Abstract
Background
As a disease with extremely complex molecular mechanisms, many deregulated miRNAs have been identified in colon cancer. Few studies have been performed by using Ingenuity Pathways Analysis (IPA) to predict miRNAs specifically expressed in colon cancer.
Methods
A characteristic microRNA-target network of colon cancer was explored using IPA. Then the clinical significance of miR-19b-3p was evaluated in 211 colon cancer patients. The roles of miR-19b-3p and its candidate target gene, SMAD4, in colon cancer progression were examined both in vitro and in vivo.
Results
Bioinformatics analysis showed that 15 microRNAs screened by IPA were significantly correlated with malignant biological behaviors of colon cancer. miR-19b-3p was the most significantly upregulated candidate based on the validation experiment using 211 colon cancer samples. High expression of miR-19b-3p was significantly associated with high N stage (P < 0.001), high AJCC stage (P < 0.001), poor histologic grade (P = 0.032), frequent venous and lymphatic invasion (P = 0.027), and liver metastasis (P < 0.001). Survival analysis revealed that miR-19b-3p was an independent prognostic factor associated with colon cancer patient’s overall survival (OS) and disease-free survival (DFS). miR-19b-3p promoted proliferation and chemoresistance of colon cancer cells, but had no effect on invasion in vitro, along with tumorigenesis in vivo. In addition, we confirmed that miR-19b-3p mediates resistance to oxaliplatin-based chemotherapy via SMAD4.
Conclusions
Our findings demonstrate the role of miR-19b-3p-SMAD4 axis in colon cancer progression, which may become a potential therapeutic target against chemotherapy resistance.
Electronic supplementary material
The online version of this article (10.1186/s13046-017-0602-5) contains supplementary material, which is available to authorized users.
Keywords: miR-19b-3p, SMAD4, Proliferation, Oxaliplatin, Chemoresistance
Background
Colon cancer is the third most common cancer with high cancer-related deaths worldwide [1]. Currently, recurrence and metastasis are the principal causes of death, despite improvements in the multidisciplinary and comprehensive treatment based on surgical operation of colon cancer [2]. In spite of the in-depth studies of the molecular mechanisms underlying colon cancer for the last decades, chemoresistance remains a crucial challenge for the treatment of colon cancer.
MicroRNAs (miRNAs) are a class of short, non-coding RNAs [3]. miRNAs bind to the 3′-untranslated regions (UTR) of partially complementary target messenger RNAs (mRNAs) by base pairing mode, thereby suppressing the expression of downstream target genes [4, 5]. More than 30% of mRNAs are regulated by miRNAs, which can act as tumor suppressor genes or oncogenes, depending on the microenvironment within the cell and the specific downstream target genes they regulate [6]. Therefore, deregulation of miRNA expression influences many biological functions of tumors such as angiogenesis, differentiation, cell proliferation, and apoptosis [7]. As a disease with extremely complex molecular mechanisms, many deregulated miRNAs have been implicated in the pathogenesis of colon cancer [8, 9].
The powerful data analysis and search capabilities of Ingenuity Pathways Analysis (IPA) contribute to the understanding of existing data, target discovery, and validation as well as analysis of biological networks [10]. The application of IPA software has been widely used in cancer research, and its powerful database makes it easier to identify potential biomarkers for cancer diagnosis and therapy [11, 12]. Although many miRNAs have been confirmed as biomarkers in colon cancer in a number of studies, limited experimental research has been performed by using IPA to better understand the miRNA/mRNA network of colon cancer.
In this study, we found that miR-19b-3p was significantly upregulated in colon cancer using IPA. To figure out its role in colon cancer, we performed the current study to evaluate the associations of miR-19b-3p dysregulation with colon cancer progression in vitro and in vivo, and further investigate its relationship with prognosis of colon cancer patients.
Methods
Bioinformatics analysis
IPA database is used for analyze and understand the complex biological and chemical systems in life science research. IPA provides exploratory investigation of genes, proteins, and biological functions, creating customized pathways or molecular interaction networks focused on drug targets and identifying potential biomarkers. The microRNA Target Filter associates microRNAs from a dataset with experimentally observed mRNA targets which is used to overlay microRNA data onto networks and pathways, to add molecules to networks, and to compare molecules from different experimental observations. In this study, we used IPA 9.0 to screen miRNAs specifically expressed in colon cancer to better understand the biology around potential mRNA targets/diseases and to identify the most biologically relevant targets.
The latest release of the miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw/) was used to collect experimentally validated target genes of miRNAs, which were screened by IPA. The miRNA-gene regulatory network was based on the interactions of miRNAs and predicted target genes. Functional annotations of the predicted target genes of these miRNAs were obtained from R annotation packages. Function enrichment analyses were performed by Fisher’s exact test. P-values were adjusted for the multiple testing corrections by R function “p.adjust”. We chose functional terms whose adjusted P-value was smaller than 0.05 as significantly enriched functions.
Patients and tissue samples
A total of 211 tissue samples from colon cancer patients with pathologically confirmed diagnosis were obtained immediately after surgery and frozen at −80 °C in liquid nitrogen before being deposited in the Anal-Colorectal Surgery Department, General Hospital of Ningxia Medical University. For each case, paired tumor and distal mucosae were collected. All patients provided informed consent and the study was approved by the institutional review boards of General Hospital of Ningxia Medical University. The pathologic verification of diagnosis and staging is summarized according to the National Comprehensive Cancer Network (NCCN) Practice guidelines. The follow-up of this cohort ended on July 14, 2014 and the median duration of follow-up was 59 (range, 12–83) months. Disease-free survival (DFS) and overall survival (OS) rates were defined as the interval from the initial surgery to clinically or radiologically proven recurrence/metastasis and death, respectively.
Cell culture and transfection
The human colon cancer cell lines, SW620, LoVo, HCT 116, SW480, Caco-2, HT-29, RKO, DLD1, and human normal colonic epithelial cells, NCM460, were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Hsa-miR-193a-3p inhibitor and negative control were purchased from Biomics Biotech (Biomics Biotech, Nantong, China). Their sequences were as follows: miR-19b-3p inhibitors, 5′ -UCAGUUUUGCAUGGAUUUGCACA-3′; negative control, 5′ -CAGUACUUUUGUGUAGUACAA-3′. The coding sequence of miR-19b-3p inhibitors or negative control was cloned into pCDH-CMV lentivectors (SBI, Mountain View, CA, USA) by BioLink Biotechnology (Shanghai, China). To package the construct, 293 T cells were co-transfected with both pPackH1 packaging plasmid mix (SBI, Mountain View, CA, USA) and the lentivectors. Then the virus particles were collected 48 h later. For transfection, cells were plated in 6-well plates at a density of 2 × 105 cells/well and cultured until they reached 80% confluence. Then RKO and SW480 cells were infected with viruses.
