ABSTRACT
Induction of interferon and proinflammatory cytokines is a hallmark of the infection of many different viruses. However, hepatitis B virus (HBV) does not elicit a detectable cytokine response in infected hepatocytes. In order to investigate the molecular mechanism underlying the innate immune evasion, a functional cyclic GMP-AMP (cGAMP) synthase (cGAS)-stimulator of interferon genes (STING) pathway was reconstituted in a human hepatoma cell line supporting tetracycline-inducible HBV replication. It was demonstrated that induction of HBV replication neither activated nor inhibited this cytosolic DNA sensing pathway. However, human hepatoma cells, as well as immortalized mouse hepatocytes, express low levels of STING, which upon activation by cGAMP, the natural ligand of STING, led to induction of a proinflammatory cytokine response. Treatment of immortalized mouse hepatocytes supporting HBV replication with either cGAMP or a small molecule pharmacologic STING agonist significantly reduced viral DNA in a STING- and Janus kinase 1-dependent manner. Moreover, cGAMP treatment was able to induce inflammatory cytokine gene expression and inhibit the transcription of covalently closed circular DNA in HBV-infected human hepatoma cells expressing sodium taurocholate cotransporting polypeptide, an essential receptor for HBV infection of hepatocytes. The studies reported here and previously (F. Guo et al., Antimicrob Agents Chemother 59:1273–1281, 2015, https://doi.org/10.1128/AAC.04321-14) thus support the notion that pharmacological activation of STING in macrophages and hepatocytes induces host innate responses that can efficiently control HBV replication. Hence, despite not playing a significant role in host innate immune response to HBV infection of hepatocytes, STING is potentially a valuable target for immunotherapy of chronic hepatitis B.
KEYWORDS: STING, hepatitis B virus, interferons
INTRODUCTION
Hepatitis B virus (HBV) chronically infects 240 million people worldwide (1) and 686,000 people die annually due to hepatitis B and its related cirrhosis and hepatocellular carcinoma (2). HBV is one of the smallest DNA viruses and contains a 3.2-kb, partially double-stranded, relaxed circular DNA (rcDNA) genome. However, unlike other DNA viruses, HBV replicates its genomic DNA via reverse transcription (RT) of an RNA intermediate, the pregenomic RNA (pgRNA) (3). Briefly, binding to sodium taurocholate cotransporting polypeptide (NTCP) on hepatocytes triggers HBV entry and subsequent delivery of nucleocapsid into the cytoplasm (4, 5). The genomic rcDNA is transported into the nucleus upon nucleocapsid uncoating at the nuclear pore complex and subsequently converted into covalently closed circular DNA (cccDNA) (6). The cccDNA serves as the template for transcription of viral RNAs that specify seven viral proteins. Viral DNA replication begins when the core protein dimers assemble the pgRNA-viral DNA polymerase complex to form nucleocapsids where viral DNA polymerase converts pgRNA first to a single-stranded DNA (ssDNA) and then to rcDNA. The rcDNA-containing nucleocapsids can either be enveloped and secreted as virions or deliver rcDNA into the nucleus to amplify cccDNA pool.
Intriguingly, unlike many other viruses that are recognized upon infection by host cellular pattern recognition receptors (PRRs) to induce interferon (IFN) and proinflammatory cytokines, HBV infection of hepatocytes does not induce a detectable cytokine response in vivo and in cultures (7, 8). Because the early cytokine response not only restricts viral replication but also plays an important role in induction of adaptive immune responses essential for resolving viral infections, evasion of the innate immune response by HBV has been considered to play a critical role in viral pathogenesis. In search for the mechanism underlying the innate immune evasion, many studies have suggested that expression of HBV core protein, polymerase, HBx, or envelope proteins inhibits TLR3 (9, 10), TLR9 (11, 12), RIG-I (13, 14), and stimulator of interferon genes (STING) (15) signal transduction pathways. Others have speculated that HBV escapes the innate immune recognition owing to its unique replication strategy (8, 16). First, HBV cccDNA exists as a minichromosome within the nucleus of infected hepatocyte and is therefore camouflaged to escape the detection of cellular DNA sensing machinery. Second, cccDNA transcribes polyadenylated viral RNAs that resemble the normal cellular mRNA and thus cannot activate cellular RNA sensors. Finally, viral DNA synthesis occurs within nucleocapsids in the cytoplasm and thus evades the detection of cytoplasmic DNA sensors. Moreover, a recent study has suggested that hepatocytes may lack an functional DNA sensing mechanism and thus favor the persistent infection of HBV (17).
