ABSTRACT
Hepcidin, an antimicrobial peptide, was discovered to integrate diverse signals from iron status and an infection threat and orchestrate a series of host-protective responses. Several studies have investigated the antimicrobial role of hepcidin, but the results have been controversial. Here, we aimed to examine the role of hepcidin in bacterial adherence and invasion in vitro. We found that porcine hepcidin could decrease the amount of the extracellular pathogen enterotoxigenic Escherichia coli (ETEC) K88 that adhered to cells because it caused the aggregation of the bacteria. However, addition of hepcidin to macrophages infected with the intracellular pathogen Salmonella enterica serovar Typhimurium enhanced the intracellular growth of the pathogen through the degradation of ferroportin, an iron export protein, and then the sequestration of intracellular iron. Intracellular iron was unavailable by use of the iron chelator deferiprone (DFO), which reduced intracellular bacterial growth. These results demonstrate that hepcidin exhibits different functions in extracellular and intracellular bacterial infections, which suggests that different defense strategies should be taken to prevent bacterial infection.
KEYWORDS: bacterial infection, hepcidin, iron regulation
INTRODUCTION
Iron is a critical determinant of the interplay between the host and the pathogen because it not only affects cell-mediated immune pathways but also represents a nutrient essential for the microbe (1, 2). This is because iron is vital for both metabolic processes, on the basis of its ability to transfer electrons (3). The expression of the iron acquisition system of pathogens has been linked to their pathogenicity (4, 5) and is essential for microbial growth and proliferation (6, 7). Thus, the control of iron homeostasis is a central battlefield determining the course of an infection.
Systemic iron homeostasis is controlled by the hormone hepcidin, which is mainly produced by hepatocytes (8). Hepcidin binds the sole known mammalian iron exporter, ferroportin (FPN), causing its internalization and degradation, thereby inhibiting iron efflux (9). The mature bioactive hepcidin peptide consists of 20 to 25 amino acids, 8 of which are cysteines, and it has four internal disulfide bonds (8, 10). Hepcidin exerts control over the trafficking of systemic iron by regulating the transfer of dietary irons and recycling and storing iron from intracellular compartments to extracellular fluid (11, 12). Given the association of iron with the outcome of infection (2, 13), determination of how hepcidin itself affects the progression of infection is clearly important.
The synthesis of hepcidin is regulated by both iron and inflammation (14). Interleukin-6 (IL-6), IL-22, and type I interferon stimulate the transcription of hepcidin through STAT3 signaling (15–17), and several microbe-derived Toll-like receptor ligands induce hepcidin expression (18, 19). However, the degree to which hepcidin can directly affect infection is not known. In the study described here, we determined the different effects of porcine hepcidin (pHepc) on extracellular and intracellular bacterial infection in vitro. Further, we observed that pHepc decreased the adherence of Escherichia coli K88 bacteria to cells because it caused the aggregation of the bacteria, while pHepc increased the cellular infectivity of Salmonella enterica serovar Typhimurium through the inhibition of iron export.
RESULTS
pHepc exhibits different effects on E. coli K88 and S. Typhimurium infections.
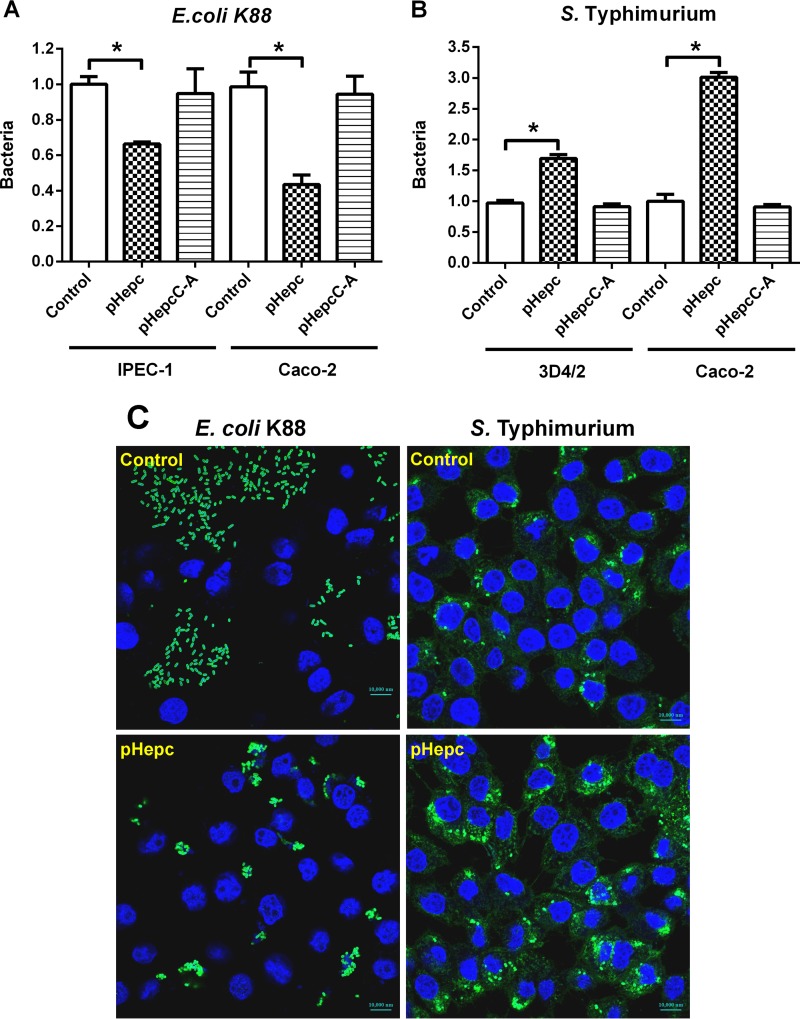
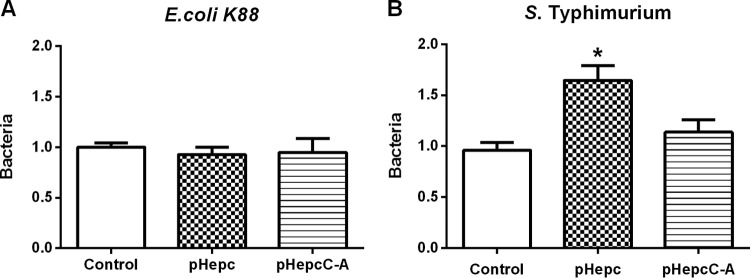
