ABSTRACT
In this study, we characterize the impact of antioxidative enzymes in amphotericin B (AmB)-resistant (ATR) and rare AmB-susceptible (ATS) clinical Aspergillus terreus isolates. We elucidate expression profiles of superoxide dismutase (SOD)- and catalase (CAT)-encoding genes, enzymatic activities of SODs, and superoxide anion production and signaling pathways involved in the oxidative stress response (OSR) in ATS and ATR strains under AmB treatment conditions. We show that ATR strains possess almost doubled basal SOD activity compared to that of ATS strains and that ATR strains exhibit an enhanced OSR, with significantly higher sod2 mRNA levels and significantly increased cat transcripts in ATR strains upon AmB treatment. In particular, inhibition of SOD and CAT proteins renders resistant isolates considerably susceptible to the drug in vitro. In conclusion, this study shows that SODs and CATs are crucial for AmB resistance in A. terreus and that targeting the OSR might offer new treatment perspectives for resistant species.
KEYWORDS: amphotericin B, resistance, Aspergillus terreus, oxidative stress response, MAPK signaling, catalases, reactive oxygen species, superoxide dismutases
INTRODUCTION
Despite intensive research, neither the molecular mechanisms of nor underlying resistance traits against amphotericin B (AmB) are fully understood in detail. The main target of AmB is ergosterol in the fungal cell membrane (1, 2). The mechanism of AmB action involves pore formation in the fungal lipid bilayer, polar interactions with phospholipid groups of the membrane, and ergosterol sequestration and/or extraction (1–4). Pore formation by AmB increases the permeability for small mono- and divalent cations and larger electrolytes, thereby depleting the intracellular ion pool.
Molecular investigations of acquired AmB resistance in Candida albicans proved that the deletion of ergosterol biosynthesis genes increased the AmB MIC >3-fold due to ergosterol depletion (5). However, these deletions conferring AmB resistance resulted in reduced tolerance to external stresses (such as high temperatures), killing by neutrophils, tissue invasion, or filamentation, and the deletion strains were avirulent in a murine infection model (5).
Several studies support the model that AmB induces reactive oxygen species (ROS) in the cell, thereby exerting a major fungicidal impact rather than ergosterol sequestration (6–9). ROS, which are key substances in the process of oxidative stress, are signaling molecules that regulate many cellular functions under physiological conditions. The cellular antioxidation system, once damaged, results in excessive ROS accumulation, which causes damage of macromolecules, such as lipids, enzymes, and nucleic acids (10).
In support of the model that AmB induces ROS, clinical AmB-resistant (ATR) and rare AmB-susceptible (ATS) Aspergillus terreus isolates did not exhibit any alterations in ergosterol content, indicating that ergosterol is not the major target in the intrinsic AmB resistance of A. terreus (11). In a recent study, we illustrated that coapplication of AmB with anti- and prooxidants significantly affected AmB efficacy in an antithetic manner. Furthermore, we showed that mitochondria, the major source of intracellular ROS formation, are involved in AmB's fungicidal activity (12). Previous work described that ATR strains exhibited increased catalase activity under untreated conditions (11). Altogether, these findings underpin a crucial role of the oxidative stress response (OSR) in AmB resistance.
Therefore, in the present study, we characterized the roles of two ROS-detoxifying systems, catalases (CATs) and superoxide dismutases (SODs), in more detail at the transcriptional and functional levels. Catalases can be classified into two subgroups, i.e., mono- and bifunctional catalases. For Aspergillus fumigatus, three functional catalases have been described (13, 14): two monofunctional catalases—Cat1 and CatA—and one bifunctional catalase—the peroxidase Cat2 (14, 15). CatA is expressed in conidia, while Cat1 is found in mycelia. In Candida albicans, one catalase is expressed (16).
SODs comprise the major antioxidant defense system against superoxide anions (O2·−) and consist of two isoforms in fungi: (i) Cu/Zn-SODs and (ii) Mn/Fe-SODs. Both SOD groups require catalytic metal (Cu or Mn) for their activation (12). For Aspergillus fumigatus, SOD functions were characterized in detail by creating single- and multiple-gene deletion mutants (17). SOD1 and SOD2 displayed SOD activity, whereas no biochemical proof for SOD activity was obtained for SOD3 and SOD4.
In addition to depicting the SOD expression and activity patterns of ATS and ATR strains, we studied mitogen-activated protein kinase (MAPK) activation initiated by AmB-mediated ROS production. MAPKs are activated in response to environmental changes, upon which they modulate osmotic stress responses, cell wall biosynthesis, and mating in fungi. We demonstrate here the crucial role, in particular, of CAT expression and activity in AmB resistance of A. terreus, as in vitro coapplication of CAT inhibitors and AmB rendered ATR strains susceptible to the drug.
RESULTS
Identification of putative SODs and CATs in A. terreus and other fungal species.
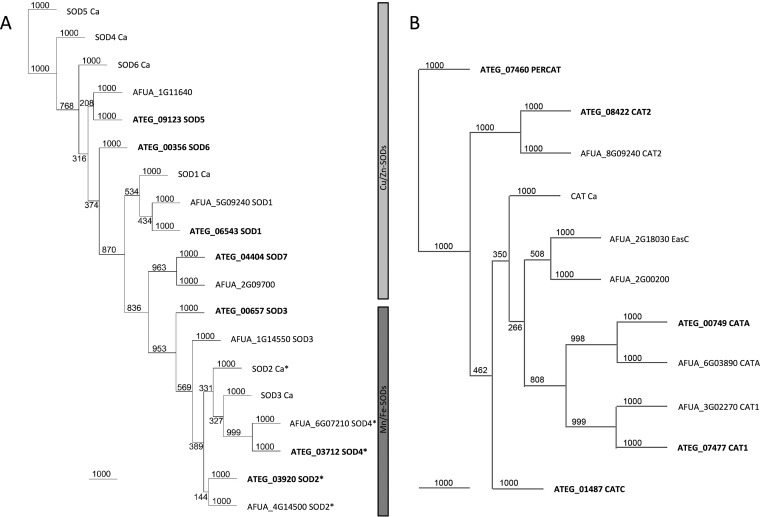
We first searched for homologs of A. fumigatus functional superoxide dismutases (SODs) and catalases (CATs) by performing InterPro (18) scans of A. terreus sequences. The in silico studies revealed that A. terreus has seven putative SOD family members: four Cu/Zn-SODs and three Mn/Fe-SODs. Similar to the Sods in A. fumigatus, we identified homologs of the previously described Sods (Sod1 to Sod4) in A. terreus, but we additionally found three other protein-encoding genes harboring Cu/Zn-SOD domains (Fig. 1A; see Table S3 in the supplemental material): ATEG_09123, termed Sod5; ATEG_00356, termed Sod6; and ATEG_04404, termed Sod7. Sod5 (ATEG_09123) shares 34.12% and 30.11% identities with the Cu/Zn-SODs Sod4 and Sod6, respectively, of C. albicans. The putative SOD proteins ATEG_09123 and ATEG_00356 share 40.5% and 41.4% identities, respectively, with AFUA_1G11640 and 34.1% identity with C. albicans Sod4. Two additional Cu/Zn-SOD proteins were detected by these in silico analyses of A. fumigatus: both are annotated as open reading frames (ORFs) and uncharacterized proteins. AFUA_2G09700 shares homology with ATEG_04404 (Sod7) (85.83% identity) and also with the superoxide dismutase chaperone Lys7/CCS1 of Saccharomyces cerevisiae (42.2% identity), required for copper incorporation in Sod1p (19–21). Phylogenetic trees of A. terreus SOD family members compared to those of C. albicans and A. fumigatus are shown in Fig. 1A and Table S3.
FIG 1.
Unrooted phylogenetic neighbor-joining tree of SOD and CAT homologs of C. albicans (Ca), A. terreus (ATEG), and A. fumigatus (AFUA). (A) C. albicans has 6, A. terreus 7, and A. fumigatus 6 SOD homologs. Proteins are denoted according to their names in the literature or according to the gene locus. Proteins marked with an asterisk harbor a mitochondrial import sequence. The two distinct subfamilies of Cu/Zn-SODs and Mn/Fe-SODs are highlighted with bars on the right. (B) Phylogenetic relationships of C. albicans catalase and the catalases and bifunctional catalases of A. terreus and A. fumigatus. Bootstrap values are depicted at the nodes, and branch lengths are shown.
