Abstract
To the astrobiologist, Enceladus offers easy access to a potential subsurface biosphere via the intermediacy of a plume of water emerging directly into space. A direct question follows: If we were to collect a sample of this plume, what in that sample, through its presence or its absence, would suggest the presence and/or absence of life in this exotic locale? This question is, of course, relevant for life detection in any aqueous lagoon that we might be able to sample. This manuscript reviews physical chemical constraints that must be met by a genetic polymer for it to support Darwinism, a process believed to be required for a chemical system to generate properties that we value in biology. We propose that the most important of these is a repeating backbone charge; a Darwinian genetic biopolymer must be a “polyelectrolyte.” Relevant to mission design, such biopolymers are especially easy to recover and concentrate from aqueous mixtures for detection, simply by washing the aqueous mixtures across a polycharged support. Several device architectures are described to ensure that, once captured, the biopolymer meets two other requirements for Darwinism, homochirality and a small building block “alphabet.” This approach is compared and contrasted with alternative biomolecule detection approaches that seek homochirality and constrained alphabets in non-encoded biopolymers. This discussion is set within a model for the history of the terran biosphere, identifying points in that natural history where these alternative approaches would have failed to detect terran life. Key Words: Enceladus—Life detection—Europa—Icy moon—Biosignatures—Polyelectrolyte theory of the gene. Astrobiology 17, 840–851.
1. Introduction
To the astrobiologist, Enceladus offers a “gimmie,” a plume of water emerging from a subsurface salty, organics-laced ocean, in contact with a rocky core (Porco et al., 2006), on an alien object that requires no landing to collect. This makes especially pressing the question: What should we seek in this plume by way of molecules that might indicate whether that ocean contains life of some kind (Lovelock, 1965; Davila and McKay, 2014)? Alternatively, we might ask what molecules would, through their absence, suggest the presence or absence of life in this ocean? These questions can be generalized to cover life in any aqueous environment, of which the Solar System is known to hold many [e.g. Europa and asteroids as well as Earth and likely Mars (Marion et al., 2012)]. Extrasolar systems may also soon be known to hold many such lagoons (Seager et al., 2012). From possible answers to these questions, we may constrain mission architectures to collect samples from the plume (or other aqueous samples).
Here, we must recognize that life on Enceladus [or in other extraterran water lagoons, or even in exotic locales on Earth (Cleland and Copley, 2005)] may have originated by a process very different from the process by which life emerged on Earth. Alternatively, alien life may have originated in the same way as terran life but did not evolve as fast or as far as the terran biosphere. For example, if we accept the “RNA world” (Gilbert, 1986) as part of a model for natural history on Earth, visits to the terran biosphere on early Earth would not have found encoded proteins, and possibly not straight-chain fatty acids (Benner et al., 1989). The same might be so on Europa or Enceladus today, even if life there began in the same way as life on Earth, if the evolution of molecular structures in those alien biospheres has not progressed as fast or as far as the terran biosphere, or has progressed in different directions.
More generally, we must be concerned about the possibility that even if terran-similar proteins, nucleic acids, and lipids are found in the biosphere that we encounter on Enceladus, Europa, or any other extraterrestrial water lagoon, they might be built from components different from those found in the analogous biopolymers today on Earth. Several alternative sets of components for proteins and nucleic acids were recently discussed by Rezzonico (2014), together with the potential use of nanopore-based devices to detect them (Sarathy et al., 2017).
The most forthright answers to any or all these questions, of course, are “We do not know.” However, experiments in the area of synthetic biology over the past three decades offer some suggestions for how we might proceed.
2. Core Needs for Biology
A decade ago, NASA commissioned the Space Science Board of the National Academies of Science to survey what was known (and what might be speculated) about the limits of organic life in the Solar System (Baross et al., 2007). The survey did not constrain our search for alien biospheres as much as we perhaps would have liked. But it did offer some suggestions.
For example, by theory as secure as the second law of thermodynamics, a biosphere requires an environment that is at thermodynamic disequilibrium, disequilibrium that is sometimes called “a source of free energy.” The Enceladus plume, for example, comes from a tens-of-kilometers-thick global ocean that sits beneath an ice shell that could be tens of kilometers thick (Thomas et al., 2015). Therefore, it seems unlikely that photosynthesis stands at the bottom of the food chain for a hypothetical biosphere on Enceladus. However, like all bodies containing rocky material, Enceladus incorporated radioactive elements as it accreted. The energy from the decay of these elements undoubtedly contributed to a subsurface disequilibrium in its early history, as on Earth. Today, tidal forces are more important sources of energy for many of these locales. In either case, it seems reasonable to assume that the thermodynamic requirement for a biosphere is not a serious constraint for this or many other Solar System locales.
The report suggested another constraint: a solvent. Chemical reactions can occur in solid phases; a field of organic chemistry (“mechanochemistry”) (James and Friščić, 2013) studies these. Further, reacting species can diffuse within solid matrices (e.g., within a frozen ice) to encounter and react with each other. Reactions of this type are especially conceivable in an environment encountering high-energy photons or cosmic particles, or both.
However, reactions in the solid phase are orders of magnitude slower than reactions in solution. For example, in water at standard temperature and pressure, molecules encounter each other with rate constants on the order of 106 M−2. This means that an enzyme that metabolizes glucose will encounter a glucose molecule 1000 times per second, if glucose is present at millimolar concentration (i.e., 0.18 grams of glucose per liter, a reasonable physiological concentration). This allows life in water to live on the millisecond timescale, with ca. 100 million opportunities for a reaction for each enzyme in a living bacterial cell in the time between cell divisions. Thus, the report recommended a fluid rather than a solid as a host for a biosphere.
The report considered liquid solvents other than water, as many such non-aqueous liquids are found throughout our solar system. Titan, for example, has large bodies of surface liquid methane at ∼95 K. Saturn itself has fluids, including supercritical dihydrogen-helium mixtures that are found in regions of this and other gas giants.
However, water remains the preferred solvent as we explore the Solar System looking for life. In part, this is because water is the liquid most likely to host a biosphere familiar to us. Further, a liquid supports intermolecular reactions between molecules only if those molecules are dissolved, and the high temperature of the liquid range of water makes it an excellent solvent.
Thus, as we survey the likelihood of biology in other liquids, it is appropriate to ask: What molecules do they dissolve? For example, efforts by Malaska and Hodyss (2014) and McLendon et al. (2015) to find molecules that might dissolve in methane at 95 K have been largely unsuccessful, especially for genetic molecules (see below).
The report offered a third constraint: Darwinism. Darwinism is believed to be the only way that organic matter can organize itself to give, if not “life” (which the community has long had difficulty defining), then at least the properties that we value in the life that we know. Indeed, a quarter century ago, a panel of NASA advisers suggested that life was “a self-sustained chemical system capable of undergoing Darwinian evolution” (Joyce, 1994).
This definition has been the target of some curious criticisms, especially with respect to its value in space exploration. For example, Chyba and Phillips (2002) allege that although “this definition may be sufficiently broad so as to be universal, operationally it is of little utility.” “How long do we wait to determine if a candidate entity is ‘capable of undergoing Darwinian evolution’?” they asked, suggesting that this was a compelling criticism of the definition or its value in space exploration.
