Abstract
Background
Anopheles (An.) coluzzii, one of Africa’s primary malaria vectors, is highly anthropophilic. This human host preference contributes greatly to its ability to transmit malaria. In contrast, the closely related An. quadriannulatus prefers to feed on bovids and is not thought to contribute to malaria transmission. The diverged preference for host odor profiles between these sibling species is likely reflected in chemosensory gene expression levels in the olfactory organs. Therefore, we compared the transcriptomes of the antennae and maxillary palps between An. coluzzii and An. quadriannulatus, focusing on the major chemosensory gene families.
Results
While chemosensory gene expression is strongly correlated between the two species, various chemosensory genes show significantly enhanced expression in one of the species. In the antennae of An. coluzzii the expression of six olfactory receptors (Ors) and seven ionotropic receptors (Irs) is considerably enhanced, whereas 11 Ors and 3 Irs are upregulated in An. quadriannulatus. In the maxillary palps, leaving aside Irs with very low level of expression, one Ir is strongly enhanced in each species. In addition, we find divergence in odorant binding protein (Obp) gene expression, with several highly expressed Obps being enhanced in the antennae and palps of An. coluzzii. Finally, the expression of several gustatory receptors (Grs) in the palps appears to be species-specific, including a homolog of a sugar-sensing Drosophila Gr.
Conclusions
A considerable number of Ors and Irs are differentially expressed between these two closely related species with diverging host preference. These chemosensory genes could play a role in the human host preference of the malaria vector An. coluzzii. Additionally, divergence in Obp expression between the two species suggests a possible role of these odor carrier proteins in determining host preference. Finally, divergence in chemosensory expression in the palps may point towards a possible role for the maxillary palps in host differentiation.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4122-7) contains supplementary material, which is available to authorized users.
Keywords: Olfaction, Chemosensory genes, Host seeking, Malaria vector, RNAseq, Antennae, Maxillary palp
Background
Several mosquitoes of the Anopheles gambiae complex are among the most important vectors of human malaria in sub-Saharan Africa. A major determinant for the effectiveness of these species as malaria vectors is their degree of human hosts preference. The species in the complex vary considerably in their host preference, with the highly anthropophilic An. gambiae s.s. and An. coluzzii, collectively referred to as An. gambiae s.l. hereafter, at one end of the spectrum, and the zoophilic An. quadriannulatus, which rarely feeds on humans, at the other [1]. The high preference of An. gambiae s.l. for human hosts translates into a high vectorial capacity for human malaria. In contrast, An. quadriannulatus, a competent vector, albeit less so than An. gambiae s.l. [2, 3], is not thought to contribute to malaria transmission because it rarely feeds on humans in the field [1], although it readily does so in the lab [4].
Host seeking in mosquitoes is primarily modulated by the olfactory system. The antennae and the maxillary palps are the two main olfactory appendages of mosquitoes. Olfactory sensory neurons that express olfactory receptors (ORs) or ionotropic receptors (IRs) are housed inside hair-like sensilla that cover these olfactory appendages [5, 6]. A transduction cascade that sends a signal to the olfactory lobes in the cerebral ganglion of the insect is triggered when odorants bind to the ORs and IRs. Odorants interact directly with these receptors, and therefore differences in host preference between species could be reflected in differences in the expression or molecular structure of the receptors.
Each OR is a heteromeric ligand-gated ion channel that is encoded by the conserved co-receptor Orco, as well as a specific Or [7]. ORs differ in their tuning breadth with some ORs responding to a few odorants, whereas others respond to a wide variety of volatiles [8, 9]. IRs are also heteromeric ligand-gated ion channels, containing up to three different subunits that include one or two of the broadly expressed co-receptors Ir25a, Ir76b and Ir8a [10, 11].
In addition to the receptors, odorant binding proteins (OBPs) play an important role in olfaction, and like the receptors, also interact directly with odorants. OBPs are small, water-soluble carrier proteins that are highly expressed in the sensilla lymph. They transport hydrophobic odorants through the hemolymph to the receptors (reviewed in [12]). Currently, 63 putative AgObps have been identified [13], but not all AgObps are expressed in female antennae [14]. Some OBPs almost certainly play a role in the transport of molecules outside the olfaction system, as some AgObps are expressed elsewhere in the body [13]. In addition, seven chemosensory proteins (CSPs), which are thought to have a similar function as OBPs, have been identified in An. gambiae s.l. [15].
The antennae express by far the largest repertoire of chemosensory genes in mosquitoes [16]. By contrast, the maxillary palps express a much smaller number of chemosensory genes [16, 17]. In mosquitoes, the palps are also responsible for CO2 detection, which in An. gambiae s.l. is modulated by three gustatory receptors; AgGr22–24 [6]. Two Ors are highly expressed in mosquito palps; AgOr8 and AgOr28 in An. gambiae s.l. and their homologs in Aedes aegypti [17] and Culex quinquefasciatus [18]. In An. gambiae s.l. AgOR8 detects the non-human specific host odorant 1-octen-3-ol, whereas AgOR28 is broadly tuned [6]. Therefore, it is clear that the palps play an important role in mosquito host detection, but it is unknown if they could play a role in differentiating between host species’ odor profiles.
An. gambiae s.l. females are attracted to volatiles emanating from human sweat and breath, as well as CO2. Volatiles present on the human skin are believed to be responsible for the human-specific odor profile that attracts An. gambiae s.l. [19, 20]. Human sweat consists of over 350 volatiles, and several of these have been identified as mosquito attractants [21]. For example, An. gambiae s.l. females are attracted to a synergistic blend of ammonia, lactic acid and carboxylic acids [22, 23]. However, the kairomones that An. gambiae s.l. uses to differentiate between humans and alternative hosts remains unknown.
Olfaction gene expression divergence is known to be correlated to differences in host preference between closely related insect species. For example, the expression level of as many as 53% of the Ors and 55% of the Obps differs in the antennae of the generalists D. melanogaster, D. simulans, and their sibling-species D. sechellia, a specialist on the toxic Morinda citrifolia. This number is significantly more than expected based on other gene families [24]. Although neutral evolution may explain some of these changes, the expression level of several genes have undergone a major change along the D. sechellia branch, suggesting an association with the host shift [24]. For example, in D. sechellia the olfactory receptor Or22a is highly up-regulated. Or22a detects a compound emitted by the fruit of D. sechellia’s host plant Morinda citrifolia [25]. Additionally, D. sechellia lost six Or genes compared to its generalist sister-species D. simulans, which did not lose any [26]. Furthermore, accelerated olfactory receptor loss has also been associated with host specialization in D. erecta [27].
A link between differential olfaction gene expression and host preference has been observed in a mosquito species as well. A recent study of Aedes aegypti tied expression differences of AeOr4 between the domestic Ae. aegypti aegypti and the sylvatic Ae. aegypti formosus to their differential host preference [28]. Furthermore, a previous comparison between the antennal transcriptome of An. coluzzii and An. quadriannulatus identified divergence in olfaction gene expression that could be related to their respective host preference [29]. This previous study was conducted during the light phase of the circadian cycle, which could have affected the results as it has been shown that olfactory gene expression can fluctuate across the circadian cycle [30]. Because Anopheles mosquitoes are night time feeders, we compared the transcriptome of the female antennae of this anthropophilic An. colluzii and zoophilic An. quadriannulatus, during the early dark cycle when both species are actively seeking hosts [31, 32]. In addition, we expand on the earlier work by comparing chemosensory gene expression between these species in the second olfactory organ, the maxillary palps.
Results
Host choice assay
We measured the preference of An. coluzzii and An. quadriannulatus laboratory strains to human odor vs cow odor in a dual choice olfactometer. Consistent with the host preference of these species in the field and previous studies, An. coluzzii significantly preferred human odor (77%, N = 770, p < 0.0001), Whereas An. quadriannulatus was significantly more attracted to cow odor (67%, N = 330, p = 0.0029). Therefore, the laboratory colonies of these species have largely preserved their natural host preference.
