ABSTRACT
Although the de novo folate biosynthesis pathway has been well studied in bacteria, little is known about its regulation. In the present study, the sigB gene in Mycobacterium tuberculosis was deleted. Subsequent drug susceptibility tests revealed that the M. tuberculosis ΔsigB strain was more sensitive to para-aminosalicylic acid (PAS) and sulfamethoxazole. Comparative transcriptional analysis was performed, and downregulation of pabB was observed in the ΔsigB strain, which was further verified by a quantitative reverse transcription-PCR and Western blot assay. Then, the production levels of para-aminobenzoic acid (pABA) were compared between the sigB deletion mutant and wild-type strain, and the results showed that sigB deletion resulted in decreased production of pABA. In addition, SigB was able to recognize the promoter of pabB in vitro. Furthermore, we found that deleting pabC also caused increased susceptibility to PAS. Taken together, our data revealed that, in M. tuberculosis, sigB affects susceptibility to antifolates through multiple ways, primarily by regulating the expression of pabB. To our knowledge, this is the first report showing that SigB modulates pABA biosynthesis and thus affecting susceptibility to antifolates, which broadens our understanding of the regulation of bacterial folate metabolism and mechanisms of susceptibility to antifolates.
KEYWORDS: Mycobacterium tuberculosis, pabB, para-aminobenzoic acid, para-aminosalicylic acid, sigB
INTRODUCTION
Tuberculosis (TB), caused by one of the toughest human pathogens, Mycobacterium tuberculosis, remains a serious public health concern in the world. The World Health Organization (WHO) estimated that in 2015 there were 10.4 million new TB cases worldwide, of which 580,000 cases were multidrug-resistant TB (MDR-TB) (1). Traditional effective anti-tubercular drugs, such as rifampin (RFP) and isoniazid (INH), were challenged by the emergence of MDR-TB and extensively drug-resistant TB, which have been major threats to global public health security due to the crisis of their detection and treatment (2, 3). Worst of all, the design of new and repurposed antimicrobial drugs has been slow (4). Thus, a better understanding of the molecular mechanisms mediating resistance or susceptibility in M. tuberculosis to preexisting antitubercular drugs is urgently required (5).
Folate is essential for all sorts of life, but mammals are unable to synthesize it, which makes the bacterial de novo folate biosynthesis pathway an ideal target for new antimicrobial drug design (6). As is well known, the bacterial de novo folate biosynthetic pathway has two branches: one for para-aminobenzoic acid (pABA) and the other for pterins. The pABA branch has two enzymes: the aminodeoxychorismate synthase composed of PabA and PabB and the aminodeoxychorismatelysase (PabC) (7–9). Meanwhile, the pterin branch starts with GTP and finally yields 7,8-dihydropterin pyrophosphate (DHPPP). These two branches are joined together by dihydropteroate synthase (DHPS), which synthesizes dihydropteroate (DHP) by using pABA and DHPPP as substrates. The DHP is then converted into dihydrofolate (DHF) through dihydrofolate synthase (DHFS), and is further reduced into tetrahydrofolate (THF) by dihydrofolate reductase (DHFR) (10–12). Thousands of antifolates have been designed targeting bacterial DHPS and DHFR (13). Para-aminosalicylic acid (PAS), a second-line drug used in clinical treatment of TB, was also classified as an antifolate that targets the de novo folate synthesis of M. tuberculosis As an analog of pABA, PAS acts as a prodrug activated by DHPS and DHFS and finally targets DHFR, thus leading to bactericidal effects (14, 15). As a result, folC mutation, which encodes the DHFS in M. tuberculosis, causes PAS resistance in M. tuberculosis clinical isolates by blocking drug activation (16, 17). In addition to folC mutations, mutations of the thymidylate synthase coding gene thyA or the upstream region of the ribD gene also result in PAS resistance in M. tuberculosis clinical isolates (18–20). ThyA is involved in folate transformation, which converts methylene THF into DHF (21). So far, mutations in these three genes could be identified in about two-thirds of PAS-resistant clinical isolates, but resistance mechanisms in the remaining third still need to be uncovered.
As essential components of the RNA polymerase (RNAP) holoenzyme, sigma factors provide specific recognition of promoters in bacteria. There are 13 sigma factors in M. tuberculosis, making the tough bacteria able to adapt to various environments (22). In M. tuberculosis, σB, encoded by the well-conserved gene sigB, is very close to the major sigma factor σA and identified to be positively regulated by the other three extracytoplasmic sigma factors: σE (23), σH (24), and σL (25). σB was also found to play a central role in response to various stress conditions, including heat shock in M. tuberculosis, sodium dodecyl sulfate (SDS), mild cold shock, cell envelope stress, and hypoxia in vitro (26, 27).
Although de novo folate biosynthesis has been well studied in bacteria, little is known regarding its regulation, especially in M. tuberculosis. Previously, deletion of sigB in Corynebacterium glutamicum resulted in a significant decrease of expression level of thyX and hypersensitivity to the DHFR inhibitor WR99210 (28). Very recently, sigB was also found to be an antifolate resistance determinant in Mycobacterium smegmatis through chemogenomic screening (29). These observations indicate that SigB might play a role on folate metabolism, thus affecting susceptibility to antifolates in bacteria. In this study, the sigB gene of M. tuberculosis H37Ra was deleted, and the effects on folate metabolism and susceptibility to different types of antifolates were probed to clarify the role of SigB on regulating folate metabolism in M. tuberculosis.
RESULTS
Effects of sigB deletion on bacterial growth.
Although σB is highly homologous to the primary sigma factor σA in mycobacteria, sigB is a nonessential gene in M. tuberculosis (30, 31). To investigate the role of σB on susceptibility to antitubercular drugs, we constructed a ΔsigB mutant by replacing the sigB gene with a hygromycin cassette on the H37Ra genome. A complemented strain, H37RaΔsigB(pMV261::sigB), was also constructed by reintroducing the episomal vector pMV261::sigB into the H37Ra ΔsigB mutant (H37RaΔsigB).
