Abstract
Background
The emergence of antibiotic-resistant bacteria can cause serious clinical and public health problems. This study describes the possibility of using bacteriophages as an alternative agent to control multidrug-resistant Salmonella Typhimurium.
Methods
The potential lytic bacteriophages (P22-B1, P22, PBST10, PBST13, PBST32, and PBST 35) were characterized by morphological property, heat and pH stability, optimum multiplicity of infection (MOI), and lytic activity against S. Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and S. Typhimurium CCARM 8009.
Results
P22-B1 and P22 belong to Podoviridae family and PBST10, PBST13, PBST32, and PBST 35 show a typical structure with polyhedral head and long tail, belonging to Siphoviridae family. Salmonella bacteriophages were highly stable at the temperatures (< 60 °C) and pHs (5.0–11.0). The reduction rates of host cells were increased at the MOI-dependent manner, showing the highest reduction rate at MOI of 10. The host cells were most effectively reduced by P22, while P22-B1 showed the least lytic activity. The ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 were resistant to ciprofloxacin, levofloxacin, norfloxacin, and tetracycline. P22 showed the highest lytic activity against S. Typhimurium KCCM 40253 (> 5 log reduction), followed by S. Typhimurium ATCC 19585 (4 log reduction) and ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585 (4 log reduction).
Conclusion
The results would provide vital insights into the application of lytic bacteriophages as an alternative therapeutics for the control of multidrug-resistant pathogens.
Keywords: Salmonella Typhimurium, Bacteriophage, Antibiotic resistance, Lytic activity, Ciprofloxacin
Background
Over the last few decades, the repeated misuse and overuse of antibiotics has accelerated the emergence of antibiotic-resistant bacteria, leading to serious clinical and public health problems [1, 2]. The rapid spread of antibiotic resistance has left the development of new antibiotics due to the long and expensive clinical testing [3]. The antibiotic resistance of Salmonella species is mainly acquired by the production of antibiotic-degrading enzymes, alteration in membrane permeability, and activation of multidrug efflux pumps [4]. Recently, the prevalence of multidrug-resistant (MDR) Salmonella serotypes has increased the failure in antibiotic treatments [5–7]. The MDR Salmonella infection has become a global public health concern due to the annual increase in morbidity and mortality rates [8]. The therapeutic limitation of current antibiotics has added to the difficulty in treating multidrug-resistant bacterial infections. Hence, the development of alternative therapeutic treatments over antibiotics is essential for controlling multidrug-resistant bacteria.
Bacteriophage has received much attention as a possible alternative due to the specificity and self-replicating property with no adverse effects on beneficial microflora and human cells [9]. The specificity to target bacteria is attributed to the binding ability of bacteriophages to host cell surface receptors such as flagella, capsule, slime layer, lipopolysaccharides, and outer membrane proteins, resulting in the lysis of bacteriophage-infected bacteria expressed as lytic activity [10, 11]. The bacteriophage-binding receptors on the host cell surface can be altered though the modification of outer membrane components [12–14]. However, there is relatively little knowledge on the interaction between bacteriophages and multidrug-resistant bacteria in terms of the alteration in host cell surface receptors. Therefore, the bacteriophages are not directly applicable to multidrug-resistant bacteria. For the successful application of bacteriophage, this study was aimed to evaluate the lytic activity of potential Salmonella bacteriophages (P22-B1, P22, PBST10, PBST13, PBST32, and PBST 35) against Salmonella enterica serovar Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and S. Typhimurium CCARM 8009, having different antibiotic resistance profiles.
Methods
Bacterial strains and culture conditions
Strains of S. Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, and S. Typhimurium CCARM 8009 were purchased from Korean Culture Center of Microorganism (KCCM, Seoul, Korea), American Type Culture Collection (ATCC, Manassas, VA, USA), and Culture Collection of Antibiotic Resistant Microbes (CCARM, Seoul, Korea), respectively. Escherichia coli ATCC 25922 was used as control strain to evaluate the antibiotic susceptibility. All strains were cultured in trypticase soy broth (TSB) (BD, Becton, Dickinson and Co., Sparks, MD, USA) at 37 °C for 20 h. The cultured cells were harvested by centrifugation at 3000×g for 20 min at 4 °C, washed twice with phosphate-buffered saline (PBS, pH 7.2), and diluted to 108 CFU/ml prior to use.
