Abstract
A meta-analysis of CD4+ T cell epitope maps reveals clusters and gaps in envelope-protein (E protein) immunogenicity that can be explained by the likelihood of epitope processing, as determined by E protein three-dimensional structures. Differential processing may be at least partially responsible for variations in disease severity among arbo-flaviruses and points to structural features that modulate protection from disease.
Keywords: : helper T cell, antigen processing, class II MHC protein
Introduction
The emergence of Zika virus (ZV) has renewed attention on arthropod-borne flaviviruses, or arbo-flaviviruses. The development of vaccines and management of disease, possibly complicated by immunopathology, face major challenges in the basic science of arbo-flaviviruses. This article seeks to highlight features of CD4+ T cell immunology that could be modulated by the major structural protein (the envelope protein, E) and offers new interpretations of existing data, based on the analysis of E structure from the standpoint of antigen processing.
Arbo-flaviviruses are responsible for some of the world's most devastating infectious diseases. Typical clinical features include pain, fever, rash, and nausea; and more serious infections cause seizures, bleeding, and mortality. The mosquito-borne flavivirus diseases take enormous tolls in tropical and subtropical countries. Yellow fever was one of the most severe before effective mosquito control reduced exposure and a highly effective live attenuated vaccine became available, but it still causes tens of thousands of deaths annually (24,57). Dengue fever has become a major threat with occurence of 100 million infections and tens of thousands of deaths worldwide each year (2). The high-mortality hemorrhagic form of dengue fever is associated with sequential infections by different dengue virus types, with four types in circulation. Japanese encephalitis approaches 100,000 cases and tens of thousands of deaths annually (9). Most transmissions of flaviviruses cause subclinical infections, and thus the frequency of transmission is vastly greater than the frequency of disease (2,51). ZV has emerged in the last year with possibly more than a million persons infected and has caused several thousand cases of microcephaly by infection in utero in Brazil alone (5). Tick-borne encephalitis cases number in the thousands, again, with much larger numbers of unreported infections (19). Multiple inactivated, recombinant, and live vaccines have been deployed for Japanese encephalitis virus (JEV) (37); inactivated vaccines are available for tick-borne encephalitis virus (TBEV) (22); and a tetravalent live recombinant virus for types 1–4 of dengue virus has recently become available (12). However, much work still remains in the development of vaccines for ZV and West Nile virus (WNV), and in the improvement of the existing ones for dengue virus and JEV.
Although they cause distinct disease entities, the arbo-flaviviruses are remarkably similar. When comparing the entire genome polyprotein, the viruses share at least 42% pairwise amino acid sequence identity. The identity ranges from 39% to 55% even for the E glycoproteins. For comparison, the influenza E protein, hemagglutinin, exhibits a similar level of identity across subtypes (38–46% identity).
The arbo-flaviviruses cycle between arthropod and mammalian hosts. For yellow fever virus (YFV), dengue fever virus (DFV), and possibly ZV, humans are the preferred mammalian host, which amplifies the virus and makes it available for a new cycle of transmission when the host is bitten by another arthropod (5). With other arbo-flaviviruses that infect humans, the preferred host is another animal, such as a bird or rodent. ZV appears to be exceptional in that it can be transmitted by multiple routes, including directly from mother to fetus and by sexual contact (8,35).
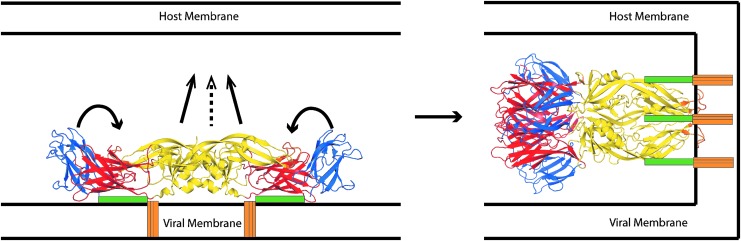
The flavivirus positive-strand RNA encodes a single genome polyprotein that includes the capsid (C), premembrane and membrane (M/M), and E proteins, and seven nonstructural proteins with enzymatic or regulatory activities. The E protein covers the viral surface as “rafts” of three E dimers that assemble into an icosahedron (53). Before activation of the fusogenic activity, the elongated monomers are arranged head-to-tail, with the long axis of the monomers oriented parallel to the viral membrane (Fig. 1). The E monomer is composed of three globular domains, stem domains, and the transmembrane domain. The polypeptide chain traverses domains I and II twice before connecting to domain III. The fusion peptide is located in domain II, projecting from the tip of one monomer into a cleft between domains I and III of the other monomer. Protonation of certain histidine residues at low pH triggers a conformational change that reorients the long axis of the monomer so that the fusion peptide projects outward from the viral membrane and toward the target cell membrane (11). The repositioning of domain III and assembly of the postfusion E trimers drive fusion of the viral and target membranes.
FIG. 1.
Flavivirus E protein conformational transition associated with membrane fusion [adapted from Stiasny and Heinz (53)]. Before fusion (at left), E forms a dimer oriented parallel to the viral envelope membrane. Low pH induces a conformational change, including the repositioning of domain III (blue) alongside domain 1 (red) and rotation of domain II (yellow), including the fusion peptide, toward the host cell membrane. After fusion, E forms a trimer oriented perpendicular to the membrane (at right). E, envelope.
Three-dimensional structures of the E proteins from different flaviviruses are very similar when representing the same conformational state (prefusion/postfusion). For examples, prefusion crystallographic structures for WNV, TBEV, and JEV have been reported (20,28,44). A well-distributed two-thirds of the backbone atoms of these structures superimpose with 1.3 Å root-mean-square deviation (RMSD). Likewise, postfusion crystallographic structures for dengue fever virus type 1 (DFV1), dengue fever virus type 2 (DFV2), and TBEV have been reported (3,21,36). Most backbone atoms of these structures superimpose with 1.5 Å RMSD. In contrast, the superimposition of prefusion and postfusion structures is essentially limited to one domain at a time. The conversion from dimer to trimer involves substantial changes in the interfaces between domains within the monomer and between monomers (29).
Immunity to flaviviruses is thought to be mediated by both neutralizing antibodies and cellular responses (43). Direct neutralization of virus is a well-established mechanism of vaccine efficacy, but antibodies protect against viruses by multiple mechanisms, including antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity (54). The inactivated TBEV and JEV vaccines are administered in multiple doses to engage affinity maturation and raise protective antibodies. CD4+ T cells contribute to these mechanisms by promoting antibody affinity maturation and isotype switching in the B cells. The cytolytic activity and other direct mechanisms by CD4+ T cells against virus may also contribute to protection. Natural DFV infection induces DFV-specific cytolytic CD4+ T cells that express the CX3CR1 chemokine receptor, which directs T cell migration from the bloodstream to peripheral tissue through the CX3CL1 ligand on endothelial cells (58). The live attenuated JEV vaccine appears to be more protective than the inactivated vaccine, possibly due to induction of more effective cellular responses (26). The recent demonstration that herpes virus-specific CD4+ T cells regulate antibody and T cell access to nervous tissue highlights another potential role in viral control (16). Protection against dengue infection of the brain required both antibodies and CD4+ T cells in a mouse model (13).