RNA extraction and quantitative real-time PCR
Total RNA, including miRNAs, was isolated from clinical tissue specimens and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The first strand cDNA was synthesized with the RevertAid First Strand cDNA Synthesis Kit (MBI Fermentas, Vilnius, Lithuania) using 1 μg of total RNA as the template. miRNAs were prepared with the High-Specificity miRNA qRT-PCR Detection Kit (Stratagene, Santa Clara, CA, USA) and U6 was used as an endogenous control. The relative miR-19b-3p level was calculated from this eq. 2-ΔCT[ΔCT = Ct (miR-19b-3p) - Ct (control)]. Real-time PCR of SMAD4 and miRNAs was carried out using ViiA™ 7 system (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. GAPDH was used as endogenous control to normalize the expression of SMAD4. The qRT-PCR primers are shown in Additional file 1: Table S1.
Cell proliferation and invasion
Cell Counting Kit-8 (CCK8) assays (Dojindo, Kumamoto, Japan) were used to evaluate the cell proliferation ability according to the manufacturer’s protocol. The invasion assays were implemented by using the transwell system. The pore size of invasion chambers is 8.0 μm coated with matrigel (Millipore, Billerica, MA, USA). The colon cancer cells were suspended after being transfected for 48 h, and then seeded in the upper chambers of 24-well transwell plates with serum and calcium-free solution supplemented with 0.2% BSA. The lower chamber was filled with 500 μL of medium containing 10% FBS. Cells were incubated for 24 h at 37 °C with 5% CO2. Next, a cotton swab was used to wipe off the matrigel and cells remaining in the upper chambers. Cells that migrated to the lower chambers were fixed with methanol for 20 min and then stained with 1% crystal violet for 10 min. The number of migrated cells was counted under a light microscope with a 200× magnification. Five fields were randomly selected on each membrane.
Cell viability and apoptosis
The Annexin V-PE and 7-AAD (BD Biosciences, San Jose, CA, USA) double staining method was used to examine cell viability. The frequency of apoptosis was measured using the BD FACSCalibur™ Flow Cytometer (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions. Before assessing the chemosensitivity of colon cancer cells to oxaliplatin, the optimal drug concentration of oxaliplatin inducing cell death was determined on untransfected cells. Cells were treated with oxaliplatin (MedChem express, Monmouth Junction, NJ, USA) at a final concentration of 50 μg/mL for 24 h. The percentage of oxaliplatin-treated and untreated apoptotic cells was calculated according to the number of cells positive or negative for Annexin V-PE and 7-AAD. Results are presented as the percentage of total cells that were living cells (Ann−/7-AAD-), early apoptotic cells (Ann+/7-AAD-), late apoptotic cells, and dead cells (Ann+/7-AAD +).
Western blot
Western blot was performed to assess SMAD4 expression in miR-19b-3p inhibitor and negative control transfected cells and tumors induced in SCID mice. SMAD4 was detected with anti-SMAD4 (Abcam, Cambridge, UK) at a 1:5000 dilution. The level of SMAD4 was normalized to the level of β-actin protein, which was detected by using anti-β-actin (Abcam, Cambridge, UK) at a 1:2000 dilution. Then horseradish peroxidase (HRP)-tagged anti-rabbit or anti-mouse immunoglobulin (Abgent, San Diego, CA, USA), at a dilution of 1:2000, was used to detect the primary antibody. Enhanced chemiluminescence reagent (Merck Millipore, Temecula, CA, USA) was applied to reveal the protein bands. The Image J software (National Institutes of Health, Bethesda, MD, USA) was used to quantify the band intensity. To further explore the effects of the interactions between miR-19b-3p and several mRNA at the protein level, a series of western blot analyses was performed following the aforementioned experimental protocols. Primary antibodies included PRKACB, ATM, CREB3L2, EGLN3, JUN, NR3C1, WEE1, RASSF1 and TGFBR2 (Abcam, Cambridge, UK).
Tumorigenesis
Twenty 6- to 8-week-old SCID mice were obtained from the Shanghai Jiaotong University and maintained in specific pathogen-free (SPF) conditions. Approximately 5 × 106 SW480 cells transfected with miR-19b-3p inhibitor or a negative control were injected subcutaneously into the opposite flanks of each mouse. Tumors grew in all animals and the tumors size was measured at 4, 8, 12, 16, and 21 days. All nude mice were euthanized by cervical dislocation at day 21 and their tumors were harvested, weighed, and photographed. All procedures followed the Shanghai Jiao tong University Affiliated Shanghai General Hospital Animal Care guidelines.
Plasmid constructs and luciferase reporter assays
Dual-Luciferase reporter assays (Promega, Madison, WI, USA) were used following the manufacturer’s instructions. The putative miR-19b-3p complementary site in the 3′-UTR of SMAD4 mRNA (NM_005395; 3′-UTR: 1352–1358) or its mutant sequence were cloned into the pmirGLO luciferase reporter vector. The novel combined plasmid was named pmirGLO-SMAD4 3′’-UTR-wt (wild type). A mutation of the 3′-UTR of SMAD4 was created by using a site-directed mutagenesis kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and designated as pmirGLO-SMAD4 3′-UTR-mut (mutant). SW480 cells were plated at 4000 cells per well in 100 μL DMEM in a 96-well microplate (BD Biosciences, USA). Twenty-four hours after plating, cells were co-transfected with 0.5 μg of pmirGLO-SMAD4 3′-UTR-wt or pmirGLO-SMAD4 3′-UTR-mut, 0.01 μg of the pMirGLO-Vector (Promega, Madison, WI, USA), 50 nM miR-19b-3p mimics (Mimics), and miRNA mimics Negative Control (NC) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The firefly and Renilla luciferase activities were measured 24 h after transfection.
In situ hybridization (ISH) and immunohistochemistry on tissue microarray
The tissue microarray (TMA) including 120 pairs of colon cancer and corresponding normal mucosa was obtained from Outdo Biotech (Shanghai, China). The TMA slides were dewaxed by xylene for 15 min twice. After being dehydrated by immersion in 100% ethanol for 5 min, the slides were air-dried and then incubated with pepsin at 37 °C for 15 min. Then the slides were fixed in 4% paraformaldehyde, dehydrated in 90% ethanol, and incubated with the digoxigenin-labeled probe (Exiqon, Denmark) complementary to miR-19b-3p at 37 °C overnight, according to the manufacturer’s instructions. The slides were washed twice with 2× Saline-Sodium Citrate Bufferat room temperature, and incubated with mouse anti-digoxigenin monoclonal antibody according to the manufacturer’s protocol. miR-19b-3p expression in the TMA was assessed by 2 independent pathologists. The proportion of positively stained tumor cells was graded as follows: 0 (no positive cells), 1 (<10% positive cells), 2 (10–50% positive cells), 3 (>50% positive cells). The intensity of the staining was recorded on a scale of 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The staining index (SI) was defined as the proportion of positively stained tumor cells × staining intensity.