Stepwise selection method
The strain of S. Typhimurium ATCC 19585 was exposed to ciprofloxacin to induce antibiotic-resistant isogenic strain according to a serial passage assay [15]. Salmonella Typhimurium ATCC 19585 strain was cultured repeatedly in TSB and TSA by increasing ciprofloxacin concentrations from 0.03 to 1 µg/ml. The ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585 were stable for more than 10 passages in antibiotic-free TSB. The strain was cultured in ciprofloxacin-free TSB at 37 °C for 20 h prior to use.
Bacteriophage propagation
Salmonella bacteriophages, P22-B1 and P22, were purchased from ATCC and PBST10, PBST13, PBST32, and PBST 35 were obtained from Bacteriophage Bank at Hankuk University of Foreign Studies (Yongin, Gyeonggi, Korea). The bacteriophages were propagated at 37 °C for 24 h in TSB containing S. Typhimurium KCCM 40253. The propagated bacteriophages were centrifuged at 5000×g for 10 min, filtered through a 0.2-μm filter to eliminate bacterial lysates, and further purified using a polyethylene glycol (PEG) precipitation assay. The bacteriophage titers were determined by using a soft-agar overlay method [16]. In brief, the collected bacteriophages were serially (1:10) diluted with PBS and gently mixed with the host cells (107 CFU/ml) in TSB (0.5% agar). The mixture was poured over the pre-warmed base agar lawn. The plates were incubated at 37 °C for 24 h to enumerate the bacteriophages expressed as plaque-forming unit (PFU).
Morphological assay
The morphological properties of Salmonella bacteriophages were determined by transmission electron microscope (TEM, LEO 912AB Omega; Carl Zeiss NTS GmbH, Oberkochen, Germany), located at the Korea Basic Science Institute (KBSI; Gangwon, Korea). The bacteriophages were transferred to the surface of carbon-coated copper film and negatively stained with 5% aqueous uranyl acetate (pH 4.0). After air-drying, the stained bacteriophages were observed under TEM (120 kV; 125,000× magnification).
Heat and pH stability of bacteriophages
The susceptibility of bacteriophages to heat was evaluated at the range of 30–80 °C for 30 min. For isothermal treatment, 0.1 ml of each Salmonella bacteriophage (5 × 105 PFU/ml) was inoculated into a pre-heated tube containing 9.9 ml of TSB and then treated at each target temperature for 30 min. After heat treatment, each tube was immediately cooled in an ice-bath. The pH stability of bacteriophages was evaluated in phosphate buffered saline (PBS, pH 7.2) adjusted to pHs ranged from 2 to 12. Each Salmonella bacteriophage (5 × 105 PFU/ml) was exposed to different pH levels at 37 °C for 30 min. The pH-treated bacteriophages were immediately diluted with PBS to avoid further inactivation. After heat or pH treatment, viable bacteriophages were enumerated by using a soft-agar overlay assay.
Determination of optimum multiplicity of infection
The lytic ability of Salmonella bacteriophages against S. Typhimurium KCCM 40253 (105 CFU/ml) was evaluated at different multiplicity of infections (MOIs) ranging from 0.01 to 10. The host cells infected by each Salmonella bacteriophage were incubated at 37 °C for 6 h. After incubation, the host cells were enumerated using an Autoplate® Spiral Plating System (Spiral Biotech Inc., Norwood, MA, USA) and a QCount® Colony Counter (Spiral Biotech Inc.). The reduction rate was estimated by comparing with the number of the control cells treated without bacteriophages.
Antibiotic disc susceptibility assay
The susceptibility of host strains, S. Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and S. Typhimurium CCARM 8009, was evaluated by using an agar disc diffusion assay. Each host strain was diluted to the turbidity of 0.5 McFarland standard and spread on Mueller–Hinton agar plate. Antibiotic discs used in this study were ampicillin (10 µg), cefotaxime (30 µg), cephalothin (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), kanamycin (30 µg), levofloxacin (5 µg), meropenem (10 µg), nalidixic acid (30 µg), norfloxacin (10 µg), sulfamethoxazole/trimethoprim (1.25/23.75 µg), and tetracycline (30 µg). The antibiotic discs were placed on Mueller–Hinton agar plate and then incubated at 37 °C for 20 h. The clear zone was measured to evaluate the antibiotic susceptibility. The susceptibility (S) and resistant (R) strains were defined as compared with the susceptibility of the control strain, E. coli ATCC 25922.