It is not. One can, for example, from the structure of a pair of rabbits, infer that the system is capable of undergoing Darwinian evolution without needing to wait for that event to transpire. Thus, this universal theory-definition of life is important because it drives us to ask what structures, which are observable, are necessary for Darwinism.
In its most general form, Darwinism requires a molecular system that can reproduce, with imperfections, where those imperfections are themselves reproducible. Further, Darwinism requires that imperfections in replication not be prospective with respect to fitness. The processes that create variation through imperfect reproduction cannot not be presumed to predict future genetic needs.
Darwinism is presumed to be the mechanism that delivers a key valued property in life: the ability to prevent the devolution of organic matter into increasingly complex tars. Organic matter, if given energy and left on its own, devolves intrinsically. Indeed, the conversion of organized matter that is a characteristic of life into increasingly “tarry” mixtures is one of the most common observations in organic chemistry. It is seen in kitchens where an oven has been left on too long, in first-year organic chemistry laboratories where students have been insufficiently attentive, and in petroleum deposits where material that used to be quite alive has devolved in a process that will end as anthracite coal.
Indeed, the organic material in carbonaceous chondrites appears to be the consequence of exactly this kind of devolution, perhaps by high-energy particles, converting material that originally had the atomic and molecular composition of the cold part of the Universe into material with more chemical diversity, more chemical cross-links, fewer hydrogen atoms, and lower solubility. Notwithstanding the amount of (and interest in) the dissolvable organic molecular repertoire of carbonaceous chondrites, the bulk of the carbon in meteorites is seemingly useless diversity, diverse to the point where no single high-molecular-weight molecular structure is likely to be found in the sample more than once.
3. Darwinism and Biology
Access to Darwinism appears to allow organic material to escape devolution. For example, on Earth, no matter where we look, if a locale holds a thermodynamic disequilibrium and a solvent, Darwinism has apparently found a way to populate that locale with what we call “life” (Navarro-González et al., 2003). Indeed, Darwinism appears to have been able to populate on Earth all such locales with a life-form whose core molecular biology is not fundamentally different from that found in humans. The genetic molecule is, as far as we know, always DNA with four nucleotides, the ribosomes and the messenger molecules are always RNA with four nucleotides, and the encoded proteins all have the same 20 amino acids. A role for carbohydrates is always somewhere in metabolism.
What molecular structures are required to support Darwinism? The minimal properties are axiomatic. A Darwinian system must be able to (a) generate replicates, where (b) those replicates are imperfect, where (c) those imperfections are themselves replicable.
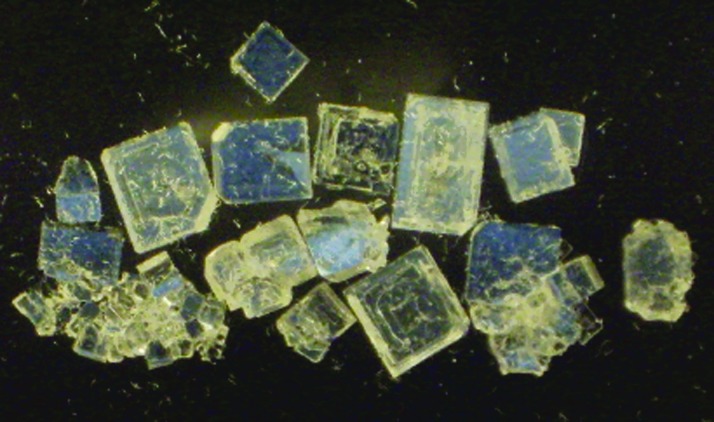
Replication of molecular order is easy. An especially common example is crystallization. For example, a crystal of sodium chlorate, once it has grown, can be powdered and used to seed the growth of more crystals of sodium chlorate (Fig. 1). This is replication.
FIG. 1.
Sodium chlorate crystallizes in two forms, similar in structure but with different chiralities; the two forms are mirror images of each other. This chirality is a single bit of information held in a crystal, and that information can be passed from a mother crystal to a daughter crystal grown from a seed. The bluer crystals in this image, created using two polarizing filters, are “left-handed” crystals; the browner crystals are “right-handed” crystals. Even though the daughter crystals seeded from the mother crystals retain the hand, they do not retain the defects in the mother crystal.
The replication of these crystals (like all crystals) is prone to error. The crystals themselves have defects, where the atoms fail to arrange themselves within the ideal crystalline lattice. The information content in a single crystal arising from those errors can be large. Thus, in any of the crystals shown in Fig. 1, specifying all the defects can easily require more information than that in the human genome. Indeed, every crystal can be said to be “different.” Further, some patterns of defects might even help crystals survive (e.g., by resisting dissolution) better than others, a basis for natural selection between crystals. However, the act of recrystallization embodies no mechanism to pass those errors and that information on, let alone that individuality, to the next generation of crystals.
The same can be said for fire and other counterexamples often offered in the “laundry list” definitions of life that are often proposed (Koshland, 2002). Fire grows, consumes food, creates descendants, and displays many of the other features on those lists. Again, the replicates are imperfect. But again, those imperfections themselves cannot be replicated as fire generates its “descendants.”
Indeed, fire as a “natural kind” has many different embodiments. That is, a fire centered at coordinates 45°12′00"N, 135°54′00"W has no microscopic similarity to a fire centered at coordinates 45°12′30"N, 135°54′30"W. What they are burning is different, the molecules being transformed at the two locations are different, and the arrangements in local space of those molecules are different. We call both “fire” simply because we, as observers, are unable to observe those differences in time and space in sufficient detail to distinguish the two.
This example captures one of the issues within the second law. “State 1” is said to have higher entropy (and therefore is more likely to emerge during time evolution) than “State 2” if it has more microstates. However, the requirement that microstates be aggregated into something called “State 1” is a function of the aggregator, not the aggregated; the requirement comes from the inability of the aggregator to distinguish microstates. Were the aggregator able to make that distinction, then State 1 would be divided into State 1′, State 1", and so on. And its entropy, and likelihood, would decrease. This problem is worth attention, but not here.
From an experimental perspective, sodium chlorate may be viewed as being more “Darwinian” than fire. Sodium chlorate crystals actually do come in two macroscopically distinguishable states, a left-handed state and a right-handed state (Fig. 1). This is a “bit” of information that can be transferred from a parent crystal to its daughter crystals. Thus, a left-handed crystal, if powdered, will seed the growth of more left-handled crystals.
This replication can, however, have errors; every now and then the daughter crystal will be right-handed. And this error is replicable; the right-handed mutant daughter crystal will seed (except for subsequent errors) the growth of (mostly) right-handed granddaughter crystals. Indeed, if left-handed sodium chlorate is placed in a chiral environment where the right-handed species is more stable, the left-handed system will evolve to give the right-handed sodium chlorate system. And if the environment is then changed to prefer sodium chlorate in its left-handed form, the system will evolve back.