Cyclic GMP-AMP synthase (cGAS) has been demonstrated to play an important role in restricting the infection of retroviruses and DNA viruses (18–21). Binding of DNA activates cGAS to synthesize 2′3′-cyclic GMP-AMP (cGAMP) from GTP and ATP. cGAMP subsequently binds to stimulator of interferon genes (STING) to induce its dimerization and translocation from the endoplasmic reticulum membrane to perinuclear membranous vesicles, where STING triggers signaling cascades leading to IRF3 phosphorylation, nuclear translocation and activation of IFN gene expression (22, 23). Recently, several reports suggested that cGAS-STING pathway may play a role in restriction of HBV replication in hepatocytes (24–27). In order to rigorously determine the interaction between HBV and the key cytoplasmic DNA sensing pathway, we reconstituted a functional cGAS-STING pathway in human hepatoma cells supporting HBV replication and demonstrated that HBV infection neither activates nor inhibits cGAS-STING pathway. However, our studies showed that human hepatoma cells and immortalized mouse hepatocytes express low levels of STING and have a functional STING signaling pathway, which is not inhibited by HBV in the context of viral replication. Furthermore, pharmacological activation of STING in those hepatocyte-derived cell lines induces a cytokine-mediated immune response that efficiently suppresses HBV replication. Together with our previous finding that activation of STING in macrophages induces a robust cytokine response to inhibit HBV replication in hepatocytes (28), our work suggests that STING is a valuable therapeutic target for pharmacological activation of host immune responses, in both hepatocytes and immune modulating cells, to cure or durably control chronic HBV infection.
RESULTS
Stable expression of cGAS and STING in HepAD38 cells reconstituted a functional DNA-sensing pathway.
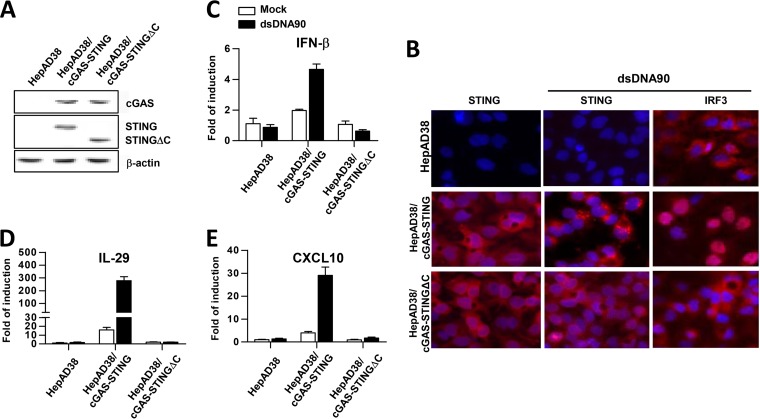
Because human hepatoma cells do not produce proinflammatory cytokines upon transfection of double-stranded DNA (dsDNA), probably due to low expression of cGAS and STING, we intended to reconstitute a functional cGAS-STING cytoplasmic DNA sensing pathway in HepAD38 cells, a HepG2-derived cell line in which viral pgRNA transcription and subsequent DNA replication are regulated by a tetracycline (TET) inducible promoter (29). To this end, HepAD38-derived cell lines constitutively expressing human cGAS and wild-type human STING or a truncated STING with deletion of 39 amino acid (aa) residues from the carboxyl terminus (STINGΔC) were established by recombinant retroviral transduction and selection with antibiotics. The resulting cell lines are designated HepAD38/cGAS-STING and HepAD38/cGAS-STINGΔC, respectively. Expression of cGAS and STING proteins in the cell lines was confirmed by a Western blot assay (Fig. 1A). Because the C-terminal domain of STING is essential for activation of IRF3 and subsequent induction of cytokines, the STINGΔC serves as a negative control (30). Indeed, functional analyses demonstrated that transfection of dsDNA90, a well-known cGAS ligand (31), only induced STING translocation, nuclear import of IRF3 (Fig. 1B) and mRNA expression of IFN-β, interleukin-29 (IL-29 [or IFN-λ1]), and CXCL10 (Fig. 1C to E) in HepAD38/cGAS-STING cells, but not in HepAD38/cGAS-STINGΔC and parental HepAD38 cells. The results thus indicate that ectopic expression of cGAS and STING can reconstitute a functional DNA sensing pathway in HepAD38 cells. Consistent with previous reports in other cell culture systems, ectopic expression of cGAS and/or STING increased the basal levels of cytokine expression, such as IL-29 and CXCL10, in HepAD38/cGAS-STING cells (32, 33).