Differential gene expression
Two replicate female antennae and two replicate maxillary palp transcriptome datasets were obtained for each of the species using RNAseq technology. For each library, between 30 and 46 million reads were generated (Table S1). After mapping of the reads to the Anopheles gambiae genome (Agam Pest Version 3.), we counted the uniquely mapped reads using the program HT-Seq [33], followed by statistical analyses using DeSeq2 package [34] on the R statistical platform. We only considered a gene to be expressed if more than 50 reads were mapped.
A total of 8857 and 9168 annotated genes were detected in the antennae of An. coluzzii and An. quadriannulatus respectively (Fig. 1a, Additional file 1). Of these, the expression of a total of 1595 genes was significantly diverged between the two species. The expression of a total of 708 genes was significantly enhanced in An. coluzzii antennae, whereas 887 genes were significantly up-regulated in An. quadriannulatus.
Fig. 1.
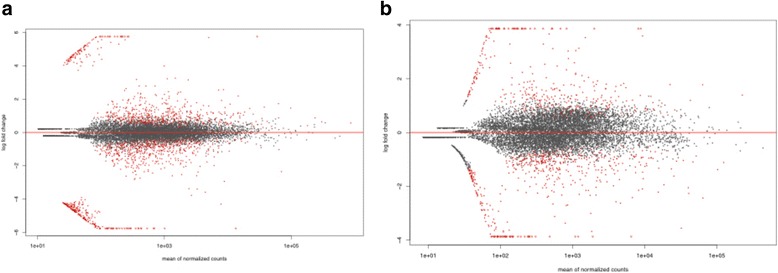
Plots of LogFC (Fold Change) against The Mean of Normalized Counts among the antennae (a) and maxillary palps (b) between An. coluzzi and An. quadriannulatus. Each dot represent a measure of abundance, with red colored dots indicating statistically significant differences in expression. logFC values above zero are those that are upregulated in An. coluzzii where as those below zero are upregulated in An. quadriannulatus
An average of 9207 and 8820 annotated genes were detected in the maxillary palps of An. coluzzii and An. quadriannulatus respectively (Fig. 1b, Additional file 1). Of these, 713 genes were differentially expressed between the two species, with 437 genes significantly enhanced in An. coluzzii and 296 in An. quadriannulatus.
Olfactory receptors
The expression of Ors was highly correlated between the two species in both the antennae (R2 = 0.99 including Orco, R2 = 0.84 excluding Orco) (Figure 2A), and the maxillary palps (R2 = 0.99 including or excluding Orco) (Fig. 3A). As expected, the co-receptor Orco was the most abundantly expressed Or in both olfactory organs, and was expressed at similar levels in both species. Of the remaining 74 known Ors, nine (Or3, 4, 5, 20, 34, 37, 40, 52 and 58) were detected (< 50 reads aligning) in the antennae or palps of neither species.
Fig. 2.
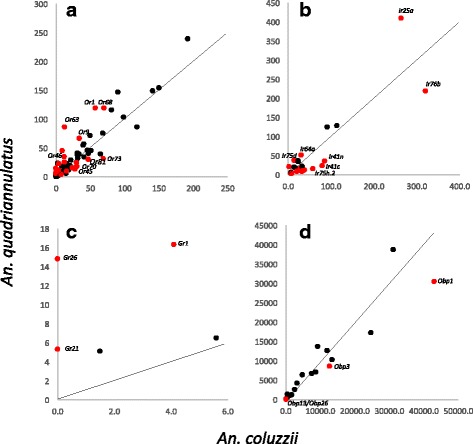
A comparison of the expression of four chemosensory gene families in the female antennae of the anthropophilic An. coluzzii and the zoophilic An. quadriannulatus. a: Ors, (b): Irs, ( c): Grs, (d): Obps. The line indicates equal expression between the two species. For the olfactory receptors Orco was excluded
Fig. 3.

A comparison of the expression of four chemosensory gene families in the female maxillary palps of the anthropophilic An. coluzzii and the zoophilic An. quadriannulatus. a: Ors, (b): Irs, ( c): Grs, (d): Obps. The line indicates equal expression between the two species. For the olfactory receptors Orco was excluded
Although it is unknown what expression level of receptor genes signifies functional relevance, it is doubtful that genes detected at very low level play a significant functional role. Therefore, we focus here on receptor genes that are expressed at >5 rpkm. Of the remaining 66 Ors, nine were expressed above 5 rpkm and significantly up-regulated in An. coluzzii antennae (Or36, 45, 57, 69, 70, 71, 72, 73, 81) (Table 1, Additional file 1). Of these, Or36, 45, 69, 70, 71, 73 and 81 stand out because they are enhanced between 1.6 and 2.4-fold in the anthropophilic An. coluzzii. All of these are expressed >14.6 rpkm in this species (Table 1).
Table 1.
Chemosensory genes significantly enhanced in the female antennae. Only receptor genes with rpkm > 5, or Obp/Csp genes with rpkm > 50 in either species are included. Fold change and log2 fold change values are presented as positive values if expression is enhanced in An. coluzzii and as negative values if expression is enhanced in An. quadriannulatus
| Gene Stable ID |
Gene name |
RPKM colluzzi |
RPKM quad |
Fold Change | log2Fold Change | padj |
|---|---|---|---|---|---|---|
| AGAP009707 | Or71 | 14.6 | 5.6 | 2.4 | 1.2 | 0.0301 |
| AGAP009719 | Or73 | 69.0 | 30.7 | 2.3 | 1.2 | 0.0000 |
| AGAP003053 | Or45 | 27.8 | 12.8 | 2.2 | 1.1 | 0.0002 |
| AGAP004357 | Or57 | 7.5 | 3.9 | 1.9 | 0.9 | 0.0259 |
| AGAP009718 | Or70 | 30.5 | 17.4 | 1.8 | 0.9 | 0.0000 |
| AGAP001012 | Or36 | 15.4 | 9.5 | 1.7 | 0.8 | 0.0087 |
| AGAP013512 | Or81 | 47.1 | 29.7 | 1.6 | 0.7 | 0.0083 |
| AGAP009705 | Or69 | 24.0 | 15.1 | 1.6 | 0.7 | 0.0326 |
| AGAP009718 | Or72 | 30.0 | 23.8 | 1.3 | 0.4 | 0.0253 |
| AGAP004354 | Or26 | 40.4 | 56.8 | −1.3 | −0.4 | 0.0407 |
| AGAP002044 | Or77 | 15.5 | 22.3 | −1.4 | −0.4 | 0.0321 |
| AGAP007797 | Or23 | 90.0 | 146.5 | −1.5 | −0.6 | 0.0081 |
| AGAP009704 | Or68 | 70.0 | 118.7 | −1.6 | −0.7 | 0.0000 |
| AGAP008333 | Or9 | 34.1 | 66.2 | −1.8 | −0.8 | 0.0081 |
| AGAP009411 | Or21 | 12.8 | 25.0 | −1.8 | −0.9 | 0.0012 |
| AGAP009640 | Or1 | 57.2 | 118.9 | −1.9 | −1.0 | 0.0000 |
| AGAP009412 | Or50 | 3.2 | 7.3 | −2.1 | −1.1 | 0.0000 |
| AGAP009390 | Or53 | 3.8 | 11.2 | −2.5 | −1.3 | 0.0022 |
| AGAP009392 | Or46 | 12.0 | 34.2 | −2.6 | −1.4 | 0.0000 |
| AGAP011991 | Or61 | 9.3 | 44.9 | −3.4 | −1.8 | 0.0028 |
| AGAP005760 | Or33 | 4.1 | 22.5 | −5.0 | −2.3 | 0.0000 |
| AGAP011990 | Or64 | 2.7 | 23.2 | −5.8 | −2.5 | 0.0000 |
| AGAP011989 | Or63 | 12.6 | 85.8 | −6.1 | −2.6 | 0.0000 |
| AGAP009410 | Or18 | 0.0 | 5.7 | −32.6 | −5.0 | 0.0000 |
| AGAP009391 | Or30 | 0.0 | 6.8 | −49.4 | −5.6 | 0.0000 |
| AGAP009720 | Or74 | 0.0 | 14.3 | −75.0 | −6.2 | 0.0000 |
| AGAP007498 | Ir75k | 32.3 | 8.0 | 4.1 | 2.0 | 0.0000 |
| AGAP001812 | Ir75h.2 | 57.7 | 15.5 | 3.7 | 1.9 | 0.0000 |
| AGAP012951 | Ir41c | 79.7 | 23.0 | 3.6 | 1.8 | 0.0000 |
| AGAP013416 | Ir7w | 35.4 | 10.2 | 3.4 | 1.8 | 0.0000 |
| AGAP000140 | Ir100a | 38.2 | 11.7 | 3.3 | 1.7 | 0.0000 |
| AGAP013285 | Ir7u | 9.0 | 3.3 | 2.8 | 1.5 | 0.0000 |
| AGAP013363 | Ir7i | 6.7 | 2.7 | 2.5 | 1.3 | 0.0000 |
| AGAP002763 | Ir7t | 20.7 | 8.5 | 2.5 | 1.3 | 0.0000 |
| AGAP003531 | Ir41n | 85.5 | 35.3 | 2.5 | 1.3 | 0.0000 |