To further confirm that SMAD4 is the target of miR-19b-3p, we detected SMAD4 protein expression levels by immunohistochemistry on the same location of the TMA. The scoring systems were similar to those used for ISH.
Statistics
Statistical analyses were performed using the SPSS statistical software program version 20 (SPSS Inc., Chicago, IL, USA). Student’s t-test was applied to compare two groups of quantitative data. The χ2 test or Fisher’s exact test for enumeration data was used to analyze the relationship between miR-19b-3p and clinicopathological features. Kaplan-Meier method was used to analyze the survival rates, and the differences between the survival curves were examined by the log-rank test. Univariate and multivariate survival analyses were performed using Cox proportional hazards models. P < 0.05 was considered statistically significant.
Results
IPA and gene function analysis
We used IPA 9.0 to identify miRNAs specifically expressed in colon cancer. A total of 15 miRNAs were identified in the Ingenuity Knowledge Base by experimental evidence (Additional file 2: Table S2). The 7 upregulated miRNAs are: miR-19b-3p, miR-155-5p, miR-17-5p, miR-183-5p, miR-25-3p, miR-21-5p, and miR-196a-5p. The 8 downregulated miRNAs are: miR-29c-3p, miR-34a-5p, miR-542-5p, let-7a-5p, miR-126-3p, miR-143-3p, miR-192-5p, and miR-194-5p. The experimentally validated target genes of the 15 significant miRNAs were searched with the miRTarBase database.
The predicted target genes in enriched functional terms were used to construct a miRNA-target network. The network was plotted by using R package “igraph” (Fig. 1). The biological function analysis of the predicted target genes based on IPA and GO annotation system enabled us to comprehensively understand their functional roles in colon cancer progression. The predicted target genes were significantly involved in many biologic processes relevant to cancer such as apoptosis, cell proliferation, and cell cycle arrest (Fig. 2). Evidence indicated that these malignant biological behaviors may lead to tumor progression [13, 14]. These bioinformatics analysis suggested that the predicted target genes may control broad biological functions associated with colon cancer.
Fig. 1.
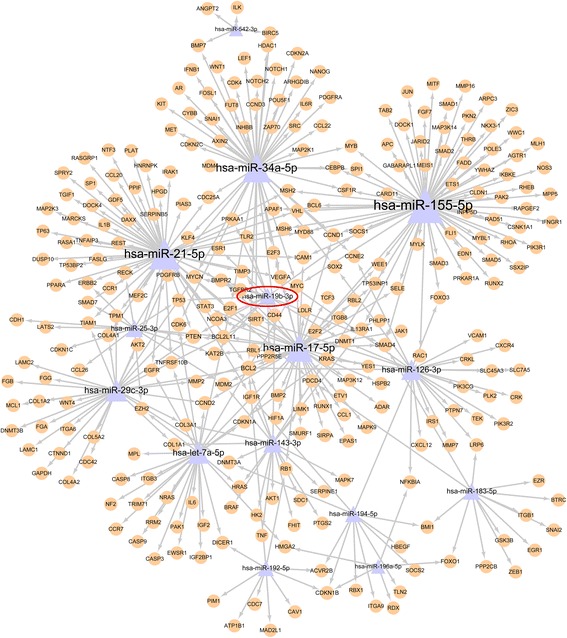
miRNA-target gene network analysis. The arrows go from miRNA to their target genes. In total, this network contains 15 miRNAs and 252 target genes
Fig. 2.
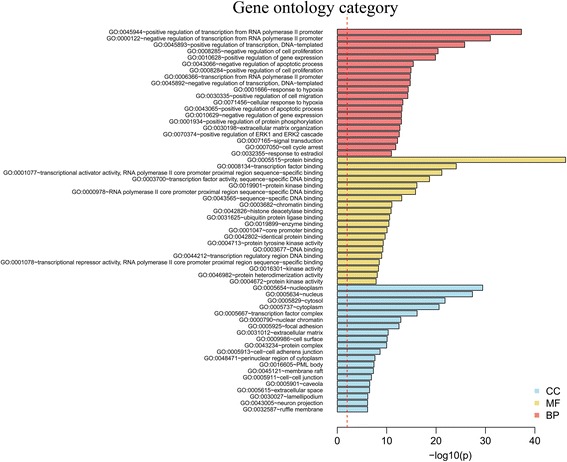
Functional annotations of the predicted target genes. Significant GO terms of miRNA targets. The vertical axis represents GO category and the horizontal axis represents the negative logarithm of P value (−Log P value), which indicates the significant level of GOs.CC (Cellular Component), MF (Molecular Function), BP (Biological Process)
miRNA expression profiling in colon cancer patients
Since gene therapy requires high expression levels of the target gene, we selected the 7 upregulated miRNAs to validate their expression patterns in the 211 colon cancer samples using qRT-PCR. The expression profiles of the 7 miRNA are presented in Fig. 3. miR-19b-3p was the most significantly upregulated candidate (P < 0.001).
Fig. 3.
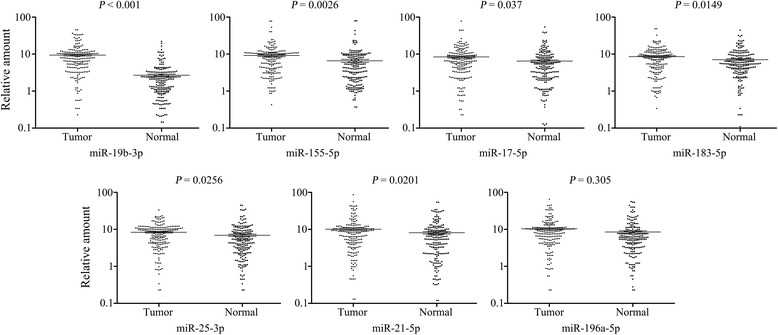
Validation of the 7 up-regulated miRNAs screened by IPA using qRT-PCR. The expression of miR-19b-3p, miR-155-5p, miR-17-5p, miR-183-5p, miR-25-3p, miR-21-5p and miR-196a-5p were significantly higher in tumor tissues compared with normal mucosa in cases from General Hospital of Ningxia Medical University
miR-19b-3p expression is upregulated in colon cancer and predicts poor prognosis of patients with colon cancer
Using qRT-PCR, we investigated miR-19b-3p expression patterns in colon cancer specimens and paired normal tissues. Results showed that miR-19b-3p expression levels were associated with the clinicopathological data (Table 1), which included high N stage (P < 0.001), high AJCC stage (P < 0.001), poor histologic grade (P = 0.032), and liver metastasis (P < 0.001).