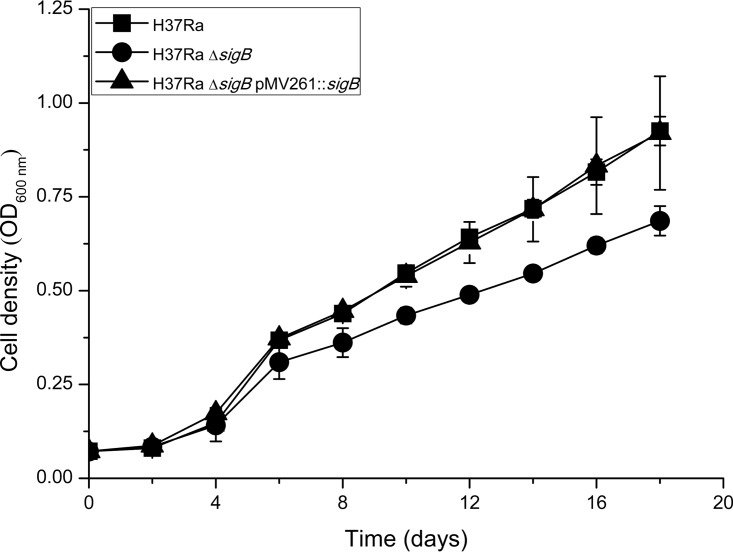
A previous study showed that inactivation of sigB in H37Rv did not affect the survival of H37Rv in macrophages but could make bacteria more sensitive to SDS stress and hypoxia in vitro (27). To probe the effect of sigB deletion on the bacterial growth of H37Ra, growth curves of H37Ra, H37RaΔsigB, and H37RaΔsigB(pMV261::sigB) were compared. As shown in Fig. 1, the ΔsigB mutant did show an obvious lag of growth compared to the wild-type and complemented strains.
FIG 1.
Growth curves of H37Ra, H37RaΔsigB, and H37RaΔsigB(pMV261::sigB) in liquid culture at 37°C. The OD600 was measured by using a SynergyH1 Hybrid reader (BioTek, USA) every 48 h. Data represent the means of three biological replicates, and error bars denote the standard deviations.
Deletion of sigB led to increased susceptibility to various antitubercular drugs.
The susceptibility of H37Ra, H37RaΔsigB, and H37RaΔsigB(pMV261::sigB) to various antitubercular drugs was tested. The results showed that sigB deletion led to increased sensitivities to most of the antitubercular drugs tested, as determined by MIC tests (Table 1). As shown in Table 1, the MICs of four drugs (RFP, streptomycin [SM], ethambutol [EMB], and norfloxacin) of the ΔsigB mutant were about 2- to 4-fold lower than for the wild-type strain, whereas MICs of two drugs (INH and ofloxacin) did not show any difference. Interestingly, we observed a remarkable decrease of MICs for the two antifolates (PAS and sulfamethoxazole [SMX]) in the ΔsigB mutant (16-fold lower than that of the wild-type strain). Moreover, changes in susceptibility to those drugs for the ΔsigB mutant could be completely reversed in the complemented strain.
TABLE 1.
MICs of various antitubercular drugs against H37Ra, H37RaΔsigB, and H37RaΔsigB(pMV261::sigB) strains
| Drug | MIC (μg ml−1) |
Fold change | ||
|---|---|---|---|---|
| H37Ra | H37RaΔsigB | H37RaΔsigB(pMV261::sigB) | ||
| para-Aminosalicylic acid | 0.04 | 0.0025 | 0.04 | 16 |
| Sulfamethoxazole | 25 | 1.5625 | 25 | 16 |
| Rifampin | 0.04 | 0.01 | 0.04 | 4 |
| Isoniazid | 0.1 | 0.1 | 0.1 | 1 |
| Streptomycin | 1.0 | 0.5 | 1.0 | 2 |
| Ofloxacin | 0.4 | 0.4 | 0.4 | 1 |
| Norfloxacin | 3.2 | 1.6 | 3.2 | 2 |
| Ethambutol | 1.25 | 0.625 | 1.25 | 2 |
| WR99210 | 25.62 | 6.405 | 25.62 | 4 |
Effect of sigB deletion on the bactericidal effect of PAS against M. tuberculosis.
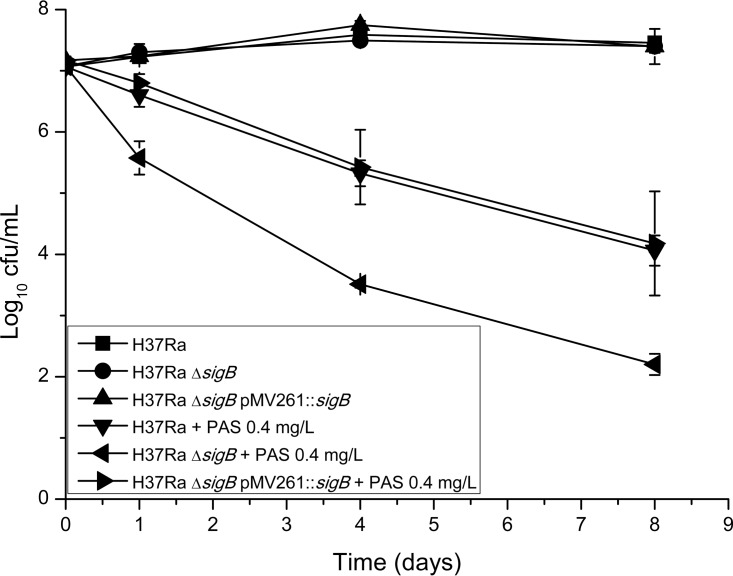
To further investigate the effect of sigB deletion on the bactericidal effect of PAS against M. tuberculosis, PAS kill kinetics were compared between the sigB knockout mutant and the wild-type strain. As shown in Fig. 2, we found that a dramatic decrease of viable bacterial cell number (from 7 to 5.5 log10 CFU/ml) was observed for the ΔsigB mutant after 24 h of PAS treatment but that only a slight decrease (∼0.5 log10 CFU/ml) was observed for the wild-type strain. After treatment for 4 days, the viable bacterial cell number of the ΔsigB mutant decreased to 3.5 log10 CFU/ml, while that of the wild-type strain fell to 5.5 log10 CFU/ml. The viable cell number of the ΔsigB mutant continued decreasing sharply from 8 days and onward, and no viable bacterial cells could be detected in 10-μl cultures dropped on the 7H10 medium after 16 days of treatment (data not shown). However, viable cell numbers of the wild-type strain could still be detected after 16 days of PAS treatment (data not shown). During the course of PAS treatment, survival of the complemented strain was very similar to that of the wild-type strain.
FIG 2.
Killing curves of different M. tuberculosis strains after exposure to PAS at 0.4 μg ml−1 in liquid medium (7H9 plus OADC). Experiments were performed in three biological replicates. Standard deviations are indicated by error bars.
Transcriptome analysis of the ΔsigB mutant.