To determine whether pHepc inhibits bacterial invasion, intestinal epithelial cells or macrophages were challenged with pHepc-treated or untreated E. coli K88 or S. Typhimurium bacteria, respectively. The ability of pHepc-treated E. coli K88 to invade intestinal porcine epithelial J1 (IPEC-1) cells was impaired (Fig. 1A). Treatment with pHepc could reduce the amount of E. coli K88 which adhered to cells by 65%. The same pattern of pHepc-mediated inhibition of E. coli K88 infection was seen in another human epithelial colorectal adenocarcinoma cell line, Caco-2. However, treatment with pHepc significantly increased the ability of S. Typhimurium to invade both Caco-2 cells and 3D4/2 macrophages (Fig. 1B). Immunofluorescence microscopy confirmed that pHepc treatment blocked E. coli K88 invasion of IPEC-1 cells but elevated the level of S. Typhimurium invasion of 3D4/2 macrophages (Fig. 1C). The cysteine mutant form of pHepc (pHepcC-A) was supposed to have no antimicrobial activity. When the peptide pHepcC-A was used at the same concentration as pHepc, pHepcC-A had no effect on bacterial invasion. No significant difference in E. coli K88 adherence to pHepc-treated and untreated intestinal epithelial cells was observed, suggesting that pHepc does not protect against E. coli K88 infection by altering the host cells (Fig. 2A).
FIG 1.
pHepc-treated bacteria exhibited different infection activities. E. coli K88 and S. Typhimurium were pretreated for 1 h with either pHepc or pHepcC-A (16 μg/ml). The bacteria were allowed to infect cells (MOI, 10) for 1 h. (A and B) Relative bacterial adherence or invasion (number of CFU) for E. coli K88 (A) and S. Typhimurium (B) infections. For E. coli K88 infection, cells were washed with PBS to remove nonadherent bacteria, while for S. Typhimurium infection, cells were washed with PBS containing gentamicin. Cell lysates were then used to quantitate adhering or intracellular bacteria (expressed on the y axis as a ratio of the level for the control group). Error bars indicate SEs. *, P < 0.05. (C) Representative images of bacterium-infected IPEC-1 cells. Cells were assayed for infection by detection of the reporter gene (green) by epifluorescence microscopy. Cell nuclei were labeled with DAPI (4′,6-diamidino-2-phenylindole; blue).
FIG 2.
Bacteria exhibited different infection activities on pHepc-treated IPEC-1 cells. Epithelial cells were preincubated with 16 μg/ml of pHepc or pHepcC-A for 1 h. The bacteria were then allowed to infect the cells (MOI, 10) for 1 h. The relative bacterial adherence or invasion (number of CFU) for E. coli K88 (A) and S. Typhimurium (B) infections is shown. For E. coli K88 infection, cells were washed with PBS to remove nonadherent bacteria, while for S. Typhimurium infection, cells were washed with PBS containing gentamicin. Cell lysates were then used to quantitate the adhering or intracellular bacteria (expressed on the y axis as a ratio of the level for the control group). Cells were then washed and lysed to quantitate adhering and intracellular bacteria (expressed as a percentage of the value for the control group). Error bars indicate SEs. *, P < 0.05.
pHepc nanonets entrap E. coli K88 in vitro.
Previous immunofluorescence assays showed that pHepc-treated E. coli K88 bacteria seemed to sit on or assemble outside of cells (Fig. 1C). In order to confirm this phenomenon, scanning electron microscopy (SEM) was used. SEM observations showed that pHepc-treated E. coli K88 bacteria were aggregated (Fig. 3A to D), and a higher magnification revealed a net-like meshwork of fibrils emanating from the bacterial surface that entangled the bacteria (Fig. 3E and F). In contrast, no nanonet was observed for pHepc-treated S. Typhimurium bacteria (Fig. 3G and H), which indicated that the mesh structures were not made up of hepcidin itself. Meanwhile, the formation of similar nanofibrils was observed in E. coli K88 only with pHepc treatment and not with pHepcC-A treatment, indicating that cysteine-mediated interactions are critical for pHepc to induce net-like structures (Fig. 3B).
FIG 3.
pHepc nanonets entrap E. coli K88 in vitro. (A to F) SEM images of in vitro nanonet formation for E. coli K88 incubated with H2O (A), pHepcC-A (B), or pHepc (64 μg/ml [C to F]) in PBS buffer. Magnifications, ×5,000 (E) and ×10,000 (F). The white rectangle (E) and the asterisks (E and F) highlight a prominent nanonet. (G and H) SEM images of S. Typhimurium incubated with H2O (G) or pHepc (H) in DMEM buffer.
pHepc inhibits iron export and promotes the replication of S. Typhimurium in macrophages.