Bacteriophage spotting assay
The lytic ability of Salmonella bacteriophages against S. Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and S. Typhimurium CCARM 8009 was determined using a spotting test. The serially (1:10) diluted bacteriophages (2 µl each) were spotted onto the soft-agar plates and incubated at 37 °C for 24 h. The formation of clear zone was determined to express the lytic capability.
Lytic activity of bacteriophage
Salmonella bacteriophages were used to evaluate the lytic activity against S. Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and S. Typhimurium CCARM 8009. The host strains (105 CFU/ml each) were mixed with bacteriophage (105 PFU/ml) at MOI of 1 and incubated at 37 °C for 12 h. After 12-h incubation, the cultures were centrifuged at 3000×g for 20 min. The collected cells were serially diluted (1:10) with PBS. The proper dilutions were plated on TSA using an Autoplate® Spiral Plating System (Spiral Biotech Inc.). After 24-h incubation, the viable cells were enumerated using a QCount® Colony Counter (Spiral Biotech Inc.). The lytic activity was expressed as log Nc/Np; Nc and Np denote the counts of bacterial host cells treated without and with bacteriophages, respectively, after 12-h incubation.
Statistical analysis
All analyses were performed in duplicate on three replicates. Data were analyzed using the Statistical Analysis System software (SAS 9.4; SAS Institute Inc., Cary, NC, USA). The general linear model (GLM) and least significant difference (LSD) procedures were used to compare means at p < 0.05.
Results
Morphological and physiological properties of various Salmonella bacteriophages
The morphological characteristics of Salmonella bacteriophages (P22-B1, P22, PBST10, PBST13, PBST32, and PBST 35) were evaluated using TEM (Fig. 1). P22-B1 (Fig. 1a) and P22 (Fig. 1b) have regular icosahedral heads of 50 and 77 nm with short tails, respectively, belonging to the Podoviridae family. PBST10 (Fig. 1c), PBST13 (Fig. 1d), PBST32 (Fig. 1e), and PBST35 (Fig. 1f) show a regular polyhedral structure with heads of 45–66 nm and long tails of 94–186 nm, belonging to Siphoviridae family.
Fig. 1.
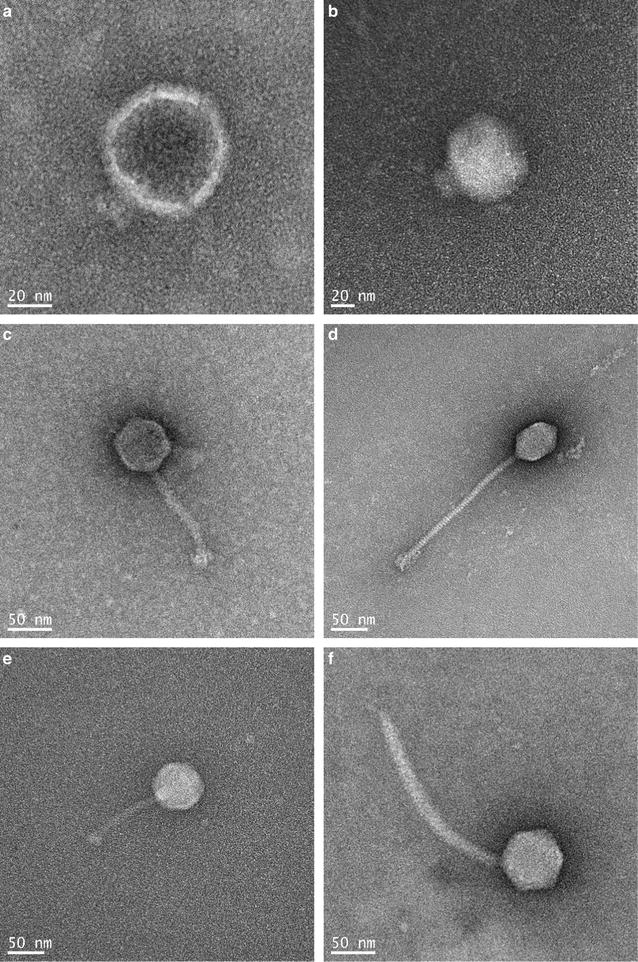
Morphology of bacteriophages; P22-B1 (a), P22 (b), PBST10 (c), PBST13 (d), PBST32 (e), and PBST35 (f)
The heat sensitivity of Salmonella bacteriophages was evaluated at the temperature ranged from 30 to 80 °C (Fig. 2a). All bacteriophages were stable to heat up to 60 °C, but the numbers of bacteriophages were reduced to below the detection limit at 80 °C. The sensitivity of Salmonella bacteriophages to pH was evaluated at different pH values ranged from 2 to 12 (Fig. 2b). P22-B1, P22, PBST32, and PBST 35 were stable at pHs ranged from 4 to 11, showing no noticeable reduction in the viable bacteriophage counts.