To obtain further insights on how sigB deletion led to increased susceptibility to multiple antitubercular drugs, we compared the global transcription profile between the ΔsigB mutant and wild-type strain using transcriptome sequencing (RNA-seq) technology. Consequently, 175 genes were identified to be significantly regulated, including 33 upregulated genes and 142 downregulated genes (see Table S1 in the supplemental material). Based on gene ontology (GO) pathway enrichment analysis, changes in the expression level of many genes involved in stress responses and pathogenesis could be observed (see Table S2 in the supplemental material), which was in consistent with the fact that σB plays a central role in stress responses (27). We also found that 37 genes involved in bacterial cell wall biosynthesis and 26 genes involved in bacterial plasma membrane biosynthesis were downregulated, which might affect the permeability of the bacteria. In addition, several transcriptional regulators were found to be up- or downregulated in the ΔsigB mutant (Table 2). For example, the expression of whiB6, which plays a role on regulating the ESX-1 secretion system and the Dos dormancy regulon in Mycobacterium marinum (32), was downregulated in H37RaΔsigB.
TABLE 2.
Genes differentially regulated in the H37RaΔsigB strain versus the wild-type H37Ra strain
| Category and gene | Descriptiona | Fold change | Tendency |
|---|---|---|---|
| Growth | |||
| yrb1B | Membrane protein | 1.54 | Up |
| yrbE4B | ABC transporter permease | 1.50 | Up |
| pabB | Aminodeoxychorismate synthase component I | 0.54 | Down |
| MRA_0320 | Hypothetical protein | 0.54 | Down |
| MRA_0637 | Hypothetical protein | 0.57 | Down |
| MRA_0881 | Hypothetical protein | 0.58 | Down |
| mprB | Two-component sensor histidine kinase | 0.65 | Down |
| cysD | Sulfate adenylyltransferase subunit 2 | 0.60 | Down |
| cysN | Adenylyl-sulfate kinase | 0.65 | Down |
| MRA_1350 | Membrane protein | 0.67 | Down |
| MRA_2389 | Hypothetical protein | 0.61 | Down |
| MRA_2684 | Antitoxin | 0.21 | Down |
| MRA_2056 | Sugar ABC transporter substrate-binding lipoprotein | 0.63 | Down |
| MRA_0067 | RNase VapC1 | 0.59 | Down |
| MRA_3842 | Membrane protein | 0.60 | Down |
| sigB | RNA polymerase sigma factor SigB | 0.00 | Down |
| Metabolic process | |||
| MRA_1007 | Acetyltransferase | 1.64 | Up |
| msrA | Peptide-methionine (S)-S-oxide reductase | 0.56 | Down |
| fabD2 | Malonyl CoA-ACP transacylase | 0.49 | Down |
| pncA | Bifunctional pyrazinamidase nicotinamidase | 0.59 | Down |
| Transcriptional regulator | |||
| MRA_0471 | Transcriptional regulator | 0.53 | Down |
| MRA_1735 | Transcriptional regulator | 0.46 | Down |
| MRA_1828 | Transcriptional regulator | 1.76 | Up |
| MRA_2649 | Transcriptional regulator | 0.54 | Down |
| MRA_2671 | Transcriptional regulator | 0.62 | Down |
| MRA_2909 | Transcriptional regulator | 1.67 | Up |
| whiB6 | Transcriptional regulator | 0.43 | Down |
CoA, coenzyme A.
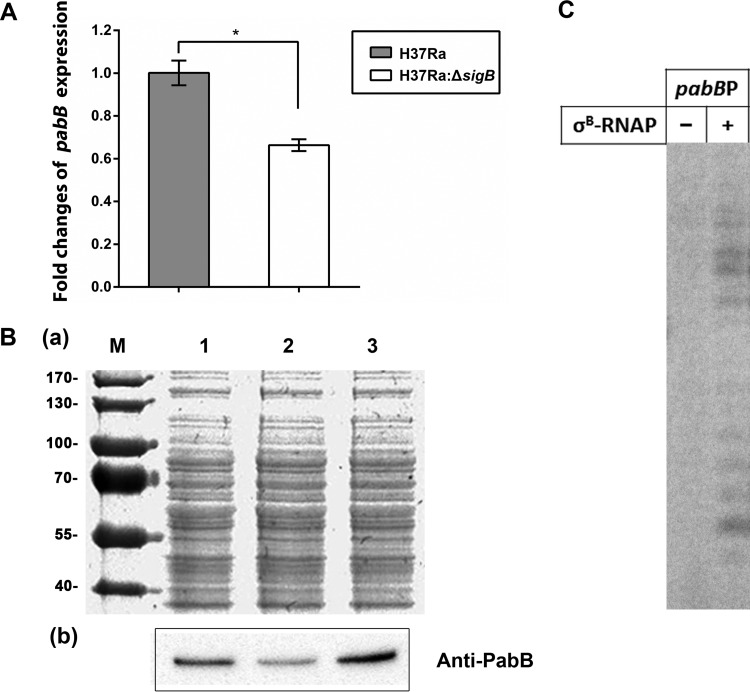
Very importantly, we found that pabB, an essential component of the pABA synthesis machinery in M. tuberculosis, was also downregulated in the ΔsigB mutant (Table 2), and this was further proved by the subsequent quantitative real-time PCR (qRT-PCR) analysis (Fig. 3A).
FIG 3.
PabB was regulated directly by σB in M. tuberculosis H37Ra. (A) Comparison of the transcriptional level of the gene pabB during the exponential phase in H37Ra (WT) and H37RaΔsigB (KO) strains by qRT-PCR. The expression levels of GAPDH mRNA were normalized as an endogenous control. Data are representative of three experiments, and the statistical significance is indicated by an asterisk: *, P < 0.01. (B) Comparison of the expressional level of PabB during the exponential phase in H37Ra, H37RaΔsigB, and H37RaΔsigB(pMV261::sigB) as determined by a Western blot assay. Experiments were repeated at least three times, and representative results are shown. (a) Total protein was normalized to 20 μg of each strain and then electrophoresed by SDS-PAGE and stained by Coomassie brilliant blue. Lane M, the prestained protein marker; lane 1, total protein of H37Ra; lane 2, total protein of H37RaΔsigB; lane 3, total protein of H37RaΔsigB(pMV261::sigB). (b) Western blot analysis of total protein immunoblotted with mice antisera of anti-PabB. Lane 1, anti-PabB immunoblotted to the total protein of H37RaΔsigB; lane 2, anti-PabB immunoblotted to the total protein of H37RaΔsigB; lane 2, anti-PabB immunoblotted to the total protein of H37RaΔsigB(pMV261::sigB). (C) [32P]RNA products synthesized in the in vitro transcription assay from the indicated promoter of pabB. Transcription was performed by RNAP holoenzyme containing σB.
Deletion of sigB led to decreased expression level of PabB.
A Western blot assay was also performed to compare the expression level of PabB between different strains, and the results showed that expression was significantly decreased in H37RaΔsigB compared to the wild-type strain, which could be restored in the complemented strain (Fig. 3B).
Overexpression of pabB in M. tuberculosis led to PAS resistance.