4. Genetic Biopolymers for Darwinism
What molecular structures are required to support Darwinism that involves more than a single bit of information? Schrödinger addressed this question by proposing the existence of a macromolecule that could give an “aperiodic crystal” (Schrödinger, 1943). Despite not knowing the structure of DNA, Schrödinger's view ended up being correct with respect to the structure of the Watson-Crick A:T and G:C pairs. Because of their similar sizes (due to the size complementarity of the pairing, big purines pairing with small pyrimidines), the A:T, T:A, G:C, and C:G pairs all fit into a size-regular double helix, notwithstanding their carrying different information. This allows the double helix of DNA to be crystal-like. The formation of the double helix can thus benefit from the reliability of a phase transition, just like the crystallization of sodium chlorate. The size uniformity of the information-holding units in the genetic biopolymer, regardless of what information they hold, is a structural feature that allows DNA to support Darwinism.
However, other molecular features are also needed to support Darwinism. Replication with errors, where those errors are subsequently themselves replicable, requires that the physical properties of a genetic biopolymer also be largely insensitive to change in the encoded information. If the physical properties of a biopolymer were to change dramatically as a consequence of changes in encoding structure, those changes could easily defeat the mechanisms for biopolymer replication.
What physical changes might we consider? At the lowest level, solubility is one. If a change in the sequence of information-encoding units in DNA were to change the solubility of the encoding DNA molecule, replication would fail at the next cycle. Darwinism would cease. Therefore, as important as Schrödinger's structural constancy is with respect to changes in encoded information, constancy in solubility in a biopolymer system is essential to support Darwinism.
Constancy of physical behavior upon changes in molecular structure is not a common feature of molecular systems (Benner, 2004). In most biopolymeric systems, even very small changes in molecular structure can lead to large changes in properties. In proteins, for example, even a single amino acid replacement can cause a protein to become insoluble in water (such a change causes the disease sickle cell anemia) (Pauling et al., 1949). For this reason, most biopolymeric systems, including proteins, cannot serve as versatile genetic molecules (Lee et al., 1996).
What molecular feature allows DNA, our Darwinian genetic molecule, to suffer nucleotide replacements, and therefore change its genetic information without materially changing its physical properties? We have suggested that the repeating negative charge in the backbone of the DNA molecule is this molecular feature (Benner and Hutter, 2002).
Operating in water, the repeating negative charge carried by the backbone phosphates in terran DNA and RNA is certainly important for many reasons, solubility in water being one (Westheimer, 1987). However, relevant to this discussion, the repeating backbone charge appears to be necessary to allow these molecules to support Darwinism. The repeating backbone charge so dominates the physical properties of DNA/RNA that any replacement of the nucleobase, a replacement that changes the genetic information of the molecule, creates only a small perturbation of those properties. Because of this repeating charge, essentially all DNA sequences, regardless of the information that they carry, dissolve in water, precipitate from ethanol, form duplexes following Watson-Crick pairing rules, and interact with DNA polymerases, more or less in the same way, regardless of their sequences and the consequent genetic information that those sequences hold.
Interestingly, natural nucleic acids offer an “exception that proves the rule.” For example, an RNA molecule with many consecutive guanines forms non-Watson-Crick structures; some are insoluble (Kim et al., 1991). This interferes with the ability of RNA to support Darwinism, and lab evolution experiments can detect examples where RNA falls into a G-rich non-Darwinian “trap” in its sequence space. The properties of DNA are less influenced by G-richness; this may be a reason why DNA replaced RNA in natural history on Earth. However, these are exceptions amid a remarkable constancy of physical behavior that the polyelectrolyte molecular structure allows.
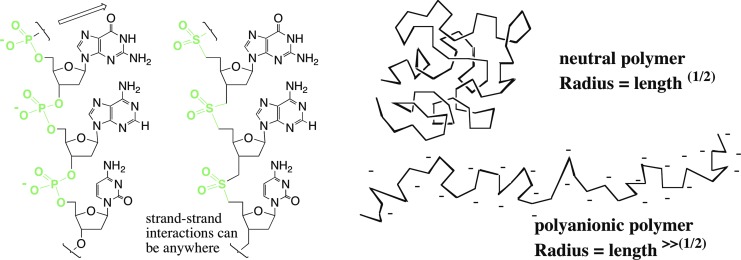
These considerations led to the “polyelectrolyte theory of the gene” (Fig. 2) (Navarro-González et al., 2003; Benner, 2004). This theory holds that any linear genetic biopolymer operating in water anywhere in the Cosmos, to support Darwinism, must have a repeating charge, either negative (as on Earth) or positive (Linkletter et al., 2001; Reddy and Bruice, 2003). According to this theory, these are the only biopolymeric systems that can robustly support Darwinian evolution, as these are the only systems that can change their structure by rearranging their building blocks without dramatically changing their overall physical behavior. Thus, life universally will have a polyelectrolyte biopolymer at its core, if it has any biopolymer at all.
FIG. 2.
The polyelectrolyte theory of the gene notes that a repeating charge in the backbone causes strand-strand interactions to be as far away from the backbone as possible; this is key to Watson-Crick pairing in DNA and RNA (Hoogsteen, 1962). Further, they cause the polymer to unfold, essential for a genetic molecule to template the synthesis of its complement. Synthetic biologists have made many variants of DNA and RNA where the repeating negatively charged phosphate groups in the backbone of natural DNA are replaced by uncharged linking groups, here exemplified by the neutral uncharged sulfone groups. This replacement generally causes the system to lose its ability to support Darwinism. Such outcomes suggest that a universal biopolymer able to support Darwinism in water must have a repeating charge in its backbone; it must be a polyelectrolyte.
Supporting this theory are many efforts by synthetic biologists to create non-ionic analogues of DNA and RNA (Miller et al., 1981). For example, replacing the anionic phosphate diester linker with the uncharged dimethylenesulfone linker generates DNA and RNA analogues that are approximately the same shape as their phosphate analogue (Fig. 2) (Huang et al., 1991). Short sulfone-linked DNA analogues (SNAs) display molecular recognition of the Watson-Crick type (Roughton et al., 1995). In longer oligosulfones, however, the loss of the repeating charge damages rule-based molecular recognition (Richert et al., 1996). Further, SNAs differing by only one nucleobase displayed different solubility, aggregation, folding, and reactivity (Steinbeck and Richert, 1998; Eschgfaeller et al., 2003).
These results suggest three hypotheses for why charged phosphate linkages are important to molecular recognition in DNA. First, the repeating charges in the backbone force interstrand interactions away from the backbone, causing strands to contact at the Watson-Crick edge of the heterocycles (Fig. 2). Without the polyanionic backbone, interstrand contacts can be anywhere. Next, the repeating charges in the backbone keep DNA strands from folding. A flexible polyanion is more likely to adopt an extended conformation suitable for templating than a flexible neutral polymer, which is more likely to fold (Flory, 1953). However, according to the polyelectrolyte theory, the repeating charges are most important because they dominate so much the physical properties of a biopolymer that they hardly change as the information content of the biopolymer changes.
5. What Does This Mean for Finding Life in Enceladus, Europa, and Other Aqueous Lagoons?
This theory offers a molecular feature that should be sought in a mission to detect life in water emerging from Enceladus or, indeed, any other aqueous sample: We must look for polymers that have a repeating charge in their backbones. We add to this the Schrödinger criterion, that these biopolymers must be able to support aperiodic crystal structures. The second criterion requires that the biopolymers be built from a small set of molecular building blocks, where the building blocks all have the same chirality (they are “homochiral”) and have similar sizes. Both features are, we suggest, essential to support Darwinism, which is presumably essential for biology.