Table 1.
Clinical characteristics of the 211 colon cancer patients according to High or Low miR-19b-3p
| Variable | Total (n = 211) | miR-19b-3p expression | P value | |
|---|---|---|---|---|
| Low (n = 115) | High (n = 96) | |||
| Age | ||||
| < 65 y | 89 | 47 | 42 | 0.721 |
| ≥ 65 y | 122 | 68 | 54 | |
| Gender | ||||
| Male | 72 | 45 | 27 | 0.637 |
| Female | 139 | 70 | 69 | |
| Location | ||||
| Left | 148 | 77 | 71 | 0.521 |
| Others | 63 | 38 | 25 | |
| T stage | ||||
| T1 + T2 | 17 | 9 | 8 | 0.764 |
| T3 + T4 | 194 | 106 | 88 | |
| N stage | ||||
| N0 | 115 | 80 | 35 | <0.001* |
| N1 + N2 | 96 | 35 | 61 | |
| AJCC stage | ||||
| I + II | 114 | 86 | 28 | <0.001* |
| III + IV | 97 | 29 | 68 | |
| Histologic grade | ||||
| Well Differentiated | 104 | 66 | 38 | 0.032* |
| Poorly differentiated | 107 | 49 | 58 | |
| Venous & Lymphatic invasion | ||||
| No | 197 | 110 | 87 | 0.247 |
| Yes | 14 | 5 | 9 | |
| Peritoneal Dissemination | ||||
| Absent | 184 | 97 | 87 | 0.359 |
| Present | 27 | 12 | 15 | |
| Liver Metastsis | ||||
| Negative | 164 | 96 | 68 | |
| Positive Multinodular | 47 | 8 | 39 | <0.001* |
P value are based on chi-square test
AJCC: American Joint Committee on Cancer
*P < 0.05 was considered significant
Kaplan-Meier and Cox proportional hazard regression analyses were performed and showed that miR-19b-3p expression level was significantly associated with patient’s survival. The results showed that high miR-19b-3p expression correlated with lower OS (P < 0.001) and DFS (P < 0.001) in 211 cases colon cancer patients (Fig. 4). Similarly, univariate analysis showed that miR-19b-3p expression was associated with OS (Table 2, P < 0.001) and DFS (Table 3, P < 0.001). Using the parameters that were significant in univariate analyses as covariates, miR-19b-3p expression level was significantly associated with OS (HR = 2.23, P = 0.008, Table 2) and DFS (HR = 2.73, P = 0.016, Table 3). It demonstrates that miR-19b-3p is an independent prognostic factor of colon cancer patients.
Fig. 4.
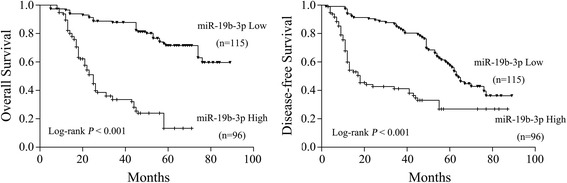
Kaplan-Meier analysis of overall survival (OS) and disease-free survival (DFS) based on the expression of miR-19b-3p. High expression of miR-19b-3p correlated with poor OS (P < 0.001) and DFS (P < 0.001) in patients from General Hospital of Ningxia Medical University
Table 2.
Univariate and multivariate Cox proportional hazard models for overall survival (OS)
| Overall survival (OS) | ||||
|---|---|---|---|---|
| Univariate | Multivariate | |||
| HR (95% CI) | P | HR (95% CI) | P | |
| Age | ||||
| < 65 y | 1 | |||
| ≥ 65 y | 1.65 (0.93–2.27) | 0.126 | ||
| Gender | ||||
| Male | 1 | |||
| Female | 1.27 (0.83–2.01) | 0.149 | ||
| Location | ||||
| Left | 1 | |||
| Others | 0.82 (0.56–1.41) | 0.632 | ||
| T stage | ||||
| T1 + T2 | 1 | |||
| T3 + T4 | 1.72 (0.52–5.29) | 0.391 | ||
| N stage | ||||
| N0 | 1 | 1 | ||
| N1 + N2 | 6.71 (3.91–11.68) | <0.001* | 0.35 (0.04–1.92) | 0.163 |
| AJCC stage | ||||
| II | 1 | 1 | ||
| III | 5.47 (4.25–10.27) | <0.001* | 18.38 (2.93–152.17) | 0.008* |
| Histologic grade | ||||
| Well Differentiated | 1 | 1 | ||
| Poorly differentiated | 3.42 (1.98–5.24) | <0.001* | 2.34 (1.36–3.57) | 0.316 |
| Venous & Lymphatic invasion | ||||
| No | 1 | |||
| Yes | 0.75 (0.37–0.95) 0.271 | |||
| Peritoneal Dissemination | ||||
| Absent | 1 | |||
| Present | 0.52 (0.24–0.82) | 0.493 | ||
| Liver Metastsis | ||||
| Negative | 1 | 1 | ||
| Positive Multinodular | 0.48 (0.29–0.81) | 0.001* | 0.59 (0.36–0.98) 0.024* | |
| MiR-19b-3p | ||||
| Low | 1 | 1 | ||
| High | 2.79 (1.80–4.67) | <0.001* | 2.23 (1.42–3.58) | 0.008* |
HR: Hazard ratio; CI: Confidence interval
*P < 0.05 was considered significant
Table 3.