FIG 1.
Stable expression of cGAS and STING in HepAD38 cells successfully reconstitutes a functional DNA-induced cytokine response pathway. (A) The expression of cGAS and STING in the indicated cells was detected by Western blotting assays with antibodies against cGAS and STING. β-Actin served as a loading control. (B) The indicated cells were mock transfected or transfected with dsDNA90 and fixed 7 h posttransfection. STING and IRF3 were visualized by indirect immunofluorescence staining (red). Cell nuclei were stained with DAPI (blue). (C to E) The indicated cells were mock transfected (open bar) or transfected with dsDNA90 (filled bar) and harvested after 7 h of transfection. The levels of IFN-β (C), IL-29 (D), and CXCL10 (E) mRNA were determined by qRT-PCR assays and normalized to β-actin mRNA and are presented as the fold induction over that in mock-transfected parental HepAD38 cells. Means and standard deviations are presented (n = 3).
HBV replication in hepatocytes neither activates nor inhibits cGAS-STING signaling pathway.
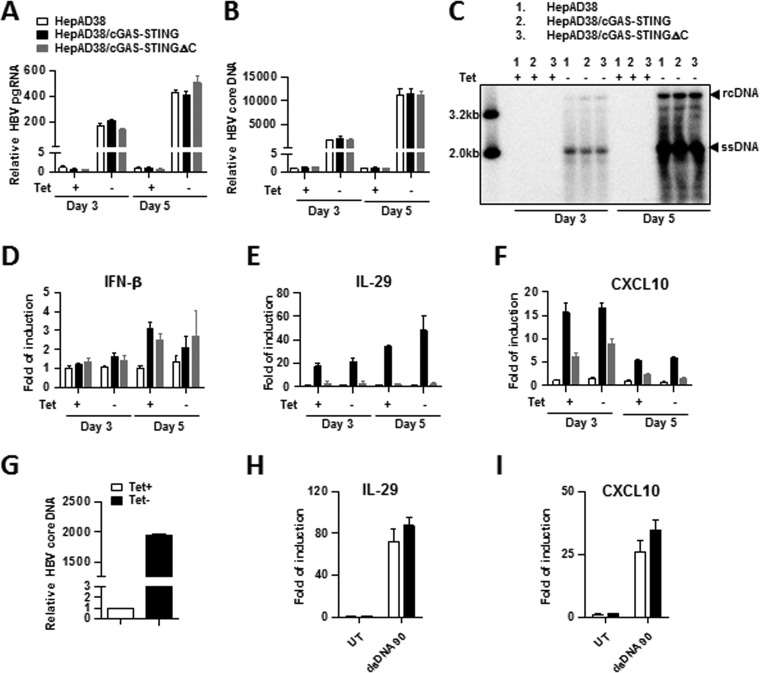
In order to investigate whether cGAS is able to sense HBV DNA replication in hepatocytes and mount a proinflammatory cytokine response, parental HepAD38, HepAD38/cGAS-STING, and HepAD38/cGAS-STINGΔC cells were cultured in the presence or absence of TET for 3 and 5 days. The removal of TET from the culture medium induced the increase of HBV pgRNA (Fig. 2A), as well as core DNA (Fig. 2B and C), in all the three cell lines in a time-dependent manner. However, the levels of HBV pgRNA and core DNA in either HepAD38/cGAS-STING or HepAD38/cGAS-STINGΔC cells did not significantly differ from that in parental HepAD38 cells, implying that the presence of a functional cGAS-STING pathway does not affect HBV replication. As anticipated, induction of HBV replication (TET-off) did not induce a detectable cytokine response in parental HepAD38 and HepAD38/cGAS-STINGΔC cells. Interestingly, induction of HBV replication in HepAD38/cGAS-STING cells also did not significantly upregulate the expression of IFN-β, IL-29, and CXCL10 (Fig. 2D to F). The results thus imply that HBV replication is unable to activate cGAS-STING pathway under the selected experimental condition. To investigate whether this is due to HBV inhibition of cGAS-STING pathway, we examined dsDNA90 induction of cytokine response in HepAD38/cGAS-STING cells cultured in the presence or absence of TET for 5 days. As expected, culturing of cells in the absence of TET for 5 days induced high levels of HBV replication (Fig. 2G); however, HBV replication had no effect on the magnitude of dsDNA90-induced upregulation of IL-29 and CXCL10 (Fig. 2H and I). Taken together, our results support the notion that HBV replication in hepatocytes neither activates nor inhibits cGAS-STING signaling pathway.