We expect such genetic polymers to arise rarely if ever without Darwinism, and essentially never in organic material devolving to give tar. Further, even in the presence of Darwinism, they will be sparse in a sample, just as they are in a sample of life on Earth.
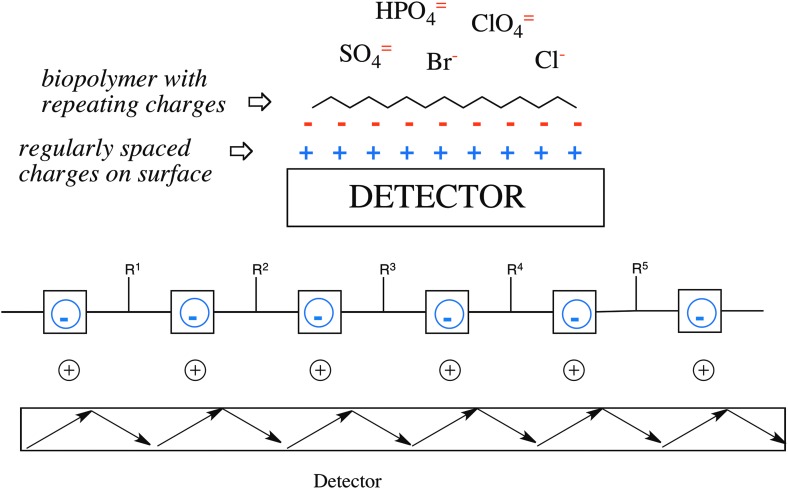
We now experience an example of good fortune. Polymers that meet the Schrödinger criterion are not, in general, easy to concentrate from a dilute aqueous solution. However, polymers having repeating charges are. A solid support presenting regularly spaced positive charges will strongly adsorb from that solution a biopolymer with repeating negative charges. Further, that adsorption is preferential over the adsorption of simple anions such as chloride, sulfate, or phosphate (Fig. 3). Conversely, a solid support presenting regularly spaced negative charges will strongly adsorb from aqueous solution a biopolymer with repeating positive charges, here preferentially over the adsorption of simple cations, such as sodium, potassium, or ammonium. This generates an easily implementable architecture to detect this universal signature of Darwinism, even if it is present in a sample only in small amounts.
FIG. 3.
A polyanion binds to a surface detector that is covered with positive charges in preference to negatively charged salt complements. Binding can be detected by total internal reflection spectroscopy.
To detect genetic biopolymers universally, both charges must be considered, as origins and subsequent evolution may have been different in encountered alien environments. It is possible that natural history on Enceladus, for example, proceeded differently than on Earth, generating a genetic biopolymer with repeating positive charges (e.g., repeating linking ammonium groups) rather than repeating negative charges (on Earth, the repeating phosphates).
Further, the sorts of surfaces able to concentrate for detection biopolymers that can support Darwinism are likely robust to space travel. Conceptually, they can be as simple as a waveguide with charges imprinted on its surface. Passing fluid from Enceladus (for example) containing the (hoped-for) Darwinian polymer over that surface will concentrate the Darwinian biopolymer molecules on that surface. The adsorption of these can be detected by total internal reflection spectroscopy with no moving parts; no reagents need be delivered. Once concentrated, the biopolymer can be recovered and analyzed for Schrödinger-type structural regularity.
Would the sensitivity of the system be adequate to detect the genetic biopolymer in the Enceladus plume or a sample of Europa? Here, the answer requires estimates about the density of putative life in the respective oceans. On Earth, the oceans contain perhaps 109 to 1010 genetic molecules per liter, perhaps 104 to 105 building blocks in length (Steward et al., 1996). Carolyn Porco (Porco et al., 2017) has provided some estimates, suggesting that “bubble scrubbing” might concentrate this material substantially. It may be possible to assemble this liter of material using a lander collecting for more than a day (Porco et al., 2017). Even if the same density is seen on Enceladus, and if the genetic polyelectrolyte is captured from a liter of sample, surface plasmon resonance detection alone would likely be too insensitive to detect the polyelectrolyte Darwinian biopolymer there.
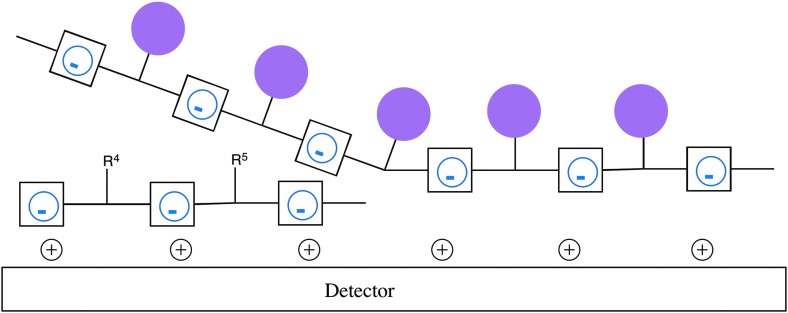
However, the ease with which Darwinian biopolymers are concentrated by Coulombic interactions supports alternative detection architectures that are more sensitive. For example, Fig. 4 shows an architecture where a fluorescently labeled strand is displaced by the adsorption of the hypothetical genetic polyelectrolyte from Enceladus. The loss of fluorescent molecules from the surface of the waveguide, or their capture on a separate detection element, also gives a signal. The detection of the movement of fluorescent species is, of course, considerably more sensitive than a simple surface plasmon resonance detection architecture. Further, electroactive moieties built into the displaced polymers can make detection of ca. 103 displaced molecules possible. This would support detection of Darwinian biopolymers in enceladan liquids at bioloads perhaps one million times lower than in an ocean on Earth.
FIG. 4.
A detector with a repeating positive charge on the surface could detect anionic Darwinian biopolymers in the enceladan geyser, by specific adsorption of the polyelectrolyte polymers onto that surface. A corresponding surface with a repeating negative charge could detect any cationic Darwinian biopolymers that might be in the geyser. Here, detection would be by displacement from the surface of a fluorescently tagged (magenta circles) short polyanionic biopolymer.
The system shown in Fig. 4 is simple to construct and even simpler to operate. To construct a device for one architecture, all that is necessary is a waveguide (a clear glass slide is adequate) illuminated by an LED emitting at a wavelength appropriate to excite a fluor attached to a short polyelectrolyte that is displaced by the universal genetic polyelectrolyte. To operate, one needs simply to flow the aqueous sample over the slide.
Once a polyelectrolyte is captured, we can then ask whether it meets the Schrödinger requirement for Darwinism: an ability to form an aperiodic crystal. An Earth-based laboratory would apply matrix-assisted laser desorption/ionization (MALDI) mass spectrometry to the adsorbed sample to address this question. Here, fragmentation patterns would allow the experimenter to estimate the diversity of building blocks within the adsorbed biopolymer, and their ability to form an aperiodic crystal. Subsequent experiments, more complex, would be needed to assess the homochirality of those building blocks.