In A. terreus, CAT1, CAT2, and CATA homologs were identified together with two other putative proteins containing monofunctional catalase domains (ATEG_01487 and ATEG_07460). Comparing the gene organization of the escA catalase gene-containing fumigaclavine C biosynthetic gene cluster of A. fumigatus (22), a homologous gene cluster is present in A. terreus, encoding the putative catalase ATEG_01487. ATEG_07460 is also annotated as a putative peroxisomal catalase and forms an outgroup in the phylogenetic tree (Fig. 1B; Table S4).
SOD- and CAT-encoding genes are transcriptionally upregulated by AmB treatment.
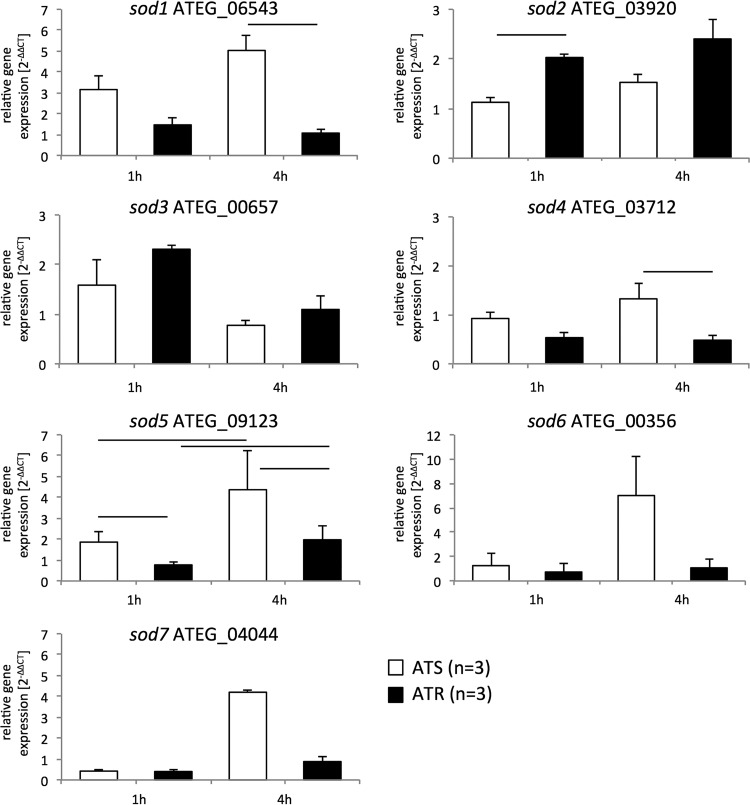
The transcriptional expression profiles of SODs and CATs of A. terreus were investigated after 1 h and 4 h of AmB treatment (1 μg/ml) of ATS (n = 3) and ATR (n = 3) strains (Fig. 2; Table S5).
FIG 2.
AmB treatment differentially affects expression of SOD-encoding genes in ATS and ATR strains. Cultures (ATS strains, white columns [n = 3]; ATR strains, black columns [n = 3]) were preincubated in RPMI 1640 liquid medium for 24 h at 37°C and 200 rpm before AmB (1 μg/ml) was added for 1 h and 4 h. Gene expression during AmB treatment was determined by RT-qPCR, and expression ratios were normalized according to the 2−ΔΔCT method, using beta-tubulin as a housekeeping gene and the untreated condition as a reference. Data are means and standard deviations for three independent experiments with ATS strains (white; 3 isolates per experiment) and ATR strains (black; 3 isolates per experiment). Lines over columns display significant changes between the respective columns (P = 0.05; two-way ANOVA and the Fisher LSD test).
Upon AmB exposure, sod1, sod4, sod5, sod6, and sod7 transcript levels increased in ATS strains, in a time-dependent manner, and were found to be highest after 4 h (Fig. 2). In contrast, ATR strains were able to better cope with AmB treatment and downmodulated these sod genes to nearly basal levels after 4 h (Fig. 2). sod3 mRNA expression levels did not differ between the resistant and susceptible strains, independent of the time point (Fig. 2), while sod2 levels were found to be significantly higher in ATR strains than in ATS strains after 1 h of AmB treatment and remained elevated 4 h after AmB addition (Fig. 2).
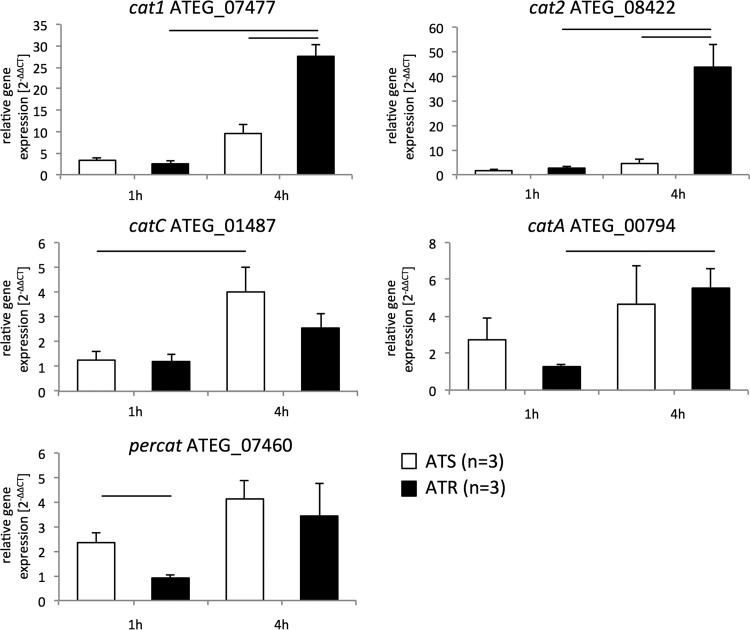
A different picture was observed respecting catalase expression profiles. cat1, cat2, and catA were significantly upregulated in ATR strains after 4 h of AmB exposure (Fig. 3). No significant upregulation of these cat transcripts was observed in ATS strains at this time point (Fig. 3). In ATS strains, only catC showed a significant increase after 4 h of AmB treatment compared to the level at the 1-h time point, but expression of catC was not significantly higher than that detected in ATR strains after 4 h (Fig. 3).
FIG 3.
AmB treatment increases transcripts of CAT-encoding genes in ATS and ATR strains. Cultures (for ATS strains, n = 3; for ATR strains, n = 3) were incubated in RPMI 1640 liquid medium for 24 h at 37°C and 200 rpm. AmB (1 μg/ml) was added for 1 h and 4 h. Gene expression during AmB treatment was determined by real-time RT-qPCR, and expression was normalized to that of beta-tubulin according to the 2−ΔΔCT method. Data are presented as means ± standard deviations for three independent experiments with ATS (white; n = 3) and ATR (black; n = 3) isolates, with technical duplicates. Lines over columns display significant changes between the respective columns (P = 0.05; two-way ANOVA and the Fisher LSD test).
This differential regulation of sod and cat genes in ATS and ATR strains points to an important regulatory function of cat genes in particular regarding AmB resistance mechanisms.
Distinct SODs are induced by AmB treatment in ATS and ATR strains.