| AGAP013085 | Ir75g | 28.6 | 15.2 | 1.9 | 0.9 | 0.0272 |
| AGAP011968 | Ir76b | 320.0 | 219.0 | 1.5 | 0.6 | 0.0283 |
| AGAP010272 | Ir25a | 263.6 | 409.4 | −1.5 | −0.6 | 0.0000 |
| AGAP004923 | Ir64a | 30.9 | 51.1 | −1.6 | −0.6 | 0.0013 |
| AGAP004969 | Ir75d | 14.1 | 37.2 | −2.4 | −1.3 | 0.0000 |
| AGAP001811 | Ir75h.1 | 2.6 | 20.1 | −6.7 | −2.7 | 0.0000 |
| AGAP004114 | Gr1 | 4.1 | 16.3 | −3.6 | −1.8 | 0.0000 |
| AGAP003260 | Gr21 | 0.0 | 5.3 | −43.9 | −5.5 | 0.0000 |
| AGAP006717 | Gr26 | 0.0 | 14.8 | −105.6 | −6.7 | 0.0000 |
| AGAP002905 | Obp13 | 140.6 | 13.7 | 9.3 | 3.2 | 0.0000 |
| AGAP012321 | Obp26 | 150.8 | 57.4 | 2.4 | 1.3 | 0.0218 |
| AGAP001409 | Obp3 | 12,763.8 | 8566.4 | 1.6 | 0.6 | 0.0000 |
| AGAP003309 | Obp1 | 43,007.8 | 30,392.5 | 1.5 | 0.6 | 0.0375 |
Similarly, a total of 17 Ors were expressed above 5 rpkm and significantly up-regulated in An. quadriannulatus antennae (Or1, 9, 18, 21, 23, 26, 30, 33, 46, 50, 53, 61, 63, 64, 68, 74 and 77) (Table 1). Of these, all but Or23, 26 and 77 are enhanced between 1.6 and 75.0 fold in this species, and all but Or18, 30, 50, and 53 are expressed >14.3 rpkm. Or18, 30 and 74 are detected only in the antennae of this species, although at relatively low levels; between 5.7 and 14.3 rpkm.
Because some genes listed in Table 1 have relatively low fold change values (<2), the RPKM and fold change values for the replicate samples are reported in Additional file 2: Table S3. For all these Ors, the expression differences between species is consistent between replicate samples, although in the case of Or69 the fold change for the second replicate of each species is considerably lower than for the first. For the majority of these Ors, including Or69, Rinker et al. [29] also reported considerable enhancement, although we should not necessarily expect their results to be the same because their sample represents day time expression.
As reported previously [6, 16], only two specific Ors are expressed at high level in the palps of An. coluzzii; Or8 and Or28. This is true for An. quadriannulatus as well (Figure 3A, Table 2). No significant differences in the expression of these genes was observed. Several other Ors were detected at very low levels; < 2.8 rpkm in An. coluzzii and <5.0 rpkm in An.quadriannulatus (Additional file 1). However, as these correspond to the most highly expressed antennal Ors, their detection is likely due to a small amount of antennal contamination during dissection.
Table 2.
Chemosensory genes significantly enhanced in the female maxillary palps. Only receptor genes with rpkm > 5, or Obp/Csp genes with rpkm > 50 in either species are included. Fold change and log2 fold change values are presented as positive values if expression is enhanced in An. coluzzii and as negative values if expression is enhanced in An. quadriannulatus
| GENE stable ID | Gene name | RPKM coluzzi | RPKM quad | Fold change | Log2 Fold Change | padj |
|---|---|---|---|---|---|---|
| AGAP013416 | Ir7w | 6.4 | 0.0 | 27.7 | 4.8 | 0.0000 |
| AGAP003531 | Ir41n | 5.8 | 0.0 | 19.0 | 4.2 | 0.0000 |
| AGAP000140 | Ir100a | 31.9 | 6.7 | 3.8 | 1.9 | 0.0001 |
| AGAP002904 | Ir41a | 6.9 | 27.1 | −3.1 | −1.6 | 0.0011 |
| AGAP001173 | Gr52 | 9.5 | 0.0 | 22.7 | 4.5 | 0.0000 |
| AGAP006717 | Gr26 | 0.0 | 16.1 | −62.3 | −6.0 | 0.0000 |
| AGAP003260 | Gr21 | 0.0 | 62.9 | −198.7 | −7.6 | 0.0000 |
| AGAP012320 | Obp25 | 2798.6 | 766.8 | 3.1 | 1.6 | 0.0010 |
| AGAP011368 | Obp57 | 10,799.9 | 3525.1 | 2.6 | 1.4 | 0.0141 |
| AGAP012321 | Obp26 | 240.5 | 74.4 | 2.5 | 1.3 | 0.0390 |
| AGAP008054 | Sap3 | 1117.4 | 2619.4 | −2.3 | −1.2 | 0.0000 |
Ionotropic receptors
The expression of Irs was highly correlated between the two species if the co-receptors Ir8a, Ir25a and Ir76b were included (R2 = 0.801 for antennae, R2 = 0.842 for palps, Figure 2B). However, if the co-receptors were excluded, Ir expression diverged considerably, especially in the palps (R2 = 0.53 for antennae, R2 = 0.157 for palps, Figure 3B). Of the 44 annotated Irs in the Anopheles gambiae genome, 11 were expressed (<50 reads) in the antennae of neither species (Additional file 1). The co-receptors, Ir76b, Ir25a and Ir8a are the most highly expressed Irs in both species: between 114.3 and 409.4 rpkm (Additional file 1). The use of these co-receptors appears to have diverged somewhat between the two species, with Ir76b significantly 1.5-fold enhanced in An. coluzzii versus Ir25a expressed 1.5-fold more in An. quadriannulatus. These differences were consistent within replicates (Additional file 2: Table S3).
Of the remaining Irs, ten (Ir7i, 7t, 7u, 7w, 41c, 41n, 75g, 75h.2, 75k, and 100a) are significantly higher expressed in An. coluzzii and expressed >5 rpkm (Table 1). Of these, all but Ir7u and 7i are expressed at >20 rpkm and between 1.9 and 4.1-fold enhanced in this species. The enhancement of Ir75g, the only Ir with fold change <2, was consistent between replicates (Additional file 2: Table S3).
In contrast, only three of the remaining Irs (64a, 75d, and 75h.1) are expressed above 5 rpkm and significantly enhanced in An. quadriannulatus. All these are expressed >20 rpkm and are between 1.6 and 6.7-fold enhanced in An. quadriannulatus (Table 1). The enhancement of Ir64a the only Ir with fold change <2, was consistent between replicates (Additional file 2: Table S3).
A total of 14 Irs were detected in the maxillary palps of An. coluzzii, but only Ir7w, 25a, 41a, 41n, 76b and 100a are expressed at >5 rpkm (Additional file 1). As in the antennae, the co-receptor Ir25a was the most highly expressed ionotropic receptor in both species. Three Irs (7w, 41n, and 100a), are expressed above 5 rpkm and significantly enhanced in the maxillary palps of An. coluzzii (Table 2). Both Ir7w and 41n were detected only in An. coluzzii, although at low level (6.4 and 5.8 rpkm respectively). Ir100a however is both the most highly expressed ionotropic receptor in An. coluzzii palps (31.9 rpkm), and is 3.8- fold enhanced in this species. This receptor is also significantly 3.3-fold enhanced in the antennae of this species, with a similar expression level (38.2 rpkm), but was barely detected in the body [16].