At Enceladus, the surface that is used to concentrate by adsorption the Darwinian biopolymer would be recuited to also serve as the support from which mass spectra are obtained. If that support contained a matrix that could be desorbed by impact of a laser beam, a MALDI mass spectrum could be obtained by direct desorption of the polymers from the surfaces that are used in the first step to gather to concentrate the biopolymers from the Enceladus plume.
These thoughts guide the design of the capture mission, which must recover the Darwinian biopolymers intact. Terran DNA is fragmented by physical shock. Sonication, for example, generates fragments ca. 10,000 nucleotides long, long enough to be recognized as products of Darwinism. However, ultrasonication gives fragments of only 100 nucleotides long, approaching the limit where a Darwinian analysis would be conclusive. Likewise, bombardment by energetic particles destroys terran DNA, and likely biopolymers in general. This constrains the mission architecture; sampling must be be sufficiently low in energy that the impact does not destroy Darwinian biopolymers that might be present, and must protect the sample from radiation.
6. The Importance of Homochirality
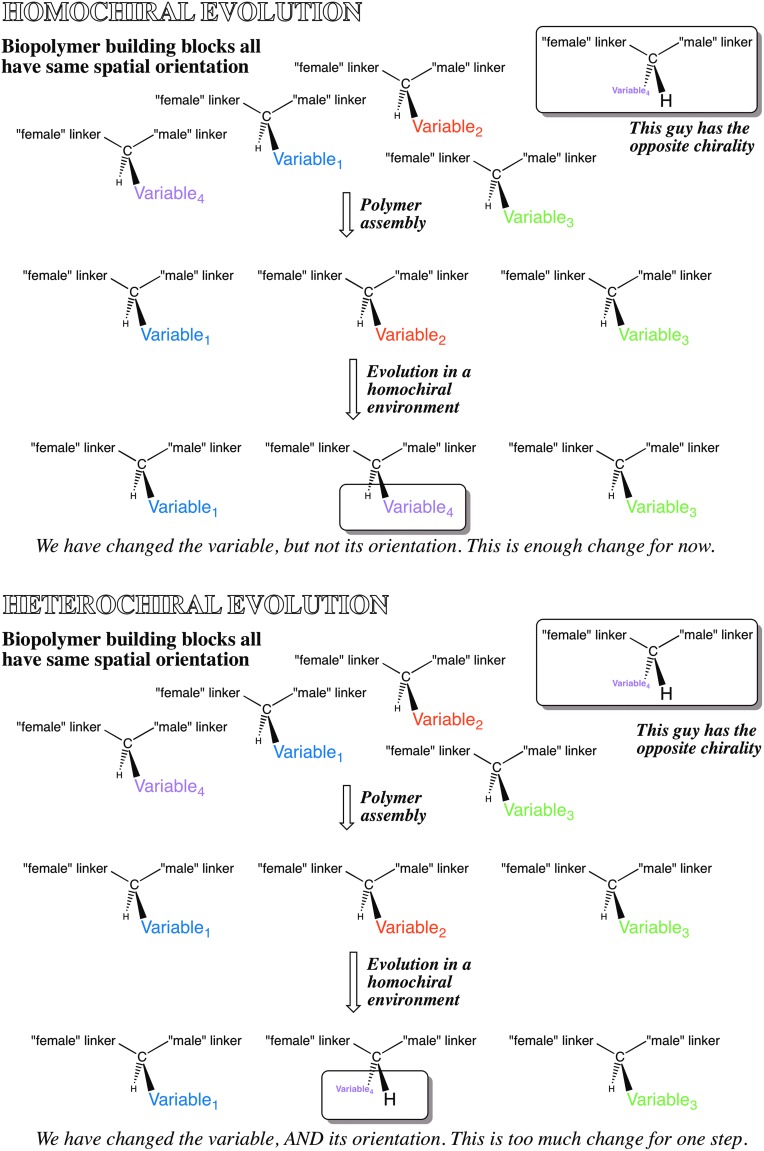
We have noted that for genetic biopolymers, homochirality is important to meet the Schrödinger criterion of an aperiodic crystal structure. It is also needed to support the evolvability of encoded biopolymers. However, homochirality is not needed for non-encoded biopolymers.
Making reference to Fig. 5, consider a set of (e.g., 20) building blocks that serve as the components of a biopolymeric system. Each of the building blocks has a side chain carrying a different functional group, chosen among the universe of possible functional groups. If the building blocks in this system are homochiral, then replacement of one during replication with error by another changes the functional group, but not its orientation. Thus, this replacement does not risk disrupting the fold (for nucleic acids, the double helix; for proteins, the alpha helix).
FIG. 5.
(Top) Mutation with a homochiral set of building blocks simply changes the side-chain information as a consequence of an error during replication. (Bottom) With a heterochiral set of building blocks, both the side-chain information and its orientation change as a consequence of a replication error. This creates discontinuity in the physical properties of the biopolymer, something that obstructs Darwinism.
However, a non-homochiral set of building blocks allows for the possibility that replacement of a functional group can also demand simultaneous change in the orientation of a functional group within the fold. For the perspective of evolvability, this requires that the system manage two different issues at the same time, the optimal selection of a functional group and the optional placement of that group. This makes the heterochiral polymeric system less evolvable than a homochiral system. For a homochiral system, the functional group is optimized by one process (here, mutation to replace one amino acid by a second with the same chirality) and its placement by another (replacement of amino acids at other sites, jostling the fold around the group).
7. Homochirality Is Not Needed in Non-Encoded Biopolymers
Under this analysis, homochirality is needed only for systems that are directly connected to Darwinism, either to support it or as a directly encoded product of it. In contrast, homochirality is not required for a non-encoded biopolymer to fold or function.
Many examples are known that illustrate this. For example, the natural antibiotic gramicidin has 15 amino acids all linked together as a biopolymer, where approximately half have the “right-handed” D-chirality and the rest have the “left-handed” L-chirality. Despite its heterochirality, gramicidin folds in a beta-helical structure and functions in that fold in a bacterial membrane. Gramicidin is, of course, not an encoded peptide; it is not created by ribosome-based translation.
The ability of heterochiral systems to fold and function is occasionally denied (Hand et al., 2017). However, it should not be. Indeed, over a century ago, Wallach and Liebisch speculated that, in general (but with exceptions), crystals composed of enantiomers (the left-handed and right-handed molecules together) are more dense than crystals composed of a single enantiomer; that is, heterochiral assemblages are better at assembling than homochiral assemblages (the “Wallach-Liebisch rule”) (Liebisch, 1894; Wallach, 1895; Ernst, 2017). This is specifically true for the amino acid alanine, but a statistical analysis based on modern structure databases finds this difference to be significant (Brock et al., 1991).
Thus, the search for homochirality in a set of building blocks as a biosignature requires that the relevant biopolymer be encoded, either as the Darwinian biopolymer itself or by a rule-based physical encoding system that changes digitally as the genetic biopolymer changes. It is not expected to be general for non-encoded biopolymers.