Univariate and multivariate Cox proportional hazard models for disease free survival (DFS)
| Disease free survival (DFS) | ||||
|---|---|---|---|---|
| Univariate | Multivariate | |||
| HR (95% CI) | P | HR (95% CI) | P | |
| Age | ||||
| < 65 y | 1 | |||
| ≥ 65 y | 1.76 (0.97–2.68) | 0.079 | ||
| Gender | ||||
| Male | 1 | |||
| Female | 1.53 (0.93–2.31) | 0.372 | ||
| Location | ||||
| Left | 1 | |||
| Others | 0.74 (0.52–1.39) | 0.354 | ||
| T stage | ||||
| T1 + T2 | 1 | |||
| T3 + T4 | 1.85 (0.56–5.37) | 0.482 | ||
| N stage | ||||
| N0 | 1 | 1 | ||
| N1 + N2 | 5.26 (2.42–10.46) | <0.001* | 4.21 (2.50–7.94) | <0.001* |
| AJCC stage | ||||
| II | 1 | 1 | ||
| III | 6.82 (3.72–10.93) | <0.001* | 2.29 (1.21–3.82) | 0.381 |
| Histologic grade | ||||
| Well Differentiated | 1 | 1 | ||
| Poorly differentiated | 2.83 (1.79–4.29) | <0.001* | 2.20 (1.35–3.62) | 0.282 |
| Venous & Lymphatic invasion | ||||
| No | 1 | |||
| Yes | 0.72 (0.56–1.27) | 0.413 | ||
| Peritoneal Dissemination | ||||
| Absent | 1 | |||
| Present | 0.84 (0.68–1.53) | 0.281 | ||
| Liver Metastsis | ||||
| Negative | 1 | 1 | ||
| Positive Multinodular | 0.52 (0.29–0.83) | 0.003* | 0.69 (0.36–1.23) | 0.032* |
| MiR-19b-3p | ||||
| Low | 1 | 1 | ||
| High | 3.34 (1.97–5.23) | <0.001* | 2.73 (1.76–3.89) | 0.016* |
HR: Hazard ratio; CI: Confidence interval
*P < 0.05 was considered significant
miR-19b-3p is overexpressed in colon cancer cell lines and promotes proliferation and chemoresistance in vitro
To further determine the potential function of miR-19b-3p in promoting colon cancer progression, we first investigated miR-19b-3p expression pattern in a panel of human colon cancer cell lines and normal colonic epithelial cells. The expression of miR-19b-3p was higher in the 8 colon cancer cells than in normal colonic epithelial cells. Two out of eight colon cancer cell lines, including SW480 and RKO, reached the highest level of statistical significance versus the control (Fig. 5a). We constructed a lentiviral vector harboring a RNAi sequence targeting the human miR-19b-3p and investigated the effects of miR-19b-3p inhibitor on a series of cancer-relevant in vitro assays, including proliferation, invasion, apoptosis, and chemosensitivity in SW480 and RKO cells. miR-19b-3p downregulation suppressed the proliferation of SW480 cells (Fig. 5b, left) and RKO cells (Fig. 5b, right) but had no significant effect on invasion using matrigel-coated transwell assays (Fig. 5c and d). miR-19b-3p downregulation had little effects on cell viability in the absence of oxaliplatin. However, the ability to promote cell apoptosis was observed when cells were treated with oxaliplatin (Fig. 5e and f). These in vitro findings showed that miR-19b-3p promotes proliferation and chemoresistance in colon cancer cells and plays an important role in colon cancer progression.
Fig. 5.
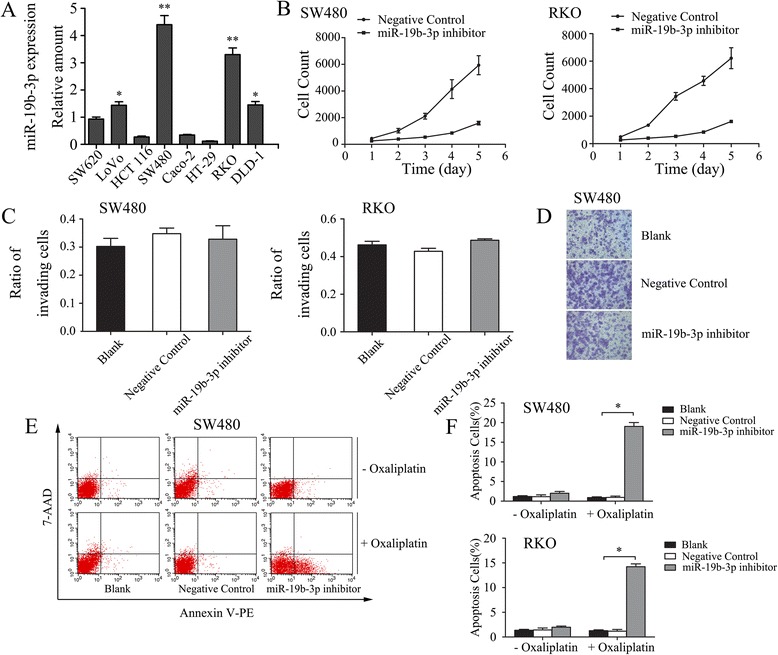
The expression of miR-19b-3p promotes proliferation and chemoresistance but has no effect on invasion of SW480 and RKO cells in vitro. a Endogenous miR-19b-3p expression by qRT-PCR in colon cancer cell lines and normal colonic epithelium cells (*, P < 0.05; **, P < 0.01). b Down-regulation of miR-19b-3p suppressed proliferation phenotype in SW480 cells (left) and RKO cells (right). a and d Matrigel-coated transwell assays showed that down-regulation of miR-19b-3p had no effect on invasive abilities of SW480 (c, left) and RKO (c, right) cells. Representative of the images were shown in (d). Statistical analysis was performed with at least three independent experiments. e and f Down-regulation of miR-19b-3p had little effect on viability in the absence of oxaliplatin but promoted cell apoptosis when treated with oxaliplatin in SW480 (f, top) and RKO cells (f, bottom). Representative images of flow cytometry were shown (e) Statistical analysis was performed with at least three independent experiments
miR-19b-3p promotes proliferation of colon cancer in vivo
To evaluate the effect of miR-19b-3p on tumorigenesis in vivo, SW480 cells were subcutaneously implanted in SCID mice. Cells transfected with the miR-19b-3p inhibitor or a negative control were injected into the opposite flanks of each animal. miR-19b-3p inhibition resulted in less spheroid tumors with clean edges, and the tumor growth rate was decreased compared to negative control tumors with more local proliferation phenotypes. The mice were euthanized at day 21 and the tumors were immediately harvested (Fig. 6a). The average tumor size of miR-19b-3p inhibitor expressing tumors was significantly reduced when compared to that of control tumors, especially at day 21 (0.237 ± 0.091 cm3 versus 1.533 ± 0.231 cm3, respectively, P < 0.05, Fig. 6b). Moreover, the average weight of miR-19b-3p inhibitor expressing tumors was also significantly reduced when compared to that of negative controls (1.28 ± 0.23 g versus 2.74 ± 0.45 g, respectively, P < 0.05, Fig. 6c). Furthermore, we performed western blotting assays to investigate SMAD4 protein levels in tumors induced in SCID mice. Results indicated that miR-19b-3p inhibitor group presented a significantly higher SMAD4 expression pattern (Fig. 6d). These results indicated that miR-19b-3p downregulation led to a significant reduction in the proliferation ability of colon cancer cells in vivo and miR-19b-3p was inversely correlated with SMAD4 during tumorigenesis.
Fig. 6.