Only six Irs are expressed in An. quadriannulatus palps, five of which are detected at >5 rpkm. Of these, only Ir41a is significantly enhanced in the palp of An. quadriannulatus. It is also the most highly expressed specific Ir in the palps of this species (27.1 rpkm) and is 3.1-fold enhanced compared to An. coluzzii. This gene is not significantly enhanced in the antennae of this species, although expressed at a similar level (33.7 rpkm, 1.3 fold). Its mRNA is also mostly absent from the body, at least in An. coluzzii [16].
Gustatory receptors
The expression of Grs showed little correlation between the two species in the antennae (R2 = 0.233, Figure 2C) (Fig. 2D). Of the 59 annotated Grs in the Anopheles gambiae genome, 39 were detected (<50 reads mapping) in the antennae of neither species, with the remaining Grs generally expressed <5 rpkm, (Additional file 1). In An. coluzzii, Gr55 is the most highly expressed (5.6 rpkm). In An. quadriannulatus, Gr1, Gr21 and Gr26 are significantly enhanced and expressed >5 rpkm. Interestingly, Gr26 is only detected in An. quadriannulatus, being expressed at 16.3 rpkm. (Table 1).
In contrast, nine Grs were detected in the palps of An. coluzzii and An. quadriannulatus, although these do not fully overlap (Additional file 3: Table S1, Figure 3c). Gr22, 23 and 24, which together encode the CO2 receptor, are of course by far the most highly expressed Grs in both species (262.9 < rpkm <729.8). Their expression level is similar between the two species, suggesting a similar sensitivity to CO2. The expression of Grs in the palps was therefore highly correlated between the two species (R2 = 0.901) if the CO2 receptor genes were included. However, this correlation disappeared completely if only the remaining Grs were examined (R2 = 0.001). A single Gr, Gr52, is specific to An. coluzzii, being detected only in this species, although in the lower range (9.5 rpkm, Table 2). None-the-less, its enhancement in An. coluzzii is highly significant (q < 0.0001). Gr52 is expressed at low levels in the antennae of this species as well (3.3 rpkm), but not in An. quadriannulatus. Two Grs (Gr21, 26) are expressed >5 rpkm and significantly enhanced in the palps of An. quadriannulatus, while not detected in An. coluzzii. Of these, Gr21 stands out by being both specific to An. quadriannulatus, and the most highly expressed Gr (62.9 rpkm) besides the CO2 receptor genes. Gr26 is also specific to An. quadriannulatus, although with a lower expression level (16.1 rpkm). In the antennae both Gr21 and Gr26 are also expressed only in An. quadriannulatus, although at a much lower level (5.0 and 11.6 rpkm, respectively), suggesting a primarily palp-specific role.
Odor-binding proteins
The expression of Obps was highly correlated between the two species (R2 = 0.915 for antennae, R2 = 0.990 for palps, Figure 2D and 3D). Thirty-four of the 63 annotated Obp s were detected in neither species in our antennae dataset (<50 reads), with another 11 expressed <50 rpkm in both species (Additional file 1). Obps comprise nine of the 10 most highly expressed genes in the antennae. Because of their high overall expression level relative to the receptor genes, we focus here only on those with expression >50 rpkm. Four Obps (Obp1, 3, 13, and 26) are significantly enhanced in An. coluzzii antennae and expressed >50 rpkm (Table 1). However, of these only Obp1 and 3 are highly expressed with rpkm values of 43,008 and 12,764 respectively. These two genes are 1.5 to 1.6-fold enhanced in An. coluzzii, with consistent expression differences between species across replicates (Additional file 2: Table S3). The expression level of Obp13 and 26 is below 151 rpkm, which is in the very low range for Obps. No Obp expressed >50 rpkm was enhanced in An. quadriannulatus antennae.
Despite the high correlation between Obp expression in the maxillary palps of the two species, the expression of several Obps has diverged substantially. Four Obps expressed >50 rpkm (Obp25, 26, 57 and 67) are significantly enhanced in Anopheles coluzzii. Of these, Obp25 and Obp57 stand out. They are among the four most highly expressed Obps in the maxillary palps of An. coluzzii (2798 and 10,800 rpkm respectively), and are considerably enhanced (3.1 and 2.6-fold respectively). Both Obp25 and Obp57 are expressed at much lower level in the antennae (407.1 and 63.0 rpkm, respectively) and therefore appear to be mostly palp-specific. In contrast, no Obp expressed at >50 rpkm is significantly enhanced in An. quadriannulatus palps. Interestingly, overall Obp expression in the An. quadriannulatus palps is 1.8-fold lower compared to An. coluzzii (33,148 vs 58,482 rpkm). This is not the case for Obp expression in the antennae.
Chemosensory proteins
Like the Obps, Csp genes probably function as odorant carrier molecules and are expressed at levels considerably higher than the receptors (as high as 4992 rpkm in the antennae and 15,504 rpkm in the maxillary palps, Additional file 1). In both the palps and antennae, the expression of these genes was highly correlated between species (R2 = 0.995 and R2 = 0.977, respectively). All seven annotated Csp genes, following Iovinella et al. [15], were detected in the antennae of both species, although several at very low level. No Csp showed enhanced expression in the antennae of either species. All seven Csps were detected in the palps of both species as well, with six expressed at >50 rpkm. One of these, Sap3, is significantly enhanced (2.3-fold) in the palp of An. quadriannulatus (Table 2). This gene is expressed at more similar levels (1.4 fold) in the antennae of the two species. The Sap3 gene is the second most highly expressed Csp in the palp of both species.
Overall Csp expression is approximately 3-fold higher in the palps of An. coluzzii than its antennae, and 2-fold higher in the palps vs antennae of An. quadriannulatus. Interestingly, Sap1 is the 3rd and 6th most highly expressed gene overall in the palps of An. coluzzii and An. quadriannulatus (15,404 and 11,762.6 rpkm, respectively), whereas it is expressed at much lower levels in the antennae (658 and 655 rpkm respectively). Vice versa, Csp3 appears to be largely antennae specific (3719 and 4992 rpkm in An. coluzzii and An. quadriannulatus respectively), with very low expression in the palps (92,7 and 64,7 rpkm in An. coluzzii and An. quadriannulatus respectively).
Gene ontology analysis
A gene ontology analysis (GO) was conducted using the online gene ontology database pantherdb (www.pantherdb.org) to recover descriptions of molecular function. We performed the GO analyses only on genes that were upregulated in the antennae or the maxillary palps of each species. This resulted in 708 antennal genes in An. coluzzii and 887 antennal genes in An. quadriannulatus. For the maxillary palps, 437 and 296 genes met these criteria in the two species, respectively. Not surprisingly, the molecular functions of the significantly enhanced genes are connected to olfaction (e.g., “protein binding”, “receptor activity”) and signal transduction (e.g “transporter activity”, “transmembrane transporter activity”). The Panther GO Biological Process classification returned “Sensory perception of smell” and “Chromatin Assembly” as the top two terms. Furthermore, we tested for enrichment of specific protein families, which showed that, based on number of genes hit against protein class, the top three protein families enriched in An. coluzzii antennae were Hydrolase (PC00121), Receptor (PC00192), and Nucleic Acid Binding (PC00171). In contrast, based on genes upregulated in An. quadriannulatus antennae showed that the protein families enriched were Oxidoreductase (PC00176), Hydrolase (PC00121), and Transferase (PC00220).
In the maxillary palps, the enriched protein classes were as follows. In An. coluzzii, they were Oxidoreductase (PC00176), Hydrolase (PC00121) and Nucleic Acid Binding (PC00171). In An. quadriannulatus, the top three protein families were Hydrolase (PC00121), Transporter (PC00227), and Oxidoreductase (PC00176). The entire list of protein families for each tissue is shown in Additional file 4: Table S4.