Indeed, if we consider only amino acids in peptides, the lack of generality extends even to the biosphere on early Earth. One “standard model” for the natural history of life on Earth includes an “RNA world,” an episode when RNA was the only encoded component of biological catalysis. That episode ended with the invention of translation that gave encoded proteins, but non-encoded proteins (like gramicidin) could have been present in the RNA world. Thus, a mission to Earth 4 billion years ago, sampling the terran biosphere, could have found non-encoded proteins but not encoded proteins. Thus, the amino acid building blocks in the proteins that that mission encountered need not have been homochiral.
Thus, using homochirality in the building blocks of proteins as a biosignature presumes that the hypothetical life being sought evolved in the same direction and at the same tempo as life on Earth. Specifically, this search strategy assumes that life universally achieved, by the time that we encounter it, the most difficult accomplishment that terran life achieved after gaining access to Darwinism: creating encoded proteins by translation. In different words, seeking amino acid homochirality on Enceladus and elsewhere must assume that life there also gained access to translation to create encoded proteins.
8. Alternative Molecular Biosignatures That Assume That Alien Natural History Proceeded in the Same Direction and with the Same Tempo as Terran Natural History
A similar discussion applies for the straight-chain lipids that appear in terran eubacteria. These have the formula C2n, where n is an integer. Thus, a clear biosignature for terran eubacteria (but not, incidentally, archaebacteria, which obtain their dominant lipid in a different way) is the observation of (for example) fatty acids with 14 carbon atoms but not 15 carbon atoms, 16 carbon atoms but not 17 carbon atoms, and so on. This pattern arises because C2n lipids are made in eubacteria by addition to a growing lipid chain of two carbons from a 3-carbon species, malonyl-ACP (ACP = acyl carrier protein). This is made by the biotin-dependent carboxylation of acetyl-ACP.
Here, the natural history has been well argued (Gilbert, 1986). Briefly, it is believed that biotin (not an RNA cofactor) emerged after ribosome-based translation to give encoded proteins (Visser and Kellogg, 1978) and that straight-chain fatty acids also postdate ribosome-based translation (Benner et al., 1989). In part, this argument is based on the observation that straight-chain lipid biosynthesis on Earth requires a ribosome-biosynthesized protein, ACP. It is perhaps buttressed by the absence of this pathway in Archaea.1
Nevertheless, the RNA appendages are widely conserved in all branches of modern terran life. Thus, the standard model interprets these appendages as handles created at a time where the only things available to grab them were RNA enzymes, which were well suited to grab RNA handles. The palimpsest model (Benner et al., 1989) for the RNA world argues that because of the abundance of RNA cofactors having these RNA appendages, the RNA world was able to catalyze a correspondingly broad range of metabolic reactions.
Again under that model (Benner et al., 1989), translation was superimposed on top of an already complex RNA world metabolism that already had multiple metabolic reactions using multiple RNA cofactors. Translation allowed translated proteins to stepwise replace the RNA enzymes in this already complex RNA-catalyzed metabolism. However, under the “constraint of multiple users,” as RNA cofactors were used throughout metabolism in multiple and consecutive steps, the proteins needed to evolve to use those the RNA handles as well; to do otherwise would have disrupted the existing RNA world metabolism.
Only when the cofactor participates in a single enzymatic reaction as a coenzyme, and not as a transferred substrate, was it possible to dispense with the RNA handle. Acyl carrier protein (ACP) as a species that makes C2n lipids and dispenses with the handle (using biotin and carbon dioxide, see below), is the archetypal example. ACP carries a stripped-down version of coenzyme A. Its predecessor, CoA, is a “ribocofactor” having a thiol (-SH) group as the active moiety; again, the RNA appendage on CoA does not participate in the reactivity of this cofactor, which is used to degrade C2n lipids and other molecules. Here, it is a cosubstrate, moving from enzyme to enzyme, requiring that the RNA handle be conserved.
For the synthesis of C2n lipids, the thiol need not move from enzyme to enzyme, and does not move. Thus, in a process that can be followed by paleogenetics reconstructions, the RNA handle has been lost, stepwise, where the steps are retained in the modern molecular biological record.
These facts are well known in the literature. However, Visser and Kellogg (1978) took the reasoning several steps further. Many organic chemists had attempted to reproduce, in small molecule systems, the reactivity of the RNA cofactors. By 1978, a large literature showed multiple successes.
The one exception with failure was biotin. Biotin is a cofactor that manages carbon dioxide, including the carbon dioxide that is used in the metabolic pathway that synthesizes C2n lipids. Even today, 40 years later, efforts to model the reactivity of biotin have been largely unsuccessful. Nor are we today entirely certain how biotin works.
Visser and Kellogg (1978) advanced their argument by pointing out that biotin is not an RNA cofactor. They then engaged in a brilliant example of chemical reasoning. RNA enzymes, they suggested, were primitive in their intrinsic catalytic potential relative to protein enzymes. Therefore, the RNA world was constrained in the kinds of reactions that it could catalyze. Visser and Kellogg suggested that chemists attempting to reproduce enzyme reactions were likewise primitive and constrained.
Those constraints, they suggested, are reflected in the kinds of reactions that RNA cofactors can participate in. For example, S-adenosylmethionine is an RNA cofactor, and its chemistry is easily modeled by primitive biomimetic organic chemists. Thus, the RNA world was able to transfer methyl groups. The RNA world was able to do oxidation-reduction reactions using niacin and flavin-containing cofactors; these reactions were also easily modeled by primitive biomimetic organic chemists, including work notably done in the Kellogg laboratory. The RNA world was able to transfer phosphate groups; ATP is an RNA cofactor, and its reactivity is easily modeled by primitive biomimetic organic chemists.
But not biotin. It is not an RNA cofactor and is not modeled by primitive organic chemists. Biotin needs sophisticated protein catalysts to function. And these emerged only after translation emerged.
This picture expands if we consider how CO2 is managed in modern terran biology when biotin is too expensive to use. For example, when fixing atmospheric CO2 in photosynthesis, the flux required is so overwhelmingly large that biotin is too expensive to be used, according to the model. Instead, ribulose-bisphosphate carboxylase is used. It is a problematic enzyme, especially in an O2 atmosphere. As it turns over, it destroys nearly half its substrate via an oxygenase reaction. The inefficiency of the carboxylase underscores the intrinsic difficulty of managing CO2 as a metabolic substrate. It is poorly soluble in water. When it does dissolve, it ionizes to form bicarbonate, shielding the electrophilicity of the central carbon atom. This makes it difficult to form a C-C bond to the central carbon of dissolved and ionized CO2.
A further dimension comes from the thermodynamics of C2n lipid biosynthesis. Condensation of two 2-carbon units (a “Claisen condensation”) lacks a large driving force. Its energetics are not conducive to addition of many 2-carbon fragments, again and again, to create a “polymeric” fatty acid, especially not at neutral pH. This is why biotin and CO2 are used in C2n fatty acid biosynthesis. It starts with a 2-carbon fragment (acetyl). It then uses an ATP and a biotin to attach a CO2 to give a malonyl (3-carbon) ACP thioester. Then, it prepares to form the bond that links two more carbon atoms to a growing lipid chain. To do this, it ejects the CO2 that it just attached using ATP, driving the formation of an “enol,” which has the energy to form the Claisen product.