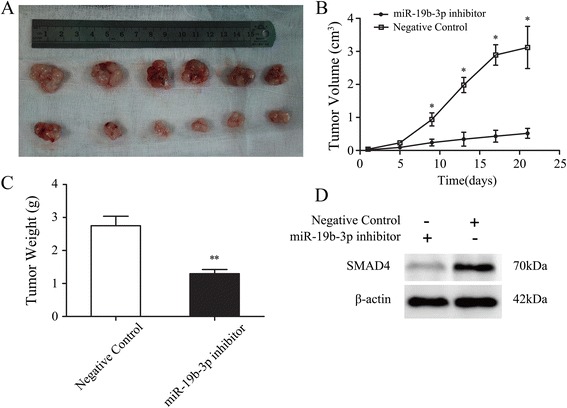
Down-regulation of miR-19b-3p suppresses colon cancer growth in vivo. a Cells transfected with the miR-19b-3p inhibitor and negative control of SW480 cells were subcutaneously implanted into the flank of nude mice (n = 6). At day 21, the mice were euthanized. b and c The average tumor size (b) and weight (c) of miR-19b-3p inhibitor tumors was reduced significantly as compared to negative control tumors. The tumor sizes were measured at 5-day intervals as soon as the tumors were measurable. The tumor weight were measured at day 21 after the mice being killed. *, P < 0.05 and **, P < 0.01 by Student’s t-test. d Representative Western blot plots of miR-19b-3p inhibitor and SMAD4 expression pattern from tumor-bearing mice
SMAD4 is targeted by miR-19b-3p in colon cancer
In order to investigate the direct target genes of miR-19b-3p that could explain the phenotype resulting from miR-19b-3p upregulation in colon cancer, the miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw/), which provides the most current and comprehensive information of experimentally validated miRNA-target interactions was utilized. SMAD4, PRKACB, ATM, CREB3L2, EGLN3, JUN, NR3C1, WEE1, RASSF1, and TGFBR2 were validated as candidate targets of miR-19b-3p in replicate experiments in SW480 and RKO cells by using qRT-PCR. SMAD4 was the top candidate gene, which was significantly upregulated in SW480 cells transfected with the miR-19b-3p inhibitor when compared with negative control SW480 cells (Fig. 7a, top). The same pattern was observed in RKO cells (Fig. 7a, bottom). Subsequent western blotting analysis were performed and confirmed that SMAD4 protein levels were significantly overexpressed in transfected miR-19b-3p inhibitor colon cancer cells, consistent with the results of qRT-PCR (Fig. 7b). In addition, miRTarBase database was queried to search for binding sites of miR-19b-3p in the 3′-UTR of SMAD4 (Fig. 7c). Results indicated that SMAD4 possesses a putative miR-19b target sequence, which matched at 7mer-m8. To verify that SMAD4 is directly targeted by miR-19b-3p, the 3′-UTR of SMAD4 containing a potential binding site for miR-19b-3p was cloned into a luciferase reporter plasmid and a dual luciferase reporter assay was performed. A segment of pmirGLO-SMAD4 3′-UTR-wt (wild type), pmirGLO-SMAD4 3′-UTR-mut (mutant), and pMirGLO-Vector were transfected together with miR-19b-3p mimics or miRNA mimics negative control into SW480 cells. Results indicated that miR-19b-3p significantly inhibited luciferase activity in the SMAD4 3′-UTR-wt transfected cells. Meanwhile, no change in luciferase activity was observed when the cells were transfected with the pMirGLO-Vector lacking the miR-19b-3p binding sequence (Fig. 7d). Furthermore, we carried out ISH and immunohistochemistry using the TMA to investigate whether miR-19b-3p inversely correlated with SMAD4 expression. Results indicated that patients whose localized colon tumors were miR-19b-3p positive presented a significantly lower SMAD4 expression pattern (Fig. 7e and Additional file 3: Table S3).
Fig. 7.
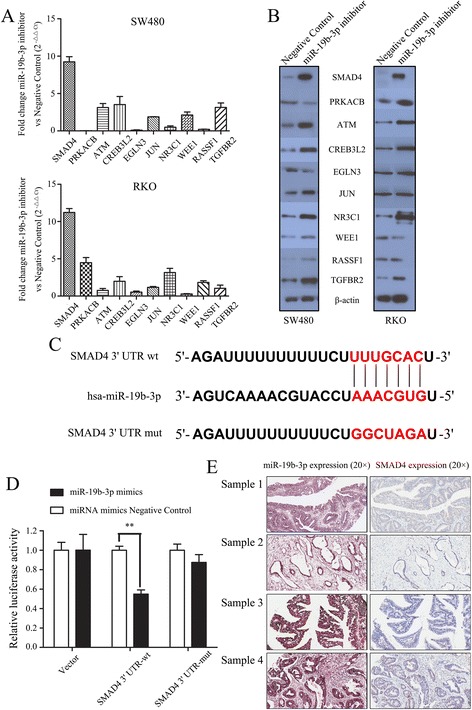
miR-19b-3p downregulates SMAD4 expression by directly targeting its 3′ UTR. a and b Endogenous candidate target genes expression were significantly upregulated in SW480 (a, qRT-PCR and b, western blotting) and RKO (a, qRT-PCR and b, western blotting) cells transfected with miR-19b-3p inhibitor compared with negative control ones. c The 3′ UTR of SMAD4 mRNA contains a complementary binding site for the seed region of miR-19b-3p. SMAD4 mut is a mutant with substitutions in the complementary region as a negative control. d The luciferase activity was detected after transfection of pmirGLO luciferase reporter vector (pmirGLO-Vector, pmirGLO-SMAD4 3’UTR-wt or pmirGLO-SMAD4 3’UTR-mut) into the miR-19b-3p mimics or miRNA mimics negative control transfected SW480 cells. **, P < 0.01. e Representative images from in situ hybridization and immunohistochemistry both in the same location of tissue microarray, showing that miR-19b-3p expression was significantly negatively correlated with SMAD4 expression. See also Additional file 3: Table S3
miR-19b-3p mediates proliferation and resistance to oxaliplatin-based chemotherapy via SMAD4
A significant increase of SMAD4 protein levels was detected upon miR-19b-3p inhibition in SW480 and RKO cell lines (Fig. 8a). In comparison with negative control cells, miR-19b-3p inhibitor-transfected SW480 and RKO cells had an impaired proliferation ability. Importantly, this phenotype could be reversed by transfection of SMAD4 shRNA (P < 0.05, Fig. 8b and c). In addition, miR-19b-3p downregulation promoted apoptosis when cells were treated with oxaliplatin compared with negative control cells. Cell apoptosis could also be reversed by transfection of SMAD4 shRNA (P < 0.05, Fig. 8d–f). Together, these data suggest that miR-19b-3p-mediated proliferation and resistance to oxaliplatin-based chemotherapy are dependent on reduced SMAD4 expression levels.
Fig. 8.