Immunopathology has long been suspected in severe cases of dengue fever (dengue hemorrhagic fever and dengue shock syndrome), which are associated with secondary infections by a different dengue virus type. Alternative mechanisms have been put forward (50). Antibody-dependent enhancement of infection occurs when antibody avidity falls below the level of neutralization, but remains high enough for opsonization, which facilitates viral entry into Fc receptor-bearing cells. This mechanism can also explain the occurrence of severe disease in infants with a primary infection, but who have acquired cross-reactive antibodies from the mother. However, severe disease has also been observed in the absence of any known source of cross-reactive antibodies (25). The alternative (and not exclusive) mechanism for severe disease is based on T cell-mediated immunopathology (50). Severe disease coincides with clearance of the virus and correlates with a cytokine storm, characterized by high levels of type 1 cytokines, including interferon-γ and tumor necrosis factor-α. The association with secondary infection is thought to derive from original antigenic sin, characterized by a robust, but less effective, response by T cells raised in the primary infection and cross-reacting with reduced avidity to the virus in the secondary infection.
CD4+ T cells differentiate from naive T cells into multiple effector and regulatory types, characterized by expression of particular cytokines and surface markers that define immune functions and tissue homing. The distribution of T cells among the various subsets is modulated by the conditions of immunization and influences the protectiveness of the response (32). Thus, the mechanisms that determine this distribution are under intense investigation, with major efforts on the role of innate immune activation, route of exposure, and persistence of antigen. Moreover, how the secondary infection triggers and then modulates a memory response adds another layer of complexity (50).
In challenge experiments, JEV vaccines demonstrated cross-protection against WNV and DFV. In the case of JEV-immunized Balb/c mice, strikingly distinct patterns of antibody cross-reactivity were detected against the different DFV types (27). Nevertheless, the vaccine provided comparable protection for all four DFV types. These results highlight the importance of cellular immunity in protection against flaviviruses. Conclusions about cross-protection by CD4+ T cells in inbred mice have uncertain relevance for humans because humans are heterogeneous in the content of polymorphic major histocompatibility class II antigen-presenting proteins (MHCIIs). Nevertheless, to the extent that antigen structure directs T cell responses in human populations, we can assume the mechanisms are independent of MHCII polymorphism.
To prime or recall a CD4+ T cell response, the flavivirus proteins must be proteolytically processed and presented to the T cells. The overall process can be thought of in terms of two colliding waves of molecular ontogeny. A wave of antigen moves inward from the endosome (or out of the cytoplasm for the infected antigen-presenting cell), and a wave of MHCII complexed with an invariant chain (MHCII-Ii) moves outward from the endoplasmic reticulum. Wave formation begins with exposure of the APC to danger signals, which activates antigen processing and blocks the turnover of MHCII (10). Wave formation terminates with the inactivation of MHCII gene expression (46). The two waves meet in the antigen-processing compartment, which is itself developing toward a more acidic and hydrolytic state. The magnitude and speed of the waves and activation of the processing compartment depend on the type of APC and level of danger signal received. There may also be multiple phases of presentation or alternative circumstances for the same APC, such as at the site of infection and then again in the lymph node. The advantage of the wave concept is that it emphasizes the kinetics of each mechanism. Few, if any, processes approach equilibrium because components are rapidly converted or sequestered. For example, it unlikely that the on-rate and off-rate for peptide binding to the MHCII have the opportunity to reach equilibrium before one or other species is destroyed or sequestered.
The dominance hierarchy of CD4+ T cell epitopes within a given antigen is a product of the naive T cell repertoire, antigen processing, and peptide-MHCII affinity (more likely, on-rate or off-rate, but not both). Clonal abundance in the naive T cell repertoire has been revealed to be a significant influence on the strength of T cell responses (31). One source of variation in clonal abundance was identified as similarity of the target epitope to epitopes of self or normal flora (39). The more similar the epitope, the lower that abundance, presumably due to mechanisms of peripheral tolerance. Variation in naive T cell abundance ranged over several fold, and the amplified populations generated by priming preserved the difference. Nevertheless, the absolute levels of T cell populations correlated only weakly with naive T cell levels, suggesting that antigen processing and peptide-MHCII affinity could be the major players in epitope dominance.
For epitope-mapping studies in humans utilizing ELISpot and natural APC, the restricting MHCII molecule is unidentified. Although the subject's human leukocyte antigen (HLA) makeup can be determined (composed of as many as five different alleles), the particular molecule(s) responsible for presenting a given epitope usually cannot. Thus, it is not currently possible to rigorously disentangle the contributions of naive T cell repertoire, antigen processing, and MHCII affinity. In this work, we evaluated the potential contributions of antigen processing and MHCII affinity.
Methods
Amino acid sequences for E proteins of YFV, TBEV, DFV2, and JEV were extracted from polyprotein sequences with Uniprot accession numbers Q6DV88, P14336, P12823, and P32886, respectively. E from WNV was from Genbank accession number YP_001527880.
Epitope likelihoods for each antigen were computed using the antigen processing-based scoring approach (34). This approach utilizes conformational stability data from sequence conservation, crystallographic B-factors, solvent-accessible surface area, and the COREX measure (15). Shannon sequence entropy was computed using BioEdit (14). Crystallographic B-factors were obtained using the protein data bank (PDB) structure for each protein. Solvent-accessible surface area was computed using MolMol with the PDB structure as input (23). The COREX metric was computed locally using source code obtained from the authors. These data were combined into an aggregate stability measure using z-scores, and were then modified using a model of antigen processing in which peptides that flank unstable regions were scored as being more likely to be processed and made available for MHC binding. Crystal structures used for prefusion conformations of TBEV, JEV, and DFV2 were PDB files 1SVB, 3P54, and 4UTC, respectively. Crystal structures for postfusion conformations of TBEV and DFV2 were 1URZ and 1OK8, respectively. The postfusion conformation of E from JEV was generated by automated homology modeling using Swiss-model and 1OK8 as template.
Results and Discussion
Available CD4+ T cell epitope maps
Using search tools provided by the PubMed interface and the Immune Epitope Database, we examined a total of 42 published reports that identified CD4+ T cell epitopes for 10 arbo-flaviviruses (DFV1, DFV2, DFV3, DFV4, JEV, TBEV, YFV, WNV, SLEV, and MVEV). For the purposes of evaluating the contribution of antigen processing to epitope dominance, we limited our analysis to eight studies on five viruses (DFV2, JEV, TBEV, YFV, and WNV) that employed a set of overlapping peptides spanning the complete E protein and that examined T cell responses following exposure to intact protein by infection or vaccination (Table 1). Many of the unused studies focused on certain epitopes for various reasons, such as strong binding to MHCII proteins or conservation across viruses. Two systematic epitope-mapping studies in mice employed T cell priming by peptides, rather than the native antigen; and thus, an epitope pattern generated by priming with the intact antigen was not determined (38,47). In the studies compiled here, specific T cell responses or T cells themselves were detected in the peripheral blood mononuclear cells (PBMCs) of immunized human subjects or the splenocytes of immunized mice. For most studies in humans and the YFV study in BALB/c mice, the T cell responses were restimulated for 16–18 h with single peptides or small pools of peptides and detected by interferon-γ or interleukin (IL)-2 ELISpot. For YFV and WNV in mice, the T cell responses were stimulated with peptide pools for 6 h and detected by intracellular staining for interferon-γ. For YFV and WNV in humans, the T cells were stimulated for 2 weeks with small pools of peptides and then detected by MHC class II tetramers loaded with the same pools of peptides.