9. Detecting Molecular Signatures of Life
Thus, again, under the “standard model” for the natural history of life on Earth, which includes an “RNA world,” a mission to Earth 4 billion years ago would not likely have encountered the regularity of C2n lipids; the standard model logically precludes the discovery of C2n lipids made by the terran pathway. However, that mission to early Earth may have encountered the regularity of C5n lipids made by terpene biosynthetic pathways. Thus, life-detection strategies to detect molecular regularities in lipids on early Earth may have detected them, just not the ones that arise from C2n lipids.
In general, in water, compelling arguments suggest that hydrophobic species of some kind are needed for Darwinism, often presumed to require compartmentalization. A hydrophobic molecule of some kind appears to be useful to obtain compartmentalization, and biosynthetic regularities that manifest themselves in the distribution of masses in these lipids might well be a biosignature. However, even on Earth today, C2n lipids and C5n lipids can be found, and one can conceive of a pathway that generates C3n lipids as well. This gives the potential to have lipids with two carbons (from malonyl-ACP), three carbons, four carbons (2 + 2), five carbons (from the terpene pathway), six carbons (either 2 + 2 + 2 or 3 + 3), eight carbons, and so on. A confusing signal, but not necessarily an uninterpretable one.
10. Conclusions
Table 1 captures these issues in summary. A theory supported by experiments on Earth can guide our search for Darwinian biosignatures in the Enceladus plumes, Jupiter's moons, and other aqueous lagoons throughout the Solar System. This assumes that the universal biopolymer (in water) able to support Darwinism is a polyelectrolyte.
Table 1.
Issues in Life Detection by Seeking Molecular Features
| Molecular target | Solution concentration principle | Comments |
|---|---|---|
| Polyelectrolyte genetic molecule | Adsorption on polyelectrolyte support | Molecular feature proposed to be universally necessary to support Darwinism, regardless of other molecular details of the building blocks. |
| Homochirality of amino acids | None | Homochirality needed only in encoded peptides; not an effective biosignature even for terran life in an RNA world (that is, pretranslation). |
| Patterns in lipids recovered | Extraction | Patterns dependent on biosynthetic pathway. On Earth, the path to C2n straight-chain lipids likely emerged after protein translation. C3n and C5n lipids all conceivable. |
Further, this polyelectrolyte structure is easily captured and concentrated, and a straight line connects these steps to its detection and, from there, to analysis and conclusions about its ability to support Darwinism. If a polyelectrolyte is found, and if it also meets Schrödingerian constraints, these two would make a compelling argument for a biosphere in any aqueous medium where it is encountered, including the Enceladus plume and Europa.
Instrument design would determine the lower limits for detection of such a biopolymer. Given the ability of the charged supports to concentrate scarce Darwinian polymers from large volumes, this lower limit of detection would be closely tied to the volume of sample that could be retrieved.
Of course, none of these considerations are unique to water coming from Enceladus. The polyelectrolyte theory of the gene is applicable to Darwinism in water, whereever it is found. Thus, the same device architecture can be used on Europa, in the terran Antarctic, or wherever liquid water is encountered.
However, for clarity, this proposal not only suggests that NASA missions to the ocean worlds of the outer Solar System should search for polyelectrolyte genetic molecules, but also suggests that the search for such polymers should be the first tool used in the search for life there. It further suggests that the search for homochirality in amino acids, or mass patterns in lipids, will not be general, applying only to life that followed the same path as life on Earth, at a similar tempo. The designs for such missions are not yet finalized, but preliminary concepts focus on the search for homochirality in amino acids and the mass spectrum in biogenic lipids (Steinbeck and Richert, 1998; Reh et al., 2016), not on the structures suggested here to be universal in Darwinian systems.
Acknowledgment
This project/publication was made possible through the support of a grant from Templeton World Charity Foundation, Inc., 0092/AB57. The opinions expressed in this publication are those of the author and do not necessarily reflect the views of Templeton World Charity Foundation, Inc. The material is also based upon work supported by NASA under awards NNX14AK37G and NNX15AF46G. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Aeronautics and Space Administration. I am indebted to discussions and comments provided by Christopher McKay, Carolyn Porco, Jack Dunitz, and several anonymous reviewers.
The persuasiveness of this model (Visser and Kellogg, 1978; Benner et al., 1989) for the natural history of the biotin cofactor is best appreciated by joining physical organic chemistry, molecular reactivity, cofactor structures, and biomimetic chemistry; all play roles in building the “big picture.” First, most cofactors in modern metabolism have pieces of RNA appended to them. These RNA appendages make absolutely no contribution to the reactivity of the cofactor. Indeed, in isolated portions of the modern terran biosphere, enzymes have emerged to perform the same reactions at essentially the same rates using substrates that lack the RNA appendages.
References
- Baross J., Benner S.A., Cody G.D., Copley S.D., Pace N.R., Scott J.H., Shapiro R., Sogin M.L., Stein J.L., Summons R., and Szostak J.W. (2007) The Limits of Organic Life in Planetary Systems, National Academies Press, Washington, DC [Google Scholar]
- Benner S.A. (2004) Understanding nucleic acids using synthetic chemistry. Acc Chem Res 37:784–797 [DOI] [PubMed] [Google Scholar]
- Benner S.A. and Hutter D. (2002) Phosphates, DNA, and the search for nonterran life. A second generation model for genetic molecules. Bioorg Chem 30:62–80 [DOI] [PubMed] [Google Scholar]
- Benner S.A., Ellington A.D., and Tauer A. (1989) Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci USA 86:7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock C.P., Schweizer W.B., and Dunitz J.D. (1991) On the validity of Wallach's rule: on the density and stability of racemic crystals compared with their chiral counterparts. J Am Chem Soc 113:9811–9820 [Google Scholar]
- Chyba C.F. and Phillips C.B. (2002) Europa as an abode of life. Orig Life Evol Biosph 32:47–67 [DOI] [PubMed] [Google Scholar]
- Cleland C.E. and Copley S.D. (2005) The possibility of alternative microbial life on Earth. International Journal of Astrobiology 4:165–173 [Google Scholar]