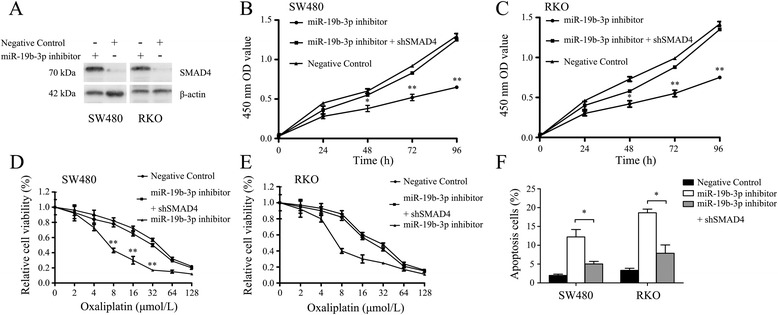
miR-19b-3p mediates proliferation and resistance to oxaliplatin-based chemotherapy via SMAD4. a Western blotting assays showed the inverse relationship between miR-19b-3p and SMAD4 in both SW480 and RKO cell lines. β-actin was acted as internal control. b and c CCk8 assays demonstrated that miR-19b-3p inhibitor-transfected SW480 and RKO cells had an impaired proliferation ability which can be reversed by transfection of SMAD4 shRNA. *, P < 0.05, **, P < 0.01. d, e and f miR-19b-3p inhibitor-transfected SW480 and RKO cells were significantly more sensitive to oxaliplatin in comparison with negative control cells,which can be reversed by transfection of SMAD4 shRNA. The ratio of apoptotic cells in every group was shown *, P < 0.05, **, P < 0.01
Discussion
Colon cancer is initiated by aberrant processing of genetic information due to genetic changes involving tumor suppressor genes and oncogenes, or altered epigenetic mechanisms manifested in global or local changes of chromatin structure [15]. Despite the continuous development of novel tumor targeting therapeutics, chemoresistance remains a challenge for the treatment of colon cancer [16]. At present, researchers have made great progress in the area of chemoresistance-associated miRNAs in colon cancer. However, the identification of novel miRNAs is still pivotal in colon cancer therapy [17, 18].
Our research used the IPA database to identify miRNAs specifically expressed in colon cancer. The 7 upregulated miRNAs were selected for further study, including miR-19b-3p, miR-155-5p, miR-17-5p, miR-183-5p, miR-25-3p, miR-21-5p, and miR-196a-5p. Several of them are associated with various human cancers. For example, miR-155-5p is deregulated in colorectal cancer, osteosarcoma, and triple-negative breast cancer [19–21]. Functional annotations of the predicted target genes of these miRNAs were obtained from R annotation packages, including GO biological process. Genes in enriched functional terms were used to construct a miRNA-target network. In this network diagram, many genes were closely related with colon cancer progression such as PTEN, STAT3, FOXO1, and SMAD4 [22–25]. The biological function analysis of the predicted target genes based on IPA and GO annotation enabled us to comprehensively understand their functional roles in colon cancer progression. The predicted target genes were significantly involved in many biologic processes relevant to cancer such as apoptosis, cell proliferation, and cell cycle arrest. These bioinformatics analyses suggested that the predicted target genes may control broad biological functions associated with colon cancer. miR-19b-3p was identified as the pivotal oncogenic component of the miR-17-92 cluster of miRNAs because of its role in promoting sustained cell survival and recognized as an onco-miR in some types of cells and tissues [26, 27]. Upregulated miR-19b-3p plays a critical role in the tumorigenesis of Em-Myc-driven B-cell lymphomas [28]. We found that high expression of miR-19b-3p in colon cancer tissues was significantly associated not only with tumorhistologic grade, but also with AJCC stage. Furthermore, Kaplan-Meier and univariate Cox proportional hazard regression analyses indicated that patients with high miR-19b-3p expression displayed a lower five-year OS and DFS. Multivariate analyses indicated that miR-19b-3p expression in colon cancer was an independent prognostic factor of survival. We also observed that miR-19b-3p downregulation had no effect on invasion, but correlated with reduced cell proliferation and decreased oxaliplatin-based chemoresistance. The proliferation ability is associated with tumor size and the invasion ability is correlated with distant metastasis. In other words, miR-19b-3p does not act as an enhancer of cancer metastasis, but of tumorigenesis in vitro. We speculate that this may be because of the complex tumor microenvironment in vivo. Recent molecular and cellular biology studies indicated that tumor growth and metastasis are not only determined by cancer cells, but are also affected by various stromal cells [29]. During metastasis, tumor cells need to go through a series of complex steps, during which they continually adapt to different microenvironments [30]. Thus, a stable in vivo model of colon cancer metastasis will be necessary to further understand the underlying mechanisms in our future study.
In this study, we demonstrated that SMAD4 is the direct target of miR-19b-3p in colon cancer. As an important mediator of the TGF-β signaling pathway, SMAD4 plays an important role in suppressing tumor progression and promoting apoptosis of tumor cells [23]. Reduced expression of SMAD4 is associated with attenuated sensitivity to chemotherapy and poor prognosis of patients with colon cancer in several clinical studies [23, 31]. The inverse relationship between miR-19b-3p and SMAD4 in colon cancer cell lines and tumor samples was investigated in our study. Importantly, since SMAD4 is targeted by this miRNA, we showed that the proliferation ability and resistance to oxaliplatin-based chemotherapy is reversed by the transfection of SMAD4 shRNA.
Conclusions
In summary, our study shows that miR-19b-3p overexpression plays an important role in promoting colon cancer progression through enhanced proliferation and reduced chemosensitivity. miR-19b-3p may be a clinical useful prognostic molecular biomarker for prognosis and a target for chemotherapy in colon cancer.
Additional files
Oligonucleotide Sequence for the primers used in the study. (DOCX 25 kb)
miRNAs specifically expressed in colon cancer after being screened by IPA. (DOCX 26 kb)
The association between miR-19b-3p and SMAD4 expression. (DOCX 19 kb)
Acknowledgments
Funding
This project was supported by the National Natural Science Foundation of China (No. 81272744, 81302111, 81560394 and 81460369) and the Scientific Research Project grant funded by Ningxia high school (NGY2015096).
Abbreviations
- AJCC
American Joint Committee on Cancer
- CCK8
Cell counting kit-8
- DFS
Disease-free survival
- GO
Gene Ontology
- IPA
Ingenuity Pathways Analysis
- OS
overall survival
- TMA
Tissue microarray
Authors’ contributions
Conception and design: TJ, LY, ZH, JF. Development of methodology: TJ, LY, ZH, YL. Acquisition of data: TJ, ZH, YL, YY. Analysis and interpretation of data: TJ, ZH, YL, YY. Writing, review, and/or revision of the manuscript: TJ, LY, ZP, JF. All authors read and approved the final manuscript.
Ethics approval
The study was approved by the ethics committee of the General Hospital of Ningxia Medical University. All animal procedures were performed according to national guidelines and approved by the Institutional Committee of Shanghai Jiao Tong University School of Medicine for Animal Research.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13046-017-0602-5) contains supplementary material, which is available to authorized users.
Contributor Information
Yinxue Yang, Email: 18895007660@163.com.