To further verify that decreased expression of pabB was responsible for increased susceptibility to both PAS and SMX in the ΔsigB strain, pabB was overexpressed in both H37RaΔsigB and the wild-type strain. The results showed that the overexpression of pabB resulted in PAS resistance (Table 3).
TABLE 3.
PAS MICs against different mycobacterial strains
| Strain | Description | MIC (μg ml−1) |
|---|---|---|
| H37Ra(pMV261) | H37Ra transformed with pMV261 | 0.04 |
| H37Ra(pMV261::pabB) | H37Ra transformed with pMV261::pabB | 0.64 |
| H37RaΔsigB(pMV261::pabB) | H37RaΔsigB transformed with pMV261::pabB | 0.16 |
σB specifically recognized the promoter of pabB in vitro.
To further investigate whether σB could recognize the promoter of pabB directly, we performed an in vitro transcriptional assay (Fig. 3C). We found that σB in combination with mycobacterial RNAP core enzymes successfully initiated transcription from the promoter of the pabB gene.
Deletion of sigB caused decreased the production of pABA.
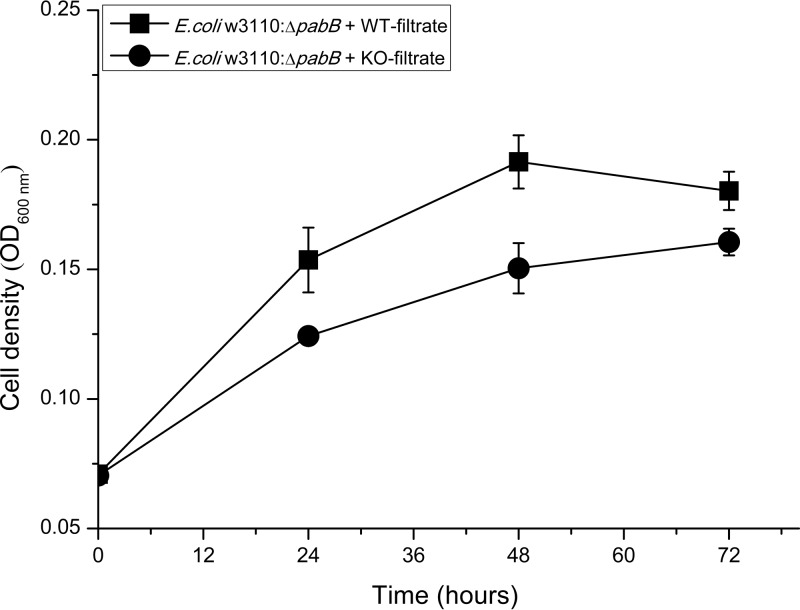
The pabB (MRA_1014) gene product was characterized to be the aminodeoxychorismate synthase, an indispensable enzyme for the biosynthesis of pABA from chorismate in folate de novo synthesis in bacteria. Consequently, decreased expression of PabB may lead to decreased production of pABA. To determine production levels of pABA in H37Ra and H37RaΔsigB mutant strains, an Escherichia coli W3110 ΔpabB mutant strain (W3110ΔpabB) was constructed, which needed exogenous pABA to support its growth both in E minimal medium and 7H9 plus oleic acid-albumin-dextrose-catalase (OADC) medium (see Fig. S1 in the supplemental material). We also found that the in vitro growth of E. coli W3110ΔpabB was pABA dose dependent (see Fig. S1A in the supplemental material). When E. coli W3110ΔpabB was cultured in E minimal medium plus culture filtrates from different H37Ra strains, there was obviously less growth when culture filtrates from H37RaΔsigB were used (Fig. 4), suggesting a decreased production of pABA in the H37RaΔsigB mutant.
FIG 4.
The synthesis of pABA was impaired in H37RaΔsigB. The growth curves of E. coli W3110ΔpabB were determined in the presence of filtrates from culture of H37Ra (WT-filtrate) and H37Ra ΔsigB (KO-filtrate) strains separately. The means of three biological replicates are shown, and error bars denoted the standard deviations.
Disruption of pABA synthesis caused increased susceptibility to PAS.
In M. tuberculosis, pabA, pabB, and pabC are all essential genes for bacterial in vitro growth. To confirm that decreased production of pABA finally caused increased susceptibility to SMX and PAS, the pABA biosynthesis pathway was disrupted by deleting pabC in H37Ra. We found that although pabC was predicted to be essential for in vitro growth of M. tuberculosis, the first generation of the H37RaΔpabC mutant did grow on 7H10 solid plates without exogenous pABA and continued growing when inoculating into liquid medium in the absence of pABA but stopped growing after it was subcultured into liquid medium. Although the H37RaΔpabC mutant did not grow as well as the wild-type strain in the absence of pABA on solid medium (see Fig. S2 in the supplemental material), it could be restored by the addition of a rather low concentration of pABA (0.01 μg ml−1). Then, the susceptibility to PAS of the H37RaΔpabC mutant were determined by MIC tests. The results showed that deletion of pabC led to increased susceptibility to PAS in the presence of a limited amount of exogenous pABA (0.01 and 0.05 μg ml−1) (Table 4).
TABLE 4.
PAS MICs against H37Ra and H37RaΔpabC strains supplemented with pABA
| pABA concn (μg ml−1) | MIC (μg ml−1) |
|
|---|---|---|
| H37Ra | H37RaΔpabC | |
| 0.01 | 0.32 | 0.02 |
| 0.05 | >0.32 | 0.08 |
DISCUSSION
Previously, Thiede et al. reported that disruption of pABA biosynthesis potentiates the antitubercular effect of anti-folates by up to 1,000-fold (33). Here, we showed that SigB regulates the expression of pabB gene in M. tuberculosis and, as a result, deletion of sigB results in decreased expression of pabB, impaired pABA production, and increase susceptibility to antifolates.
Although the folate de novo biosynthesis pathway has been well studied in bacteria, very little is known about its regulation. In 2010, a eubacterial riboswitch class that selectively binds derivatives of folate was discovered, indicating a role of riboswitches for the regulation of folate biosynthesis (34). However, further evidence is required to prove this. Very recently, a vitamin B12-binding light sensing transcriptional regulator (PhrR) was found in Halomonas, which modulates the expression of three genes related to folate biosynthesis (folE, folK, and folM) and shows the existence of transcriptional regulation of folate biosynthesis in bacteria (35). Further studies are necessary to see whether there would be any homologue of PhrR in other bacteria beyond gammaproteobacteria.