FIG 2.
HBV replication neither activates nor inhibits cGAS-STING signaling pathway. (A to F) Each of the indicated cell lines was cultured in the presence or absence of TET for 3 and 5 days. The levels of HBV pgRNA (A), cytoplasmic HBV core DNA (B), IFN-β mRNA (D), IL-29 mRNA (E), and CXCL10 mRNA (F) were determined by qPCR assays and are presented as the fold change compared to that in HepAD38 cells cultured in the presence of TET. Means and standard deviations are shown (n = 4). Proper induction of HBV DNA replication upon TET removal was further confirmed by Southern blotting hybridization with a [32P]UTP-labeled riboprobe specific for minus-strand of HBV DNA (C). Relaxed circular DNA (rcDNA), single-stranded DNA (ssDNA), and 3.2- and 2.0-kb DNA size markers are indicated. (G to I) HepAD38/cGAS-STING cells were cultured in the presence (open bar) or absence (filled bar) of TET for 5 days. The cells were then reseeded at a density of 5 × 105 cells per well of a 12-well plate and mock transfected or transfected with dsDNA90 using Lipofectamine-3000 at 12 h after seeding. The levels of HBV core DNA in pretransfected cells (G), as well as IL-29 mRNA (H) and CXCL10 mRNA (I), at 12 h posttransfection were determined by qPCR assays and are presented as the fold change compared to that in HepAD38/cGAS-STING cells cultured in the presence of TET. Means and standard deviations are shown (n = 6).
Hepatocyte-derived cell lines express low levels of STING that can be activated by cGAMP to induce a cytokine response.
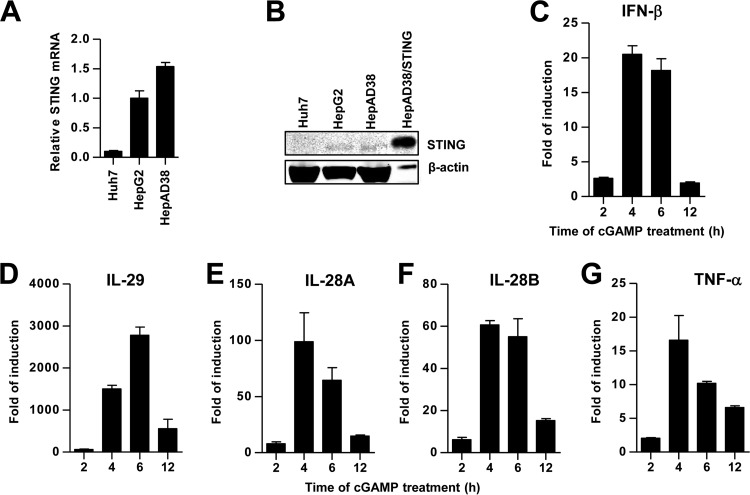
As an adaptor protein for various cytoplasmic and nuclear DNA sensors, STING plays a crucial role in antimicrobial defense and the maintenance of tissue homeostasis. Consistent with previous reports (15), we found that STING mRNA and protein levels were ∼10-fold higher in HepG2 and HepAD38 cells than in Huh7 cells (Fig. 3A and B). Treatment of HepG2 cells with cGAMP induced a type III IFN-dominant cytokine response and the cytokine mRNA levels peaked at 2 to 4 h posttreatment (Fig. 3C to G).
FIG 3.