Discussion
The host seeking behavior of the sister species An. gambiae and An. coluzzii is primarily modulated by their olfactory system [1]. It is therefore expected that their preference for human hosts has a strong basis in the genes that make up the most important components of this system. The olfactory system is primarily housed in the antennae, which express a large number of olfactory genes. However, the maxillary palps also play an important role in host detection as they contain the receptor for CO2, as well as two olfactory receptors that detect the host kairomone 1-octen-3-ol and a range of other chemicals [7]. Additionally, olfactory receptors are expressed in the labellum of mosquito proboscises which may play a role in discrimination host at short range [35, 36].
Here we compared the transcriptomes of the antennae and maxillary palps of two species of malaria mosquitoes with diverging host preference during the dark cycle when they are actively seeking hosts: the anthropophilic An. coluzzii and the zoophilic An. quadriannulatus. We focused on five chemosensory gene families that interact directly with odorants. The expression profiles of these chemosensory gene families generally show a high correlation between the two species, but considerable differences in expression of individual chemosensory genes are present. These could point to genes responsible for the diverging host preference between these closely related species.
Divergence in olfaction gene expression associated with host specialization has been observed in other insects. Recently, it was shown that divergent expression of AaOr4, an olfactory receptor without an ortholog in An. gambiae s.l., between an anthropophilic and zoophilic form of Aedes aegypti contributes to differences in host preference [28]. Similarly, specialization to host plants has been accompanied by differential expression of olfactory receptors between closely related Drosophila species. Or expression in the antennae differs markedly between the generalists D. melanogaster, D. simulans, and their specialist sister-species D. sechellia, with the expression of up to 53% of Ors differing between species pairs [24]. Therefore, the enhanced expression of some of the chemosensory genes in An. coluzzii could very well be linked to their preference for human odor.
In the antennal transcriptome of An. coluzzii, seven Ors (Or36, 45, 69, 70, 71, 73 and 81) and eight Irs (Ir7w, 7t, 75h.2, 75g, 75k 41n, 41c, and 100a) stand out by being among the more highly expressed receptor genes, while also being considerably up-regulated compared to An. quadriannulatus (1.6 to 4.8-fold). Previous studies have reported the response of 56 AgORs to a range of odorants [8, 9]. Of the seven Ors enhanced in An. coluzzii that are highlighted here, only OR36 showed a positive response to the tested odorants. In comparison with other tested ORs it appears to be narrowly tuned, and it responded strongly to only two odorants, neither of which are of human origin [9]. However, 346 volatiles have been identified in human sweat [21], and only a subset of these volatiles have been tested.
The relative role of ORs vs IRs in host differentiation is not entirely clear. IRs are thought to detect several important human kairomones; lactic acid, carboxylic acids and amonia [37–40]. Although IRs play a role in the detection of host odors, this role may depend on the presence of CO2, as Aedes aegypti females in which Orco is knocked-out are attracted to human hosts, but can no longer distinguish between different vertebrate hosts [41]. This suggests that in Ae. aegypti IRs do not play a role in host differentiation. However, it is not clear that this rules out a potential role of IRs in human host preference in Anopheles. The divergence in Ir expression in the olfactory tissues observed in this study suggests some species-specific role of these receptors.
OBPs carry odorants through the hemolymph to olfactory receptor neurons and are essential for the proper function of the olfactory system [12]. For example, Drosophila melanogaster that carry a mutant OBP called LUSH, do not detect an aggregation pheromone [42], and Obp57d and Obp57e are linked to diverging oviposition behavior between Drosophila species [43]. In An. gambiae RNAi knock-down of AgObp1 abolishes the antennal response to indole [44]. Interestingly, knocking down AgObp4, which is expressed primarily in the antennae, also suppresses blood feeding from a membrane feeder which does not rely on odor cues [45]. In Culex quinquefasciatus knockdown of CqObp1 results in lowered response to an oviposition pheromone [46]. The importance of some OBPs to the olfactory system of insects is therefore well established, but it is not clear if they contribute to host selectivity [12, 17].
We note the divergence in the expression of several AgObps between the two species. In the antennae, the highly expressed Obp1 and 3 are enhanced 1.5 to 1.6 -fold in An. coluzzii. More remarkable however is the enhancement of the highly expressed Obp25 and 57 in the palps of this species. These two Obps are enhanced 2.6 to 3.1–fold in this species compared to An. quadriannulatus. In addition, Obp25 is expressed in the antennae of An. coluzzii at only appr. 0.15-fold level compared to the palps, and Obp57 is expressed at very low level in the antennae (63.0 rpkm). Additionally, both these Obps are detected only at very low levels in the body of An. coluzzii [16]. This suggests a mostly palp-specific function of these genes.
Although the maxillary palps obviously have an important role in host seeking by detecting CO2 [47], to our knowledge no one has examined if these organs function in host differentiation in mosquitoes. The small and similar AgOr repertoire in the palps suggests that they do not [6]. However, the considerable difference in the expression of several palp-specific Obps, as well as Ir100a and Ir41a, does suggest a species-specific role of the maxillary palps in olfaction.
In addition, several Grs showed a species-specific expression pattern in the maxillary palps. Gr52 is specific to An. coluzzii, and Gr21 and Gr26 are both specific to An. quadriannulatus palps in this study, although Gr21 was detected at low levels in An. coluzzii palps in a previous study [16]. We found an approximately 3-fold higher expression of Gr52 in the palps vs the antennae in this species. However, a previous study found that the transcript of this gene is present in the whole bodies, antennae and palps of this species at similar levels, with only approximately 1.5-fold enhancement in the palps [16]. Therefore, this gene likely has a function other than a gustatory or olfactory one.
One the An. quadriannulatus specific Grs, AgGr21, is homologous to DmGr64a and DmGr61a [48], which function as sugar receptors in Drosophila melanogaster [49, 50]. Interestingly, it was recently found that several sugar receptors are expressed in olfactory neurons in Drosophila antennae and/or palps, and one of these is co-expressed with Orco. This could suggest a hitherto unknown role of these receptors in olfaction [51]. The other An. quadriannulatus specific Gr, Gr26, has a relatively low expression level and it is unknown if this level of expression for a receptor gene (approx. 12 rpkm) could indicate a species-specific role in olfaction. Neither AgGr21 nor AgGr26 has a known ortholog in Aedes or Drosophila.
Although the An. quadriannulatus strain examined in this study preferred bovine odor, this was not the case in a previous study in which this species did not distinguish between human and cow odor [3]. Furthermore, in an additional study it blood fed equally on a human and equal sized calf [52]. This suggests that An. quadriannulatus is more of a generalist than An. gambiae with a wider host preference. The number of expressed and enhanced olfaction genes in An. quadriannulatus indicates it likely also responds to a complex blend of odors during host seeking, rather than focusing on a few universal mammalian kairomones.
Rinker et al. [29] previously compared the daytime transcriptomes of the antennae of An. coluzzii and An. quadriannulatus. When comparing that study with our results, we observe considerable agreement. Specifically, when looking at the antennal Ors, Irs and Obps highlighted in our study, Rinker et al. [29] also found most of these genes to be enhanced in either An. coluzzii (Or36, Or45, Or69, Or70, Or73, Or81, Ir7w, Ir7u, Ir75g, Ir75h.2, Ir41n, Ir41c, Ir100a, Obp1 and Obp3) or An. quadriannulatus (Or1, Or9, Or33, Or46, Or50, Or63, Or64, Or68, Ir75d and 75h.1) [29] (Additional file 4: Table S4 and Additional file 5: Table S5). The expression differences of these genes can therefore be interpreted as representing real consistent biological differences between the species. Differences observed between the two studies could be due to fluctuations of gene expression during the circadian cycle, differences in rearing conditions, or genetic drift in laboratory colonies.
Our study used a laboratory strain of An. coluzzii. Until recently this species was considered the An. gambiae M form, but was elevated to species level on the basis of ecological differences in larval breeding, premating isolation, and several small highly differentiated genomic regions [53] that encode for premating isolation between it and the An. gambiae S form, now known as An. gambiae s.s. [54]. Both species are highly anthropophilic and have very limited genomic divergence. Therefore, their human host preference is almost certainly an ancestral trait with a shared genetic basis. We therefore expect that the results from our study will be largely applicable to An. gambiae s.s. as well.