Fig. 2.
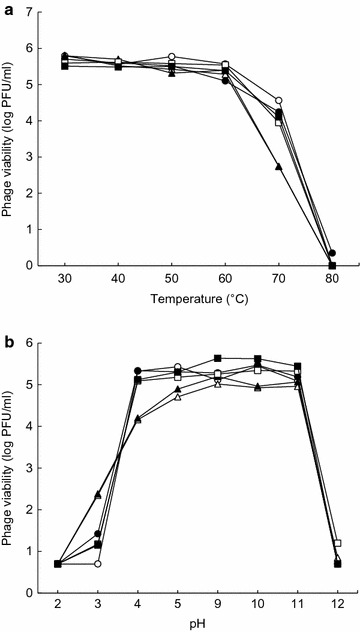
Susceptibility of Salmonella bacteriophages (○, P22-B1; ●, P22; □, PBST10; ■, PBST13; ∆, PBST32; ▲, PBST35) to heat (a) and pH (b)
The ability of Salmonella bacteriophages to lyse the host cells (S. Typhimurium KCCM 40253) was evaluated at different MOIs of 0.01, 01, 1, and 10 (Fig. 3). The reduction rates of S. Typhimurium KCCM 40253 were increased with increasing the MOI. The highest reduction rate of host cells was 54% for P22 at MOI of 10, followed by PBST35, PBST13, and PBST32. P22 was most effective on the reduction of host cells, whereas the least lytic activity was observed for P22-B1 at all MOIs.
Fig. 3.
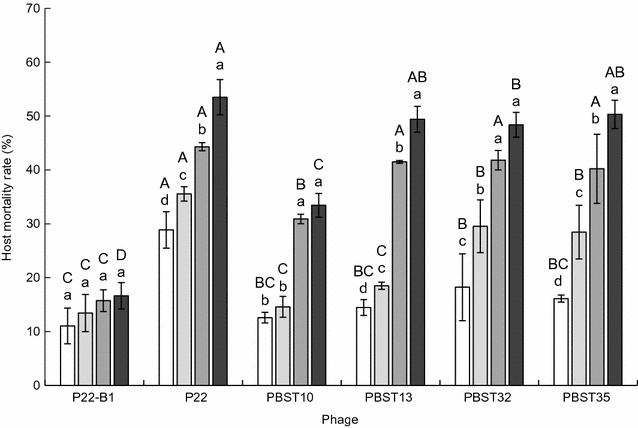
Reduction rate (%) of Salmonella Typhimurium KCCM 40253 treated with bacteriophages at different multiplicity of infections (MOIs) of 0.01 (□), 0.1 ( ), 1 (
), 1 ( ), and 10 (■). Bars with different uppercase (A–D) among bacteriophages and lowercase (a–d) within MOIs are significantly different at p < 0.05
), and 10 (■). Bars with different uppercase (A–D) among bacteriophages and lowercase (a–d) within MOIs are significantly different at p < 0.05
Antibiotic susceptibility and bacteriophage host ranges of S. Typhimurium
The antibiotic susceptibility of S. Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 was determined using a disc diffusion assay (Table 1). Salmonella Typhimurium KCCM 40253 and S. Typhimurium ATCC 19585 were sensitive to most antibiotics. The susceptibility of S. Typhimurium ATCC 19585 to cephalothin, ciprofloxacin, levofloxacin, nalidixic acid, norfloxacin, and tetracycline was decreased after ciprofloxacin induction. The clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 was resistant to ampicillin, ciprofloxacin, kanamycin, levofloxacin, norfloxacin, sulfamethoxazole/trimethoprim, and tetracycline.