- Davila A.F. and McKay C.P. (2014) Chance and necessity in biochemistry. Implications for the search for extraterrestrial biomarkers in Earth-like environments. Astrobiology 14:534–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst K.H. (2017) On the validity of calling Wallach's rule Wallach's rule. Isr J Chem 57:24–30 [Google Scholar]
- Eschgfaeller B., Schmidt J.G., and Benner S.A. (2003) Synthesis and properties of oligodeoxynucleotide analogs with bis(methylene) sulfone-bridges. Helv Chim Acta 86:2957–2997 [Google Scholar]
- Flory P.J. (1953) Principles of Polymer Chemistry, Cornell University Press, Ithaca, NY [Google Scholar]
- Gilbert W. (1986) Origin of life: the RNA world. Nature 319:618 [Google Scholar]
- Hand K.P., Murray A.E., Garvin J.B., Brinkerhoff W.B., Christner B.C., Edgett K.S., Ehlmann G.L., German C.R., Hayes A.G., Hoehler T.M., Horst S.M., Lunine J.I., Nealson K.H., Paranicas C., Schmidt B.E., Smith D.E., Rhoden A.R., Russell M.J., Templeton A.D., Willis P.A., Yingst R.A., Phillips C.B., Cable M.L., Craft K.L., Hofmann A.E., Nordheim T.A., Pappalardo R.P., and the Project Engineering Team. (2017) Report of the Europa Lander Science Definition Team, JPL publication D-97667, Jet Propulsion Laboratory, Pasadena, CA [Google Scholar]
- Hoogsteen K. (1962) The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallographica 16:907–916 [Google Scholar]
- Huang Z., Schneider K.C., and Benner S.A. (1991) Building blocks for analogs of ribo- and deoxyribonucleotides with dimethylene-sulfide, -sulfoxide and -sulfone groups replacing phosphodiester linkages. J Org Chem 56:3869–3882 [Google Scholar]
- James S.L. and Friščić T. (2013) Mechanochemistry. Chem Soc Rev 42:7494–7496 [DOI] [PubMed] [Google Scholar]
- Joyce J. (1994) Foreword. In Origins of Life: The Central Concepts, edited by Deamer D.W. and Fleischaker G.R., Jones and Bartlett, Boston: The original reference is a 1992 internal NASA document entitled “Exobiology: Discipline Science Plan.” [Google Scholar]
- Kim J., Cheong C., and Moore P.B. (1991) Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature 351:331–332 [DOI] [PubMed] [Google Scholar]
- Koshland D.E. (2002) The seven pillars of life. Science 295:2215–2216 [DOI] [PubMed] [Google Scholar]
- Lee D.H., Granja J.R., Martinez J.A., Severin K., and Ghadiri M.R. (1996) A self replicating peptide. Nature 382:525–528 [DOI] [PubMed] [Google Scholar]
- Liebisch T. (1894) Mitgetheilt in der Sitzung der Königl. Gesellschafter der Wissenschaftern zu Göttingen, 8 Dez [Google Scholar]
- Linkletter B.A, Szabo I.E, and Bruice T.C. (2001) Solid-phase synthesis of oligopurine deoxynucleic guanidine (DNG) and analysis of binding with DNA oligomers. Nucleic Acids Res 29:2370–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock J.E. (1965) A physical basis for life detection experiments. Nature 207:568–570 [DOI] [PubMed] [Google Scholar]
- Malaska M.J. and Hodyss R. (2014) Dissolution of benzene, naphthalene, and biphenyl in a simulated Titan lake. Icarus 242:74–81 [Google Scholar]
- Marion G.M., Kargel J.S., Catling D.C., and Lunine J.I. (2012) Modeling ammonia–ammonium aqueous chemistries in the Solar System's icy bodies. Icarus 220:932–946 [Google Scholar]
- McLendon C., Opalko F.J., Illangkoon H.I., and Benner S.A. (2015) Solubility of polyethers in hydrocarbons at low temperatures. A model for potential genetic backbones on warm Titans. Astrobiology 15:200–206 [DOI] [PubMed] [Google Scholar]
- Miller P.S., McParland K.B., Jayaraman K., and Tso P.O.P. (1981) Biochemical and biological effects of nonionic nucleic acid methylphosphonates. Biochemistry 20:1874–1880 [DOI] [PubMed] [Google Scholar]
- Navarro-González R., Rainey F.A., Molina P., Bagaley D.R., Hollen B.J., de la Rosa J., Small A.M., Quinn R.C., Grunthaner F.J., Caceres L., Gomez-Silva B., and McKay C.P. (2003) Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 302:1018–1021 [DOI] [PubMed] [Google Scholar]
- Pauling L., Itano H.A., Singer S.J., and Wells I.C. (1949) Sickle cell anemia, a molecular disease. Science 110:543–548 [DOI] [PubMed] [Google Scholar]
- Porco C.C., Helfenstein P., Thomas P.C., Ingersoll A.P., Wisdom J., West R., Neukum G., Denk T., Wagner R., Roatsch T., Kieffer S., Turtle E., McEwen A., Johnson T.V., Rathbun J., Veverka J., Wilson D., Perry J., Spitale J., Brahic A., Burns J.A., GelGenio A.D., Dones L., Murray C.D., and Squyres S. (2006) Cassini observes the active south pole of Enceladus. Science 311:1393–1401 [DOI] [PubMed] [Google Scholar]
- Porco C.C., Dones L., and Mitchell C. (2017) Could it be snowing microbes on Enceladus? Assessing conditions in its plume and implications for future missions. Astrobiology 17:876–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P.M. and Bruice T.C. (2003) Solid-phase synthesis of positively charged deoxynucleic guanidine (DNG) oligonucleotide mixed sequences. Bioorg Med Chem Lett 13:1281–1285 [DOI] [PubMed] [Google Scholar]
- Reh K., Spilker L., Lunine J.I., Waite J.H., Cable M.L., Postberg F., and Clark K. (2016) Enceladus Life Finder: the search for life in a habitable moon. In 2016 IEEE Aerospace Conference, IEEE, Piscataway, NJ, doi: 10.1109/AERO.2016.7500813 [DOI] [Google Scholar]
- Rezzonico F. (2014) Nanopore-based instruments as biosensors for future planetary missions. Astrobiology 14:344–351 [DOI] [PubMed] [Google Scholar]
- Richert C., Roughton A.L., and Benner S.A. (1996) Nonionic analogs of RNA with dimethylene sulfone bridges. J Am Chem Soc 118:4518–4531 [Google Scholar]
- Roughton A.L., Portmann S., Benner S.A, and Egli M. (1995) Crystal structure of a dimethylene sulfone linked ribodinucleotide analog. J Am Chem Soc 117:7249–7250 [Google Scholar]
- Sarathy A., Qiu H., and Leburton J.P. (2017) Graphene nanopores for electronic recognition of DNA methylation. J Phys Chem B 121:3757–3763 [DOI] [PubMed] [Google Scholar]
- Schrödinger E. (1943) Was ist Leben, Serie Piper, Vol. 1134 [Google Scholar]
- Seager S., Schrenk M., and Bains W. (2012) An astrophysical view of Earth-based metabolic biosignature gases. Astrobiology 12:61–82 [DOI] [PubMed] [Google Scholar]
- Steinbeck C. and Richert C. (1998) The role of ionic backbones in RNA structure: an unusually stable non-Watson-Crick duplex of a nonionic analog in an apolar medium. J Am Chem Soc 120:11576–11580 [Google Scholar]
- Steward G.F., Smith D.C., and Azam F. (1996) Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. Mar Ecol Prog Ser 131:287–300 [Google Scholar]
- Thomas P., Tajeddine R., Tiscareno M., Burns J., Joseph J., Loredo T., Helfenstein P., and Porco C. (2015) Enceladus's measured physical libration requires a global subsurface ocean. Icarus 264:37–47 [Google Scholar]
- Visser C.M. and Kellogg R.M. (1978) Biotin. Its place in evolution. J Mol Evol 11:171–178 [DOI] [PubMed] [Google Scholar]
- Wallach O. (1895) Zur Kenntniss der Terpene und der ätherischen Oele. Ueber gebromte Derivate der Carvonreihe. Liebigs Ann Chem 286:119–143 [Google Scholar]
- Westheimer F.H. (1987) Why Nature chose phosphates. Science 235:1173–1178 [DOI] [PubMed] [Google Scholar]