The olfactory system of Anopheles mosquitoes plays an important role in other aspects of their biology. It is used to locate sugar sources such as nectar, as well as oviposition sites. We do not know from what source or how often or An. quadriannulatus females obtain sugar meals. However, it has been established that An. coluzzii stops responding to honey volatiles five days after emergence, and starts being strongly attracted to human odor at that point [55]. Observations of An. quadriannulatus strains kept in our laboratory indicates that they also switch to host seeking around this time. The two species appear to have a similar larval ecology, as larvae of both species can be found in shallow, open, sunlit fresh water pools [56, 57]. In any case the search for oviposition-sites does not begin until 48 h after blood feeding. None-the-less, we cannot rule out that the divergence in chemosensory gene expression observed in this study are due other biological differences between the two species than host preference.
Conclusion
This comparison of gene expression in the olfactory organs of the anthropophilic An. coluzzii and the zoophilic An. quadriannulatus identified chemosensory genes that may underlie differences in host preference between the two species. Several medium high expressed olfactory and ionotropic receptors show considerable enhancement in the antennae of either species. In addition, expression of several Obps in the antennae and maxillary palps is considerably diverged between the two species. Finally, a small number of gustatory receptors in the maxillary palps were found to be species-specific. It is likely that some of the chemosensory genes identified here play a role in the differential host preference between the two species.
Methods
Mosquito rearing
Study populations in this study were lab colonies of An. colluzzii and An. quadriannulatus. The laboratory strains of An. coluzzii (formerly classified as An. gambiae M form, GASUA) originate from Suakoko, Liberia, and the An. quadriannulatus (SANQUA) colony originates from female mosquitoes collected in Sangwe, Zimbabwe, and reared in the insectaries at Wageningen University, The Netherlands (host choice experiment), and Texas A&M University, College Station, TX, USA (RNAseq analyses). Colonies were maintained at 25 °C, 75–85% relative humidity and a light:dark photoperiod of 12 h. Female mosquitoes were blood fed on defibrinated rabbit blood using an artificial membrane feeding system. Larvae were fed finely ground fish food (Tetramin, Melle, Germany), and were maintained at densities of approximately 150 per 2 L container. Pupae were collected and placed into cages at densities of two cups of 150 pupae per cage.
To obtain mosquitoes for RNAseq analyses newly emerged mosquitoes were removed daily from rearing cages. To obtain mosquitoes from the same age, pupae that failed to eclose were transferred to new cages. Male and female mosquitoes were kept in the same cage and maintained on a 5–10% sucrose solution until tissue dissections. Therefore, females were allowed to mate, but not to blood-feed. Insemination rate was found to be high (82%) in six-day old females. Species identity of the mosquito colonies was confirmed using the RFLP-PCR develop by Fanello et al. [58].
Dual odor-choice assay
A total of 750 female An. colluzzii and 330 An. quadriannulatus females were used to determine their preference for human or cow odor. Seventy-five to eighty mosquitoes were placed in release cages the night before experiments with access to a wet cotton ball for hydration. The next morning the release cage was attached to the dual-choice olfactometer. The human odor sample consisted of a socks worn by volunteers for 24 h, and the cow odor sample consisted of a panty hose that had been tied around the leg of a cow for 24 h. Odor sources were switched between the left and right port of the olfactometer between runs. A single CO2 source, placed between the two odor ports was used as activator. Conditions during the experiments were as follows: temperature = 26–28 °C, humidity = 55–75% inside olfactometer, 80% in front of port holes, air-speed 018–0.22 ms−1, and released [CO2] = 4.5%. Mosquitoes were released into the olfactometer during the dark-cycle for 15 min. Experiments were conducted under semi-dark conditions. Mosquitoes that remained in the wind tunnel and did not enter an odor port during the experiment were discarded.
Olfactory tissue dissections
Shortly after dark, anophelines begin their host seeking activity following their normal circadian patterns, and Orco expression in An. coluzzii peaks at this time. Hence, to capture the olfactory gene expression in these mosquitoes during their host-seeking phase, we collected their antennae and maxillary palps soon after dark. First, we immobilized female mosquitoes shortly after the start of the dark cycle by placing them at −20 °C for about a minute, and then placed them inside petri-plates on ice. The antennae and maxillary palps were each separately removed from immobilized mosquitoes while placed under a 10× dissecting microscope, and immediately stored in RNAlater® Ice (Ambion). Between 300 and 500 six-day old females were dissected for each replicate and two replicates per species were included for a total of four samples per species. Samples were stored at −20 °C until RNA extraction, which was carried out for all samples within 48 h after dissection.
Total RNA was isolated from each sample using miRNeasy (Qiagen) columns using the protocol provided by Qiagen. A Qubit fluorometer (Life Technologies) was used to quantify RNA originaly. Subsequently, the RNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific) followed by a quality assessment with a RNA Pico LabChip analysis on an Agilent BioAnalyzer 2100 (Agilent Technologies) by the Agrilife Genomics Center at Texas A&M University. Messenger RNA was enriched from about 1 μg of total RNA using the Poly-A enrichment method and cDNA libraries were prepared using an Illumina TruSeq RNA Library kit (Illumina). All libraries were sequenced over two lanes of Illumina HiSeq 2500 platform running in high-output mode, and generating 100 bp single-end reads. Preparation and sequencing of libraries were both performed at the Agrilife Genomics core facility at Texas A&M University, College Station, Texas. Approximately 30–46 million reads with an average read of 100 bp were generated for each replicate sample and used for further analysis.
RNA sequencing analyses
Two replicate female antennae and two replicate maxillary palp transcriptome datasets were obtained for each of the two species using RNAseq technology on the Illumina HiSeq 2500 platform. For each library, between 20 M and 30 M reads were generated. The quality of raw data was first assessed using FastQC (version 0.10.0). Fastq files were trimmed and QC-filtered using NGSQCTooklit package [59], and only the reads with at least 80% of the bases at phred score = > 30 were retained, and the rest were discarded. After quality filtering and trimming, an average of 96% of the reads were retained in each library. Filtered-trimmed files were then aligned to the mtGenome from An. gambiae assembly Pest Version 4.2 (agamp4) using the short-read aligner Bowtie2 [60] to remove mitochondrial RNA and/or rRNA contamination that was not removed during library preparation. We found 6–10% of the reads were mapping to mitochondrial rRNA for each of the samples, and we subsequently excluded the reads mapping to the mitochondrial genome for differential gene expression analyses. Reads that did not map to mtDNA reference were extracted out into a new Fastq file. These reads were aligned to Anopheles gambiae reference genome (PEST strain, Assembly version 4.2, retrieved on May 13th 2015 from www.vectorbase.org), using the de novo splice assembler STAR [61, 62]. After mapping, the reads mapping to ‘exon’ features were counted using the R package HTSeq (geneset: AgamP4.2, retrieved from www.vectorbase.org on May 13th 2015). Aligned reads were counted using the tool HTSeq-Count [33]. Reads that mapped only once in the genome were used for the differential expression analyses. Characteristics of data used for analysis are provided in Table S1. Reads mapping to multiple loci were excluded from the analyses. The counts data for each annotated feature on the reference genome was used for downstream statistical comparisons.
Clustering of the variance-stabilized transformed counts indicated that there was relatively little variation between biological replicates compared to between species for both antennae and maxillary palp samples. It was therefore deemed to use them as replicates in statistical analysis.
Analysis of differential gene expression
With the ever-increasing variety of software packages to estimate differential expression, and the absence of standardized RNAseq specific methods [63], the repeatability, and the interpretation of the biological significance of expressed genes will rest on the variance structure unique to each dataset, and the statistical methods. To this end, we decided to use the popular and well-reviewed statistical [64] analysis package for DGE analysis - namely DESeq2 [34]. The DESeq2 statistical package is available as part of the Bioconductor suite on the R platform, and uses implementations of negative binomial modeling of counts data.
