ABSTRACT
There are limited therapeutic options to treat infections caused by vancomycin-resistant Enterococcus faecium (VREfm). The lipoglycopeptide oritavancin exhibits in vitro activity against this pathogen, although its utility against infections caused by VREfm has not been clinically established. In this study, the pharmacodynamic activity of free-drug levels associated with 12 mg/kg/day of daptomycin and a single 1,200-mg dose of oritavancin were determined against three VanA VREfm isolates in an in vitro pharmacokinetic/pharmacodynamic model.
KEYWORDS: daptomycin, Enterococcus faecium, oritavancin, lipoglycopeptide, vancomycin resistance
TEXT
Vancomycin-resistant enterococci (VRE) are an important cause of nosocomial infections in the United States (1, 2). The oxazolidinone linezolid remains the only agent indicated for infections caused by vancomycin-resistant Enterococcus faecium (VREfm). Although the lipopeptide daptomycin is not indicated for the treatment of VRE infections, high-dose regimens (≥8 mg/kg/day, exceeding the approved dose of 6 mg/kg/day for Staphylococcus aureus bloodstream infections) are often considered for first-line therapy (3, 4). Higher doses of daptomycin may improve outcomes by maximizing exposure to compensate for the elevated daptomycin MICs of VRE relative to S. aureus and by limiting the emergence of daptomycin nonsusceptibility (5, 6).
The long-acting lipoglycopeptide oritavancin is approved as a single 1,200-mg dose treatment of acute bacterial skin and skin structure infections caused by Gram-positive pathogens (7). Oritavancin exerts in vitro activity against VRE isolates expressing both the VanA and VanB phenotypes (8–10). It has shown efficacy in a rat model of vancomycin-resistant E. faecium bloodstream infection (11) and enhanced activity in combination with gentamicin in a rabbit model of enterococcal endocarditis (12, 13). In this study, we describe the pharmacodynamic (PD) activity of free-drug levels associated with 12 mg/kg/day of daptomycin and a single 1,200-mg dose of oritavancin against clinical isolates of VREfm in an in vitro pharmacokinetic (PK)/PD model.
(Part of this work was presented at IDWeek 2016, New Orleans, LA, 26 to 30 October 2016 [14].)
Oritavancin (The Medicines Company, Parsippany, NJ) and daptomycin (APIChem Technology Company, Hangzhou, China) broth microdilution MICs of the three VanA VREfm clinical isolates ATCC 51559, B7181440, and B7231527 were determined following CLSI M07-A10 guidelines (6) using the quality control isolate Enterococcus faecalis ATCC 29212 to assess appropriate drug and assay performance (7). MICs of the derived mutants that survived daptomycin challenge were determined before and after serial passage on nonselective medium (Mueller-Hinton agar) for 5 days to assess the stability of the susceptibility changes. Subcultures of the VREfm isolates in exponential phase were inoculated at 106 CFU/ml into a dilutional one-compartment in vitro PK/PD model (15) containing 250 ml of cation-adjusted