Zhihai Peng, Email: pengzhihai1958@163.com.
Junwei Fan, Email: drjunweifan@163.com.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Manfredi S, Bouvier AM, Lepage C, Hatem C, Dancourt V, Faivre J. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg. 2006;93(9):1115–1122. doi: 10.1002/bjs.5349. [DOI] [PubMed] [Google Scholar]
- 3.Tuschl GMT. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Garofalo M. Croce CM: microRNAs: master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 5.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiekhattar RIGaR MicroRNA Biogenesis and Cancer. Cancer Res. 2005;65(9):3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Chen Y, Zhao J, Kong F, Zhang Y. miR-203 reverses chemoresistance in p53-mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett. 2011;304(1):52–59. doi: 10.1016/j.canlet.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Shang J, Yang F, Wang Y, Wang Y, Xue G, Mei Q, Wang F, Sun S. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J Cell Biochem. 2014;115(4):772–784. doi: 10.1002/jcb.24721. [DOI] [PubMed] [Google Scholar]
- 10.Gusev Y, Schmittgen TD, Lerner M, Postier R, Brackett D. Computational analysis of biological functions and pathways collectively targeted by co-expressed microRNAs in cancer. BMC bioinformatics. 2007;8(Suppl 7):S16. doi: 10.1186/1471-2105-8-S7-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Zhao YC, Lu YD, Ma CP. Integrated bioinformatics analyses identify dysregulatedmiRNAs in lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:2270–2274. [PubMed] [Google Scholar]
- 12.YU F, Shen XY, Fan L. Genome-wide analysis of genetic variations assisted by ingenuity pathway analysis to comprehensively investigate potential genetic targets associated with the progression of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2014;18:2102–2108. [PubMed] [Google Scholar]
- 13.Millan A, Huerta S. Apoptosis-inducing factor and colon cancer. J Surg Res. 2009;151(1):163–170. doi: 10.1016/j.jss.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura Y, Miyoshi N. Cell death induction by isothiocyanates and their underlying molecular mechanisms. Biofactors. 2006;2006(26):123–134. doi: 10.1002/biof.5520260203. [DOI] [PubMed] [Google Scholar]
- 15.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 16.Grassilli E, Narloch R, Federzoni E, Ianzano L, Pisano F, Giovannoni R, Romano G, Masiero L, Leone BE, Bonin S, et al. Inhibition of GSK3B bypass drug resistance of p53-null colon carcinomas by enabling necroptosis in response to chemotherapy. Clin Cancer Res. 2013;19(14):3820–3831. doi: 10.1158/1078-0432.CCR-12-3289. [DOI] [PubMed] [Google Scholar]
- 17.Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W, Jiang X, Zhang C, Qu J. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J Cell Physiol. 2015;230(3):535–545. doi: 10.1002/jcp.24366. [DOI] [PubMed] [Google Scholar]
- 18.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28(46):4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu YL, Wang HF, Sun ZQ, Tang Y, Han XN, Yu XB, Liu K. Up-regulated miR-155-5p promotes cell proliferation, invasion and metastasis in colorectal carcinoma. Int J Clin Exp Pathol. 2015;8(6):6988–6994. [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya S, Chalk AM, Ng AJ, Martin TJ, Zannettino AC, Purton LE, Lu J, Baker EK, Walkley CR. Increased miR-155-5p and reduced miR-148a-3p contribute to the suppression of osteosarcoma cell death. Oncogene. 2016;35(40):5282–5294. doi: 10.1038/onc.2016.68. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Li C, Zhu Z, Teng Y, Che X, Wang Y, Ma Y, Wang Y, Zheng H, Liu Y, et al. miR-155-5p antagonizes the apoptotic effect of bufalin in triple-negative breast cancer cells. Anti-Cancer Drugs. 2016;27(1):9–16. doi: 10.1097/CAD.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 22.Ren W, Shen S, Sun Z, Shu P, Shen X, Bu C, Ai F, Zhang X, Tang A, Tian L, et al. Jak-STAT3 pathway triggers DICER1 for proteasomal degradation by ubiquitin ligase complex of CUL4A(DCAF1) to promote colon cancer development. Cancer Lett. 2016;375(2):209–220. doi: 10.1016/j.canlet.2016.02.055. [DOI] [PubMed] [Google Scholar]
- 23.Yan P, Klingbiel D, Saridaki Z, Ceppa P, Curto M, McKee TA, Roth A, Tejpar S, Delorenzi M, Bosman FT, et al. Reduced expression of SMAD4 is associated with poor survival in colon cancer. Clin Cancer Res. 2016;22(12):3037–3047. doi: 10.1158/1078-0432.CCR-15-0939. [DOI] [PubMed] [Google Scholar]
- 24.Yu M, Trobridge P, Wang Y, Kanngurn S, Morris SM, Knoblaugh S, Grady WM. Inactivation of TGF-beta signaling and loss of PTEN cooperate to induce colon cancer in vivo. Oncogene. 2014;33(12):1538–1547. doi: 10.1038/onc.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coomans de Brachene A, Demoulin JB. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 2016;73(6):1159–1172. doi: 10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huhn D, Kousholt AN, Sorensen CS. Sartori AA: miR-19, a component of the oncogenic miR-17 approximately 92 cluster, targets the DNA-end resection factor CtIP. Oncogene. 2015;34(30):3977–3984. doi: 10.1038/onc.2014.329. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma Y, Chen Y, Pan F, Wang K, Ni J, et al. Circulating MiR-16-5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics. 2015;5(7):733–745. doi: 10.7150/thno.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuge R, Kea Y. mTOR and PDGF pathway blockade inhibits liver metastasis of colorectal cancer by modulating the tumor microenvironment. Am J Pathol. 2015;185(2):399–408. doi: 10.1016/j.ajpath.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Davis C, Shah S, Hughes D, Ryan JC, Altomare D, Pena MM. IL-33 promotes growth and liver metastasis of colorectal cancer in mice by remodeling the tumor microenvironment and inducing angiogenesis. Mol Carcinog. 2017;56(1):272–287. doi: 10.1002/mc.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baraniskin A, Munding J, Schulmann K, Meier D, Porschen R, Arkenau HT, Graeven U, Schmiegel W, Tannapfel A, Reinacher-Schick A. Prognostic value of reduced SMAD4 expression in patients with metastatic colorectal cancer under oxaliplatin-containing chemotherapy: a translational study of the AIO colorectal study group. Clin Colorectal Cancer. 2011;10(1):24–29. doi: 10.3816/CCC.2011.n.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide Sequence for the primers used in the study. (DOCX 25 kb)
miRNAs specifically expressed in colon cancer after being screened by IPA. (DOCX 26 kb)
The association between miR-19b-3p and SMAD4 expression. (DOCX 19 kb)