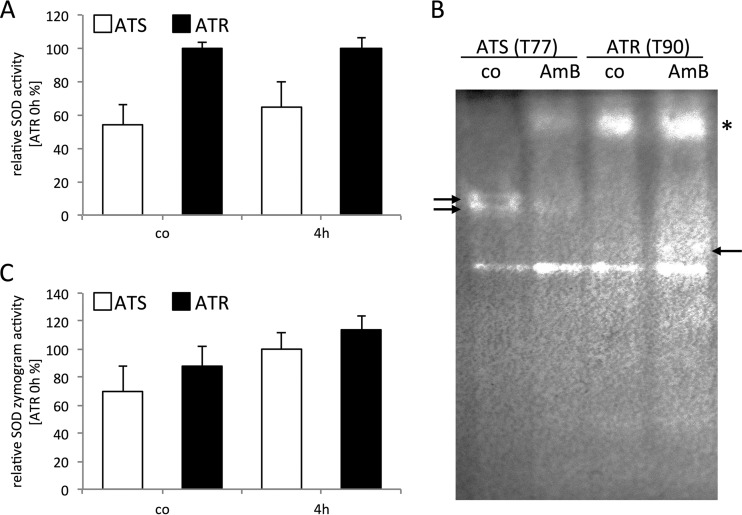
To further characterize the role of SODs in ATS and ATR strains upon AmB treatment, we next analyzed their enzymatic activity via two different assays: we used a commercially available SOD determination kit, and we examined SOD activity by using zymograms. Determining the enzymatic activity in protein extracts under control conditions (time zero) revealed that basal SOD activity was nearly twice as high in ATR strains (85% ± 3%) as that in ATS strains (46% ± 10%) (Fig. 4A). After 4 h of AmB exposure, ATR strains showed constant SOD activity levels, whereas ATS strains showed increased enzymatic activity (∼1.20-fold [to 55% ± 13%]). SOD zymograms showed a similar overall SOD activity pattern (Fig. 4B and C). Substantial differences in distinct Sod proteins were observed between ATS and ATR strains (Fig. 4B). One Mn/Fe-SOD displayed activity in both groups (Fig. 4B, lowest band), and this was increased upon AmB treatment. ATS strains exhibited additional bands (marked with arrows at the left), which decreased with AmB treatment. AmB caused induction of a high-molecular-weight SOD isoform in ATS strains which already existed in ATR strains under control conditions (Fig. 4B, asterisk). In ATR strains, an additional band appeared after AmB treatment (Fig. 4B, arrow at the right).
FIG 4.
AmB increases SOD activity and induces distinct SOD responses in ATR and ATS strains. (A) ATR strains (black bars; n = 3) exhibited higher total SOD activity than that of ATS strains (white bars; n = 3) under basal conditions and with AmB treatment (4 h, 1 μg/ml AmB). AmB caused an increase in SOD activity in ATS strains, while ATR SOD activity remained almost constant. SOD activity was determined with a SOD detection kit (Sigma-Aldrich) in these experiments. (B) Representative SOD zymogram of ATS (isolate T77) and ATR (isolate T90) strains, displaying distinct SOD activities under control conditions (co) for the ATS and ATR strains, which were modulated by AmB treatment (1 μg/ml, 4 h). Eighty-microgram aliquots of protein were loaded onto 10% native polyacrylamide gels, and gels were stained for superoxide dismutase activity. The asterisk indicates Cu/Zn-SOD, and the arrows highlight altered protein patterns seen with AmB exposure. (C) SOD zymograms of ATS and ATR strains (n = 3 per group) were evaluated with ImageJ and normalized to those of ATR strains under basal conditions. For all experiments shown, strains were cultured in RPMI 1640 medium at 37°C and 200 rpm. AmB (1 μg/ml) was added for 4 h.
Overall, these results indicate that ATR strains display a higher basal SOD activity and that AmB treatment activates differential SOD isoforms in ATR and ATS strains.
AmB exaggerates superoxide anion generation in ATS mitochondria.
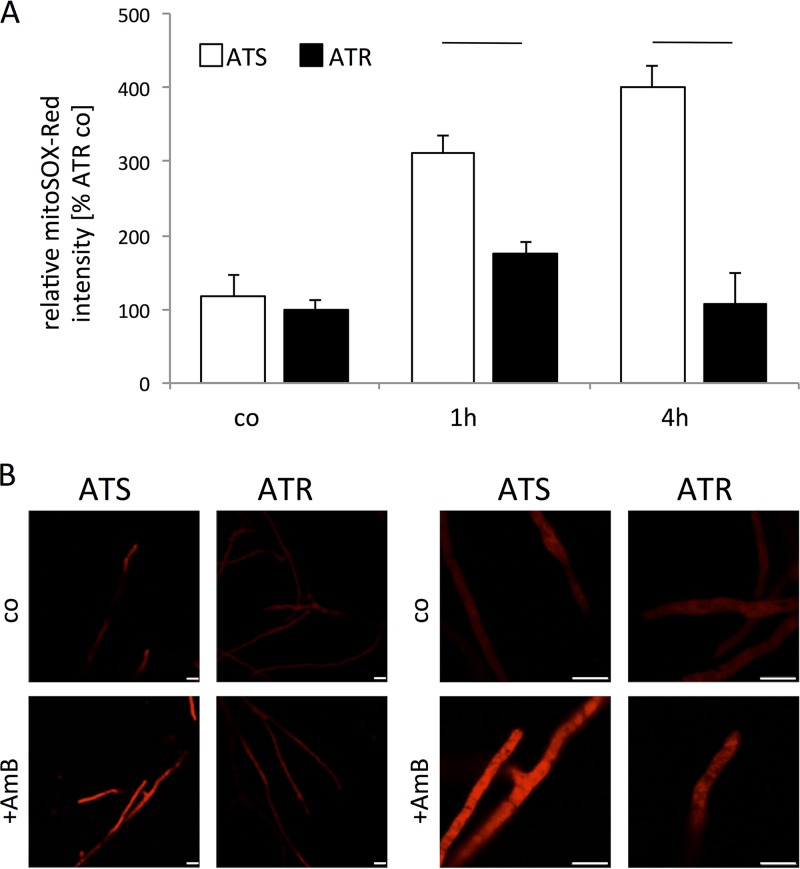
Due to the observed differences in SOD expression and activity, we next examined mitochondrial superoxide anion generation upon AmB treatment in ATS and ATR strains by using the mitochondrial superoxide anion indicator MitoSOX-Red. Under control conditions, superoxide anion levels were similar in ATS and ATR strains (Fig. 5). After 1 h of AmB exposure, superoxide anion production was nearly twice as high in ATS strains, and it was enhanced nearly 4-fold after 4 h (Fig. 5A). In contrast, mitochondrial superoxide anion production decreased to control levels in ATR strains after 4 h of AmB exposure (Fig. 5A). Also, confocal microscopic analyses revealed that ATS strains produced more superoxide anions after AmB treatment than ATR strains did (Fig. 5B).
FIG 5.
AmB induces high superoxide levels in ATS mitochondria. (A) Superoxide anions in mitochondria were measured using the mitochondrial superoxide anion indicator MitoSOX-Red. Strains (ATS, white columns [n = 3]; ATR, black columns [n = 3]) were cultured in 100 μl RPMI 1640 medium without phenol red (105 conidia/ml) for 18 h before AmB (1 μg/ml) was added for 1 h and 4 h. Fifteen minutes before measurements, 5 μM MitoSOX-Red was added to each sample. Fluorescence was detected in a microplate reader at 580 nm after excitation at 510 nm. Experiments were performed in triplicate and repeated thrice. Data were normalized to those for ATR strains under untreated conditions (control [co]) and are presented as means and standard deviations. AmB treatment resulted in increasing superoxide anion levels in ATS mitochondria, while in ATR mitochondria only slight increases were observed at the early time point. Lines over columns display significant changes between the respective columns (P = 0.05; two-way ANOVA and the Fisher LSD test). (B) Representative confocal microscopic images of AmB-treated (1 μg/ml, 4 h) and MitoSOX-Red-stained specimens of ATS (T77) and ATR (T90) strains.
These findings indicate that AmB treatment results in significantly higher mitochondrial superoxide anion levels in ATS strains, with increased mitochondrial superoxide anion accumulation. The superoxide anion reduction in ATR strains over 4 h of AmB exposure illustrated the ability of resistant isolates to reduce AmB-mediated superoxide anion accumulation in mitochondria.
Inhibition of SODs and CATs increases the AmB susceptibility of resistant strains.