Mueller-Hinton broth (CAMHB) supplemented with either 50 μg/ml CaCl2 (for daptomycin) or 0.01% polysorbate 80 (for oritavancin). Daptomycin was added as bolus daily doses and a pump flow rate (0.34 ml/min for 72 h) was used to simulate free-drug exposures expected from 12 mg/kg/day in healthy volunteers (assuming protein binding of 91.5%, a free peak concentration [ƒCmax] of 15.6 μg/ml, a half-life [t1/2] of 8 h, and an area under the concentration-time curve from 0 to 24 h [ƒAUC0–24 h] of 171 μg · h/ml) (16). For oritavancin, a single dose was infused over 3 h and flow rates (1.25 ml/min for 5 h, 0.94 ml/min for 1 h, 0.31 ml/min for 23 h, and 0.04 ml/min for 43 h) were used to approximate the mean free-drug concentration-time profile (assuming protein binding of 85% [17]; an ƒCmax of 20.7 μg/ml; alpha, beta and gamma t1/2 of 2.3 h, 13.4 h, and 245 h, respectively, and an ƒAUC0–24 of 178 μg · h/ml) observed in patients receiving a 1,200-mg dose (15, 18). After 5 h of drug exposure, cultures were transferred to new sterilized in vitro PK/PD model systems to ensure that only drug-exposed bacteria were present. For control cultures (no drug exposure), fresh media were supplied using the flow rates indicated for oritavancin over 24 h (until turbid cultures were apparent). Aliquots were sampled at the indicated times for bacterial viability as previously described (15) and then frozen at −20°C until drug concentrations were determined. Statistical differences (P < 0.05) in mean changes in bacterial viability (log CFU/ml) relative to inoculum were compared by t test. Daptomycin concentrations were quantified using a described bioassay (15). Oritavancin was quantified by fluorescence polarization using a fluorescein-labeled d-Ala-d-Ala peptide (Ac-l-Lys-Ala-d-Ala-OH; Pharmaron, Irvine, CA) (19). Oritavancin standards (0.06 to 32 μg/ml) were prepared in CAMHB containing 0.01% P80 (assay linear range of sensitivity 0.25 μg/ml to 16 μg/ml); samples from the in vitro PK/PD model were diluted 1 in 4 in CAMHB. Assays (100 μl) were performed in 96-well plates (reference number 3694; Corning Inc., Corning, NY) using a final concentration of 90 nM of the fluorescein-labeled peptide and excitation and emission wavelengths of 485 nm and 535 nm, respectively. PCR amplification of the liasFSR locus and cls gene from genomic DNA (GenElute Bacterial Genomic DNA kit; Sigma-Aldrich, Oakville, Ontario, Canada) was performed using published primer sequences (20). For amplification of liaS in ATCC 51559, the primers 5′-AAAGGGATAGGCAGAACACG-3′ (forward) and 5′-CAATACCAGCTACTCGTTCTTTGA-3′ (reverse) were used due to allelic sequence variation. Sanger sequencing of the amplicons was performed at McGill University and the Génome Québec Innovation Centre (Montreal, Québec, Canada).
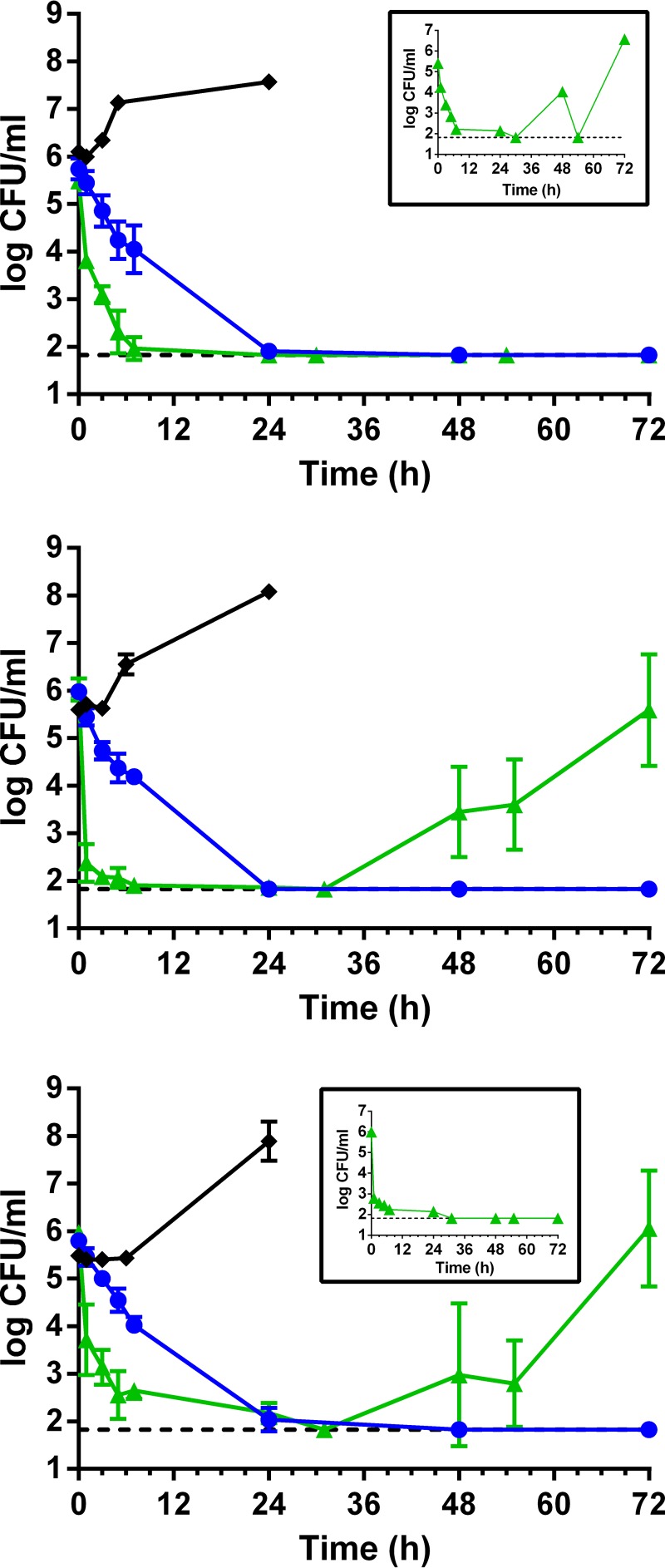