Expression and functionality of STING in human hepatoma cells. The relative levels of STING mRNA (A) and protein (B) in the human hepatoma cells were determined by qRT-PCR and Western blot assays, respectively. The amounts of STING mRNA detected in each of the cell lines were normalized to β-actin mRNA and are expressed as relative levels to that in HepG2 cells. For the Western blot assay, a HepAD38-derived stable cell line expressing human STING (HepAD38/hSTING) was used as a positive control. β-Actin served as a loading control. Compared to other cells, one-tenth of the cell lysate from HepAD38/hSTING cells was loaded. (C to G) HepG2 cells were mock treated or treated with cGAMP at 10 μg/ml for 30 min and then harvested 2, 4, 6, and 12 h posttreatment. The levels of IFN-β, IL-29, IL-28A, IL-28B, and tumor necrosis factor alpha (TNF-α) mRNA were determined by qRT-PCR assays and normalized to β-actin mRNA and are expressed as the fold of induction over that in mock-treated controls. Means and standard deviations are presented (n = 2 to 4).
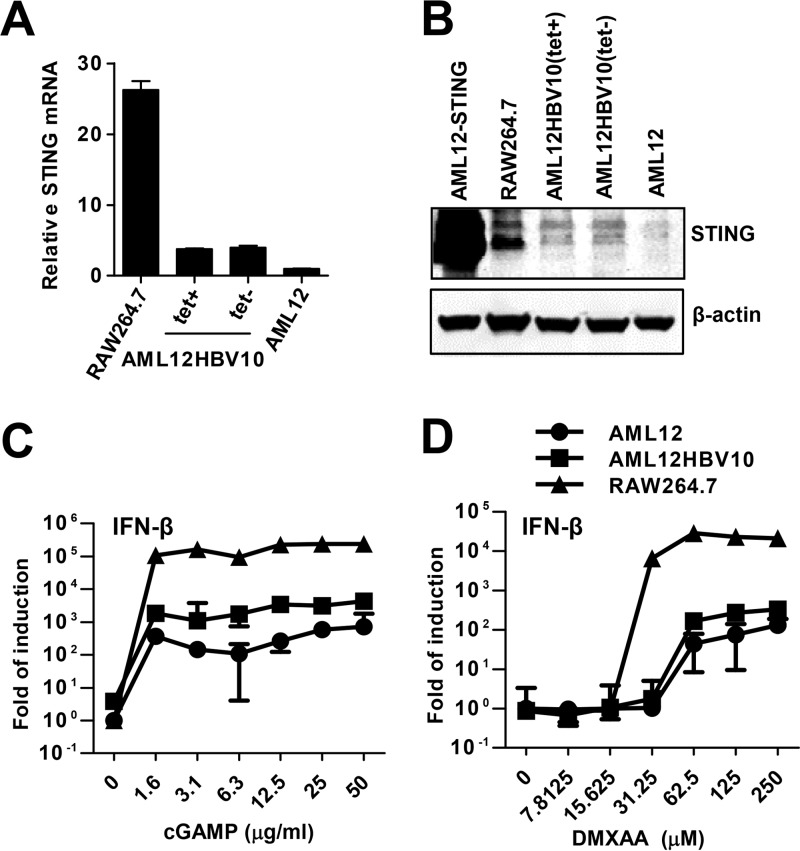
In addition, immortalized mouse hepatocyte line AML12 has been used to investigate the molecular mechanism underlying the immune control of HBV infection (27, 34). We have reported previously that STING agonist mediated activation of mouse macrophages leads to secretion of cytokines that efficiently suppress HBV replication in AML12 cells by reducing the amount of cytoplasmic viral nucleocapsids (28). To determine the expression and functionality of STING in AML12 and its derived cell line supporting TET-inducible HBV replication (AML12HBV10), the levels of STING mRNA and protein were measured by quantitative RT-PCR (qRT-PCR) and Western blot assays, respectively. AML12 cells transduced with retroviral vector expressing mouse STING (AML12-mSTING) and mouse macrophage cell line RAW246.7 were used as positive controls. As shown in Fig. 4A and B, both parental AML12 and AML12HBV10 cells expressed low but detectable levels of STING compared to RAW246.7 cells. Treatment of the cells with either cGAMP (Fig. 4C) or mouse STING agonist 5,6-dimethylxanthenone-4-acetic acid (DMXAA) (Fig. 4D) dose dependently induced the expression of IFN-β mRNA. The potency of the STING agonists to induce IFN-β expression in all three cell lines was positively correlated to their levels of STING expression.