We next performed blocking experiments using SOD and CAT inhibitors (N-N′-diethyldithiocarbamate [DDC] and 3-amino-1,2,4-triazole [3-AT], respectively) (23–25) to link enzymatic activity to AmB resistance. First, we determined concentrations of 3-AT and DDC that did not affect radial growth of the fungi (Table 1; Table S2). ATR strains displayed a more pronounced growth reduction when challenged with the CAT inhibitor 3-AT, while ATS strains were more sensitive to the SOD inhibitor DDC (Table S6). For susceptibility testing, 2 mM 3-AT and 12.5 μM DDC were found to be optimal regarding fungal growth. The MIC of ATR strains cotreated with AmB and DDC was significantly reduced (Table 1) (AmB MIC, 32.00 ± 0.00 mg/liter; AmB-DDC MIC, 1.25 ± 0.36 mg/liter). AmB-DDC cotreatment did not exert any significant effect on ATS strains (Table 1) (AmB MIC, 0.23 ± 0.03 mg/liter; AmB-DDC MIC, 0.11 ± 0.02 mg/liter). Consistent with the stronger growth inhibition of ATR strains with the CAT blocker 3-AT, an even larger reduction in the AmB MIC was observed upon AmB–3-AT cotreatment (Table 1) (AmB MIC, 32.00 ± 0.00 mg/liter; AmB–3-AT MIC, 0.44 ± 0.08 mg/liter). However, no changes in MICs were observed for coapplying voriconazole or caspofungin with DDC or 3-AT (data not shown). As proof of principle that CAT and SOD inhibition increases AmB susceptibility, clinical isolates of A. flavus were cotreated with DDC or 3-AT and AmB. A. flavus clinical isolates (MICs, >2 μg/ml for AmB) also showed a decrease in AmB MICs when they were cotreated with 3-AT or DDC, albeit a smaller one than that seen with ATR strains.
TABLE 1.
Effects of 3-AT and DDC on radial growth and AmB susceptibility in ATS and ATR strains and A. flavus strainsa
| Inhibitor | Radial growth (% of control level) |
AmB MIC (mg/liter) |
||||
|---|---|---|---|---|---|---|
| ATS strains | ATR strains | A. flavus strains | ATS strains | ATR strains | A. flavus strains | |
| Control | 100.00 ± 8.18 | 100.00 ± 6.90 | 100.00 ± 5.96 | 0.23 ± 0.03 | 32.00 ± 0.00 | 24.33 ± 13.28 |
| 3-AT (2 mM) | 85.49 ± 18.44 | 79.31 ± 18.43 | 89.92 ± 1.68 | 0.22 ± 0.04 | 0.57 ± 0.26 | 13.33 ± 2.31 |
| DCC (12.5 μM) | 94.54 ± 3.35 | 100.00 ± 4.83 | 98.34 ± 3.73 | 0.11 ± 0.02 | 1.20 ± 0.36 | 4.67 ± 3.06 |
For radial growth assays, 100 conidia were point inoculated onto RPMI plates supplemented with the respective inhibitors and incubated at 37°C for 48 h. For MIC determination, conidial suspensions with 1 × 106 conidia/ml were streaked onto RPMI plates supplemented with the respective inhibitors before AmB Etest strips were applied. Experiments (for ATS strains, n = 3; for ATR strains, n = 3; and for A. flavus strains, n = 3) were always performed with freshly harvested spores and repeated at least three times per strain.
These results highlight the importance of antioxidative enzymes for AmB susceptibility in A. terreus and emphasize that CAT activity especially is indispensable for AmB resistance in A. terreus.
AmB exposure causes differential MAPK activation in ATS and ATR strains.
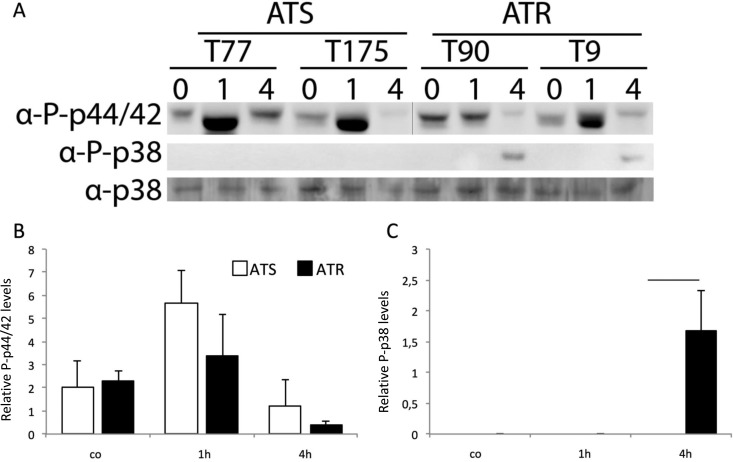
MAPK signaling is involved in oxidative stress responses in A. fumigatus (26, 27). Therefore, we also studied phosphorylation of the MpkA (p44/p42 MAPK in humans) and SakA/Hog1 (p38 MAPK in humans) orthologs in ATS and ATR strains upon AmB exposure over time (Fig. 6). While MpkA was highly activated in ATS strains upon 1 h of AmB treatment (Fig. 6A, upper panel, and B), SakA/Hog1 phosphorylation was induced only in AmB-treated ATR strains after 4 h (Fig. 6A, middle panel, and C). ATR strains displayed a higher basal MpkA phosphorylation level, which was not affected upon AmB treatment after 1 h and was downmodulated after 4 h (Fig. 6A, upper panel). SakA/Hog1 protein levels were used as loading controls (Fig. 6A, lower panel). These findings illustrate that MAPKs are alternatively regulated in ATS and ATR strains upon AmB treatment and that basal MpkA phosphorylation levels are elevated in ATR strains.
FIG 6.
Immunoblot analysis of ATS and ATR strains treated with AmB reveals distinct phosphorylation statuses of SakA and MpkA. (A) Western blot analysis of phosphorylated MpkA (using an anti-phospho-p44/42 antibody) and SakA (using an anti-phospho-p38 antibody) in ATS and ATR strains treated with AmB (1 μg/ml) for 1 and 4 h. The anti-p38 antibody was used as a standard loading control in all experiments. The vertical line in the blot for phosphorylated MpkA indicates splicing of the image to give the same order of samples. (B and C) Summaries of two independent Western blot experiments (as shown in panel A) with three ATS and three ATR strains. Expression levels of phosphorylated MpkA and SakA were normalized to the level of SakA. Lines over columns display significant changes between the respective columns (P = 0.01; two-way ANOVA and the Fisher LSD test).
DISCUSSION
A. terreus has intrinsic resistance to AmB, and only a few susceptible isolates are known so far (12, 28). We and others recently depicted that AmB induces oxidative stress in fungal cells rather than targeting ergosterol content in the fungal plasma membrane. In this study, the roles of ROS-detoxifying SODs and CATs in AmB resistance were investigated in A. terreus. Little is known of A. terreus SODs and CATs, and these enzymes have not been studied in detail regarding their relevance to AmB resistance (29–32). The cytosolic Cu/Zn-SOD1 is found at high levels in conidia and germinated conidia of A. fumigatus. We illustrated here that upon AmB treatment, the sod1, sod4, sod5, sod6, and sod7 genes were elevated in ATS but not ATR strains.
At the transcriptional level, sod4 expression displayed opposite regulation in the two groups. Transcripts increased in ATS strains but decreased in ATR strains and resulted in a significant difference at the 4-h time point; this antithetic regulation of sod4 underlines the distinct mitochondrial functions in ATS and ATR strains (12). Also, transcription of sod5, sod6, and sod7 was strongly enhanced upon AmB treatment in ATS strains, while ATR strains downregulated these genes and reached control levels after 4 h.
The mitochondrial Mn/Fe-SOD, encoded by sod2 in A. fumigatus, is strongly expressed in conidia and at later growth stages. The SOD2 homolog of A. terreus (88.55% identity) also contains a mitochondrial import sequence, and transcription of sod2 is induced mainly in ATR strains by AmB treatment. Sod4 was proven to be essential in A. fumigatus. Similar to Sod2, the Sod4 ortholog contains a mitochondrial import sequence.