The three VREfm isolates were susceptible to daptomycin (MIC ≤ 4 μg/ml), exhibiting daptomycin MICs of 2 to 4 μg/ml (Table 1); oritavancin MICs ranged from 0.06 to 0.5 μg/ml. Exposure of the VREfm isolates to daptomycin at free-drug concentrations expected from dosing with 12 mg/kg/day (obtained PK parameters shown in Table 2) resulted in rapid bactericidal activity (≥3 log kill relative to the starting inoculum) within 5 h and sustained suppression of regrowth for 24 h (Fig. 1 and Table 3). However, each isolate exhibited instances of bacterial regrowth by 48 h following exposure to the second dose of daptomycin (in 1 of 4 replicates of ATCC 51559 [Fig. 1A, inset], 4 of 4 replicates of B7181440 [Fig. 1B], and 3 of 4 replicates of B7231527 [Fig. 1C]). Regrowth among the three VREfm isolates was coincident with stable increases in daptomycin MICs ranging from 4- to 8-fold (daptomycin MICs of 16 μg/ml) relative to their corresponding parental isolates (Table 1). In one derived mutant (1440-141-1), the oritavancin MIC also increased 4-fold above its parental MIC (Table 1) and therefore it will be of interest to elucidate the genetic changes that provoke cross-reduced susceptibility to oritavancin in VREfm. Other studies have also shown the development of reduced susceptibility to daptomycin in VREfm (including ATCC 51559) when modeling free-drug exposures associated with ≤10 mg/kg/day (16, 21). In contrast, modeling of total drug exposures (in the presence of 3.5 g/dl of human albumin) associated with ≥10 mg/kg/day prevented the development of reduced susceptibility (5, 6) and established a total AUC0–24/MIC (area under the concentration-time curve from 0 to 24 h divided by the MIC) value of 781 as a cutoff to prevent the emergence of reduced susceptibility for the tested VREfm isolate. The projected total AUC0–24/MIC ratio for VREfm ATCC 51559 and B7181440 is 953 (ƒAUC0–24 of 162, 91.5% protein binding, and daptomycin MICs of 2 μg/ml), an exposure that exceeded the cutoff value but did not prevent emergence of reduced susceptibility. In immunocompetent hosts, it is plausible that the small population of daptomycin-nonsusceptible bacteria that emerge (as shown in Fig. 1) could be eliminated by the immune system. Nevertheless, case reports of emergence of reduced susceptibility in patients receiving 10 mg/kg/day have been published (22, 23) and hence the appropriateness of the current daptomycin susceptibility breakpoint and high-dose daptomycin regimens for enterococci are under scrutiny (24).
TABLE 1.
Characterization of VREfm parental isolates and derived mutants that survived daptomycin challenge in the in vitro PK/PD model
| Parental isolate or derived mutant | MIC (μg/ml) ofa: |
Mutationb | |
|---|---|---|---|
| Daptomycin | Oritavancin | ||
| ATCC 51559 | 2 | 0.25 | NAc |
| 51559-146-4d | 16 | 0.25 | Unknown |
| B7181440 | 2 | 0.06 | NA |
| 1440-140-1d | 16 | 0.12 | Unknown |
| 1440-140-2d | 16 | 0.12 | LiaF truncation |
| 1440-141-1d | 16 | 0.25 | Unknown |
| 1440-141-2d | 16 | 0.12 | LiaF L181S |
| B7231527 | 4 | 0.5 | NA |
| 1527-140-4d | 16 | 1 | LiaS V129E |
| 1527-141-3d | 16 | 0.5 | Unknown |
| 1527-141-4d | 16 | 0.25 | Unknown |
Modal MICs are presented from ≥3 independent determinations.