Table 1.
Antibiotic disc diffusion (mm) of Salmonella Typhimurium strains
| Antibiotic disc |
S. Typhimurium KCCM40253 |
S. Typhimurium ATCC19585 |
S. Typhimurium ATCC19585-CIP |
S. Typhimurium CCARM8009 |
|---|---|---|---|---|
| AMP | 27.4 ± 0.5 (S) | 24.3 ± 0.5 (S) | 22.4 ± 0.7 (S) | 6.00 (R) |
| CEF | 33.3 ± 0.9 (S) | 31.5 ± 0.4 (S) | 28.8 ± 0.6 (S) | 32.0 ± 0.5 (S) |
| CEP | 24.8 ± 0.8 (S) | 24.8 ± 0.8 (S) | 22.7 ± 0.4 (R) | 25.2 ± 0.5 (S) |
| CHL | 27.1 ± 0.4 (S) | 25.7 ± 0.3 (S) | 24.9 ± 0.7 (S) | 23.5 ± 0.3 (S) |
| CIP | 34.6 ± 0.6 (S) | 33.2 ± 0.7 (S) | 30.7 ± 1.0 (R) | 30.9 ± 0.7 (R) |
| KAN | 21.8 ± 0.9 (S) | 21.8 ± 0.3 (S) | 21.4 ± 0.6 (S) | 6.00 (R) |
| LEV | 31.0 ± 0.5 (S) | 31.6 ± 0.5 (S) | 27.4 ± 0.9 (R) | 28.3 ± 0.5 (R) |
| MER | 34.2 ± 0.5 (S) | 32.3 ± 0.2 (S) | 31.8 ± 1.2 (S) | 32.5 ± 0.7 (S) |
| NAL | 24.6 ± 0.9 (R) | 23.3 ± 0.6 (R) | 6.00 (R) | 23.6 ± 0.7 (R) |
| NOR | 32.5 ± 0.7 (S) | 31.8 ± 0.5 (S) | 28.6 ± 0.6 (R) | 29.5 ± 0.4 (R) |
| SMA/TMP | 23.6 ± 0.6 (S) | 25.7 ± 1.0 (S) | 24.4 ± 0.5 (S) | 22.7 ± 0.5 (R) |
| TET | 18.6 ± 0.8 (R) | 19.3 ± 0.6 (S) | 17.4 ± 0.2 (R) | 7.5 ± 0.1 (R) |
S and R indicate susceptible and resistant strain. AMP, ampicillin (10 µg); CEF, cefotaxime (30 µg); CEP, cephalothin (30 µg); CHL, chloramphenicol (30 µg); CIP, ciprofloxacin (5 µg); KAN, kanamycin (30 µg); LEV, levofloxacin (5 µg); MER, meropenem (10 µg); NAL, nalidixic acid (30 µg); NOR, norfloxacin (10 µg); SMA/TMP, sulfamethoxazole/trimethoprim (1.25/23.75 µg); TET, tetracycline (30 µg)
The host range of bacteriophages (P22-B1, P22, PBST10, PBST13, PBST32, and PBST 35) was determined using a spotting assay (Fig. 4). All bacteriophages tested in this study showed the specificity to S. Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, and ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585. No host specificity was observed for P22-B1 and P22 against clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009, showing no clear plaques on the soft agar.
Fig. 4.
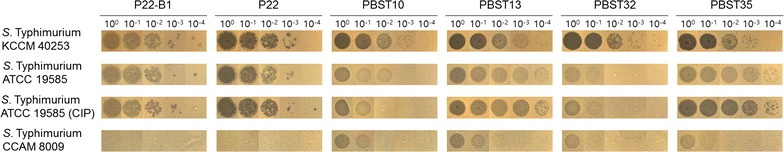
Lytic ability of bacteriophages (P22-B1, P22, PBST10, PBST13, PBST32, and PBST35) against Salmonella Typhimurium KCCM 40253, S. Typhimurium ATCC 19585, ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585, and clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009
Lytic activity of bacteriophages against antibiotic-resistant Salmonella Typhimurium
The lytic activity of bacteriophages against various host strains was evaluated as shown in Fig. 5. The highest lytic activity of P22 was observed against S. Typhimurium KCCM 40253 (> 5 log reduction), followed by S. Typhimurium ATCC 19585 and ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585. The clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 showed the highest resistance to Salmonella bacteriophages (P22-B1, P22, PBST10, PBST13, PBST32, and PBST 35).