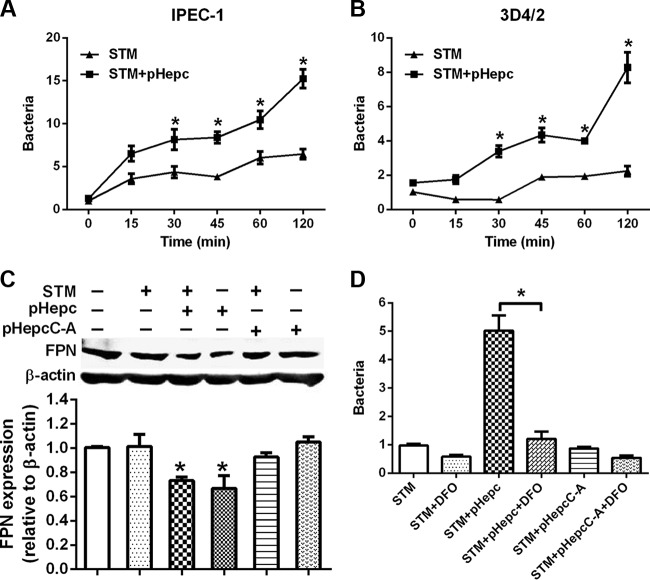
pHepc pretreatment of either bacteria (Fig. 1B) or epithelial cells (Fig. 2B) resulted in increases in the amounts of intracellular S. Typhimurium bacteria. The hypothesis that pHepc enhanced the level of invasion by promotion of the phagocytosis activity of cells was rejected by the finding that the amount of intracellular bacteria was not different from the initial amount when pHepc treatment was used (Fig. 4A and B). However, the rapid growth of pHepc-treated bacteria was observed at later times. To examine whether elevated levels of bacterial growth are induced by iron retention, we proceeded to investigate the impacts of FPN-mediated iron export on the intracellular replication of S. Typhimurium. The results showed that the addition of pHepc led to a decrease in FPN protein levels (Fig. 4C), which was expected to increase the intracellular iron content. To test this hypothesis, we used the iron chelator deferoxamine (DFO) to chelate intracellular iron. We observed that the elevated bacterial growth induced by pHepc addition was inhibited by DFO treatment (Fig. 4D). These results suggest that the increased level of invasion of macrophages by S. Typhimurium after pHepc treatment is largely through an increase in the level of iron acquisition by intracellular bacteria.
FIG 4.
pHepc induced the downregulation of FPN and increased intracellular bacterial growth. (A and B) IPEC-1 enterocytes (A) or 3D4/2 macrophages (B) were infected with pHepc-treated S. Typhimurium bacteria for 1 h, and extracellular bacteria were washed away with PBS containing gentamicin. Cell lysates were then used to quantitate the intracellular bacteria for measurement of the number of CFU (expressed on the y axis as a ratio of the level for the control group). (C) Western blot analysis showing FPN protein levels in 3D4/2 cells. (D) The iron chelator DFO removes the iron stored in macrophages, limiting intracellular S. Typhimurium growth. 3D4/2 macrophages were infected with pHepc-treated S. Typhimurium for 1 h, extracellular bacteria were washed away, and cells were incubated in the presence of DFO for 4 h. Cells were processed for immunofluorescence, and the number of bacterial foci per cell was determined as described above. Experiments were repeated a minimum of 3 times. Error bars indicate SEs. *, P < 0.05.
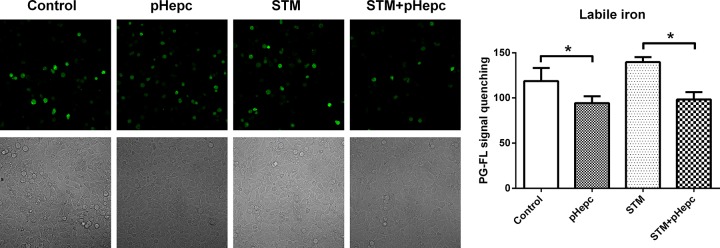
pHepc increased the intracellular iron concentration.
In order to confirm that hepcidin favors growth by increasing bacterial iron acquisition, intracellular iron stores were measured by determination of the quenching of the fluorescence signal of the green fluorescent heavy metal indicator Phen Green FL (PG-FL). The fluorescence of PG-FL is quenched upon binding to Fe2+ and Fe3+. pHepc treatment resulted in the significant suppression of the fluorescence intensity in both uninfected and S. Typhimurium-infected 3D4/2 cells (Fig. 5). This finding suggests that the intracellular iron content was increased by pHepc treatment.
FIG 5.
pHepc increased the intracellular iron concentration. (Left) 3D4/2 macrophages were infected with S. Typhimurium (STM) for 1 h with or without pretreatment with 16 μg/ml pHepc before the detection of fluorescence. (Right) Phen Green FL was added for 1 h before the amount of intracellular iron was determined by confocal laser scanning microscopy on the basis of the recorded PG-FL fluorescence intensity. Experiments were repeated a minimum of 3 times. Error bars indicate SEs. *, P < 0.05.