Table 1.
Systematic T Cell Epitope Mapping Studies of E Proteins
| Virus | Reference | Subjects | N | Peptides | Detection | Epitope densitya |
|---|---|---|---|---|---|---|
| DFV2 | (45) | Humans | 5 | 100 | Elispot, IFNγ | 1.2 |
| JEV | (55) | Humans | 26 | 62 | Elispot, IFNγ | 3.3 |
| YFV | (56) | C57BL/6 mice | N/A | 50 | Flow, IFNγ | 1 |
| YFV | (30) | BALB/c mice | 3–5 | 81 | Elispot, IFNγ | <6 |
| YFV | (18) | Humans | 40 | 83 | Tetramer flow | 3.6b |
| TBEV | (49) | Humans | 34 | 124 | Elispot, IL-2 | 5c |
| WNV | (4) | C57BL/6 mice | 2–5 | 100 | Flow, IFNγ | <2 |
| WNV | (17) | Humans | 40 | 83 | Tetramer flow | 3.6d |
Average number of epitopes in E detected per subject or tetramer allele.
Seven alleles tested.
Median reported for a subset of subjects.
Ten alleles tested.
DFV2, dengue fever virus type 2; JEV, Japanese encephalitis virus; YFV, yellow fever virus; WNV, West Nile virus; TBEV, tick-borne encephalitis virus; IFN, interferon; IL, interleukin; N/A, not available.
The total number of epitopes reported in a given study was related to the number of subjects and the number of MHC II alleles involved. The number of test peptides spanning the E protein sequence was relatively constant. The fewest epitopes (one or two) were reported for studies in small groups of C57BL/6 mice. A large number of epitopes was discovered in the tetramer-guided epitope mapping studies, using up to 10 MHC II alleles present in 40 human subjects. The largest number of epitopes was discovered using IL-2 ELISpot with a cohort of 34 TBEV-vaccinated human subjects, each of whom could have as many as five different MHC II alleles. In that study, three-fourths of the test peptides were found to contain an epitope in at least one subject. Thus, for the purposes of comparing epitope maps and scoring dominance across viruses, we considered only the nine epitope clusters identified in E from TBEV, rather than all epitopes identified.
The relative abundance of a viral protein predicts the relative intensity of that protein's CD4+ T cell immunogenicity. The structural proteins E, C, and M constitute the largest aggregate mass in the viral particle and therefore dominate the responses. In the DFV2 mapping study, CD4+ responses were found to target E and C proteins, whereas CD8+ responses were found to target the nonstructural proteins (45). Likewise, in the JEV mapping study, E, C, and M proteins were the sources of most of the CD4+ epitopes (55). For TBEV, the number of ELISpots responding to E, C, and M peptides correlated with the total mass of the respective protein in the virus particle (49). In the WNV tetramer-guided mapping, T cell epitopes were discovered in the structural proteins at roughly twice the rate of discovery for the less abundant nonstructural proteins (17). These results foreshadow a relationship between the CD4+ T cell response and the relative yield of peptides within a given antigen from antigen processing.
Heinz and coworkers generated a prediction of epitope dominance for TBEV structural proteins using MHCII-peptide affinity and the HLA allele distribution in groups of exposed individuals (49). For epitopes in E, the correlation of predicted and observed epitopes was poor, whereas for epitopes in C, the correlation was very good. The authors attributed the difference to the influence of E structure on antigen processing, whereas C is largely disordered when dissociated from the viral RNA, as it is expected to be in the acidic endolysosome. Of nine epitope clusters identified in E by Heinz and coworkers in TBEV vaccinees, only three clusters could be explained by the expected affinity of the sequences for binding to the MHCII proteins. Remarkably, the single most dominant cluster, recognized by more than 50% of vaccinees, was not predicted to bind strongly to the MHCII proteins present.
Tetramer-guided epitope mapping revealed a large number of epitopes in the E proteins of YFV and WNV (17,18). In this new approach to epitope mapping, pools of peptides that assembled onto individual MHC proteins were tested for binding to T cells from vaccinated subjects. The technique is powerful because the phenotype of T cells may be interrogated, but it potentially misses epitopes for weakly binding peptides and can identify T cells for epitopes to which the subject is actually naive. In the study of YFV epitopes, the second issue was addressed by staining the T cells for “antigen experience,” revealed by low-level expression of CD45RA. For a subset of epitopes, the authors also confirmed that the epitope could be presented from the intact antigen in an assay for T cell restimulation. Epitopes associated with low CD45RA and can be presented from an intact antigen are strong candidates for natural processing during vaccination or infection.
Epitope clusters and gaps in E
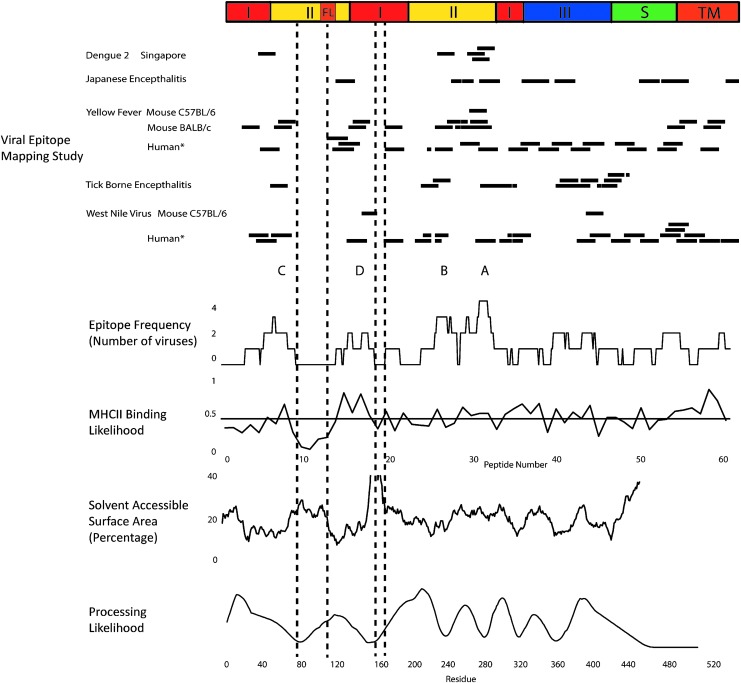
Comparison of epitope mapping results for all systematically mapped E proteins revealed considerable similarity, especially in domains I and II. Sequence positions were scored for occurrence in an epitope (or epitope cluster, in the case of TBEV). Most positions were contained in an epitope for at least two viruses. Four distinct epitope superclusters were evident (Fig. 2, labeled A–D). The concentration of epitopes into superclusters suggests a conserved pattern of epitope immunogenicity, whether through sequence composition or structural constraints. The presence of immunodominant superclusters could be a success or failure of the immune system, depending on the superclusters' relative protectiveness and contribution to immunopathology.
FIG. 2.