Except for PhrR, sigB, a well-known transcriptional regulator in bacteria, has also been shown to be related to folate metabolism. Previously, the alternative thymidylate synthase ThyX, which is involved in folate transformation, was found to be regulated by SigB (28). Since M. tuberculosis also has an alternative thymidylate synthase gene thyX (36), we wondered whether SigB could also regulate the expression of thyX in M. tuberculosis. Therefore, we deleted the sigB gene in H37Ra and tested its effect on thyX expression through Western blot analysis. Meanwhile, the susceptibility of the H37RaΔsigB mutant to different types of antitubercular drugs, including antifolates, was also tested. To our surprise, although deletion of sigB in M. tuberculosis caused increased susceptibility to PAS and SMX, it did not affect the expression of thyX (see Fig. S3 in the supplemental material). These data suggested that in M. tuberculosis SigB does not regulate the expression of thyX and that the increased susceptibility to PAS and SMX caused by sigB deletion was not related to thyX.
Although the microarray data of H37Rv ΔsigB under normal growth and stress conditions had been reported (27), we failed to find any reasonable clue as to why the sigB knockout mutant was more sensitive to antifolates. From the known action mechanisms of PAS and SMX, we can see that pABA is able to antagonize the antitubercular effects of both drugs, which means disruption of the pABA biosynthesis pathway may result in increased sensitivity to both drugs. Thus, we speculated that the deletion of sigB might affect the expression of genes involved in pABA biosynthesis. The data from our comparative RNA-seq analysis between H37Ra and its sigB deletion mutant strain showed that in M. tuberculosis the expression of pabB was regulated by SigB, which was confirmed by subsequent qRT-PCR and Western blot analysis.
In addition, our data of in vitro transcriptional analysis further confirmed that sigB is indeed able to recognize the promoter of pabB and initiate gene transcription. Although the transcription of pabB could be initiated by σB in vitro, the expression of pabB was not completely eliminated in the ΔsigB mutant (Fig. 3), suggesting the existence of other transcriptional regulators, such as SigA, for pabB in M. tuberculosis. The alternative sigma factor SigB of M. tuberculosis shows 62.9% identity to its principal sigma factor SigA in amino acid sequences (37, 38). Furthermore, a previous study showed that these two sigma factors recognize similar promoters with a conserved −10 element in TANNNT (39). In accordance, the pabB promoter contains a 5′-TAAGAT-3′ as the −10 element upstream of the transcriptional start site identified by Shell et al. (40). Thus, SigA and SigB may both be in charge of the initiation of pabB transcription in M. tuberculosis.
Since pabB is essential for pABA biosynthesis and thus essential for in vitro bacterial growth, we speculated that a decrease in pabB expression might result in decreased production of pABA in the ΔsigB mutant. Thus, we used the E. coli W3110 ΔpabB mutant to compare the pABA production level between different H37Ra strains, and the results showed that the deletion of sigB led to decreased pABA production. Considering the impairment to pABA synthesis caused by sigB deletion, SigB should not be only regarded as an environmental stress responder in M. tuberculosis. In fact, it has been reported that in C. glutamicum, SigB was involved in the positive regulation of glucose metabolism even during the aerobic exponential phase (41). In addition, the homologue of SigB in E. coli, RpoS, was also shown to play an important role in iron acquisition and other metabolism gene regulation during exponential growth (42). Our data (in combination with those previous observations) suggested that SigB also played important roles under standard physiological growth conditions, and as a result, inactivation of sigB led to a slower growth under normal in vitro growth conditions (Fig. 1).
It has been previously reported that in Pseudomonas aeruginosa, RpoS (the homologue of SigB) was involved in tolerance to multiple antibiotics (43, 44). Therefore, except for antifolates, susceptibility to other antitubercular drugs was also tested for the H37RaΔsigB mutant (Table 1). The results showed that the deletion of sigB in M. tuberculosis also led to increased susceptibility to many other antitubercular drugs. To obtain further insights into how sigB deletion affects susceptibility to multiple antitubercular drugs, we returned to our comparative RNA-seq data and found that deletion resulted in the downregulation of 37 genes involved in biosynthesis of the bacterial cell wall and 26 genes involved in biosynthesis of bacterial the plasma membrane, which might change bacteria permeability. However, further studies are required to probe the effect of sigB deletion on M. tuberculosis cell permeability.
Taken together, we found that deletion of sigB affected the in vitro growth of H37Ra and susceptibility to various antitubercular drugs, including PAS and SMX. It seemed that, in M. tuberculosis H37Ra, sigB could affect susceptibility to PAS and SMX through at least two different ways: first, by regulating the expression of pabB, which has been verified in this study, and second, by modulating the permeability of the bacterial cell, which requires further verification. So far, this is the first report showing that SigB modulates the biosynthesis of pABA by regulating the expression of pabB, which broadens our understanding of both the physiological function of SigB and the regulation mechanisms of folate biosynthesis in mycobacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
M. tuberculosis strains were cultured at 37°C in 7H9 medium consisting of Middlebrook 7H9 broth (Difco), 10% (vol/vol) OADC (Difco), 0.5% (vol/vol) glycerol, and 0.05% (vol/vol) Tween 80 (Sigma-Aldrich) or on 7H10 agar medium (Difco) supplemented with 10% (vol/vol) OADC (Difco) and 0.5% (vol/vol) glycerol. Mycobacterium smegmatis mc2155 was grown in Middlebrook 7H9 medium or on 7H10 agar medium, both without adding OADC. E. coli trains HB101 and BL21(DE3) were cultured in Luria-Bertani (LB) medium (Difco) at 37°C or in E minimal medium (citric·H2O [2 g/liter], MgSO4·7H2O [0.2 g/liter], K2HPO4·3H2O [13.09 g/liter], NaNH4HPO4·4H2O [3.5 g/liter]) supplemented with 0.5% d-(+)-glucose. Plasmid pET-21a (Novagen) and pMV261 were used for the construction of expression plasmids. The gene-specific primers used for the construction of recombinant plasmids are listed in Table S3 in the supplemental material. Where appropriate, the culture medium was supplemented with hygromycin at 75 μg ml−1 for mycobacteria and 150 μg ml−1 for E. coli, kanamycin at 25 μg ml−1 for mycobacteria and 50 μg ml−1 for E. coli, and ampicillin at 100 μg ml−1 for E. coli.
Construction of mycobacterial mutant and complemented strains.