Fig. 5.
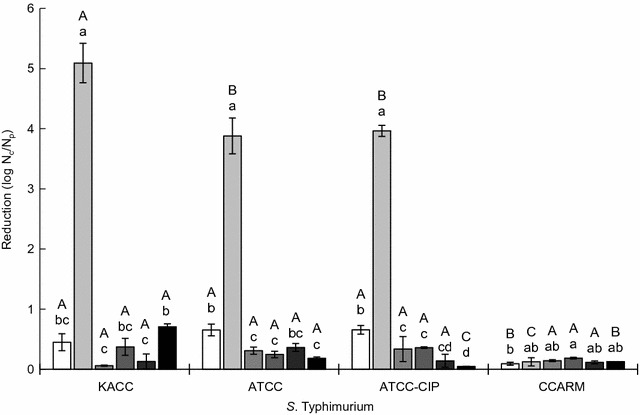
Lytic activity of Salmonella bacteriophages (□, P22-B1;  , P22;
, P22;  , PBST10;
, PBST10;  , PBST13;
, PBST13;  , PBST32; ■, PBST35) against Salmonella Typhimurium KCCM 40253 (KACC), S. Typhimurium ATCC 19585 (ATCC), ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585 (ATCC-CIP), and clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 (CCARM). Nc and Np indicate the numbers of host cells treated without and with bacteriophages, respectively, after 12-h incubation. Bars with different uppercase (A–C) among host strains and lowercase (a–d) within bacteriophages are significantly different at p < 0.05
, PBST32; ■, PBST35) against Salmonella Typhimurium KCCM 40253 (KACC), S. Typhimurium ATCC 19585 (ATCC), ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585 (ATCC-CIP), and clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 (CCARM). Nc and Np indicate the numbers of host cells treated without and with bacteriophages, respectively, after 12-h incubation. Bars with different uppercase (A–C) among host strains and lowercase (a–d) within bacteriophages are significantly different at p < 0.05
Discussion
This study demonstrates the lytic activity of selected bacteriophages against multidrug-resistant S. Typhimurium strains, including antibiotic-induced resistant and clinically isolated strains. In order to compare the host specificity depending on the interaction between bacteriophages and host receptors, the isogenic strain was used to induce the antibiotic resistance. The application of bacteriophages can be one possible approach as an alternative agent to control multidrug-resistant S. Typhimurium.
The heat stability of PBST10 and PBST13 was significantly decreased at 70 °C when compared to other bacteriophages (Fig. 2a). PBST10 and PBST13 were relatively more stable than other bacteriophages at pH 3 but less stable at pH above 4 (Fig. 2b). All bacteriophages were highly sensitive to pH 2 and pH 12 (Fig. 2b). This is in good agreement with previous study showing most bacteriophages are highly stable at pHs between 4 and 9 [17, 18]. The observations suggest that the adsorption rates of Salmonella bacteriophages tested in this study may not be changed at the broad ranges of temperature and pH, which are the important environmental factors for the lytic activity of bacteriophages [19].
The different lytic activities of P22 and P22-B1 were observed depending on MOI (Fig. 3). The interaction of bacteriophages and host cells plays an important role in practical application. The efficacy of lytic activity is associated with the optimum ratio of bacteriophage to host cells, which leads to the increase in the binding chance of bacteriophages to the host cells [20]. The primary step in bacteriophage infection process is the attachment and adsorption, which contribute to the host specificity [10, 11]. Unlike the host cell reduction, the highest phage titers were observed at low MOI (data not shown). The result is due to the lysis-from-without phenomenon that the lysis of host cells can occur without bacteriophage multiplication at high MOI [20, 21]. The bacteriophages could effectively self-replicate at the optimum MOI of 1.