Mutations in the liaFSR system and cardiolipin synthase gene cls were determined by DNA sequencing.
NA, not applicable.
Daptomycin-nonsusceptible derived mutant. One of four replicates of ATCC 51559, four of four replicates of B7181440, and three of four replicates of B7231527 showed regrowth of derived mutants with reduced susceptibility to daptomycin following daptomycin exposure in the in vitro PK/PD model. MICs of the derived mutants were unchanged following 5 days of passage on nonselective medium.
TABLE 2.
Pharmacokinetic parameters obtained for the indicated dosing regimens in the in vitro PK/PD model
| Parametera | Daptomycin, 12 mg/kg/day |
Oritavancin, 1,200-mg single dose |
||
|---|---|---|---|---|
| Targetedb | Obtained ± SD | Targetedc | Obtained ± SD | |
| ƒCmax (μg/ml) | 15.6 | 15.1 ± 0.2 | 20.7 | 20.1 ± 3.1 |
| ƒAUC0–24 (μg · h/ml) | 171 | 162 ± 7.7 | 178 | 164 ± 30.5 |
| ƒAUC0–72 (μg · h/ml) | ND | ND | 246 | 223 ± 46.2 |
| t1/2 (h) | 8 | 8.1 ± 0.5 | ND | ND |
ƒCmax, free peak concentration; ƒAUC0–24, area under the concentration-time curve from 0 to 24 h; ƒAUC0–72, area under the concentration-time curve from 0 to 72 h; t1/2, half-life; SD, standard deviation; ND, not determined.
The targeted PK values for daptomycin were derived from Benvenuto et al. (28) and the prescribing information (29) with the assumption of 91.5% protein binding. The daptomycin t1/2 in the in vitro PK/PD model was determined by nonlinear regression analysis using GraphPad Prism 6 software. The targeted ƒAUC (area under the concentration-time curve for the free, unbound fraction of a drug) values were calculated using a simulated daptomycin concentration-time profile (Prism 6) that respects the targeted PK parameters (ƒCmax of 15.6 μg/ml and t1/2 = 8 h).
FIG 1.
Pharmacodynamic activity of daptomycin and oritavancin at free-drug exposures associated with 12 mg/kg/day daptomycin (green triangles) and a single 1,200-mg dose of oritavancin (blue circles) against the clinical isolates of VanA VREfm ATCC 51559 (A), B7181440 (B), and B7231527 (C) in an in vitro PK/PD model over 72 h. Mean values ± standard deviation (SD) are from two independent experiments done in duplicate. Control cultures are shown for each isolate (black diamonds), using the flow rates for oritavancin to supply fresh drug-free medium over 24 h. The inset in panel A depicts the single occurrence of regrowth of ATCC 51559 following exposure to daptomycin. The inset in panel C depicts the single occurrence of eradication of B7231527 following exposure to daptomycin. The dashed line indicates the limit of detection (<66.7 CFU/ml).
TABLE 3.
Mean changes in bacterial viability relative to inoculum of the tested VREfm isolates exposed to daptomycin and oritavancin in the in vitro PK/PD model
| Time (h) | Mean decrease in bacterial viability relative to starting inoculum (log CFU/ml ± SD) fora: |
|||||
|---|---|---|---|---|---|---|
| VREfm ATCC 51559 |
VREfm B7181440 |
VREfm B7231527 |
||||
| Daptomycinb | Oritavancin | Daptomycin | Oritavancin | Daptomycin | Oritavancin | |
| 1 | −1.7 ± 0.2 | −0.3 ± 0.2c | −3.6 ± 0.4 | −0.5 ± 0.3c | −2.3 ± 0.8 | −0.3 ± 0.1c |
| 3 | −2.4 ± 0.1 | −0.9 ± 0.2c | −4.0 ± 0.2 | −1.2 ± 0.3c | −2.8 ± 0.4 | −0.8 ± 0.1c |
| 5 | −3.2 ± 0.4 | −1.5 ± 0.2c | −4.0 ± 0.1 | −1.6 ± 0.4c | −3.4 ± 0.5 | −1.2 ± 0.2c |
| 7 | −3.5 ± 0.1 | −1.7 ± 0.5c | −4.2 ± 0.3 | −1.7 ± 0.1c | −3.3 ± 0.1 | −1.7 ± 0.2c |
| 24 | −3.7 ± 0.1 | −3.8 ± 0.2 | −4.2 ± 0.1 | −4.2 ± 0.2 | −3.8 ± 0.1 | −3.8 ± 0.2 |
| 48 | −3.7 ± 0.1 | −3.9 ± 0.2 | −2.8 ± 1.2 | −4.2 ± 0.2 | −3.0 ± 1.5 | −4.0 ± 0.1 |
| 72 | −3.7 ± 0.1 | −3.9 ± 0.2 | −0.7 ± 1.5 | −4.2 ± 0.2c | 0.1 ± 1.2 | −4.0 ± 0.1c |
Mean ± SD values shown are from two independent experiments done in duplicate (n = 4).