FIG 4.
Expression and functionality of STING in immortalized mouse hepatocytes. (A) The levels of STING mRNA in parental AML12 and AML12HBV10 cells cultured in the presence or absence of TET were determined by a qRT-PCR assay. RAW264.7 cells served as a positive control. The amounts of STING mRNA detected in each of the cell lines were normalized to β-actin mRNA and are expressed as relative levels to that in AML12 cells. (B) The levels of STING protein in the indicated cells were determined by a Western blot assay. β-Actin served as a loading control. (C and D) RAW264.7, AML12, and AML12HBV10 cells were mock treated or treated with the indicated concentrations of cGAMP for 30 min (C) or else treated with the indicated concentrations of DMXAA for 4 h (D). The levels of IFN-β mRNA were determined by a qRT-PCR assay and normalized to β-actin mRNA and are expressed as the fold of induction over that in mock-treated AML12 cells. Means and standard deviations from a representative duplicate experiment are shown.
HBV replication does not inhibit STING agonist induced cytokine response.
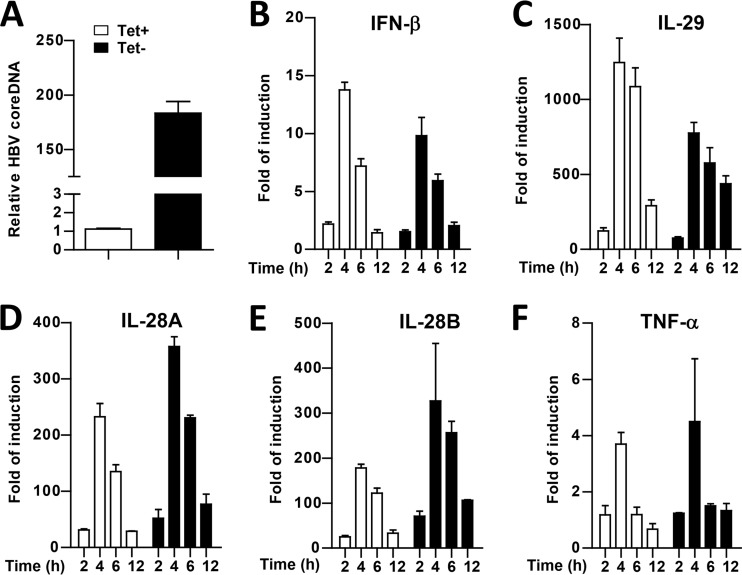
It was reported previously that HBV polymerase protein disrupts STING signaling by physical association with STING and inhibition of STING K63-linked polyubiquitination (15). However, such an effect was observed under the condition of polymerase protein and STING overexpression in human hepatoma cells. To investigate whether HBV inhibits STING signaling pathway in the context of HBV replication and at physiological level of STING expression, we compared cGAMP-induced cytokine expression in HepAD38 cells cultured in the presence or absence of TET. As expected, although the cells cultured in TET-containing medium do not have detectable amounts of HBV core DNA, HepAD38 cells cultured in TET-free medium induced the accumulation of HBV core DNA by 180-fold (Fig. 5A). However, the extent and kinetics of cGAMP-induced cytokine mRNA expression did not significantly differ in HepAD38 (Fig. 5B to F) with (TET−) or without (TET+) active HBV replication. A similar observation was obtained in AML12HBV10 cells (data not shown). Our results thus demonstrated that in the context of HBV replication and physiological level of STING expression, HBV does not interfere with STING signal transduction.
FIG 5.
HBV replication in hepatocytes does not inhibit STING agonist activation of cytokine response. HepAD38 cells were cultured in the presence (open bar) or absence (filled bar) of TET for 5 days. (A) HBV cytoplasmic core DNA levels were detected by a qPCR assay. The cells were then treated with 10 μg/ml cGAMP for 30 min and harvested at 2, 4, 6 and 12 h posttreatment. (B to F) The levels of IFN-β (B), IL-29 (C), IL-28A (D), IL-28B (E), and TNF-α (F) mRNA were determined by qRT-PCR assays and normalized to β-actin mRNA and are expressed as the fold of induction over that in mock-treated cells. Means and standard deviations are presented (n = 2 to 4).