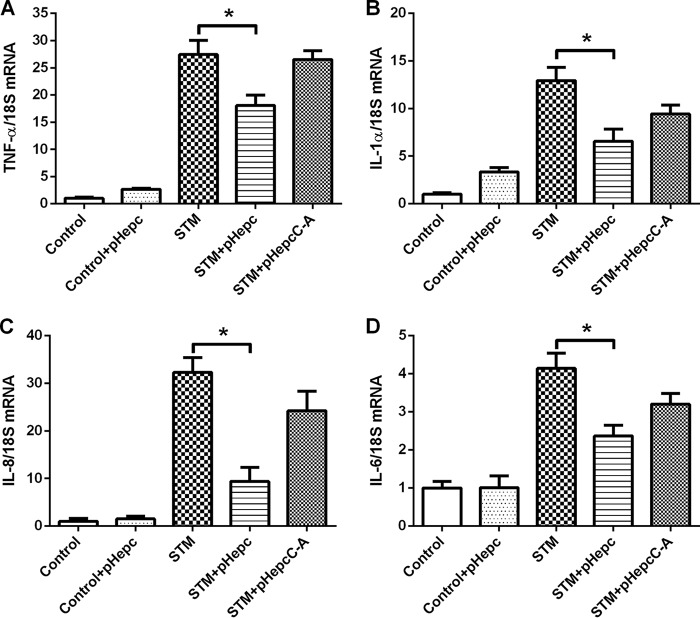
pHepc suppresses the cytokine expression induced by S. Typhimurium infection.
Hepcidin expression is altered in response to inflammation and iron loading. We determined the effects of hepcidin on the induction of IL-6, tumor necrosis factor alpha (TNF-α), IL-1α, and IL-8 resulting from S. Typhimurium infection. Bacterial infection of macrophages resulted in the induction of transcripts for the inflammatory cytokines (Fig. 6). Pretreatment of macrophages with pHepc 2 h before infection greatly reduced the levels of induction of TNF-α, IL-1α, IL-8, and IL-6. This finding suggests that hepcidin-mediated transcription is anti-inflammatory.
FIG 6.
Decreased inflammatory response in pHepc-treated macrophages. Cells were infected with Salmonella or treated with pHepc as described in the legend to Fig. 4B. The levels of TNF-α (A), IL-1α (B), IL-8 (C), and IL-6 (D) mRNA were determined by quantitative reverse transcription-PCR. 18S rRNA was used as the housekeeping gene. The mRNA expression ratio was normalized to the mean value for unstimulated control macrophages, which was equal to 1. Error bars indicate SEs. *, P < 0.05.
DISCUSSION
Hepcidin is a peptide hormone that regulates iron homeostasis and acts as an antimicrobial peptide. On the one hand, iron homeostasis in vertebrates is regulated by the binding of hepcidin to the iron transporter FPN, inducing the internalization and degradation of hepcidin (9). On the other hand, hepcidin functions as a member of the defensin family of antimicrobial peptides (10, 20). In most mammals, there is only one hepcidin gene, and it has both antimicrobial and iron-regulatory activities.
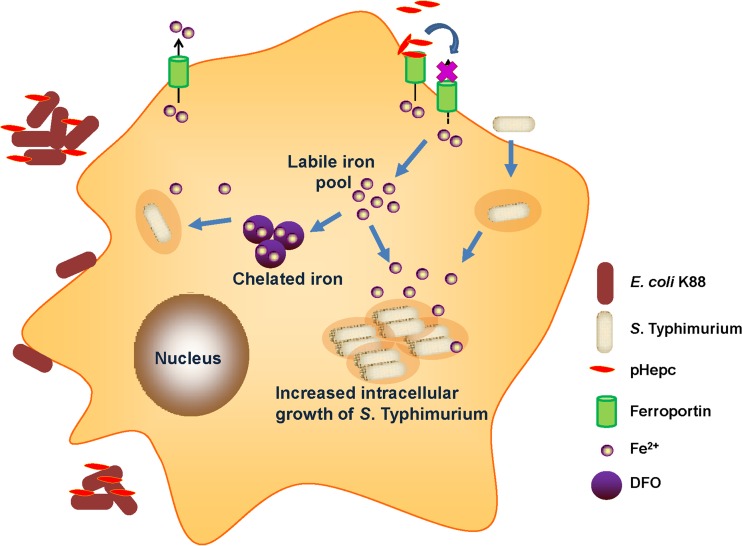
Several experiments have been performed in porcine cell lines, such as IPEC-1 and 3D4/2 cells, so we used porcine hepcidin in this study. The synthetic pHepc used in this study is 84% identical to the human ortholog, and most residues of pHepc are used to define hydrophobicity, net charge, and intramolecular disulfide bridges (21). Our previous studies showed that, like hepcidin from humans, pHepc exerts certain bacteriostatic activity (22). Here we showed that pHepc has different effects on E. coli K88 and S. Typhimurium bacterial infections in vitro. The level of adherence of pHepc-treated E. coli K88 bacteria to cells was decreased because of the aggregation of the bacteria, while pHepc increased the cellular infectivity of S. Typhimurium through the inhibition of iron export (Fig. 7).
FIG 7.
Proposed model for interference of pHepc in bacterial infection. pHepc inhibits E. coli K88 invasion by aggregation. However, pHepc binds to the iron exporter ferroportin, inducing its internalization and degradation. This causes an increase in the cytosolic iron content, which stimulates the growth of S. Typhimurium. Treatment with the iron chelator DFO reduces intracellular bacterial growth.