In DESeq2, size factors for each dataset were calculated to normalize library sizes across replicates, and overall means and variances were determined using a negative binomial distribution model. Genes were considered to be differentially expressed if q < 0.05 after correcting for multiple testing. Differentially expressed genes were tested by the GLM likelihood ratio test (after fitting negative binomial models and estimating dispersion), and considered significantly differentially expressed if the adjusted P-value, (padj) <0.05. Reported fold change values reported were calculated using the 2^(Log2FoldChange) equation in excel on the Log2 fold changes obtained from the DESeq analysis.
Gene ontology analysis
Genes that met the following criteria were used for gene ontology analysis. Genes that appeared on the consensus list by virtue of having FDR < 0.05 under the edgeR analysis and padj < 0.05 in the DESeq analysis between antennae or maxillary palps of the two species. GO Annotation was performed using the gene ontology analysis engine Pantherdb (www.pantherdb.org). The gene ids were retrieved from Ensembl release 22 via Biomart (http://metazoa.ensembl.org/index.html). GO annotation was used for assessment of the molecular function of genes that were differentially expressed between species. GO annotation associates analyzed transcripts with terms from hierarchical vocabularies describing, e.g., molecular function or biological process.
Additional files
Gene expression data for the antennae and maxillary palps of female Anopheles coluzzii and Anopheles quadriannulatus. (XLSX 3020 kb)
RPKM and fold change values for replicate samples for chemosensory genes with small (fold change <2), yet significantly different expression between species. (XLSX 45 kb)
Mapping statistics. (XLSX 27 kb)
Protein families enriched in An. coluzzii and An. quadriannulatus antennae and maxillary palps. (XLSX 33 kb)
Gene expression data for chemosensory genes in antennae reported in this paper and in Rinker et al. [29]. RPKM and fold change values between replicate antennae samples for genes listed in Table 1. with a Fold Change values <2. Also included are the expression levels for these genes reported in Rinker et al. [29]. (XLSX 80 kb)
Acknowledgements
We are thankful to Leon Westerd for assistance with colony maintenance, anonymous volunteers for providing human odor samples, to Jeroen Spitzen for provided cow odor samples, and to Dr. Craig Coates for providing rearing space. We also thank the reviewers for providing valuable comments for improving the manuscript.
Author’s Contribution
GA: Data collection, analysis and manuscript preparation; LVC: Data collection, data analysis and manuscript preparation, ZPH: Data collection; SP: Data analysis and manuscript preparation; WT: Supervision and planning of experiments, and manuscript preparation; MAS: Conceived, planned and supervised experiments, manuscript preparation. All authors read and approved the final manuscript.
Funding
This work was supported by NIH/NIAID grant 1R01 AI085079 to MAS. The funding body played no role in the design of the study and collection, analysis and interpretation of the data and in writing the manuscript.
Availability of data and materials
The sequence data generated during this study is deposited on the NCBI Sequence Read Archive, under BioProject ID PRJNA400609. The full gene expression data are available in “Additional file 1”.
Consent for publication
N/A
Abbreviations
- An
Anopheles
- Csp
Chemosensory receptor
- D
Drosophila
- GO
Gene ontology
- Gr
Gustatory receptor
- Ir
Ionotropic repector
- Obp
Odorant binding protein
- Or
Olfactory receptor
Ethics approval and consent to participate
This study did not include the use of humans, vertebrate animals of plants directly. No ethics approval is required for experimentation of the study organisms; Anopheles coluzzii and An. quadriannulatus.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4122-7) contains supplementary material, which is available to authorized users.
Contributor Information
G. Athrey, Email: giri.athrey@tamu.edu
L. V. Cosme, Email: luciano.cosme@yale.edu
Z. Popkin-Hall, Email: zpopkinh@tamu.edu
S. Pathikonda, Email: spathikonda@tamu.edu
W. Takken, Email: willem.takken@wur.nl
M. A. Slotman, Phone: 1 (979) 845 7556, Email: maslotman@tamu.edu
References
- 1.Takken W, Knols B. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu Rev Entomol. 1999;44:131–157. doi: 10.1146/annurev.ento.44.1.131. [DOI] [PubMed] [Google Scholar]
- 2.Takken W, Eling W, Hooghof J, Dekker T, Hunt R, Coetzee M. Susceptibility of Anopheles quadriannulatus Theobald (Diptera: Culicidae) to Plasmodium falciparum. Trans R Soc Trop Med Hyg. 1999;93:578–580. doi: 10.1016/S0035-9203(99)90054-8. [DOI] [PubMed] [Google Scholar]
- 3.Habtewold T, Povelones M, Blagborough AM, Christophides GK. Transmission blocking immunity in the malaria non-vector mosquito Anopheles quadriannulatus species a. PLoS Pathog. 2008;4(5):e1000070. doi: 10.1371/journal.ppat.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pates HV, Takken W, Curtis CF. Laboratory studies on the olfactory behaviour of Anopheles quadriannulatus. Entomologia Experimentalis et Applicata. 2005;114:153–159. doi: 10.1111/j.1570-7458.2005.00249.x. [DOI] [Google Scholar]
- 5.Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc Natl Acad Sci U S A. 2004;101:5058–5063. doi: 10.1073/pnas.0308146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu T, Qiu YT, Wang G, Kwon JY, Rützler M, Kwon H-W, et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr Biol. 2007;17:1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b Encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Carey AF, Wang G, Su C-Y, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. Stern DL, editor. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 13.Li ZX, Pickett JA, Field LM, Zhou J-J. Identification and expression of odorant-binding proteins of the malaria-carrying mosquitoes Anopheles gambiae and Anopheles arabiensis. Arch Insect Biochem Physiol. 2005;58:175–189. doi: 10.1002/arch.20047. [DOI] [PubMed] [Google Scholar]
- 14.Biessmann H, Nguyen QK, Le D, Walter MF. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae. Insect Mol Biol. 2005;14:575–589. doi: 10.1111/j.1365-2583.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- 15.Iovinella I, Bozza F, Caputo B, Torre Della A, Pelosi P. Ligand-binding study of Anopheles gambiae chemosensory proteins. Chem Senses. 2013;38:409–419. doi: 10.1093/chemse/bjt012. [DOI] [PubMed] [Google Scholar]
- 16.Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011;12:271. doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohbot JD, Sparks JT, Dickens JC. The maxillary palp of Aedes aegypti, a model of multisensory integration. Insect Biochem Mol Biol. 2014;48:29–39. doi: 10.1016/j.ibmb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Syed Z, Leal WS. Maxillary palps are broad spectrum odorant detectors in Culex quinquefasciatus. Chem Senses. 2007;32:727–738. doi: 10.1093/chemse/bjm040. [DOI] [PubMed] [Google Scholar]
- 19.Braks MAH, Takken W. Incubated human sweat but not fresh sweat attracts the malaria mosquito Anopheles gambiae Sensu stricto. J Chem Ecol. 1999;25:663–672. doi: 10.1023/A:1020970307748. [DOI] [Google Scholar]
- 20.Verhulst NO, Andriessen R, Groenhagen U, Bukovinszkiné Kiss G, Schulz S, Takken W, et al. Differential attraction of malaria mosquitoes to volatile blends produced by human skin bacteria. PLoS One. 2010;5:e15829. doi: 10.1371/journal.pone.0015829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernier UR, Kline DL, Barnard DR, Schreck CE, Yost RA. Analysis of human skin emanations by gas chromatography/mass spectrometry. 2. Identification of volatile compounds that are candidate attractants for the yellow fever mosquito (Aedes aegypti). Anal. Chem. 2000;72:747–756. doi: 10.1021/ac990963k. [DOI] [PubMed] [Google Scholar]
- 22.Braks MAH, Meijerink J, Takken W. The response of the malaria mosquito, Anopheles gambiae, to two components of human sweat, ammonia and l-lactic acid, in an olfactometer. Physiol Entomol. 2001;26:142–148. doi: 10.1046/j.1365-3032.2001.00227.x. [DOI] [Google Scholar]
- 23.Dekker T, Steib B, Carde RT, Geier M. L-lactic acid: a human-signifying host cue for the anthropophilic mosquito Anopheles gambiae. Med Vet Entomol. 2002;16:91–98. doi: 10.1046/j.0269-283x.2002.00345.x. [DOI] [PubMed] [Google Scholar]