A modified strategy for specialized transduction was used to construct the M. tuberculosis H37RaΔsigB mutant. Genomic regions flanking sigB, 705 bp upstream (region containing MRA_2737 and MRA_2736) and 838 bp downstream (region containing ideR), were amplified by PCR. The primers used for amplification of the upstream of sigB were sigBkoLFP and sigBkoLRP, and those for the region downstream were sigBkoRFP and sigBkoRRP. The recombinant plasmid p0004s-L+R was constructed by inserting the Van91I-digested PCR products into the plasmid p0004s digested with Van91I. Then, the p0004s-L+R was digested with PacI and ligated to the PacI-digested shuttle phasmid vector phAE159. After ligation, the recombinant cosmid phAE159-p0004s-L+R was transduced into E. coli HB101 in an in vitro λ-packaging reaction (Epicentre Biotechnologies, MaxPlax Lambda packaging extracts). Phasmid DNA prepared from confirmed selected hygromycin-resistant transductants was electroporated into M. smegmatis mc2155 to generate the specialized transducing phage. As described in a previous study (45), the transducing phage at the most efficient titer was used to infect H37Ra at a multiplicity of infection of 10. Successful specialized transduction of H37Ra was confirmed by comparing the size of the PCR-amplified product of hygromycin-resistant colonies with wild-type H37Ra using primers sigBLYZ and sigBRYZ (see Table S3 in the supplemental material). The gene sigB was amplified from M. tuberculosis H37Ra genomic DNA using the specific primers sigB-L and sigB-R (see Table S3) and then cloned into pMV261 to yield pMV261::sigB. The complemented strain was constructed by electrotransforming the recombinant plasmid pMV261::sigB into the competent cell of H37RaΔsigB and plated on 7H10 medium supplemented with hygromycin at 75 μg ml−1 and kanamycin at 25 μg ml−1.
Construction of the H37RaΔpabC mutant was performed by the same strategies described above except for the last screening step in which, screening was performed on 7H10 medium supplemented with hygromycin at 75 μg ml−1 and an additional 10 μg ml−1 pABA.
Drug susceptibility tests.
Mycobacterial cells were cultured to optical density at 600 nm (OD600) of 0.5 to 1.0 and diluted to about 105 CFU ml−1 by 10-fold serial dilutions in fresh 7H9 medium with or without 10% OADC. Bacteria were plated onto 7H10 agar solid plates containing various concentrations of different drugs: PAS (0, 0.00125, 0.0025, 0.005, 0.01, 0.02, 0.04, 0.08, 0.16, and 0.32 μg ml−1), SMX (0, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, 50, 100, and 200 μg ml−1), WR99210 (0, 1, 2, 4, 8, 16, 32, 64, and 128 μM), INH (0, 0.00625, 0.0125, 0.025, 0.05, 0.1, 0.2, 0.4, 0.8, and 1.6 μg ml−1), RIF (0, 0.000625, 0.00125, 0.0025, 0.005, 0.01, 0.02, 0.04, 0.08, and 0.16 μg ml−1), SM (0, 0.03125, 0.0625, 0.125, 0.25, 0.5, 1, 2, 4, and 8 μg ml−1), EMB (0, 0.15625, 0.3125, 0.625, 1.25, 2.5, 5, and 10 μg ml−1), ofloxacin (0, 0.025, 0.05, 0.1, 0.2, 0.4, 0.8, and 1.6 μg ml−1), and norfloxacin (0, 0.2, 0.4, 0.8, 1.6, 3.2, and 6.4 μg ml−1). All antibiotics were purchased from Sigma-Aldrich and solubilized according to the manufacturer's recommendations. Cultures were incubated at 37°C for 21 days. The MIC was defined as the lowest required concentration of antibiotics to inhibit the growth of 99% bacterial CFU. MIC tests for the H37RaΔpabC mutant were performed on the 7H10 medium in the presence of different concentrations of pABA (0.01, and 0.05 μg ml−1) with various concentrations of PAS as described above.
Kill kinetics of PAS against M. tuberculosis.
Bacteria were grown to OD600 of 0.5 to 1.0 and diluted to about OD600∼0.1 (107 CFU ml−1) in fresh 7H9 medium with OADC, and 0.4 μg ml−1 (10×MIC of WT) of PAS was used in PAS treatment. Cultures were incubated at 37°C, and aliquots of samples were taken and plated on 7H10 medium after serial dilutions at days 0, 1, 4, and 8, separately.
RNA-seq.
Total RNA was isolated using an RNeasy minikit (Qiagen, Germany). Library constructions were prepared using TruSeq stranded total RNA sample preparation kit (Illumina, USA), and RNA sequencing was conducted using an Illumina HiSeq 2500 at Shanghai Biotechnology Corporation. The insert size conformation of purified libraries was validated by an Agilent 2100 bioanalyzer (Agilent Technologies, USA). Bowtie2 v2-2.0.5 was used to map the cleaned reads to the M. tuberculosis H37Ra genome acquired from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/nuccore/148659757?report=GenBank). Then, HTSeq v2.1.1 was run with a reference annotation to generate fragments per kilobase of exon model per million mapped reads values for estimation of fold changes. Three biological replicates were used in RNA-seq and the P and q values were calculated. The differentially expressed genes were selected using the following filter criteria: a false discovery rate of ≤0.05 and a fold change of ≥1.5.
Quantitative real-time PCR assays.
Total RNA was extracted as described previously, and cDNA was synthesized with a ReverTra Ace qPCR kit (Toyobo) according to the manufacturer's instructions. Quantification of the gene expression levels was performed by real-time qPCR analysis on a 7900 HT sequence detection system (ABI, USA) with ABI Power SYBR green PCR master mix. Primers specific to pabB were designed and synthesized by Sangon Biotech (Shanghai) Co., Ltd. Expression levels of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA were normalized as an endogenous control. The gene-specific primers pabB-qRT-L and pabB-qRT-R used are listed in Table S3 in the supplemental material.
In vitro transcription assay.
The recombinant M. tuberculosis RNAP core enzyme containing 6×His tag at the C terminus of the β′ subunit was purified from E. coli BL21(DE3) carrying pMR4 plasmid as described previously (39). Mycobacterial SigB was expressed in Escherichia coli BL21(DE3) using pET28a plasmid and purified as previously described (39). In vitro transcription was performed in transcription buffer (20 mM Tris-HCl [pH 7.9], 50 mM NaCl, 5 mM MgSO4, 1 mM dithiothreitol, 0.1 mM EDTA, and 5% glycerol) in a total volume of 5 μl. RNAP holoenzyme was assembled by mixing the 600 nM SigB with 200 nM core RNAP, followed by incubation for 5 min. A 15 nM promoter DNA fragment was added, followed by incubation at 37°C for 10 min. Transcription was initiated by the addition of 50 μM ATP, GTP, and CTP and 3 μCi of [α-32P]UTP and then carried out for 10 min at 37°C. Reactions were stopped by adding 8 M urea, and the synthesized RNA products were analyzed on denaturing (7 M urea) 18% polyacrylamide gel electrophoresis (PAGE). The primers pabB-p-L and pabB-p-R listed in Table S3 in the supplemental material were designed according to the reported transcription start site of pabB analyzed by Shell et al. for in vitro transcriptional assay (40).
Purification of recombinant histidine-tagged PabB.