The calculations of mean decrease in bacterial viability for the daptomycin exposures excluded both that of the single replicate of ATCC 51559 in which reduced susceptibility to daptomycin was observed and that of the single replicate of B7231527 in which no regrowth occurred.
Decrease in log CFU/ml is significantly different (P < 0.05, t test) from the corresponding value obtained for daptomycin at the same exposure time.
Infusion of oritavancin over 3 h into the in vitro PK/PD model (obtained PK parameters shown in Table 2) resulted in bactericidal activity against the three VREfm isolates that was significantly less rapid than that of daptomycin over the first 7 h of exposure (P < 0.05) (Table 3 and Fig. 1), as bacterial counts were reduced by approximately 1.7 log. Whereas bacterial killing by oritavancin was not significantly different to that of daptomycin at 24 h (P > 0.05) (Table 3), oritavancin reduced counts of all three VREfm isolates to below the limit of detection (≤66.7 CFU/ml) between 48 and 72 h (Fig. 1) with suppression of regrowth of B7181440 and B7231527 that differed significantly from the regrowth of those isolates at 72 h following daptomycin exposure (P < 0.05) (Table 3). A limitation of this study is that the duration of oritavancin exposure was limited to 72 h and therefore it is unknown whether longer exposures that more completely represent the terminal half-life of oritavancin (245 h) would confirm the suppression of regrowth.
Mutations in the cardiolipin synthase gene cls and the three-component regulatory system operon liaFSR have been implicated in clinical development of reduced susceptibility to daptomycin in VREfm isolates (20, 25). No mutations in cls were observed in the derived daptomycin-nonsusceptible mutants. A total of 171 singlenucleotide polymorphisms (SNPs) were observed within the liaFSR operon of ATCC 51559 relative to its counterpart shared by B7181440 and B7231527 (data not shown); the SNPs accounted for 7, 12, and 3 amino acid differences in LiaF, LiaS, and LiaR, respectively. Queries of the GenBank database revealed that both allelic variations are conserved in different E. faecium isolates (data not shown). Three of the eight derived mutants had incurred mutations within the liaFSR locus (Table 1). In mutant 1440-140-2, a deletion of the thymine residue at position 24 of liaF caused a frameshift mutation, presumably truncating the resultant protein. In mutant 1440-141-2, a nonsynonymous point mutation resulted in a change of leucine to serine at position 181 of LiaF. Impairment of LiaF function in an E. faecalis isolate was shown to cause a 3-fold increase in its daptomycin MIC and abolished the bactericidal activity of the drug (26). In mutant 1527-140-4, a nonsynonymous point mutation resulted in a change of valine to glutamic acid at position 129 of LiaS. For the other five derived mutants, no changes in liaFSR were observed and consequently analysis of other genes that have been implicated in reduced susceptibility to daptomycin (27) is warranted.
In conclusion, oritavancin demonstrated sustained bactericidal activity in vitro against VREfm isolates at free-drug exposures expected to occur in patients receiving a single 1,200 mg-dose. These results support further investigation of its safety and efficacy in clinical VREfm infections.
ACKNOWLEDGMENTS
This study was funded by The Medicines Company (Parsippany, NJ).
A.B., F.A., and G.M. are employees of The Medicines Company. D.L.-S. is a former employee of The Medicines Company.
