy for Malignant Glioma. Gan to kagaku ryoho Cancer & chemotherapy. 2016;43:702–705. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.