The transcriptional activation of CAT-encoding genes was more pronounced than that of SOD-encoding genes in ATR strains after AmB treatment. CAT1 mRNA expression was significantly upregulated in ATR strains after 4 h of AmB exposure compared to that at the 1-h time point or that in ATS strains at 4 h, indicating that ATR strains display a more pronounced OSR at the transcriptional level. The active protein was recently investigated and characterized for an A. terreus environmental isolate from oil fields in India (30, 33). Significantly higher transcription of CAT2, the bifunctional catalase-peroxidase, was also induced in ATR strains by 4 h of treatment with AmB. CAT2 was also identified in a proteomics study of potato tuber colonization by A. terreus (32) and displayed a significant induction in the plant host environment. The AFUA_2G18030 gene, termed easC, was identified as a gene of the ergot alkaloid gene cluster that is required for chanoclavine I biosynthesis (22), with confirmed in vitro catalase activity required for the synthesis of chanoclavine I, rather than being involved in the OSR. This protein harbors a type I peroxisomal targeting sequence (PTS I) at the carboxy terminus (ARL) (34). A similar PTS I sequence also exists at the carboxy terminus of ATEG_01487 (CATC). catC transcripts significantly increased at the 4-h time point solely in ATS strains. The ortholog of CATA is known to be a conidium-specific CAT in A. fumigatus. catA transcripts were induced by AmB treatment in both ATS and ATR strains. Since A. terreus is known to produce aleuroconidia (35), arising from hyphae in liquid cultures, it is not surprising that catA transcripts were detected in both strains in liquid cultures. The last putative catalase identified by InterPro searches was annotated as a putative peroxisomal catalase according to UniProtKB/TrEMBL (accession no. Q0CFS4). The predicted protein contains a CAT domain from residues 23 to 369. However, the peroxisomal CAT annotation is questionable, for two reasons: a PTS I motif is missing at the carboxy terminus, and no type II PTS (PTS II) [(R/K)(L/I)xxxxx(H/Q)L] is present at the amino terminus. A type II PTS was solely found at the end of the catalase domain, at position 321.
Taken together, the data here show that all tested A. terreus clinical isolates—three susceptible and three resistant—exhibited distinct basal SOD and CAT expression levels and different patterns upon AmB treatment. Upon AmB exposure, transcription of the cytosolic Cu/Zn-SOD1 was significantly upregulated in ATS strains, while in ATR strains the mitochondrial Mn/Fe-SOD2 level was significantly increased. Levels of the cytoplasmic Mn/Fe-SOD3 did not greatly differ over time between ATS and ATR strains after AmB addition. Interestingly, the ortholog of the essential mitochondrial Mn/Fe-SOD4 enzyme illustrated an antithetic mRNA expression pattern, with AmB-induced upregulation in ATS strains and downregulation in ATR strains, emphasizing the role of mitochondrial functions in AmB resistance.
In turn, the catalase genes cat1, cat2, and catA were significantly upregulated in ATR strains after 4 h of AmB treatment. Since CAT1 and CAT2 were illustrated to be involved in H2O2 degradation in A. fumigatus (14), we suggest that cat1 and cat2 orthologs in A. terreus fulfill these major functions as well.
In our analysis to determine the consequences of altered transcription patterns in ATS and ATR strains, SOD activity assays revealed that ATS strains displayed lower SOD activity. Interestingly, Cu/Zn-SOD1 transcriptional expression was higher in ATS strains, but this increase was observed regarding its protein activity. In a recent study, we examined the effects of pro- and antioxidants on CAT and SOD activities, and after blocking with DDC, we identified the high-molecular-weight band to constitute the Cu/Zn-SOD complex (12). Holdom et al. biochemically characterized the Cu/Zn-SOD1 of A. terreus and compared it to orthologs of related species (29). In that study, only A. fumigatus SODs retained thermostability at 37°C, while Aspergillus nidulans and A. terreus Cu/Zn-SODs significantly lost activity at this temperature, and all but A. fumigatus Cu/Zn-SODs were inhibited by chelators, such as EDTA. The decreased Cu/Zn-SOD activities in ATS strains are supposed to result from inefficiently translated transcripts, from missing functional SOD homomultimers, or from missing metal cofactors. We highlighted the differential SOD activity between ATS and ATR strains already under control conditions—in zymograms, ATS strains displayed two additional bands with Mn/Fe-SOD activity which were not detected for ATR strains. Upon AmB treatment, the activity of this Mn/Fe-SOD was also decreased in ATS strains, while in ATR strains another SOD activity band appeared. Consistent with decreased SOD activity in ATS strains, superoxide anions were more abundant. Elevated mitochondrial superoxide anion levels were detected in ATS strains under basal conditions and significantly increased upon AmB exposure. ATR strains, in contrast, showed moderate accumulation of superoxide anions due to efficient detoxification of these reactive oxygen species by activation of specific Sod proteins.
To evaluate the association of SOD activity and AmB resistance, we used DDC, an inhibitor of Cu/Zn-SODs. DDC is a chelating agent which is used to mobilize metals and is also used as an antidote for nickel and cadmium poisoning. DDC is the main metabolite of disulfiram, a drug discovered in the 1920s as an antialcoholism drug inducing an acute sensitivity to ethanol. Disulfiram has been used orally for the clinical treatment of alcoholism since 1949. Additionally, DDC displays growth-inhibitory effects against bacteria, viruses, fungi, and protozoa (36–40). Upon DDC-AmB combinatorial treatment, ATR strains were rendered susceptible to AmB, while slight changes were observed for ATS strains, which are already susceptible to AmB treatment alone. Aspergillus flavus isolates (MICs of >2 μg/ml for AmB) also showed a significant decrease in MIC values upon AmB-DDC combinatorial treatment.
The use of the catalase inhibitor 3-AT in combination with AmB resulted in highly susceptible ATR strains, indicating the essential role of H2O2-detoxifying CATs with regard to overcoming AmB-induced ROS accumulation. Also, A. flavus clinical isolates showed increased susceptibility when 3-AT was applied in combination with AmB. 3-AT not only inhibits catalase activity but also inhibits histidine biosynthesis by inhibiting the imidazole-phosphate dehydratase. For this reason and the anticipated carcinogenicity of 3-AT found in animal experiments, 3-AT is disqualified for clinical applications.
Interestingly, MAPK signaling showed significant differences between ATS and ATR strains. The major physiological activities assigned to MAPKs in fungi are cell wall biosynthesis, osmoregulation, mating, carbon utilization, stress responses, and host invasion (41, 42). Similar to A. fumigatus, A. terreus encodes four putative terminal MAPKs. MpkA is central in regulating cell wall integrity and initiates responses to cell wall-disturbing agents and ROS in A. fumigatus (42, 43). So far, MpkA activation has not been investigated in A. terreus. SakA/Hog1 proteins from Aspergillus spp. share similarities with Hog1 of Saccharomyces cerevisiae, the major regulator of the high-osmolarity glycerol (HOG) pathway (44). SakA and MpkA were reported to play a role in stress related to caspofungin, a β-glucan synthase inhibitor (45, 46), and to be initiated in S. cerevisiae under stress conditions (47). In order to elucidate the roles of SakA/Hog1 and MpkA during AmB-mediated stress, we determined their phosphorylation states over time. Under control conditions, the MpkA phosphorylation levels were similar in ATS and ATR strains. MpkA phosphorylation was induced in ATS strains after 1 h of treatment with AmB, to levels 3-fold higher than those under control conditions. In ATR strains, MpkA phosphorylation levels were only 1.3-fold higher in AmB-exposed ATR strains than in the untreated controls. MpkA decreased to basal levels in ATS and ATR strains after 4 h of AmB treatment. In ATR strains, this decrease in MpkA at 4 h of AmB exposure was accompanied by phosphorylation of SakA/Hog1, indicating a role for SakA/Hog1 as a salvage pathway. In ATR strains, differential SOD activation was initiated relative to that in ATS strains, and fewer SOD anions were detected as a consequence. This inverse regulation of MpkA and SakA/Hog1 implies a regulatory cross talk of MAPKs in ATR strains which is lacking in ATS strains. A recent MAPK network modeling study investigating caspofungin stress in A. fumigatus demonstrated that HOG pathway activation occurs either as a consequence of MpkA repression or by caspofungin-induced osmotic stress (45). Our results support the first assumption. Studies of yeast link HOG1 activity to oxidative stress, and HOG1-dependent genes, such as cat1, are transcriptionally increased in AmB-resistant isolates (48). In support of HOG-mediated activation of CATs, transcripts of cat1 and cat2 increased hand in hand with SakA/Hog1 phosphorylation in ATR strains. Furthermore, in C. albicans, HOG depended on activation by HSP90, and HOG was also activated by osmotic stress (49). We earlier demonstrated that HSP90 activity is important for AmB resistance in A. terreus and that ATR strains exhibit increased CAT activity. Moreover, we recently showed that ROS are involved in AmB fungicidal activity (12). This study complements our recent studies, provides additional insights into the AmB resistance mechanisms of A. terreus, and sheds light on the differential OSR of ATS and ATR strains. Tackling SODs and CATs renders resistant isolates susceptible to AmB treatment. Thereby, targeting SOD activity with DDC, an inhibitor that displays a good safety profile and is already in clinical use, might provide novel approaches to treating AmB-resistant fungal infections.