Epitope frequency aligned with predicted MHCII binding, solvent-accessible surface area, and processing likelihood. At top, a schematic diagram indicates structural domains identified in crystallographic structures. Horizontal bars indicate peptides identified as CD4+ epitopes or immunodominant epitope clusters (TBEV study) as they appear in the corresponding aligned E protein sequences. Epitope frequency was compiled as the number of viruses for which a given amino acid residue appears in an epitope or cluster, not including those identified by tetramer-guided mapping (*). MHCII binding likelihood indicates the average normalized prediction of binding affinity (“1-log50k”) for seven MHCII proteins (HLA-DRB1*0101, HLA-DRB1*0301, HLA-DRB1*0401, HLA-DRB1*0404, HLA-DRB1*0701, HLA-DRB1*1101, and HLA-DRB1*1501). The horizontal line indicates the mean for all sequence positions. Solvent-accessible area was calculated using MOLMOL with the crystal structure of the E protein from DFV2 (PDB: 1OK8) and then smoothed with a 15-residue averaging window. Processing likelihood was calculated for DFV2 using the algorithm previously described (34). HLA, human leukocyte antigen; MHCII, major histocompatibility class II; TBEV, tick-borne encephalitis virus; DFV2, dengue fever virus type 2.
The compilation of epitope-mapping data reveals two gaps of immunogenicity, which we identify as segments of at least six residues that were not observed in any CD4+ T cell epitopes (residues 76–108 and 150–158, DFV2 numbering). We considered three alternative hypotheses for the gaps. First, the number of epitopes identified was so few that they would not cover the whole sequence even if they were distributed randomly. Second, the distribution of CD4+ T cell epitopes was strictly related to the sequence preferences of the class II MHC proteins, and the gaps were evolutionarily selected either to avoid MHCII binding or to achieve another function that simultaneously reduces MHCII binding (e.g., high flexibility). Third, the sequences in the gaps were not available for presentation due to the interaction of E protein structure with antigen-processing mechanisms. Each hypothesis is addressed below.
Since the mechanisms controlling CD4+ T cell epitope profiles are poorly understood, the shape of epitope profile expected in the absence of any peptide selection bias was important to consider. Twenty hypothetical random distributions of 65 epitopes were generated using a pool of 75 seventeen-mer peptides that spanned a 445-residue antigen in six-residue steps. Most of the hypothetical random profiles exhibited several peaks and troughs, not unlike the experimental epitope frequency presented in Figure 2. The average percentage of residues that was not found in any epitope was 9% ± 4%. In the experimental profiles, the nonimmunogenic residues amounted to 20% of the sequence, which is more than three standard deviations larger than the 9% expected from a random selection of epitopes. Thus, epitope selection is not at all random; and the gaps are most likely due to weak MHCII binding and/or processing of those peptides. (Note that each replicate hypothetical profile exhibited gaps in different segments, whereas we predict that experimental gaps would be located in the same segments if a compilation was assembled from a new set of mapping experiments due to biases in the MHCII binding and processing of peptides.)
To evaluate the influence of MHCII-peptide affinity on immunogenicity, it is necessary to know which MHCII protein presented the peptide. In the case of the tetramer-guided mapping of E from YFV, the MHC II allele for each epitope is known by the identity of its tetramer (18). Thus, it is possible to predict peptide affinity. A total of 25 epitopes were identified using seven HLA molecules. Only four peptides were identified with multiple HLA proteins, indicating relatively little overlap in peptide selectivity. The values of “1-log50k” were collected for theoretical binding of E peptides of YFV to each HLA protein, as reported by the NETMHCII website (40). Values were normalized to the range of 0 to 1 for comparison of dominance profiles across HLA molecules. Strikingly, almost all the tetramer-mapped epitope peptides were predicted to bind to their respective HLA molecules with affinity above the 80th percentile (data not shown). This is an uncommonly high level of accuracy in CD4+ T cell epitope prediction, and it contrasts with the results of Elispot-based epitope mapping for the E protein of TBEV, where a poor correlation with predicted (population weighted) MHCII affinity was found (49). CD4+ epitope mapping studies have generally found only weak predictive power of MHCII binding (7,42). Important features of the YFV study are (i) the use of tetramer-guided epitope mapping and (ii) the fact that the PBMCs were cultured with peptides for 14 days before analysis. Both features could result in the selective detection of high MHCII-affinity peptides. They could also account for the much larger number of epitopes being reported for YFV (and WNV) if some of the high MHCII-affinity peptides are poorly represented in a natural response.
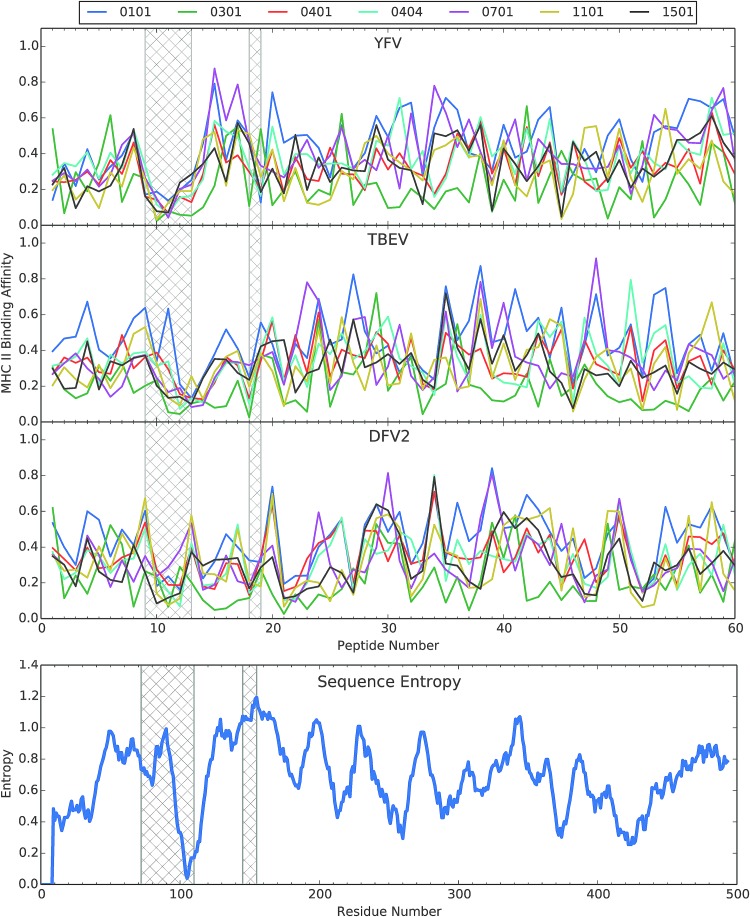
As expected for divergent peptide selectivity in the different HLA molecules, the theoretical peptide binding profiles displayed considerable variability, except for a prominent dip in affinity spanning from peptide 9 to peptide 13, corresponding to residues 72–110 (Fig. 3). The dip in affinity almost completely overlaps the largest gap in the compilation of experimental epitope profiles. The dip in MHCII affinity and epitope gap coincide with a large surface-exposed loop in the N-terminal segment of domain II that includes the highly conserved fusion peptide (Figs. 2 and 4A). Thus, the single least immunogenic segment in the protein coincides with its most conserved structural feature (the fusion loop). The coincidence of low MHCII affinity and conformational flexibility most probably derives from the high content of charged amino acids in the surface-exposed loop.
FIG. 3.
Alignment of predicted MHCII binding to selected HLA molecules of peptides from three flaviviruses and the plot of Shannon sequence entropy for an alignment of 21 flaviviruses. Hatched bars indicate the two gaps in the experimental CD4+ epitope-mapping profiles, as identified in Figure 2. MHCII binding was predicted using the NETMHCII 2.2 server (40) and then the values of “1-log50k” for each profile were normalized to the range of 0 to 1. The selected HLA molecules were those used in the tetramer-guided mapping of epitopes in the E protein for YFV (18). YFV, yellow fever virus.