We thank the staff at Mount Sinai Hospital, University of Toronto, for providing VREfm isolates B7181440 and B7231527.
REFERENCES
- 1.Munita JM, Murray BE, Arias CA. 2014. Daptomycin for the treatment of bacteraemia due to vancomycin-resistant enterococci. Int J Antimicrob Agents 44:387–395. doi: 10.1016/j.ijantimicag.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 37:1288–1301. doi: 10.1017/ice.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKinnell JA, Arias CA. 2015. Editorial commentary: linezolid vs daptomycin for vancomycin-resistant enterococci: the evidence gap between trials and clinical experience. Clin Infect Dis 61:879–882. doi: 10.1093/cid/civ449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munita JM, Arias CA, Murray BE. 2012. Enterococcal endocarditis: can we win the war? Curr Infect Dis Rep 14:339–349. doi: 10.1007/s11908-012-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall AD, Steed ME, Arias CA, Murray BE, Rybak MJ. 2012. Evaluation of standard- and high-dose daptomycin versus linezolid against vancomycin-resistant Enterococcus isolates in an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations. Antimicrob Agents Chemother 56:3174–3180. doi: 10.1128/AAC.06439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werth BJ, Steed ME, Ireland CE, Tran TT, Nonejuie P, Murray BE, Rose WE, Sakoulas G, Pogliano J, Arias CA, Rybak MJ. 2014. Defining daptomycin resistance prevention exposures in vancomycin-resistant Enterococcus faecium and E. faecalis. Antimicrob Agents Chemother 58:5253–5261. doi: 10.1128/AAC.00098-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Medicines Company. 2016. ORBACTIV (oritavancin) for injection, for intravenous use. Prescribing information. The Medicines Company, Ville St-Laurent, Quebec, Canada. [Google Scholar]
- 8.McKay GA, Beaulieu S, Arhin FF, Belley A, Sarmiento I, Parr T Jr, Moeck G. 2009. Time-kill kinetics of oritavancin and comparator agents against Staphylococcus aureus, Enterococcus faecalis and Enterococcus faecium. J Antimicrob Chemother 63:1191–1199. doi: 10.1093/jac/dkp126. [DOI] [PubMed] [Google Scholar]
- 9.Zhanel GG, Calic D, Schweizer F, Zelenitsky S, Adam H, Lagace-Wiens PR, Rubinstein E, Gin AS, Hoban DJ, Karlowsky JA. 2010. New lipoglycopeptides: a comparative review of dalbavancin, oritavancin and telavancin. Drugs 70:859–886. doi: 10.2165/11534440-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 10.Sweeney D, Stoneburner A, Shinabarger DL, Arhin FF, Belley A, Moeck G, Pillar CM. 2017. Comparative in vitro activity of oritavancin and other agents against vancomycin-susceptible and -resistant enterococci. J Antimicrob Chemother 72:622–624. doi: 10.1093/jac/dkw451. [DOI] [PubMed] [Google Scholar]
- 11.Rupp ME, Fey PD, Longo GM. 2001. Effect of LY333328 against vancomycin-resistant Enterococcus faecium in a rat central venous catheter-associated infection model. J Antimicrob Chemother 47:705–707. doi: 10.1093/jac/47.5.705. [DOI] [PubMed] [Google Scholar]
- 12.Lefort A, Saleh-Mghir A, Garry L, Carbon C, Fantin B. 2000. Activity of LY333328 combined with gentamicin in vitro and in rabbit experimental endocarditis due to vancomycin-susceptible or -resistant Enterococcus faecalis. Antimicrob Agents Chemother 44:3017–3021. doi: 10.1128/AAC.44.11.3017-3021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleh-Mghir A, Lefort A, Petegnief Y, Dautrey S, Vallois JM, Le Guludec D, Carbon C, Fantin B. 1999. Activity and diffusion of LY333328 in experimental endocarditis due to vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 43:115–120. doi: 10.1093/jac/43.suppl_1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belley A, Lalonde-Seguin D, Arhin FF, Moeck G. 2016. Antibacterial activity of oritavancin and daptomycin against clinical isolates of vancomycin-resistant Enterococcus faecium in in vitro pharmacokinetic/pharmacodynamic models, abstract 1990 IDWeek 2016, New Orleans, LA, 26-30 October 2016. [Google Scholar]