MATERIALS AND METHODS
Fungal strains and growth conditions.
In order to directly compare the AmB responses in ATS and ATR strains, clinical isolates of AmB-susceptible Aspergillus terreus from different geographic origins (ATS strains) (n = 3; T77, T164, and T175) and of AmB-resistant Aspergillus terreus (ATR strains) (n = 3; T5, T9, and T90) were used for all experiments (see Table S1 in the supplemental material), unless otherwise indicated. Additionally, for antifungal susceptibility testing, three A. flavus clinical isolates (Afl20, Afl22, and Afl25) were included. All strains were equally susceptible to azole antifungals. Fungi were grown on Aspergillus complete medium (ACM) (recipe given in reference 50) for 7 days at 37°C. Conidia were harvested, filtered through a 45-μm cell strainer (BD), and counted with a hemocytometer. For RNA and protein analyses, 1 × 106 conidia/ml of the respective strains were inoculated into 200 ml RPMI 1640 medium and incubated at 37°C and 200 rpm. After 24 h of incubation, liquid cultures were treated with 1 μg/ml AmB deoxycholate (Bristol Meyer Squibb), which reflects physiologic concentrations attainable in patients undergoing therapy.
In silico analyses of CAT and SOD genes in the A. terreus genome.
The following databases and assemblies were used to obtain protein and DNA sequences for the respective species: for Aspergillus terreus, the BROAD Institute database (http://www.broadinstitute.org/fetgoat/index.html; accessed 11 November 2014) and the Central Aspergillus Data REpository; for Aspergillus fumigatus, the Central Aspergillus Data REpository and the Aspergillus genome database (51; http://aspgd.org [accessed 14 November 2014]); and for C. albicans, the Candida genome database (52; http://www.candidagenome.org). The comparative fungal genomics platform CFGP2.0 (53) was used to query the conserved Mn/Fe-SOD and Cu/Zn-SOD families (InterPro accession numbers IPR001189 and IPR001424, respectively). In order to retrieve mono- and bifunctional catalase genes from the A. terreus genome, searches were performed with the sequences under InterPro accession numbers IPR018028 and IPR000763 (54). Multiple-protein-sequence alignments were performed with ClustalW and the Pasteur bioweb2.0 database (55). Based on this alignment, an unrooted phylogenetic tree was constructed using phylip3.67 and the Kimura formula distance model. The confidence level was analyzed using bootstrapping with 1,000 iterations. Protein identities were calculated with Clustal Omega, using default parameters (http://www.ebi.ac.uk/Tools/msa/clustalo; accessed 14 September 2016).
SOD and CAT inhibitor preparation and growth assays.
For inhibition of catalases, 3-amino-1,2,4-triazole (3-AT) was used, and for inhibition of Cu/Zn-SODs, N-N′-diethyldithiocarbamate (DDC) was used. Both chemicals were obtained from Sigma-Aldrich, and stocks were prepared in distilled water. For growth assays, the inhibitors were diluted and used to supplement RPMI 1640 plates. Five-microliter aliquots of spore suspensions (2 × 104 spores per ml) were used for point inoculation, and radial growth was monitored for 48 h at 37°C. RPMI 1640 agar plates without inhibitors served as controls. For further combinatorial susceptibility testing, inhibitor concentrations were selected such that radial growth was compromised less than 40% with the respective inhibitor alone.
Antifungal susceptibility testing.
A. terreus strains displaying AmB MICs of ≤1 mg/liter (n = 3) and >2 mg/liter (n = 3) were categorized as ATS and ATR strains, respectively (18, 56) (Table S1). The impacts of SOD and CAT inhibitors on AmB efficacy were evaluated by Etest (bioMérieux) on RPMI 1640 plates supplemented with the respective inhibitors as indicated (3-AT, 2 mM; DDC, 12.5 μM). MICs were assayed and compared to MICs obtained from RPMI plates with no supplementation (57, 58).
Extraction of nucleic acids, primer design, and RT-qPCR analyses.
RNA isolation of homogenized mycelia was performed with Tri reagent (Sigma-Aldrich) as previously described (12). Sequences for A. terreus SOD and CAT genes were retrieved from the Central Aspergillus Data REpository (51). Primers were designed to span exon-intron borders, and gene identities and primer sequences are listed in Table S2. Reverse transcription-quantitative PCR (RT-qPCR) was performed with total RNA by using SsoFast EvaGreen Supermix (Bio-Rad) and the reverse transcriptase qScript (Quanta Bioscience) as described before (12, 50). Expression levels were calculated according to the 2−ΔΔCT method (56).
Mitochondrial superoxide anion quantification.
Fungal cultures (105 conidia/ml) were grown at 37°C in black, clear-bottomed 96-well culture plates (Greiner). After 18 h, cultures were treated with AmB and incubated for an additional 4 h. All samples were stained with 5 μM MitoSOX-Red for 15 min at 37°C before fluorescence measurements. Samples were excited at 510 nm, and emission was detected at 580 nm by use of a fluorescence plate reader (Tecan, Austria). Values are presented as relative fluorescence units (RFU). Confocal microscopy was performed on a Leica SP5 confocal microscope equipped with a glycerol immersion objective (63×). Fungi (104 conidia/ml) were incubated in petri dishes on coverslips in RPMI medium without phenol red for 18 h at 37°C. AmB (1 μg/ml) was added to the respective samples for 4 h in the dark at 37°C. MitoSOX-Red (5 μM) was used to monitor mitochondrial superoxide anion production. Samples were incubated with MitoSOX-Red for 15 min, and then the coverslips were washed three times with phosphate-buffered saline (PBS) and mounted in Mowiol (Sigma-Aldrich).
SOD activity and SOD zymograms.
To determine SOD activity in ATR and ATS strains, a SOD activity determination kit (Sigma-Aldrich) was used according to the manufacturer's instructions. The relative SOD activity (%) was calculated as specified by the manufacturer. SOD zymograms were created as specified previously (12).
Western blot analysis.
Protein extraction and immunoblot analyses were performed as previously described by Blatzer et al. (50). Briefly, 40 μg protein was loaded into a 12% SDS-PAGE gel and then transferred to a nitrocellulose membrane (Bio-Rad). Anti-p38, anti-phospho-p44/42 MAPK (extracellular signaling-regulated kinase 1/2 [ERK1/2]) (Thr202/Tyr204), and anti-phospho-p38 MAPK (Thr180/Tyr182) antibodies (Cell Signaling Technologies) were used to detect the SakA ortholog and phosphorylated forms of the MpkA (ATEG_03252) and SakA (ATEG_00489) orthologs of A. terreus, respectively. After overnight incubation with the primary antibodies in Tris-buffered saline with Tween 20 (TBS-T)–3% bovine serum albumin (BSA), horseradish peroxidase (HRP)-conjugated secondary antibodies were used. Blots were visualized with an ImageQuant Las 4000 machine (GE Healthcare Life Sciences).