The specificity of P22 to antibiotic-sensitive S. Typhimurium ATCC 19585 was not different from that to ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585 (Fig. 5). The specificity of P22 to S. Typhimurium ATCC 19585 was not changed after the induction of antibiotic resistance. The observation implies that P22 can be used as biocontrol agent against multidrug-resistant S. Typhimurium. Bacteriophages have renewed attention to potential alternative over conventional antibiotics [22]. However, the lowest lytic efficacy of bacteriophages against the clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 might be due to the alteration in receptors, leading to the decrease in binding affinity [23]. The specificity of bacteriophages against host cells is highly associated with the binding affinity between the receptors in the host and receptor-binding proteins in bacteriophages [10, 24]. The alterations in host cell surface receptors are responsible for bacteriophage resistance, resulting in the reduced lytic activity [12, 25].
Conclusions
This study highlights the possibility of using bacteriophages for biocontrol of multidrug-resistant S. Typhimurium. The most significant finding of this study was that the ciprofloxacin-induced antibiotic-resistant S. Typhimurium ATCC 19585 was effectively inactivated by P22, but the clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009 was resistant to P22 due to the alteration in bacteriophage-binding receptors. PBST10, PBST13, PBST32, and PBST35 showed the lytic activity against the clinically isolated antibiotic-resistant S. Typhimurium CCARM 8009. The results would open the door for developing new bacteriophage control system against multidrug-resistant pathogens. However, in order to use bacteriophage as a potential alternative agent against multidrug-resistant S. Typhimurium, further study is needed to elucidate the alterations in bacteriophage-binding receptors in association with the lytic efficacy of bacteriophages.
Authors’ contributions
LJ conducted all experiments as a first author and TD contributed to the writing and preparation of the manuscript as a co-corresponding author. JA contributed to the experimental design, data interpretation, and manuscript writing as a corresponding author. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data supporting the conclusions are included within the manuscript.
Ethics approval and consent to participate
Not applicable.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1D1A3B01008304).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- MOI
multiplicity of infection
- TSB
trypticase soy broth
- CFU
colony-forming unit
- PFU
plaque-forming unit
- PEG
polyethylene glycol
- TEM
transmission electron microscope
- ATCC
American Type Culture Collection
- KCCM
Korean Culture Center of Microorganism
- AMP
ampicillin
- CEF
cefotaxime
- CEP
cephalothin
- CHL
chloramphenicol
- CIP
ciprofloxacin
- KAN
kanamycin
- LEV
levofloxacin
- MER
meropenem
- NAL
nalidixic acid
- NOR
norfloxacin
- SMA/TMP
sulfamethoxazole/trimethoprim
- TET
tetracycline
Contributor Information
Lae-seung Jung, Email: jls@kangwon.ac.kr.
Tian Ding, Email: tding@zju.edu.cn.
Juhee Ahn, Email: juheeahn@kangwon.ac.kr.
References
- 1.Khameneh B, Diab R, Ghazvini K, Fazly Bazzaz BS. Breakthroughs in bacterial resistance mechanisms and the potential ways to combat them. Microb Pathog. 2016;95(1):32–42. doi: 10.1016/j.micpath.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Beceiro A, Tomás M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26(2):185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hede K. Antibiotic resistance: an infectious arms race. Nature. 2014;509(7498):S2–S3. doi: 10.1038/509S2a. [DOI] [PubMed] [Google Scholar]
- 4.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119(6A):S3–S10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Mulvey MR, Finley R, Allen V, Ang L, Bekal S, El Bailey S, et al. Emergence of multidrug-resistant Salmonella enterica serotype involving human cases in Canada: results from the Canadian Integrated Program on Antimicrobial Resistance Surveillance (CIPARS) J Antimicrob Chemother. 2013;68(9):1982–1986. doi: 10.1093/jac/dkt149. [DOI] [PubMed] [Google Scholar]