PabB was amplified from M. tuberculosis H37Ra genomic DNA using the specific primers MpabB-L and MpabB-R (see Table S3) and cloned into pET21a to yield pET21a::pabB. After sequence verification, these recombinant plasmids were transformed into E. coli BL21(DE3). The transformed E. coli BL21(pET21a::pabB) cells were grown at 37°C in LB broth to an OD600 of ∼0.6. Then, 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, followed by incubation at 16°C for 20 h. The cells were harvested by centrifugation after incubation and resuspended in 50 mM Tris-HCl, 500 mM NaCl, and 20 mM imidazole (pH 8.0). Suspensions were disrupted by sonication and clarified by high-speed centrifugation. After centrifugation, the supernatants were mixed with prewashed nickel-nitrilotriacetic acid His-Trap HP affinity resin (GE Healthcare) at 4°C overnight, and nonspecifically bound protein was removed by washing the resin with 50 mM Tris-HCl, 0.5 M NaCl, and 60 mM imidazole (pH 8.0). Meanwhile, recombinant PabB was eluted with 50 mM Tris-HCl, 0.5 M NaCl, and 200 mM imidazole (pH 8.0) separately. SDS-PAGE was used to analyze the acquired elution.
Antibody preparation of PabB.
Five mice as a group were immunized with purified recombinant PabB. They were injected intramuscularly with 0.5 mg of recombinant proteins in a ratio of 1:1 (vol/vol) with Freund complete adjuvant for the first immunization. Then, every 2 weeks a similar injection was subcutaneously administered, except that Freund incomplete adjuvant was used. Immune sera were collected after four injections, and mice were bled a week after the fourth injection. The specificity of antisera against recombinant proteins was tested by Western blotting assay.
Western blot analysis.
The H37Ra, H37RaΔsigB, and H37RaΔsigB(pMV261::sigB) strains were cultured in 50 ml of 7H9 medium at 37°C and harvested at log phase by centrifugation. For Western blot analysis, pellets were resuspended in 50 mM Tris-HCl, 500 mM NaCl, and 20 mM imidazole (pH 8.0) and then lysed by using zirconium beads. Protein samples acquired from the supernatant after centrifugation were separated by SDS–10% PAGE and immediately transferred to a polyvinylidene difluoride membrane (Merck Millipore, Darmstadt, Germany) by a Bio-Rad SD device (Bio-Rad Laboratories, Hercules, CA) at 15 V for 25 min. Finally, the proteins were probed with mice antisera against PabB.
Growth curves of the E. coli W3110 ΔpabB mutant.
Filtrates from cultures of the three strains [H37Ra, H37RaΔsigB, and H37RaΔsigB(pMV261::sigB)] were collected, respectively, through sterile filters (Millipore). E. coli W3110ΔpabB was cultured to log phase in LB medium at 37°C and then harvested by centrifugation. After that, the pellet was washed twice and resuspended for inoculation (at ca. 106 to 107 CFU/ml) into fresh E medium supplemented with 0.5% d-(+)-glucose for starvation. After incubation at 37°C to consume the endogenous pABA for 72 h, the E. coli W3110ΔpabB mutant was inoculated (105 CFU/ml) into 2 ml of ETG plus 8-ml filtrates from the different strains mentioned above. The growth of bacterial cultures was measured by monitoring the OD600 every 24 h using a spectrophotometer (Bio-Rad).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Key Programs of the Chinese Academy of Sciences (ZDRW-ZS-2016-4), the Open Project Grant from the State Key Lab of Respiratory Disease (2014SKLRD-O06), the Hubei Provincial Natural Science Foundation of China (2013CFA072), and the National Natural Science Foundation of China (grant 31300050).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00551-17.
REFERENCES
- 1.World Health Organization. 2016. Global tuberculosis report 2016. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/global_report/en/. [Google Scholar]
- 2.Nguyen L. 2016. Antibiotic resistance mechanisms in Mycobacterium tuberculosis: an update. Arch Toxicol 90:1585–1604. doi: 10.1007/s00204-016-1727-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients coinfected with tuberculosis and HIV in a rural area of South Africa. Lancet 368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 4.Wallis RS, Maeurer M, Mwaba P, Chakaya J, Rustomjee R, Migliori GB, Marais B, Schito M, Churchyard G, Swaminathan S, Hoelscher M, Zumla A. 2016. Tuberculosis-advances in development of new drugs, treatment regimens, host-directed therapies, and biomarkers. Lancet Infect Dis 16:e34–e46. doi: 10.1016/S1473-3099(16)00070-0. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen L, Jacobs MR. 2012. Counterattacking drug-resistant tuberculosis: molecular strategies and future directions. Expert Rev Anti Infect Ther 10:959–961. doi: 10.1586/eri.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermingham A, Derrick JP. 2002. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24:637–648. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- 7.Ye QZ, Liu J, Walsh CT. 1990. p-Aminobenzoate synthesis in Escherichia coli: purification and characterization of PabB as aminodeoxychorismate synthase and enzyme X as aminodeoxychorismate lyase. Proc Natl Acad Sci U S A 87:9391–9395. doi: 10.1073/pnas.87.23.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green JM, Nichols BP. 1991. p-Aminobenzoate biosynthesis in Escherichia coli: purification of aminodeoxychorismate lyase and cloning of pabC J Biol Chem 266:12971–12975. [PubMed] [Google Scholar]
- 9.Nichols BP, Seibold AM, Doktor SZ. 1989. para-Aminobenzoate synthesis from chorismate occurs in two steps. J Biol Chem 264:8597–8601. [PubMed] [Google Scholar]
- 10.Richey DP, Brown GM. 1969. The biosynthesis of folic acid. IX. Purification and properties of the enzymes required for the formation of dihydropteroic acid. J Biol Chem 244:1582–1592. [PubMed] [Google Scholar]
- 11.Shiota T, Baugh CM, Jackson R, Dillard R. 1969. The enzymatic synthesis of hydroxymethyldihydropteridine pyrophosphate and dihydrofolate. Biochemistry 8:5022–5028. doi: 10.1021/bi00840a052. [DOI] [PubMed] [Google Scholar]
- 12.Blakley RL, Mcdougall BM. 1961. Dihydrofolic reductase from Streptococcus faecalis R. J Biol Chem 236:1163–1167. [PubMed] [Google Scholar]
- 13.Estrada A, Wright DL, Anderson AC. 2016. Antibacterial antifolates: from development through resistance to the next generation. CSH Perspect Med 6:a028324. doi: 10.1101/cshperspect.a028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty S, Gruber T, Barry CE III, Boshoff HI, Rhee KY. 2012. para-Aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339:88–91. doi: 10.1126/science.1228980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J, Rubin EJ, Bifani P, Mathys V, Lim V, Au M, Jang J, Nam J, Dick T, Walker JR, Pethe K, Camacho LR. 2013. para-Aminosalicylic acid is a prodrug targeting dihydrofolate reductase in Mycobacterium tuberculosis. J Biol Chem 288:23447–23456. doi: 10.1074/jbc.M113.475798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao F, Wang XD, Erber LN, Luo M, Guo AZ, Yang SS, Gu J, Turman BJ, Gao YR, Li DF, Cui ZQ, Zhang ZP, Bi LJ, Baughn AD, Zhang XE, Deng JY. 2014. Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:1479–1487. doi: 10.1128/AAC.01775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Liu L, Zhang Y, Dai G, Huang H, Jin Q. 2015. Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob Agents Chemother 59:1320–1324. doi: 10.1128/AAC.03695-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rengarajan J, Sassetti CM, Naroditskaya V, Sloutsky A, Bloom BR, Rubin EJ. 2004. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol Microbiol 53:275–282. doi: 10.1111/j.1365-2958.2004.04120.x. [DOI] [PubMed] [Google Scholar]