Statistical analysis.
Results are expressed as means ± standard deviations for n samples, obtained from at least three independent experiments. Statistical analyses were performed by two-way analysis of variance (ANOVA) and the Fisher least significant difference (LSD) test. P values of <0.05 and <0.01 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. M. Tortorano and D. Stevens for providing the AmB-susceptible A. terreus strains.
This work was supported by the Austrian Science Fund (FWF) (project number W1253-B24 for the doctoral program HOROS [C.L.-F.] and grant I 661 to C.L.-F.) and the Christian Doppler (CD) Laboratory for Invasive Fungal Infections (C.L.-F.). Additional support came from the Tiroler Wissenschaftsfonds (grant 2012-1-19 to E.J.) and the Medical University of Innsbruck (MUI start grant 2012032004 to M.B.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00670-17.
REFERENCES
- 1.Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, Nieuwkoop AJ, Comellas G, Maryum N, Wang S, Uno BE, Wildeman EL, Gonen T, Rienstra CM, Burke MD. 2014. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol 10:400–406. doi: 10.1038/nchembio.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palacios DS, Dailey I, Siebert DM, Wilcock BC, Burke MD. 2011. Synthesis-enabled functional group deletions reveal key underpinnings of amphotericin B ion channel and antifungal activities. Proc Natl Acad Sci U S A 108:6733–6738. doi: 10.1073/pnas.1015023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, Burke MD. 2012. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci U S A 109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palacios DS, Anderson TM, Burke MD. 2007. A post-PKS oxidation of the amphotericin B skeleton predicted to be critical for channel formation is not required for potent antifungal activity. J Am Chem Soc 129:13804–13805. doi: 10.1021/ja075739o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. 2013. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol 11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol-Anderson M, Sligh JE Jr, Elberg S, Brajtburg J, Kobayashi GS, Medoff G. 1988. Role of cell defense against oxidative damage in the resistance of Candida albicans to the killing effect of amphotericin B. Antimicrob Agents Chemother 32:702–705. doi: 10.1128/AAC.32.5.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokol-Anderson ML, Brajtburg J, Medoff G. 1986. Amphotericin B-induced oxidative damage and killing of Candida albicans. J Infect Dis 154:76–83. doi: 10.1093/infdis/154.1.76. [DOI] [PubMed] [Google Scholar]
- 8.Sangalli-Leite F, Scorzoni L, Mesa-Arango AC, Casas C, Herrero E, Gianinni MJ, Rodriguez-Tudela JL, Cuenca-Estrella M, Zaragoza O. 2011. Amphotericin B mediates killing in Cryptococcus neoformans through the induction of a strong oxidative burst. Microbes Infect 13:457–467. doi: 10.1016/j.micinf.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Mesa-Arango AC, Trevijano-Contador N, Roman E, Sanchez-Fresneda R, Casas C, Herrero E, Arguelles JC, Pla J, Cuenca-Estrella M, Zaragoza O. 2014. The production of reactive oxygen species is a universal action mechanism of amphotericin B against pathogenic yeasts and contributes to the fungicidal effect of this drug. Antimicrob Agents Chemother 58:6627–6638. doi: 10.1128/AAC.03570-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiseman H, Halliwell B. 1996. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem J 313:17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speth C, Blum G, Hagleitner M, Hortnagl C, Pfaller K, Posch B, Ott HW, Wurzner R, Lass-Florl C, Rambach G. 2013. Virulence and thrombocyte affectation of two Aspergillus terreus isolates differing in amphotericin B susceptibility. Med Microbiol Immunol 202:379–389. doi: 10.1007/s00430-013-0300-7. [DOI] [PubMed] [Google Scholar]
- 12.Blatzer M, Jukic E, Posch W, Schopf B, Binder U, Steger M, Blum G, Hackl H, Gnaiger E, Lass-Flörl C, Wilflingseder D. 2015. Amphotericin B resistance in Aspergillus terreus is overpowered by coapplication of pro-oxidants. Antioxid Redox Signal 23:1424–1438. doi: 10.1089/ars.2014.6220. [DOI] [PubMed] [Google Scholar]
- 13.Shibuya K, Paris S, Ando T, Nakayama H, Hatori T, Latge JP. 2006. Catalases of Aspergillus fumigatus and inflammation in aspergillosis. Nihon Ishinkin Gakkai Zasshi 47:249–255. doi: 10.3314/jjmm.47.249. [DOI] [PubMed] [Google Scholar]
- 14.Paris S, Wysong D, Debeaupuis JP, Shibuya K, Philippe B, Diamond RD, Latge JP. 2003. Catalases of Aspergillus fumigatus. Infect Immun 71:3551–3562. doi: 10.1128/IAI.71.6.3551-3562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calera JA, Paris S, Monod M, Hamilton AJ, Debeaupuis JP, Diaquin M, Lopez-Medrano R, Leal F, Latge JP. 1997. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect Immun 65:4718–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa Y. 2008. Catalase gene disruptant of the human pathogenic yeast Candida albicans is defective in hyphal growth, and a catalase-specific inhibitor can suppress hyphal growth of wild-type cells. Microbiol Immunol 52:16–24. doi: 10.1111/j.1348-0421.2008.00006.x. [DOI] [PubMed] [Google Scholar]
- 17.Lambou K, Lamarre C, Beau R, Dufour N, Latge JP. 2010. Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol Microbiol 75:910–923. doi: 10.1111/j.1365-2958.2009.07024.x. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culotta VC, Klomp LW, Strain J, Casareno RL, Krems B, Gitlin JD. 1997. The copper chaperone for superoxide dismutase. J Biol Chem 272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 20.Sturtz LA, Culotta VC. 2002. Superoxide dismutase null mutants of baker's yeast, Saccharomyces cerevisiae. Methods Enzymol 349:167–172. doi: 10.1016/S0076-6879(02)49332-9. [DOI] [PubMed] [Google Scholar]
- 21.Gamonet F, Lauquin GJ. 1998. The Saccharomyces cerevisiae LYS7 gene is involved in oxidative stress protection. Eur J Biochem 251:716–723. doi: 10.1046/j.1432-1327.1998.2510716.x. [DOI] [PubMed] [Google Scholar]
- 22.Goetz KE, Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. 2011. Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Curr Genet 57:201–211. doi: 10.1007/s00294-011-0336-4. [DOI] [PubMed] [Google Scholar]
- 23.Margoliash E, Novogrodsky A, Schejter A. 1960. Irreversible reaction of 3-amino-1:2:4-triazole and related inhibitors with the protein of catalase. Biochem J 74:339–348. doi: 10.1042/bj0740339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda M, Kinoshita H, Yoshida T, Kamasawa N, Osumi M, Tanaka A. 2003. Effect of catalase-specific inhibitor 3-amino-1,2,4-triazole on yeast peroxisomal catalase in vivo. FEMS Microbiol Lett 219:93–98. doi: 10.1016/S0378-1097(02)01201-6. [DOI] [PubMed] [Google Scholar]
- 25.Lushchak V, Semchyshyn H, Lushchak O, Mandryk S. 2005. Diethyldithiocarbamate inhibits in vivo Cu,Zn-superoxide dismutase and perturbs free radical processes in the yeast Saccharomyces cerevisiae cells. Biochem Biophys Res Commun 338:1739–1744. doi: 10.1016/j.bbrc.2005.10.147. [DOI] [PubMed] [Google Scholar]
- 26.Jain R, Valiante V, Remme N, Docimo T, Heinekamp T, Hertweck C, Gershenzon J, Haas H, Brakhage AA. 2011. The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol Microbiol 82:39–53. doi: 10.1111/j.1365-2958.2011.07778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valiante V, Heinekamp T, Jain R, Hartl A, Brakhage AA. 2008. The mitogen-activated protein kinase MpkA of Aspergillus fumigatus regulates cell wall signaling and oxidative stress response. Fungal Genet Biol 45:618–627. doi: 10.1016/j.fgb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Blum G, Kainzner B, Grif K, Dietrich H, Zelger B, Sonnweber T, Lass-Florl C. 2013. In vitro and in vivo role of heat shock protein 90 in amphotericin B resistance of Aspergillus terreus. Clin Microbiol Infect 19:50–55. doi: 10.1111/j.1469-0691.2012.03848.x. [DOI] [PubMed] [Google Scholar]