- 24.Kopp A, Barmina O, Hamilton AM, Higgins L, McIntyre LM, Jones CD. Evolution of gene expression in the Drosophila olfactory system. Mol Biol Evol. 2008;25:1081–1092. doi: 10.1093/molbev/msn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D sechellia. Current Biology. 2006;16:101–109. doi: 10.1016/j.cub.2005.11.075. [DOI] [PubMed] [Google Scholar]
- 26.McBride CS. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci U S A. 2007;104:4996–5001. doi: 10.1073/pnas.0608424104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride CS, Arguello JR, O'Meara BC. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics. 2007;177:1395–1416. doi: 10.1534/genetics.107.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride CS, Baier F, Omondi AB, Spitzer SA, Lutomiah J, Sang R, et al. Evolution of mosquito preference for humans linked to an odorant receptor. Nature. 2014;515:222–227. doi: 10.1038/nature13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinker DC, Zhou X, Pitts RJ, Consortium AGC, Rokas A, Zwiebel LJ. Antennal transcriptome profiles of anopheline mosquitoes reveal human host olfactory specialization in Anopheles Gambiae. BMC Genomics. 2013;14:749. doi: 10.1186/1471-2164-14-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rund SSC, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc Natl Acad Sci U S A. 2011;108:E421–E430. doi: 10.1073/pnas.1100584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekker T, Takken W. Differential responses of mosquito sibling species Anopheles arabiensis and An. quadriannulatus to carbon dioxide, a man or a calf. Med Vet Entomol. 1998;12:136–140. doi: 10.1046/j.1365-2915.1998.00073.x. [DOI] [PubMed] [Google Scholar]
- 32.Sharp BL, Quicke FC, Jansen EJ. Aspects of the behavior of five anopheline species in the endemic malaria area of Natal. Journal of the Entomological Society of Southern Africa. 1984;47:251–258. [Google Scholar]
- 33.Anders S, Pyl PT, Huber W. HTSeq--a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed]
- 35.Kwon H-W, Lu T, Rützler M, Zwiebel LJ. Olfactory responses in a gustatory organ of the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2006;103:13526–13531. doi: 10.1073/pnas.0601107103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sparks JT, Bohbot JD, Dickens JC. The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect Biochem Mol Biol. 2014;48:8–16. doi: 10.1016/j.ibmb.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Smallegange RC, Qiu YT, van Loon JJA, Takken W. Synergism between ammonia, lactic acid and carboxylic acids as kairomones in the host-seeking behaviour of the malaria mosquito Anopheles gambiae Sensu stricto (Diptera: Culicidae) Chem Senses. 2005;30:145–152. doi: 10.1093/chemse/bji010. [DOI] [PubMed] [Google Scholar]
- 38.Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GSXE, et al. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min S, Ai M, Shin SA, Suh GSB. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A. 2013;110:E1321–E1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride CS. Genes and odors underlying the recent evolution of mosquito preference for humans. Curr Biol. 2016;26:R41–R46. doi: 10.1016/j.cub.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeGennaro M, McBride CS, Seeholzer L, Nakagawa T, Dennis EJ, Goldman C, et al. Orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498:487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu P, Atkinson R, Jones DNM, Smith DP. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 43.Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5:e118. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E, et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One. 2010;5:e9471. doi: 10.1371/journal.pone.0009471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das S, Dimopoulos G. Molecular analysis of photic inhibition of blood-feeding in Anopheles gambiae. BMC Physiol. 2008;8:23. doi: 10.1186/1472-6793-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelletier J, Guidolin A, Syed Z, Cornel AJ, Leal WS. Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J Chem Ecol. 2010;36:245–248. doi: 10.1007/s10886-010-9762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Healy TP, Copland MJ. Activation of Anopheles gambiae mosquitoes by carbon dioxide and human breath. Med Vet Entomol. 1995;9:331–336. doi: 10.1111/j.1365-2915.1995.tb00143.x. [DOI] [PubMed] [Google Scholar]
- 48.Kent LB, Robertson HM. Evolution of the sugar receptors in insects. BMC Evol Biol. 2009;9:41. doi: 10.1186/1471-2148-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiao Y, Moon SJ, Montell C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci U S A. 2007;104:14110–14115. doi: 10.1073/pnas.0702421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto T, Chen Y, Slone J, Amrein H. Identification of a Drosophila glucose receptor using Ca2+ imaging of single chemosensory neurons. PLoS One. 2013;8:e56304. doi: 10.1371/journal.pone.0056304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujii S, Yavuz A, Slone J, Jagge C, Song X, Amrein H. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr Biol. 2015;25:621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pates HV, Takken W, Curtis CF, Huisman PW, Akinpelu O, Gill GS. Unexpected anthropophagic behaviour in Anopheles quadriannulatus. Med Vet Entomol. 2001;15:293–298. doi: 10.1046/j.0269-283x.2001.00310.x. [DOI] [PubMed] [Google Scholar]
- 53.Coetzee M, Hunt RH, Wilkerson R, Torre Della A, Coulibaly MB, Besansky NJ. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae Complex. Zootaxa. 2013;3619:246–274. doi: 10.11646/zootaxa.3619.3.2. [DOI] [PubMed] [Google Scholar]
- 54.Aboagye-Antwi F, Alhafez N, Weedall GD, Brothwood J, Kandola S, Paton D, et al. Experimental swap of Anopheles gambiae’s assortative mating preferences demonstrates key role of X-chromosome divergence island in incipient sympatric speciation. PLoS Genet. 2015;11:e1005141. doi: 10.1371/journal.pgen.1005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foster WA, Takken W. Nectar-related vs. human-related volatiles: behavioural response and choice by female and male Anopheles gambiae (Diptera: Culicidae) between emergence and first feeding. Bull. Ento. Res. 2004; 94:145–157. [DOI] [PubMed]
- 56.Coluzzi M. Heterogeneities of the malaria vectorial system in tropical Africa and their significance in malaria epidemiology and control. Bull World Health Organ. 1984; [PMC free article] [PubMed]
- 57.Gillies MT, Coetzee M. A supplement to the Anophilinae of Africa south of the Sahara. Johannesburg: Publication of the South African Institute for Medical Research; 1987. [Google Scholar]
- 58.Fanello C, Santolamazza F, Torre DA. Simultaneous identification of species and molecular forms of the Anopheles gambiae Complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- 59.Patel RK, Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A, Gingeras TR. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformatics. 2015;Suppl 51:–11.14.1, 11.14.19. [DOI] [PMC free article] [PubMed]
- 62.Dobin A, Gingeras TR. Optimizing RNA-Seq mapping with STAR. Methods Mol Biol. 2016;1415:245–262. doi: 10.1007/978-1-4939-3572-7_13. [DOI] [PubMed] [Google Scholar]
- 63.Kvam VM, Liu P, Si Y. A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am J Bot. 2012;99:248–256. doi: 10.3732/ajb.1100340. [DOI] [PubMed] [Google Scholar]
- 64.Rapaport F, Khanin R, Liang Y, Krek A, Zumbo P, Mason CE, et al. Comprehensive evaluation of differential expression analysis methods for RNA-seq data. Genome Biol. 2013;14(9):R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression data for the antennae and maxillary palps of female Anopheles coluzzii and Anopheles quadriannulatus. (XLSX 3020 kb)
RPKM and fold change values for replicate samples for chemosensory genes with small (fold change <2), yet significantly different expression between species. (XLSX 45 kb)
Mapping statistics. (XLSX 27 kb)
Protein families enriched in An. coluzzii and An. quadriannulatus antennae and maxillary palps. (XLSX 33 kb)
Gene expression data for chemosensory genes in antennae reported in this paper and in Rinker et al. [29]. RPKM and fold change values between replicate antennae samples for genes listed in Table 1. with a Fold Change values <2. Also included are the expression levels for these genes reported in Rinker et al. [29]. (XLSX 80 kb)
Data Availability Statement
The sequence data generated during this study is deposited on the NCBI Sequence Read Archive, under BioProject ID PRJNA400609. The full gene expression data are available in “Additional file 1”.