The induction of hepcidin during infection causes a drop in the amount of extracellular iron, as the withholding of iron from invading pathogens is thought to be a general defense mechanism against many infections (13). However, our findings suggest that pHepc exerts iron-independent effects on antibacterial infection by causing the aggregation of E. coli K88. Some studies have proposed that hepcidin can influence immune responses independently of its role in iron homeostasis (1). The hepcidin molecule, like defensins, is an amphipathic peptide with a net cationic charge, is rich in cysteine bonds, and has a β-sheet structure (20, 21). Consistent with this, hepcidin has microbicidal activity against many classes of microbes in vitro, leading to the hypothesis that such a direct antimicrobial effect may be relevant in vivo during infection (23). However, serum hepcidin concentrations are 1 to 2 orders of magnitude lower than those required for antimicrobial effects, making it unlikely that hepcidin directly kills pathogens in the bloodstream (20). Although pHepc has little bactericidal activity (22), pHepc provides a nidus that triggers a dynamic and deterministic process of self-assembly, building nanonets that can aggregate E. coli K88 cells and/or impede the close physical contact of E. coli K88 bacteria with epithelial cells required for bacterial attachment or invasion. Similar phenomena were also observed for some human α-defensins (HDs). HD6 affords protection against invasion by enteric bacterial pathogens in vitro and in vivo (24). HD5 binds to BK virus, leading to the aggregation of virion particles and the prevention of normal virus binding to the cell surface and uptake into cells (25). Although hepatocytes are the main source of hepcidin, experiments performed in vivo revealed hepcidin upregulation in immune cells at the site of bacterial infection and in neutrophils and macrophages in skin after subcutaneous bacterial infection (26). While hepcidin is locally produced in infected tissue, it is unknown if local hepcidin concentrations reach those required to produce antimicrobial activity (27). There is currently little evidence in the literature that hepcidin plays a directly antimicrobial role in vivo in mammalian infections (13). However, the previous research suggests that the local production of hepcidin may alter the availability of iron to pathogens at foci of infection without necessarily altering the systemic iron balance.
Hepcidin reduces iron availability in serum by inhibiting the function of FPN in macrophages and enterocytes (28), potentially limiting pathogen growth therein. While the upregulation of hepcidin in response to infection is generally considered protective, our observations indicate that the opposite is true for the intracellular pathogen S. Typhimurium. Exogenous pHepc increased the cellular infectivity of S. Typhimurium by inhibiting iron export. Hepcidin induces iron accumulation in macrophages and may be detrimental as a defense against pathogens that occupy this intracellular niche. Our data generally support the role of the hepcidin-FPN axis in the pathogenesis of S. Typhimurium infection, where pHepc causes intracellular iron accumulation and allows greater bacterial growth. The role of the hepcidin-FPN axis in the pathogenesis of S. Typhimurium infection is also supported by the finding that in vitro infection of macrophages with Salmonella resulted in lower intracellular bacterial burdens when macrophages were transfected to overexpress FPN (29). Our findings also indicate an important role for FPN in controlling the growth of S. Typhimurium inside cells and suggest that altered FPN expression and function in hemochromatosis and possibly other iron overload states may explain the susceptibility of intracellular pathogens to cells under these conditions (30, 31).
Macrophages facilitate the rapid throughput of iron and strongly express FPN, so the growth of pathogens that target and infect this cell type may be particularly influenced by hepcidin. How hepcidin and FPN might influence iron acquisition by S. Typhimurium is not yet clear, although high levels of hepcidin would increase the amount of iron stored within the macrophage. On the other hand, the generation of microbicidal molecules (including reactive oxygen species and nitric oxide) by macrophages is iron dependent. Our findings that hepcidin can suppress the increase in IL-6, TNF-α, IL-1α, and IL-8 levels confirmed that hepcidin may play an important role in affecting inflammation in bacterial infection. Hepcidin's modulation of inflammation may represent a feedback mechanism to limit the inflammatory response to bacterial infection. The downregulation of FPN immediately decreases iron export, while hepcidin's effects on transcription are delayed relative to the timing of its effects on the alteration of iron transport. Therefore, the two effects of hepcidin are temporally separated. Several lines of evidence indicate that hepcidin activation of transcription through FPN/JAK2/STAT3 and SOCS3 completes a negative-feedback loop for inflammation (32). Further, cells other than macrophages can express hepcidin, suggesting the possibility that hepcidin/FPN transcriptional responses may affect local inflammatory responses. This double-edged nature of iron, as a nutrient needed for pathogen growth and for host antimicrobial defense, may be particularly important for macrophage-tropic organisms, and how this balance is regulated by hepcidin requires careful exploration. We used only mammal cells to identify the effects of pHepc. In the absence of in vivo data, any iron-independent or iron-dependent role of hepcidin in host defense remains speculative.
In summary, our data demonstrate that hepcidin plays different roles in infections caused by the extracellular bacterium E. coli K88 and the intracellular bacterium S. Typhimurium in vitro. pHepc exerted iron-independent effects on antibacterial infection by aggregating E. coli K88 bacteria, while it increased the cellular infectivity of S. Typhimurium by inhibiting iron export. We have also shown that the chelators can reduce the intracellular growth of S. Typhimurium in pHepc-treated macrophages. These results suggest that these iron chelators might be efficacious in the treatment of persistent intracellular bacterial infections.
MATERIALS AND METHODS
Bacterial strains, cell lines, and growth conditions.