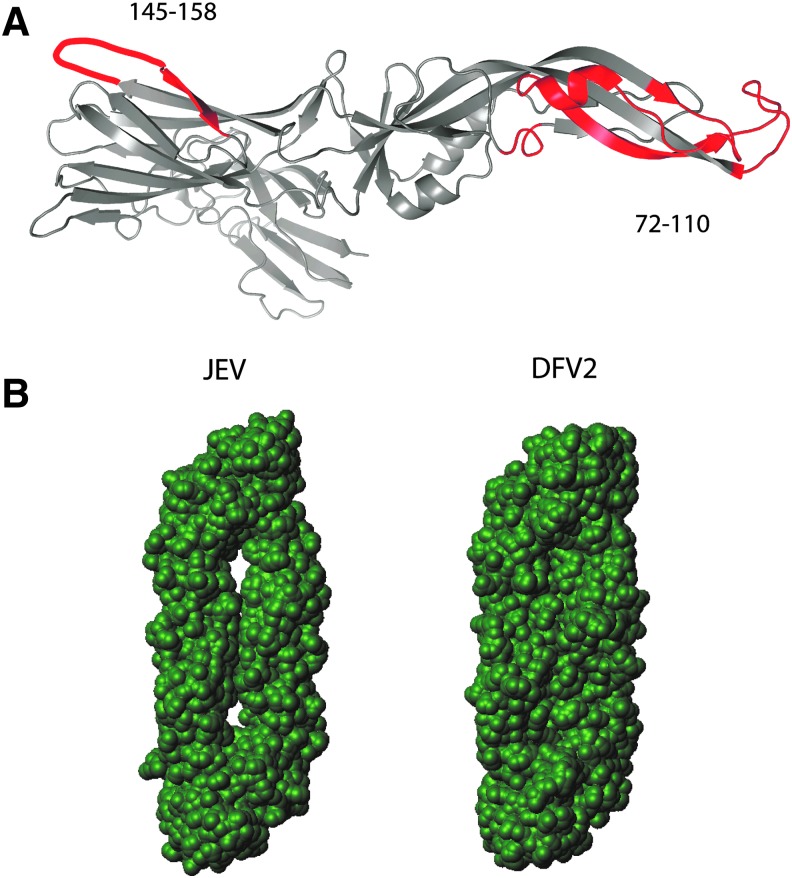
FIG. 4.
Structures of flavivirus E proteins in the prefusion conformation. In (A), the ribbon diagram of E protein from TBEV, illustrating prominent flexible loops that are poorly immunogenic for CD4+ T cells. In (B), the solvent-accessible area for E proteins from JEV and DFV2, illustrating greater solvent accessibility between monomers of the JEV dimer. JEV, Japanese encephalitis virus.
Since the flavivirus structures are well conserved, we examined the predicted binding of the same group of seven HLA molecules to E from TBEV and DFV2. For these two viruses, the dip in MHCII affinity was not so prominent (Fig. 3). From the information presently available, we cannot determine whether the seven HLA molecules in the YFV study adequately represent the human populations that yielded the epitope maps for these three viruses. On the one hand, it is possible that the collection of HLA molecules was appropriate for YFV, but not for DFV2, because the two viruses came under selective pressure to reduce MHCII binding affinity in different human populations. On the other hand, the lack of MHCII affinity may be only part of the explanation for the gap in CD4+ epitope immunogenicity. Another potential explanation is that surface exposure and conformational flexibility of the gap sequence make it a target of destructive proteolytic antigen processing.
The second gap in the CD4+ epitope map of E from YFV (residues 150–158) exhibits typical affinities for the HLA molecules tested; and thus, its poor immunogenicity cannot be attributed to weak MHCII binding (Fig. 3). This gap corresponds to a surface-exposed loop (example for TBEV in Fig. 4A) that becomes completely disordered in the postfusion E structures of TBEV, DFV1, DFV2, and SLEV. We hypothesize that potential epitopes in this segment of E were destroyed by an early proteolytic processing event.
If we limit our attention to CD4+ epitopes that were not identified by tetramer-guided mapping, the distribution of epitopes corresponds reasonably well to the profile of epitope processing likelihood generated from structural constraints on antigen processing and presentation (example of DFV2 in Fig. 2, bottom). The calculation of processing likelihood is based on association of the epitope with a nearby conformationally flexible site that may serve as a site for antigen processing (34). Epitopes were scored by the number of viruses and then the profile was tested for correlation with the profile for prediction of epitopes in the E protein of DFV2, resulting in a modest, but significant correlation of 0.4 and p < 0.001 (data not shown). Upon examination of the individual E profiles, the influence of structural considerations becomes more apparent. Of the 31 epitopes identified by experiment, 15 were predicted with at least 80% processing likelihood (Fig. 5). The accuracy of epitope prediction is indicated by the percentage of peptides with high processing likelihood, which were actually observed as epitopes. Of the peptides predicted above the 80th percentile, 27% were actually observed as epitopes (using postfusion structures to calculate processing likelihood). This is a significant enrichment over random selection (p < 0.02), although by only twofold. Approximately, the same enrichment was previously obtained with epitope predictions for a collection of nine unrelated antigens (34). We are not aware of any other approach to CD4+ T cell epitope prediction that achieves greater accuracy, and we speculate that substantial improvements in accuracy can be obtained by optimizing the structure-based prediction and combining it with MHCII binding-based prediction.
FIG. 5.