- 15.Belley A, Arhin FF, Sarmiento I, Deng H, Rose W, Moeck G. 2013. Pharmacodynamics of a simulated single 1,200-milligram dose of oritavancin in an in vitro pharmacokinetic/pharmacodynamic model of methicillin-resistant staphylococcus aureus infection. Antimicrob Agents Chemother 57:205–211. doi: 10.1128/AAC.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steed ME, Vidaillac C, Rose WE, Winterfield P, Kaatz GW, Rybak MJ. 2011. Characterizing vancomycin-resistant Enterococcus strains with various mechanisms of daptomycin resistance developed in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 55:4748–4754. doi: 10.1128/AAC.00084-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arhin FF, Belley A, McKay G, Beaulieu S, Sarmiento I, Parr TR Jr, Moeck G. 2010. Assessment of oritavancin serum protein binding across species. Antimicrob Agents Chemother 54:3481–3483. doi: 10.1128/AAC.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubino CM, Bhavnani SM, Moeck G, Bellibas SE, Ambrose PG. 2015. Population pharmacokinetic analysis for a single 1,200-milligram dose of oritavancin using data from two pivotal phase 3 clinical trials. Antimicrob Agents Chemother 59:3365–3372. doi: 10.1128/AAC.00176-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu L, Zhong M, Wei Y. 2010. Direct fluorescence polarization assay for the detection of glycopeptide antibiotics. Anal Chem 82:7044–7048. doi: 10.1021/ac100543e. [DOI] [PubMed] [Google Scholar]
- 20.Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob Agents Chemother 56:4354–4359. doi: 10.1128/AAC.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith JR, Barber KE, Raut A, Rybak MJ. 2015. β-Lactams enhance daptomycin activity against vancomycin-resistant Enterococcus faecalis and Enterococcus faecium in in vitro pharmacokinetic/pharmacodynamic models. Antimicrob Agents Chemother 59:2842–2848. doi: 10.1128/AAC.00053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casapao AM, Kullar R, Davis SL, Levine DP, Zhao JJ, Potoski BA, Goff DA, Crank CW, Segreti J, Sakoulas G, Cosgrove SE, Rybak MJ. 2013. Multicenter study of high-dose daptomycin for treatment of enterococcal infections. Antimicrob Agents Chemother 57:4190–4196. doi: 10.1128/AAC.00526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matono T, Hayakawa K, Hirai R, Tanimura A, Yamamoto K, Fujiya Y, Mawatari M, Kutsuna S, Takeshita N, Mezaki K, Ohmagari N, Miyoshi-Akiyama T. 2016. Emergence of a daptomycin-non-susceptible Enterococcus faecium strain that encodes mutations in DNA repair genes after high-dose daptomycin therapy. BMC Res Notes 9:197. doi: 10.1186/s13104-016-2003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla BS, Shelburne S, Reyes K, Kamboj M, Lewis JD, Rincon SL, Reyes J, Carvajal LP, Panesso D, Sifri CD, Zervos MJ, Pamer EG, Tran TT, Adachi J, Munita JM, Hasbun R, Arias CA. 2016. Influence of minimum inhibitory concentration in clinical outcomes of Enterococcus faecium bacteremia treated with daptomycin: is it time to change the breakpoint? Clin Infect Dis 62:1514–1520. doi: 10.1093/cid/ciw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, Reyes J, Roh JH, Lobos E, Sodergren E, Pasqualini R, Arap W, Quinn JP, Shamoo Y, Murray BE, Weinstock GM. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365:892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munita JM, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, Arias CA. 2013. A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 57:2831–2833. doi: 10.1128/AAC.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, Wollam A, Reyes J, Panesso D, Rojas NL, Shamoo Y, Murray BE, Weinstock GM, Arias CA. 2014. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 58:4527–4534. doi: 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benvenuto M, Benziger DP, Yankelev S, Vigliani G. 2006. Pharmacokinetics and tolerability of daptomycin at doses up to 12 milligrams per kilogram of body weight once daily in healthy volunteers. Antimicrob Agents Chemother 50:3245–3249. doi: 10.1128/AAC.00247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merck Sharp & Dohme Corp. 2016. CUBICIN (daptomycin for injection) for intravenous use. Prescribing information. Merck Sharp & Dohme Corp., Whitehouse Station, NJ. [Google Scholar]