- 29.Holdom MD, Hay RJ, Hamilton AJ. 1996. The Cu,Zn superoxide dismutases of Aspergillus flavus, Aspergillus niger, Aspergillus nidulans, and Aspergillus terreus: purification and biochemical comparison with the Aspergillus fumigatus Cu,Zn superoxide dismutase. Infect Immun 64:3326–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vatsyayan P, Goswami P. 2016. Highly active and stable large catalase isolated from a hydrocarbon degrading Aspergillus terreus MTCC 6324. Enzyme Res 2016:4379403. doi: 10.1155/2016/4379403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vatsyayan P, Goswami P. 2011. Acidic pH conditions induce dissociation of the haem from the protein and destabilise the catalase isolated from Aspergillus terreus. Biotechnol Lett 33:347–351. doi: 10.1007/s10529-010-0442-2. [DOI] [PubMed] [Google Scholar]
- 32.Louis B, Waikhom SD, Roy P, Bhardwaj PK, Singh MW, Chandradev SK, Talukdar NC. 2014. Invasion of Solanum tuberosum L. by Aspergillus terreus: a microscopic and proteomics insight on pathogenicity. BMC Res Notes 7:350. doi: 10.1186/1756-0500-7-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vatsyayan P, Bordoloi S, Goswami P. 2010. Large catalase based bioelectrode for biosensor application. Biophys Chem 153:36–42. doi: 10.1016/j.bpc.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 35.Lass-Florl C, Rief A, Leitner S, Speth C, Wurzner R, Dierich MP. 2005. In vitro activities of amphotericin B and voriconazole against aleurioconidia from Aspergillus terreus. Antimicrob Agents Chemother 49:2539–2540. doi: 10.1128/AAC.49.6.2539-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horita Y, Takii T, Yagi T, Ogawa K, Fujiwara N, Inagaki E, Kremer L, Sato Y, Kuroishi R, Lee Y, Makino T, Mizukami H, Hasegawa T, Yamamoto R, Onozaki K. 2012. Antitubercular activity of disulfiram, an antialcoholism drug, against multidrug- and extensively drug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 56:4140–4145. doi: 10.1128/AAC.06445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan S, Singhal S, Mathur T, Upadhyay DJ, Rattan A. 2007. Antifungal potential of disulfiram. Nihon Ishinkin Gakkai Zasshi 48:109–113. doi: 10.3314/jjmm.48.109. [DOI] [PubMed] [Google Scholar]
- 38.Bouma MJ, Snowdon D, Fairlamb AH, Ackers JP. 1998. Activity of disulfiram (bis(diethylthiocarbamoyl)disulphide) and ditiocarb (diethyldithiocarbamate) against metronidazole-sensitive and -resistant Trichomonas vaginalis and Tritrichomonas foetus. J Antimicrob Chemother 42:817–820. doi: 10.1093/jac/42.6.817. [DOI] [PubMed] [Google Scholar]
- 39.Nash T, Rice WG. 1998. Efficacies of zinc-finger-active drugs against Giardia lamblia. Antimicrob Agents Chemother 42:1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubner L, Ernst M, von Laer D, Schwander S, Flad HD. 1991. Enhancement of monocyte antimycobacterial activity by diethyldithiocarbamate (DTC). Int J Immunopharmacol 13:1067–1072. doi: 10.1016/0192-0561(91)90157-3. [DOI] [PubMed] [Google Scholar]
- 41.Rispail N, Soanes DM, Ant C, Czajkowski R, Grunler A, Huguet R, Perez-Nadales E, Poli A, Sartorel E, Valiante V, Yang M, Beffa R, Brakhage AA, Gow NA, Kahmann R, Lebrun MH, Lenasi H, Perez-Martin J, Talbot NJ, Wendland J, Di Pietro A. 2009. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol 46:287–298. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Valiante V, Macheleidt J, Foge M, Brakhage AA. 2015. The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front Microbiol 6:325. doi: 10.3389/fmicb.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valiante V, Jain R, Heinekamp T, Brakhage AA. 2009. The MpkA MAP kinase module regulates cell wall integrity signaling and pyomelanin formation in Aspergillus fumigatus. Fungal Genet Biol 46:909–918. doi: 10.1016/j.fgb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Du C, Sarfati J, Latge JP, Calderone R. 2006. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med Mycol 44:211–218. doi: 10.1080/13693780500338886. [DOI] [PubMed] [Google Scholar]
- 45.Altwasser R, Baldin C, Weber J, Guthke R, Kniemeyer O, Brakhage AA, Linde J, Valiante V. 2015. Network modeling reveals cross talk of MAP kinases during adaptation to caspofungin stress in Aspergillus fumigatus. PLoS One 10:e0136932. doi: 10.1371/journal.pone.0136932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valiante V, Monteiro MC, Martin J, Altwasser R, El Aouad N, Gonzalez I, Kniemeyer O, Mellado E, Palomo S, de Pedro N, Perez-Victoria I, Tormo JR, Vicente F, Reyes F, Genilloud O, Brakhage AA. 2015. Hitting the caspofungin salvage pathway of human-pathogenic fungi with the novel lasso peptide humidimycin (MDN-0010). Antimicrob Agents Chemother 59:5145–5153. doi: 10.1128/AAC.00683-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-Pena JM, Garcia R, Nombela C, Arroyo J. 2010. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast 27:495–502. doi: 10.1002/yea.1792. [DOI] [PubMed] [Google Scholar]
- 48.Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O'Shea EK. 2008. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet 40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diezmann S, Michaut M, Shapiro RS, Bader GD, Cowen LE. 2012. Mapping the Hsp90 genetic interaction network in Candida albicans reveals environmental contingency and rewired circuitry. PLoS Genet 8:e1002562. doi: 10.1371/journal.pgen.1002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blatzer M, Blum G, Jukic E, Posch W, Gruber P, Nagl M, Binder U, Maurer E, Sarg B, Lindner H, Lass-Florl C, Wilflingseder D. 2015. Blocking Hsp70 enhances the efficiency of amphotericin B treatment against resistant Aspergillus terreus strains. Antimicrob Agents Chemother 59:3778–3788. doi: 10.1128/AAC.05164-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mabey Gilsenan J, Cooley J, Bowyer P. 2012. CADRE: the Central Aspergillus Data REpository 2012. Nucleic Acids Res 40:D660–D666. doi: 10.1093/nar/gkr971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Binkley J, Arnaud MB, Inglis DO, Skrzypek MS, Shah P, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. 2014. The Candida Genome Database: the new homology information page highlights protein similarity and phylogeny. Nucleic Acids Res 42:D711–D716. doi: 10.1093/nar/gkt1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi J, Cheong K, Jung K, Jeon J, Lee GW, Kang S, Kim S, Lee YW, Lee YH. 2013. CFGP 2.0: a versatile web-based platform for supporting comparative and evolutionary genomics of fungi and Oomycetes. Nucleic Acids Res 41:D714–D719. doi: 10.1093/nar/gks1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neron B, Menager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics 25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 57.Subcommittee on Antifungal Susceptibility Testing of the ESCMID European Committee for Antimicrobial Susceptibility Testing. 2008. EUCAST technical note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia-forming moulds. Clin Microbiol Infect 14:982–984. doi: 10.1111/j.1469-0691.2008.02086.x. [DOI] [PubMed] [Google Scholar]
- 58.Arendrup MC, Guinea J, Cuenca-Estrella M, Meletiadis J, Mouton JW, Lagrou K, Howard SJ, Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). 22 December 2015. EUCAST definitive document E.DEF 9.3. Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia forming moulds. http://www.eucast.org/ast_of_fungi/methodsinantifungalsusceptibilitytesting/susceptibility_testing_of_moulds/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.