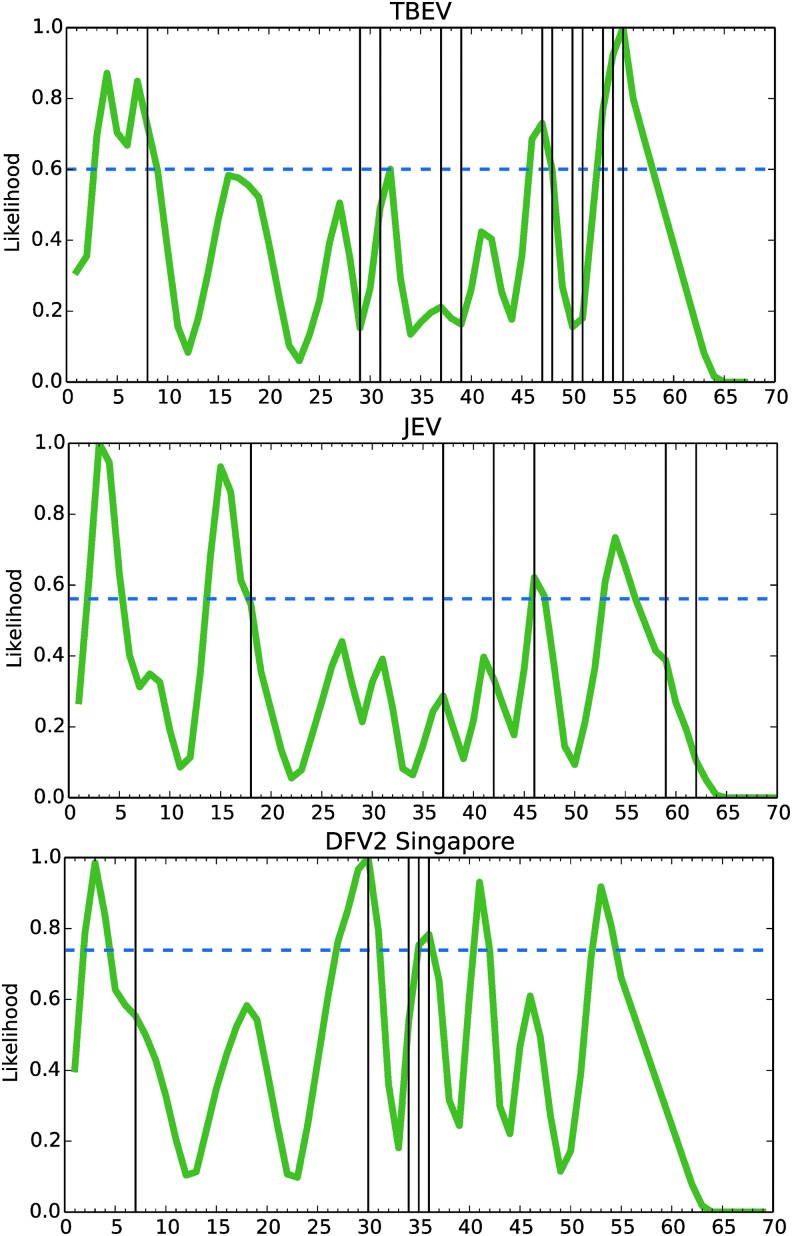
Processing likelihood compared to selected experimental T cell epitope maps. The selected studies excluded tetramer-guided mapping, but included a DFV2 study that did not discriminate CD4+ and CD8+ T cells. Experimentally observed epitopes were reassigned to the corresponding peptides of hypothetical sets (to equalize sampling across studies) and then compared to the processing likelihood. Experimental epitopes were discovered at a rate of 11% of tested peptides (31 epitopes of 276 peptides tested). Experimental epitopes were more than twofold enriched (p < 0.02) within the subset of peptides having high processing likelihood (above the 80th percentile), when based on the postfusion conformations (15 correct of 55 peptides), but not the prefusion conformations (8 correct of 55 peptides). The 80th percentile of peptides ranked by processing likelihood varies by protein (horizontal dashed line).
Differential processing of E protein domains
The different flavivirus E proteins exhibit distinct epitope dominance patterns at the level of protein domains. The CD4+ epitope mapping study of DFV2 found epitopes in domains I and II, but none in domain III or the C-terminal stem and transmembrane domains (Fig. 2). Although the observations are few, we interpret the concentration of epitopes in domains I and II as epitope dominance. This interpretation is supported by an earlier DFV2 epitope mapping study in Vietnam that did not distinguish whether the T cell responses were CD4+ or CD8+ (52). This study should not be disregarded for two reasons. Several epitopes occurred at identical positions in both studies; and, like the later Singapore study, there was a complete absence of epitopes in the C-terminal domains. Thus, we argue that domains I and II dominate the human CD4+ response to DFV2. This contrasts with the maps reported for TBEV and JEV, which have few epitopes in domains I and II and a majority of epitopes in domain III and the C-terminal domains. The YFV and WNV maps cannot be compared to the others because many of the epitopes have not been tested for natural processing. The distinct epitope dominance patterns for DFV2 versus TBEV and JEV could arise at the level of MHCII binding or antigen processing.
The subject populations could have widely different compositions of HLA alleles, which have distinct peptide binding preferences. The DFV2 populations were from Singapore, the JEV population was from South India, and the TBEV populations were European Caucasian. However, if the tetramer-guided mapping results can be taken to indicate typical epitope potential based on MHCII binding, then there would appear to be abundant MHCII binding sequences in each domain (Fig. 3). Thus, it seems unlikely that the MHCII binding preferences would account for the different dominance patterns.
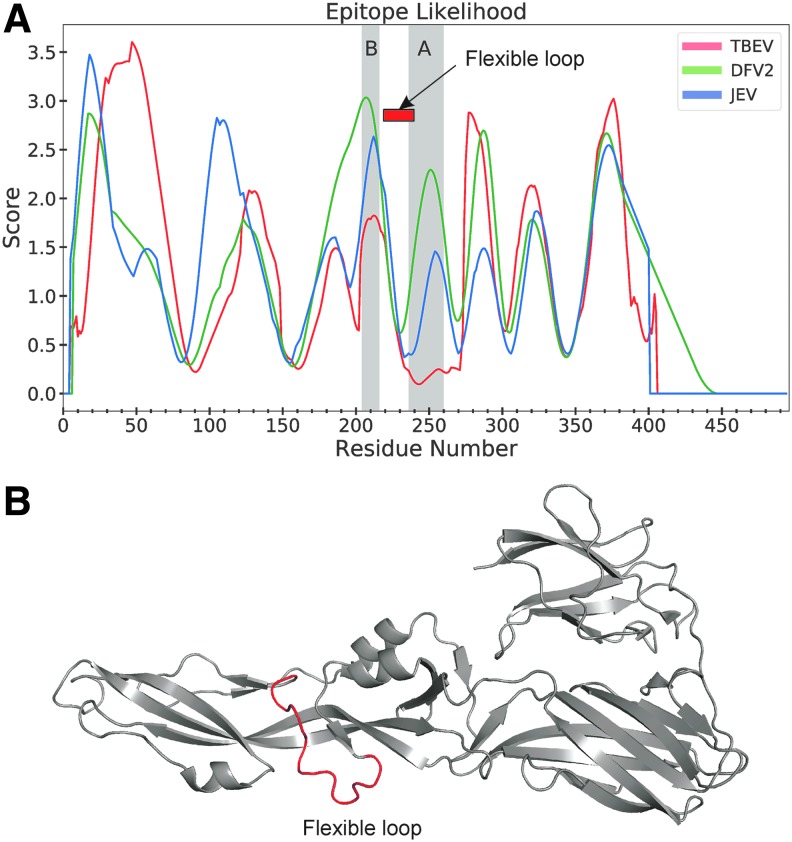
Differential processing is a possible explanation for distinct epitope dominance patterns. The crystallographic structures reveal a much smaller solvent-accessible surface in the prefusion dimer of DFV2, compared to TBEV and JEV (28) (illustrated for JEV and DFV2 in Fig. 4B). Thus, processing proteases could have much less access to domains I and II in the prefusion structure of DFV2, allowing epitopes in those domains to avoid proteolytic destruction. These trends are also reflected in the structure-based predictions, which indicated a greater likelihood of epitopes for DFV2 in the N-terminal domains, especially superclusters A and B, compared to the same regions of E proteins from TBEV and JEV (Fig. 6A). These regions of elevated epitope likelihood coincide with regions of above-average conformational stability on the flanks of the 220–240 loop (Fig. 6A, B).
FIG. 6.
Greater immunogenicity of superclusters A and B in the DFV2 E protein, compared to in E proteins of TBEV or JEV, coincides with greater processing likelihood (A) and conformational stability on the flanks of the flexible loop at residues 220–240, highlighted in the ribbon diagram of E from DFV2 (B).