Enterotoxigenic Escherichia coli (ETEC) K88 and Salmonella enterica serovar Typhimurium CMCC 50013 were obtained from the China General Microbiological Culture Collection Center (Beijing, China) and grown in Mueller-Hinton broth (MHB) under aerobic conditions. 3D4/2 porcine alveolar macrophages, intestinal porcine epithelial J1 (IPEC-1) cells, and human epithelial colorectal adenocarcinoma (Caco-2) cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). They were cultured in RPMI 1640 medium or Dulbecco modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS; non-heat inactivated; HyClone), penicillin (100 IU/ml), and streptomycin (100 μg/ml). Cells in mid-logarithmic phase were used for bacterial infection assays; no antibiotic was added 24 h prior to infection. Cells were grown at 37°C in 5% CO2. At the cell density used (1 × 106 cells/ml), the proportion of dead cells was <1% according to the results of trypan blue dye exclusion tests.
Peptide synthesis.
The naturally secreted form of porcine hepcidin (pHepc; DTHFPICIFCCGCCRKAICGMCCKT) and the cysteine mutant form of pHepc (pHepcC-A; DTHFPIAIFAAGAARKAIAGMAAKT) were acquired through synthesis, refolding, and purification by reversed-phase high-performance liquid chromatography (HPLC) as previously described (22). Peptides were diluted to the appropriate concentration in phosphate-buffered saline (PBS) prepared with endotoxin-free water before addition to cultured cells.
Infection assay.
Fresh cultured bacteria were collected, washed twice with sterile PBS (pH 7.0), and dispersed into cell culture medium without FBS in the absence or presence of 16 μg/ml pHepc for 1 h. The final concentration of hepcidin (16 μg/ml) was determined as described previously (22). The multiplicity of infection (MOI) was adjusted to 10 using a standardized calibration curve of the optical density at 600 nm/number of CFU. For E. coli K88 infection, bacteria were added to IPEC-1 and Caco-2 cells at 70% confluence in 12-well dishes. After 1 h at 37°C, the cells were repeatedly washed with warm PBS to remove nonadherent bacteria. Cell lysates were prepared by hypotonic lysis of the cell monolayers with sterile water. Serial dilutions were then plated on Mueller-Hinton agar and incubated at 37°C overnight. For S. Typhimurium infection, 3D4/2 cells were incubated with the bacteria for 1 h at 37°C. Extracellular bacteria were removed by washing 3 times with warm PBS containing antibiotics (0.1 mg/ml gentamicin; Sigma-Aldrich), and the cells were plated on Mueller-Hinton agar and incubated at 37°C for another 24 h. Infected cells were fixed and processed for immunofluorescence assay or lysed in 150 mM NaCl, 10 mM EDTA, 10 mM Tris (pH 7.4), 1% Trion X-100, and a protease inhibitor cocktail (Roche) for Western blot analysis.
Scanning electron microscopy.
Bacteria were grown to mid-logarithmic phase in MHB. Bacteria (2 × 106 CFU) in growth medium were centrifuged (5,000 × g for 5 min) and resuspended in 1 ml of 50 mM Tris-maleate buffer (pH 6.4). The bacteria were then incubated with 64 μg/ml of pHepc for 1 h at room temperature and then centrifuged (5,000 × g for 5 min) to pellet the bacteria. A higher dose of hepcidin (64 μg/ml) was used to exaggerate the effects. The bacterial pellets were resuspended in Karnovsky's fixative (2.5% paraformaldehyde, 2.5% glutaraldehyde in 0.06 M Sorensen's phosphate buffer [0.2 M sodium phosphate, pH 7.2]) and left overnight at room temperature. Specimens were dehydrated in graded ethanol (twice in 30% ethanol and twice in 100% ethanol for 15 min each time) and then transferred to a critical point dryer (Pelco CPD-2) and dried with CO2. The samples were then mounted onto 12-mm aluminum stubs followed by sputter coating with gold. Samples were photographed using a Philips XL-30 scanning electron microscope.
Western blot analysis and immunofluorescence assay.
Cellular proteins were extracted with lysis buffer, and total protein concentrations were determined using the bicinchoninic acid reagent (KeyGen, China). Protein samples were separated on 4% to 20% acrylamide gels (Bio-Rad, USA) and transferred onto a polyvinylidene difluoride membrane (Millipore, USA). FPN was detected using rabbit anti-FPN antibody (1:400; Bioss, China) with peroxidase-conjugated goat anti-rabbit IgG as the secondary antibody (1:5,000; Abcam, USA). Actin was detected using a mouse anti-beta-actin antibody (1:2,500; Proteintech, USA). The chemiluminescent method was used for detection.
For immunofluorescence assay, IPEC-1 cells were fixed by use of a methanol flash for 5 min at −20°C, permeabilized in PBS containing 1% bovine albumin serum and 0.1% saponin, and incubated with rabbit anti-Salmonella or anti-E. coli antibody (1:100; Abcam, USA) for 1 h at room temperature, followed by incubation with Alexa Fluor 594-conjugated goat anti-rabbit immunoglobulin antibody for 1 h at room temperature. Cells were visualized using an epifluorescence microscope (Olympus) with a 60× oil immersion objective. Images were acquired using Magnifier analysis software.
Intracellular iron measurement.
Intracellular iron levels were measured using green fluorescent heavy metal indicator Phen Green FL (PG-FL; Invitrogen) fluorescence signal quenching (33). 3D4/2 macrophages were treated with PBS (control), 16 μg/ml pHepc, S. Typhimurium, and pHepc plus S. Typhimurium for 1 h before detection of fluorescence. After incubation, the cells were twice washed with warm PBS (37°C) and then incubated with 30 μM PG-FL in PBS for 1 h at 37°C. The excess PG-FL was twice washed off with PBS, and the PG-FL fluorescence intensity was recorded (excitation and emission, 490 and 528 nm, respectively) under a confocal laser scanning microscope (LSM 780; Zeiss).