- 6.Dionisi AM, Lucarelli C, Benedetti I, Owczarek S, Luzzi I. Molecular characterisation of multidrug-resistant Salmonella enterica serotype Infantis from humans, animals and the environment in Italy. Int J Antimicrob Agent. 2011;38(5):384–389. doi: 10.1016/j.ijantimicag.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, Zhao H, Sun J, Liu Y, Zhou X, Beier RC, et al. Characterization of multidrug-resistant Salmonella enterica serovars Indiana and Enteritidis from chickens in Eastern China. PLoS ONE. 2014;9(5):e96050. doi: 10.1371/journal.pone.0096050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eng S-K, Pusparajah P, Ab Mutalib N-S, Ser H-L, Chan K-G, Lee L-H. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015;8(3):284–293. doi: 10.1080/21553769.2015.1051243. [DOI] [Google Scholar]
- 9.Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, et al. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J Infect Chemother. 2005;11(5):211–219. doi: 10.1007/s10156-005-0408-9. [DOI] [PubMed] [Google Scholar]
- 10.Le S, He X, Tan Y, Huang G, Zhang L, Lux R, et al. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS ONE. 2013;8(7):e68562. doi: 10.1371/journal.pone.0068562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol. 2010;59(3):145–155. [PubMed] [Google Scholar]
- 12.Qimron U, Marintcheva B, Tabor S, Richardson CC. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc Natl Acad Sci. 2006;103(50):19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madaras-Kelly KJ, Daniels C, Hegbloom M, Thompson M. Pharmacodynamic characterization of efflux and topoisomerase IV-mediated fluoroquinolone resistance in Streptococcus pneumoniae. J Antimicrob Chemother. 2002;50(2):211–218. doi: 10.1093/jac/dkf095. [DOI] [PubMed] [Google Scholar]
- 14.Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob Agent Chemother. 2000;44(5):1223–1228. doi: 10.1128/AAC.44.5.1223-1228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michéa-Hamzehpour M, Kahr A, Pechère JC. In vitro stepwise selection of resistance to quinolones, β-lactams and amikacin in nosocomial Gram-negative bacilli. Infection. 1994;22(2):S105–S110. doi: 10.1007/BF01793574. [DOI] [PubMed] [Google Scholar]
- 16.Bielke L, Higgins S, Donoghue A, Donoghue D, Hargis BM. Salmonella host range of bacteriophages that infect multiple genera. Poult Sci. 2007;86(12):2536–2540. doi: 10.3382/ps.2007-00250. [DOI] [PubMed] [Google Scholar]
- 17.Chow MS, Rouf MA. Isolation and partial characterization of two Aeromonas hydrophila bacteriophages. Appl Environ Microbiol. 1983;45(5):1670–1676. doi: 10.1128/aem.45.5.1670-1676.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo J, Ahn J. Assessment of synergistic combination potential of probiotic and bacteriophage against antibiotic-resistant Staphylococcus aureus exposed to simulated intestinal conditions. Arch Microbiol. 2014;196(10):719–727. doi: 10.1007/s00203-014-1013-z. [DOI] [PubMed] [Google Scholar]
- 19.Fischetti VA. Bacteriophage lysins as effective antibacterials. Curr Opin Microbiol. 2008;11(5):393–400. doi: 10.1016/j.mib.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong CL, Sieo CC, Tan WS, Abdullah N, Hair-Bejo M, Abu J, et al. Evaluation of a lytic bacteriophage, Φ st1, for biocontrol of Salmonella enterica serovar Typhimurium in chickens. Int J Food Microbiol. 2014;172(2):92–101. doi: 10.1016/j.ijfoodmicro.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 21.Huff WE, Huff GR, Rath NC, Donoghue AM. Evaluation of the influence of bacteriophage titer on the treatment of colibacillosis in broiler chickens. Poult Sci. 2006;85(8):1373–1377. doi: 10.1093/ps/85.8.1373. [DOI] [PubMed] [Google Scholar]
- 22.Jassim SAA, Limoges RG. Natural solution to antibiotic resistance: bacteriophages ‘The Living Drugs’. World J Microbiol Biotechnol. 2014;30(8):2153–2170. doi: 10.1007/s11274-014-1655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller-Merbach M, Kohler K, Hinrichs J. Environmental factors for phage-induced fermentation problems: replication and adsorption of the Lactococcus lactis phage P008 as influenced by temperature and pH. Food Microbiol. 2007;24(7–8):695–702. doi: 10.1016/j.fm.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Javed MA, Poshtiban S, Arutyunov D, Evoy S, Szymanski CM. Bacteriophage receptor binding protein based assays for the simultaneous detection of Campylobacter jejuni and Campylobacter coli. PLoS ONE. 2013;8(7):e69770. doi: 10.1371/journal.pone.0069770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Jo A, Ding T, Lee H-Y, Ahn J. Assessment of altered binding specificity of bacteriophage for ciprofloxacin-induced antibiotic-resistant Salmonella Typhimurium. Arch Microbiol. 2016;198(6):521–529. doi: 10.1007/s00203-016-1210-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the conclusions are included within the manuscript.