Destructive processing may account for the lack of epitopes in domains I and II in the CD4+ T cell response of TBEV-infected subjects. The type of exposure, for example, vaccine versus infection, affects innate immune activation and the types of cells present when the dendritic cell (DC) collects, processes, and presents the antigen. DFV2 infection induces innate immune responses through reactive oxygen species (ROS)-dependent signaling, mediated by interferon regulatory factor 3 (IRF3) or signal transducer and activator of transcription 1 (STAT1) and NF-κB (41). ROS also directly activate transcription factor EB (TFEB), the master regulator of lysosome function (46,59). Thus, infection could accelerate the acidification of lysosomes and activate proteolysis, resulting in the proteolytic destruction of epitopes. Although we are unaware if TBEV infection induces ROS as does DFV2 infection, this mechanism could be responsible for distinct epitope dominance patterns in TBEV-infected subjects, compared to TBEV vaccinees. TBEV-infected subjects failed to prime epitopes in E domains I and II, which is consistent with the activation-dependent destruction of those epitopes when E was processed by DC (49).
The implications of prefusion and postfusion conformations for antigen processing and immunity of E protein have been largely unexplored. From the standpoint of protective antibody, the prefusion conformation is the more important target for immunity. However, this need not be the case for T cell responses. Given that the postfusion conformation is induced at lysosomal pH, it seems likely that T cells raised against epitopes derived from the postfusion conformation would be able to provide help to B cells specific for the prefusion conformation. A key requirement for this situation is that processing in the B cell yields at least some of the same peptides as generated by processing in the DC. Although both cell types employ similar proteolytic activities and an acidified antigen-processing compartment, antigen processing is substantially slower in DC and positively regulated by innate immune signals from the environment (33). Given that the immunogenicity of specific epitopes, and even whole proteins, can be determined by individual proteolytic cleavage sites, there appears to be enormous potential for pathogens to evade immune responses, as well as for vaccine design to optimize protection, by modulating the accessibility of proteolytic cleavage sites (1,48).
The CD4+ T cell epitope dominance pattern in flavivirus E is most likely a product of the interaction of E protein conformation and DC proteolytic activities, as modulated by innate immune activation. Theoretically, the striking difference in dominance patterns observed for DFV2 versus JEV and TBEV could be due to enhanced proteolysis and acidification in DCs experiencing increased activation in DFV2. Likewise, the DCs in TBEV-infected patients could be more activated than those in TBEV vaccinees. A difference in acidification could be particularly important because it affects both the activity of the proteases and the conformation of E protein. For example, the postfusion conformation exhibits a drastic increase in disorder at residues 150–158, which could promote destruction of epitopes in that segment. We analyzed epitope processing likelihood for both the prefusion and postfusion conformations of DFV2, TBEV, and JEV. Analysis of only the postfusion conformations produced a significant level of accuracy in epitope prediction (Table 2). We are optimistic that future studies will directly assess the presentation of flavivirus epitopes from multiple cell types in various states of activation, and any shift in epitope dominance will be revealed.
Table 2.
Fraction of Epitopes in E Predicted by Processing Likelihood
| E conformational state | ||
|---|---|---|
| Virus | Prefusion | Postfusion |
| TBEV | 3/12 | 5/12 |
| JEV | 1/7 | 2/7 |
| DFV2 (Singapore) | 2/5 | 3/5 |
| DFV2 (Vietnam) | 2/7 | 5/7 |
| Total | 8/31 | 15/31 |
E, envelope.
It is intriguing that JEV has arguably the lowest frequency of clinical disease and the most solvent-accessible domain I and II of the known flavivirus structures. However, it is unclear whether the prefusion dimer or postfusion trimer is the source of peptides that prime CD4+ T cells. Are peptides from the different structures presented by different cell types? Were the E protein structures under selective pressure to maintain the dimer structure in DC, but convert to the trimer conformation in macrophages? Are responses to the C-terminal epitopes in JEV more effective, or do the N-terminal responses to DFV2 cause excessive immunopathology? The molecular and immunological basis of dengue virus infection enhancement remain under investigation. One possible mechanism is an ineffective or inappropriate T cell response, such as might arise from differentially processed E proteins from different dengue types. Antibodies could also play a role in shaping the T cell response by binding to E and affecting antigen processing, as has been proposed for antibodies that bind to HIV Env glycoprotein (6).
General considerations for CD4+ T cell targeting in flavivirus vaccines
Vaccine designs based on epitope-specific targeting of CD4+ T cells have not found wide application for a number of reasons. Although numerous studies report that CD4+ T cells directly participate in viral clearance, many more studies have documented the importance of CD4+ T cells in promoting antibody and CD8+ T cell responses, which also play important roles in protection. Thus, the most effective vaccine is likely to take the form of a virus, virus subunit, or nucleic acid that displays proteins in a native-like conformation that induces neutralizing antibodies. A native-sized protein subunit is also likely to carry one or more CD8+ T cell epitopes. Our cognizance and manipulation of the CD4+ T cell response under these constraints demand a more complete understanding of antigen processing and presentation.
Acknowledgments
This research was funded by a grant (R21-AI122199) to RRM from the U.S. National Institutes of Health and an NSF training grant (DGE-1144646) fellowship to M.L.F.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Antoniou AN, Blackwood SL, Mazzeo D, et al. Control of antigen presentation by a single protease cleavage site. Immunity 2000;12:391–398 [DOI] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, et al. The global distribution and burden of dengue. Nature 2013;496:504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressanelli S, Stiasny K, Allison SL, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J 2004;23:728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brien JD, Uhrlaub JL, and Nikolich-Zugich J. West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. J Immunol 2008;181:8568–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JF, Choi GK, Yip CC, et al. Zika fever and congenital Zika syndrome: an unexpected emerging arboviral disease. J Infect 2016;72:507–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien PC, Jr, Cohen S, Tuen M, et al. Human immunodeficiency virus type 1 evades T-helper responses by exploiting antibodies that suppress antigen processing. J Virol 2004;78:7645–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowe SR, Miller SC, Brown DM, et al. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine 2006;24:457–467 [DOI] [PubMed] [Google Scholar]
- 8.D'Ortenzio E, Matheron S, Yazdanpanah Y, et al. Evidence of sexual transmission of zika virus. N Engl J Med 2016;374:2195–2198 [DOI] [PubMed] [Google Scholar]
- 9.Daep CA, Muñoz-Jordán JL, and Eugenin EA. Flaviviruses, an expanding threat in public health: focus on dengue, West Nile, and Japanese encephalitis virus. J Neurovirol 2014;20:539–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiebiger E, Meraner P, Weber E, et al. Cytokines regulate proteolysis in major histocompatibility complex class II-dependent antigen presentation by dendritic cells. J Exp Med 2001;193:881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritz R, Stiasny K, and Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol 2008;183:353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gessner BD, and Wilder-Smith A. Estimating the public health importance of the CYD-tetravalent dengue vaccine: vaccine preventable disease incidence and numbers needed to vaccinate. Vaccine 2016;34:2397–2401 [DOI] [PubMed] [Google Scholar]