- 19.Moradigaravand D, Grandjean L, Martinez E, Li H, Zheng J, Coronel J, Moore D, Torok ME, Sintchenko V, Huang H, Javid B, Parkhill J, Peacock SJ, Koser CU. 2016. dfrA thyA double deletion in para-aminosalicylic acid-resistant Mycobacterium tuberculosis Beijing strains. Antimicrob Agents Chemother 60:3864–3867. doi: 10.1128/AAC.00253-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng YS, Sacchettini JC. 2016. Structural insights into Mycobacterium tuberculosis Rv2671 protein as a dihydrofolate reductase functional analogue contributing to para-aminosalicylic acid resistance. Biochemistry 55:1107–1119. doi: 10.1021/acs.biochem.5b00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carreras CW, Santi DV. 1995. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem 64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol Rev 30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 23.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor σE: role in global gene expression and survival in macrophages. Mol Microbiol 41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 24.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. 2002. Role of the extracytoplasmic-function sigma factor σH in Mycobacterium tuberculosis global gene expression. Mol Microbiol 45:365–374. doi: 10.1046/j.1365-2958.2002.03005.x. [DOI] [PubMed] [Google Scholar]
- 25.Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palu G, Manganelli R. 2006. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor sigma L and roles in virulence and in global regulation of gene expression. Infect Immun 74:2457–2461. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol Microbiol 31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 27.Fontan PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, Fattorini L, Schoolnik GK, Smith I. 2009. The Mycobacterium tuberculosis sigma factor σB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol 191:5628–5633. doi: 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho S, Yang S, Rhie H. 2012. The gene encoding the alternative thymidylate synthase ThyX is regulated by sigma factor SigB in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol Lett 328:157–165. doi: 10.1111/j.1574-6968.2011.02494.x. [DOI] [PubMed] [Google Scholar]
- 29.Guzzo MB, Nguyen HT, Pham TH, Wyszczelska-Rokiel M, Jakubowski H, Wolff KA, Ogwang S, Timpona JL, Gogula S, Jacobs MR, Ruetz M, Krautler B, Jacobsen DW, Zhang GF, Nguyen L. 2016. Methylfolate trap promotes bacterial thymineless death by sulfa drugs. PLoS Pathog 12:e1005949. doi: 10.1371/journal.ppat.1005949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez M, Doukhan L, Nair G, Smith I. 1998. sigA is an essential gene in Mycobacterium smegmatis. Mol Microbiol 29:617–628. doi: 10.1046/j.1365-2958.1998.00960.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Karakousis PC, Bishai WR. 2008. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J Bacteriol 190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen ZK, Hu YB, Cumming BM, Lu P, Feng LP, Deng JY, Steyn AJC, Chen SY. 2016. Mycobacterial WhiB6 differentially regulates ESX-1 and the Dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Rep 16:2512–2524. doi: 10.1016/j.celrep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 33.Thiede JM, Kordus SL, Turman BJ, Buonomo JA, Aldrich CC, Minato Y, Baughn AD. 2016. Targeting intracellular p-aminobenzoic acid production potentiates the anti-tubercular action of antifolates. Sci Rep 6:38083. doi: 10.1038/srep38083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ames TD, Rodionov DA, Weinberg Z, Breaker RR. 2010. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem Bio 17:681–685. doi: 10.1016/j.chembiol.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romine MF, Rodionov DA, Maezato Y, Anderson LN, Nandhikonda P, Rodionova IA, Carre A, Li X, Xu C, Clauss TR, Kim YM, Metz TO, Wright AT. 2017. Elucidation of roles for vitamin B12 in regulation of folate, ubiquinone, and methionine metabolism. Proc Natl Acad Sci U S A 114:E1205–E1214. doi: 10.1073/pnas.1612360114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fivian-Hughes AS, Houghton J, Davis EO. 2012. Mycobacterium tuberculosis thymidylate synthase gene thyX is essential and potentially bifunctional, while thyA deletion confers resistance to p-aminosalicylic acid. Microbiology 158:308–318. doi: 10.1099/mic.0.053983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu YM, Coates ARM. 1999. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol 181:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manganelli R, Proveddi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol 186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu YB, Morichaud Z, Perumal AS, Roquet-Baneres F, Brodolin K. 2014. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res 42:10399–10408. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA. 2015. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet 11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehira S, Shirai T, Teramoto H, Inui M, Yukawa H. 2008. Group 2 sigma factor SigB of Corynebacterium glutamicum positively regulates glucose metabolism under conditions of oxygen deprivation. Appl Environ Microbiol 74:5146–5152. doi: 10.1128/AEM.00944-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong T, Kirchhof MG, Schellhorn HE. 2008. RpoS regulation of gene expression during exponential growth of Escherichia coli K-12. Mol Genet Genomics 279:267–277. doi: 10.1007/s00438-007-0311-4. [DOI] [PubMed] [Google Scholar]
- 43.Murakami K, Ono T, Viducic D, Kayama S, Mori M, Hirota K, Nemoto K, Miyake Y. 2005. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol Lett 242:161–167. doi: 10.1016/j.femsle.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Kayama S, Murakami K, Ono T, Ushimaru M, Yamamoto A, Hirota K, Miyake Y. 2009. The role of rpoS gene and quorum-sensing system in ofloxacin tolerance in Pseudomonas aeruginosa. FEMS Microbiol Lett 298:184–192. doi: 10.1111/j.1574-6968.2009.01717.x. [DOI] [PubMed] [Google Scholar]
- 45.Bardarov S, Bardarov S Jr, Pavelka MS Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, and Jacobs WR Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG, and M. smegmatis. Microbiology 148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.