Effect of pHepc on S. Typhimurium-induced cytokine mRNA expression.
The total RNA of 3D4/2 cells infected with S. Typhimurium was extracted by using the TRIzol agent (Sigma, China) according to the manufacturer's recommendations. Reverse transcription and quantitative PCR were conducted as described previously (34). Inflammatory cytokines TNF-α, IL-1α, IL-8, and IL-6 were detected, and the 18S rRNA housekeeping gene was used as an internal standard. The primers used for amplification are listed in Table 1.
TABLE 1.
Primer sequences for real-time PCR amplification
| Gene | Orientation | Primer sequence (5′–3′) | GenBank accession no. |
|---|---|---|---|
| TNF-α | Forward | CCAATGGCAGAGTGGGTATG | NM_214022.1 |
| Reverse | TGAAGAGGACCTGGGAGTAG | ||
| IL-1α | Forward | CCCGTCAGGTCAATACCTC | NM_214029.1 |
| Reverse | GCAACACGGGTTCGTCTTC | ||
| IL-8 | Forward | TTCGATGCCAGTGCATAAATA | M86923.1 |
| Reverse | CTGTACAACCTTCTGCACCCA | ||
| IL-6 | Forward | TGGCTACTGCCTTCCCTACC | NM_001252429.1 |
| Reverse | CAGAGATTTTGCCGAGGATG | ||
| 18S rRNA | Forward | CCCACGGAATCGAGAAAGAG | AY265350.1 |
| Reverse | TTGACGGAAGGGCACCA |
Statistics.
The difference between groups was determined by Student's t test or, if there were more than two groups, by one-way analysis of variance followed by Duncan's test, using SPSS software (version 18.0). All of the data are presented as the mean ± standard error of the mean, and a P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the Natural Science Foundation for Distinguished Young Scholars of Zhejiang Province of China (no. LR16C170001) and the National Natural Science Foundation of China (no. 31272455 and 31572411).
None of the authors has a conflict of interest.
REFERENCES
- 1.Cassat JE, Skaar EP. 2013. Iron in infection and immunity. Cell Host Microbe 13:510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soares MP, Weiss G. 2015. The iron age of host-microbe interactions. EMBO Rep 16:1482–1500. doi: 10.15252/embr.201540558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papanikolaou G, Pantopoulos K. 2005. Iron metabolism and toxicity. Toxicol Appl Pharmacol 202:199–211. doi: 10.1016/j.taap.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Clark MA, Goheen MM, Fulford A, Prentice AM, Elnagheeb MA, Patel J, Fisher N, Taylor SM, Kasthuri RS, Cerami C. 2014. Host iron status and iron supplementation mediate susceptibility to erythrocytic stage Plasmodium falciparum. Nat Commun 5:4446. doi: 10.1038/ncomms5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiester SE, Nwugo CC, Penwell WF, Neary JM, Beckett AC, Arivett BA, Schmidt RE, Geiger SC, Connerly PL, Menke SM, Tomaras AP, Actis LA. 2015. Role of the carboxy terminus of SecA in iron acquisition, protein translocation, and virulence of the bacterial pathogen Acinetobacter baumannii. Infect Immun 83:1354–1365. doi: 10.1128/IAI.02925-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Othman R, Flannery AR, Miguel DC, Ward DM, Kaplan J, Andrews NW. 2014. Leishmania-mediated inhibition of iron export promotes parasite replication in macrophages. PLoS Pathog 10:e1003901. doi: 10.1371/journal.ppat.1003901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paradkar PN, De Domenico I, Durchfort N, Zohn I, Kaplan J, Ward DM. 2008. Iron depletion limits intracellular bacterial growth in macrophages. Blood 112:866–874. doi: 10.1182/blood-2007-12-126854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganz T. 2003. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood 102:783–788. doi: 10.1182/blood-2003-03-0672. [DOI] [PubMed] [Google Scholar]
- 9.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. 2004. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 10.Park CH, Valore EV, Waring AJ, Ganz T. 2001. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 11.Drakesmith H, Prentice AM. 2012. Hepcidin and the iron-infection axis. Science 338:768–772. doi: 10.1126/science.1224577. [DOI] [PubMed] [Google Scholar]
- 12.Ganz T. 2011. Hepcidin and iron regulation, 10 years later. Blood 117:4425–4433. doi: 10.1182/blood-2011-01-258467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michels K, Nemeth E, Ganz T, Mehrad B. 2015. Hepcidin and host defense against infectious diseases. PLoS Pathog 11:e1004998. doi: 10.1371/journal.ppat.1004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganz T. 2006. Hepcidin—a peptide hormone at the interface of innate immunity and iron metabolism. Curr Top Microbiol 306:183–198. [DOI] [PubMed] [Google Scholar]
- 15.Ichiki K, Ikuta K, Addo L, Tanaka H, Sasaki Y, Shimonaka Y, Sasaki K, Ito S, Shindo M, Ohtake T, Fujiya M, Torimoto Y, Kohgo Y. 2014. Upregulation of iron regulatory hormone hepcidin by interferon alpha. J Gastroenterol Hepatol 29:387–394. doi: 10.1111/jgh.12348. [DOI] [PubMed] [Google Scholar]
- 16.Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, Ernst M, Klein C, Trautwein C. 2007. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology 132:294–300. (Erratum, 132:1208.) [DOI] [PubMed] [Google Scholar]