- 13.Goncalves AJS, Oliveira ERA, Costa SM, et al. Cooperation between CD4+T cells and humoral immunity is critical for protection against dengue using a DNA vaccine based on the NS1 antigen. Plos Negl Trop Dis 2015;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95–98 [Google Scholar]
- 15.Hilser VJ, and Freire E. Structure-based calculation of the equilibrium folding pathway of proteins. Correlation with hydrogen exchange protection factors. J Mol Biol 1996;262:756–772 [DOI] [PubMed] [Google Scholar]
- 16.Iijima N, and Iwasaki A. Access of protective antiviral antibody to neuronal tissues requires CD4 T-cell help. Nature 2016;533:552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James EA, Gates TJ, LaFond RE, et al. Neuroinvasive West nile infection elicits elevated and atypically polarized T cell responses that promote a pathogenic outcome. PLoS Pathog 2016;12:e1005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James EA, LaFond RE, Gates TJ, et al. Yellow fever vaccination elicits broad functional CD4+ T cell responses that recognize structural and nonstructural proteins. J Virol 2013;87:12794–12804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser R. Tick-borne encephalitis. Infect Dis Clin North Am 2008;22:561–575, x. [DOI] [PubMed] [Google Scholar]
- 20.Kanai R, Kar K, Anthony K, et al. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J Virol 2006;80:11000–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein DE, Choi JL, and Harrison SC. Structure of a dengue virus envelope protein late-stage fusion intermediate. J Virol 2013;87:2287–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kollaritsch H, Paulke-Korinek M, Holzmann H, et al. Vaccines and vaccination against tick-borne encephalitis. Expert Rev Vaccines 2012;11:1103–1119 [DOI] [PubMed] [Google Scholar]
- 23.Koradi R, Billeter M, and Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 1996;14:51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- 24.Kupferschmidt K. Infectious diseases. Yellow fever outbreak triggers vaccine alarm. Science 2016;352:128–129 [DOI] [PubMed] [Google Scholar]
- 25.L'Azou M, Moureau A, Sarti E, et al. Symptomatic dengue in children in 10 Asian and latin American countries. N Engl J Med 2016;374:1155–1166 [DOI] [PubMed] [Google Scholar]
- 26.Li J, Chen H, Wu N, et al. Characterization of immune responses induced by inactivated, live attenuated and DNA vaccines against Japanese encephalitis virus in mice. Vaccine 2013;31:4136–4142 [DOI] [PubMed] [Google Scholar]
- 27.Li J, Gao N, Fan D, et al. Cross-protection induced by Japanese encephalitis vaccines against different genotypes of dengue viruses in mice. Sci Rep 2016;6:19953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luca VC, AbiMansour J, Nelson CA, et al. Crystal structure of the Japanese encephalitis virus envelope protein. J Virol 2012;86:2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luca VC, Nelson CA, and Fremont DH. Structure of the St. Louis encephalitis virus postfusion envelope trimer. J Virol 2013;87:818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maciel M, Kellathur SN, Chikhlikar P, et al. Comprehensive analysis of T cell epitope discovery strategies using 17DD yellow fever virus structural proteins and BALB/c (H2d) mice model. Virology 2008;378:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malhotra D, Linehan JL, Dileepan T, et al. Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat Immunol 2016;17:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martins KA, Cooper CL, Stronsky SM, et al. Adjuvant-enhanced CD4 T cell responses are critical to durable vaccine immunity. EBioMedicine 2016;3:67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellman I. Dendritic cells: master regulators of the immune response. Cancer Immunol Res 2013;1:145–149 [DOI] [PubMed] [Google Scholar]
- 34.Mettu RR, Charles T, and Landry SJ. CD4+ T-cell epitope prediction using antigen processing constraints. J Immunol Methods 2016;432:72–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mlakar J, Korva M, Tul N, et al. Zika virus associated with microcephaly. N Engl J Med 2016;374:951–958 [DOI] [PubMed] [Google Scholar]
- 36.Modis Y, Ogata S, Clements D, et al. Structure of the dengue virus envelope protein after membrane fusion. Nature 2004;427:313–319 [DOI] [PubMed] [Google Scholar]
- 37.Muangchana C, Henprasertthae N, Nurach K, et al. Effectiveness of mouse brain-derived inactivated Japanese encephalitis vaccine in Thai National Immunization Program: a case–control study. Vaccine 2012;30:361–367 [DOI] [PubMed] [Google Scholar]
- 38.Nascimento EJ, Mailliard RB, Khan AM, et al. Identification of conserved and HLA promiscuous DENV3 T-cell epitopes. PLoS Negl Trop Dis 2013;7:e2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson RW, Beisang D, Tubo NJ, et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity 2015;42:95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen M, and Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics 2009;10:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olagnier D, Peri S, Steel C, et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 2014;10:e1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul S, Lindestam Arlehamn CS, Scriba TJ, et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods 2015;422:28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol 2010;17:1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rey FA, Heinz FX, Mandl C, et al. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 1995;375:291–298 [DOI] [PubMed] [Google Scholar]
- 45.Rivino L, Kumaran EA, Jovanovic V, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol 2013;87:2693–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roche PA, and Cresswell P. Antigen processing and presentation mechanisms in myeloid cells. Microbiol Spectr 2016;4 [Epub ahead of print]; DOI: 10.1128/microbiolspec.MCHD-0008-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roehrig JT, Risi PA, Brubaker JR, et al. T-helper cell epitopes on the E-glycoprotein of dengue 2 Jamaica virus. Virology 1994;198:31–38 [DOI] [PubMed] [Google Scholar]
- 48.Schneider SC, Ohmen J, Fosdick L, et al. Cutting edge: introduction of an endopeptidase cleavage motif into a determinant flanking region of hen egg lysozyme results in enhanced T cell determinant display. J Immunol 2000;165:20–23 [DOI] [PubMed] [Google Scholar]
- 49.Schwaiger J, Aberle JH, Stiasny K, et al. Specificities of human CD4+ T cell responses to an inactivated flavivirus vaccine and infection: correlation with structure and epitope prediction. J Virol 2014;88:7828–7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Screaton G, Mongkolsapaya J, Yacoub S, et al. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 2015;15:745–759 [DOI] [PubMed] [Google Scholar]
- 51.Sharmin S, Glass K, Viennet E, et al. A Bayesian approach for estimating under-reported dengue incidence with a focus on non-linear associations between climate and dengue in Dhaka, Bangladesh. Stat Methods Med Res 2016. DOI: 10.1177/0962280216649216 [DOI] [PubMed]
- 52.Simmons CP, Dong T, Chau NV, et al. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J Virol 2005;79:5665–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stiasny K, and Heinz FX. Flavivirus membrane fusion. J Gen Virol 2006;87:2755–2766 [DOI] [PubMed] [Google Scholar]
- 54.Tan GS, Leon PE, Albrecht RA, et al. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog 2016;12:e1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turtle L, Bali T, Buxton G, et al. Human T cell responses to Japanese encephalitis virus in health and disease. J Exp Med 2016;213:1331–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Most RG, Harrington LE, Giuggio V, et al. Yellow fever virus 17D envelope and NS3 proteins are major targets of the antiviral T cell response in mice. Virology 2002;296:117–124 [DOI] [PubMed] [Google Scholar]
- 57.Wasserman S, Tambyah PA, and Lim PL. Yellow fever cases in Asia: primed for an epidemic. Int J Infect Dis 2016;48:98–103 [DOI] [PubMed] [Google Scholar]
- 58.Weiskopf D, Bangs DJ, Sidney J, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci U S A 2015;112:E4256–E4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Cheng X, Yu L, et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat Commun 2016;7:12109. [DOI] [PMC free article] [PubMed] [Google Scholar]