- 17.Smith CL, Arvedson TL, Cooke KS, Dickmann LJ, Forte C, Li HY, Merriam KL, Perry VK, Tran L, Rottman JB, Maxwell JR. 2013. IL-22 regulates iron availability in vivo through the induction of hepcidin. J Immunol 191:1845–1855. doi: 10.4049/jimmunol.1202716. [DOI] [PubMed] [Google Scholar]
- 18.Guida C, Altamura S, Klein FA, Galy B, Boutros M, Ulmer AJ, Hentze MW, Muckenthaler MU. 2015. A novel inflammatory pathway mediating rapid hepcidin-independent hypoferremia. Blood 125:2265–2275. doi: 10.1182/blood-2014-08-595256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koening CL, Miller JC, Nelson JM, Ward DM, Kushner JP, Bockenstedt LK, Weis JJ, Kaplan J, De Domenico I. 2009. Toll-like receptors mediate induction of hepcidin in mice infected with Borrelia burgdorferi. Blood 114:1913–1918. doi: 10.1182/blood-2009-03-209577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. 2000. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 480:147–150. doi: 10.1016/S0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 21.Sang Y, Ramanathan B, Minton JE, Ross CR, Blecha F. 2006. Porcine liver-expressed antimicrobial peptides, hepcidin and LEAP-2: cloning and induction by bacterial infection. Dev Comp Immunol 30:357–366. doi: 10.1016/j.dci.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Liu D, Pu YT, Xiong HT, Wang YZ, Du HH. 2015. Porcine hepcidin exerts an iron-independent bacteriostatic activity against pathogenic bacteria. Int J Pept Res Ther 21:229–236. doi: 10.1007/s10989-014-9451-7. [DOI] [Google Scholar]
- 23.Maisetta G, Petruzzelli R, Brancatisano FL, Esin S, Vitali A, Campa M, Batoni G. 2010. Antimicrobial activity of human hepcidin 20 and 25 against clinically relevant bacterial strains: effect of copper and acidic pH. Peptides 31:1995–2002. doi: 10.1016/j.peptides.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Chu HT, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B, Salzman NH, Underwood MA, Tsolis RM, Young GM, Lu WY, Lehrer RI, Baumler AJ, Bevins CL. 2012. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337:477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dugan AS, Maginnis MS, Jordan JA, Gasparovic ML, Manley K, Page R, Williams G, Porter E, O'Hara BA, Atwood WJ. 2008. Human alpha-defensins inhibit BK virus infection by aggregating virions and blocking binding to host cells. J Biol Chem 283:31125–31132. doi: 10.1074/jbc.M805902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motley ST, Morrow BJ, Liu XJ, Dodge IL, Vitiello A, Ward CK, Shaw KJ. 2004. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell Microbiol 6:849–865. doi: 10.1111/j.1462-5822.2004.00407.x. [DOI] [PubMed] [Google Scholar]
- 27.Peyssonnaux C, Zinkernagel AS, Datta V, Lauth X, Johnson RS, Nizet V. 2006. TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood 107:3727–3732. doi: 10.1182/blood-2005-06-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivera S, Nemeth E, Gabayan V, Lopez MA, Farshidi D, Ganz T. 2005. Synthetic hepcidin causes rapid dose-dependent hypoferremia and is concentrated in ferroportin-containing organs. Blood 106:2196–2199. doi: 10.1182/blood-2005-04-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chlosta S, Fishman DS, Harrington L, Johnson EE, Knutson MD, Wessling-Resnick M, Cherayil BJ. 2006. The iron efflux protein ferroportin regulates the intracellular growth of Salmonella enterica. Infect Immun 74:3065–3067. doi: 10.1128/IAI.74.5.3065-3067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordeuk VR, McLaren CE, MacPhail AP, Deichsel G, Bothwell TH. 1996. Associations of iron overload in Africa with hepatocellular carcinoma and tuberculosis: Strachan's 1929 thesis revisited. Blood 87:3470–3476. [PubMed] [Google Scholar]
- 31.Wanachiwanawin W. 2000. Infections in E-beta thalassemia. J Pediatr Hematol Oncol 22:581–587. doi: 10.1097/00043426-200011000-00027. [DOI] [PubMed] [Google Scholar]
- 32.De Domenico I, Zhang TY, Koening CL, Branch RW, London N, Lo E, Daynes RA, Kushner JP, Li D, Ward DM, Kaplan J. 2010. Hepcidin mediates transcriptional changes that modulate acute cytokine-induced inflammatory responses in mice. J Clin Invest 120:2395–2405. doi: 10.1172/JCI42011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X, Wang H, Shi W, Shen Z, Shen H, Li M. 2014. Hyperinsulinemia induces hepatic iron overload by increasing liver TFR1 via the PI3K/IRP2 pathway. J Mol Endocrinol 53:381–392. doi: 10.1530/JME-14-0122. [DOI] [PubMed] [Google Scholar]
- 34.Gao Y, Han F, Huang X, Rong Y, Yi H, Wang Y. 2013. Changes in gut microbial populations, intestinal morphology, expression of tight junction proteins, and cytokine production between two pig breeds after challenge with Escherichia coli K88: a comparative study. J Anim Sci 91:5614–5625. doi: 10.2527/jas.2013-6528. [DOI] [PubMed] [Google Scholar]