STING agonist treatment induces a Janus kinase 1 (JAK1)-dependent antiviral response against HBV in immortalized mouse hepatocytes.
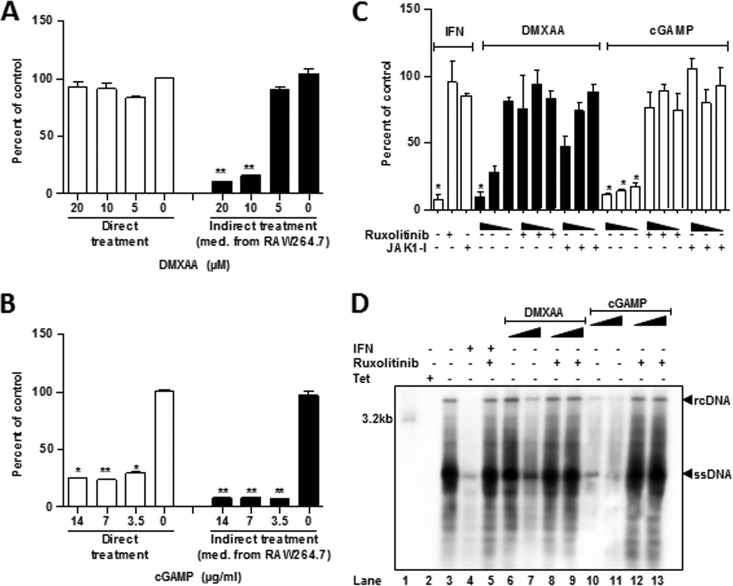
In order to determine whether activation of STING in hepatocytes inhibits HBV replication, we initially compared the indirect and direct effects of STING agonists on HBV replication in AML12HBV10 cells. As shown in Fig. 6A and B, in agreement with our previous report (28), treatment of AML12HBV10 cells with conditioned media of cGAMP- or DMXAA (5,6-dimethylxanthenone-4-acetic acid)-treated RAW246.7 cells efficiently inhibited HBV DNA replication. Interestingly, direct treatment of AML12HBV10 cells with cGAMP, but not DMXAA, reduced HBV DNA. Because DMXAA only induced IFN-β gene expression in AML12HBV10 cells at concentrations higher than 30 μM (Fig. 4D), we further examined its antiviral effects at higher concentrations and showed that direct treatment of AML12HBV10 cells with 500, 165, and 55 μM DMXAA dose dependently reduced HBV DNA (Fig. 6C and D).
FIG 6.
DMXAA inhibition of HBV replication in hepatocytes can be abolished by JAK inhibitors. (A and B) AML12HBV10 cells were cultured in the absence of TET and treated with the indicated concentrations of DMXAA (A) or cGAMP (B) (direct treatment) for 2 days. Alternatively, RAW264.7 cells were treated with the indicated concentrations of DMXAA (A) or cGAMP (B) for 12 h. The culture medium from the treated cells was diluted with fresh medium at 1:1 and then applied to AML12HBV10 cells for 2 days (indirect treatment). The levels of intracellular HBV core DNA were quantified by a qPCR assay and are presented as the percentages of that in untreated cells cultured in the absence of TET. (C) AML12HBV10 cells were cultured in the absence of TET and treated with mouse IFN-α at 10 U/ml or serial concentrations of cGAMP (10, 3, and 1 μg/ml) or DMXAA (500, 165, and 55 μM) in the absence or presence of the JAK inhibitor Ruxolitinib (20 μM) or JAK1-I (20 μM) for 2 days. Intracellular HBV core DNA levels were quantified by a qPCR assay and are presented as the percentages of that in untreated cells cultured in the absence of TET. Means and standard deviations are presented (n = 3). *, P < 0.01; **, P < 0.001. (D) AML12HBV10 cells were cultured in the absence of TET and treated with mouse IFN-α at 10 U/ml or serial concentrations of cGAMP (10 and 1 μg/ml) or DMXAA (500 and 165 μM) in the absence or presence of JAK inhibitors Ruxolitinib (20 μM) for 2 days. Intracellular HBV core DNA were detected by Southern blotting hybridization with a [32P]UTP-labeled riboprobe specific for minus-strand of HBV DNA.
We demonstrated previously that activation of MAVS and TRIF-mediated innate immune response in human hepatoma cells induces cytokine-independent intracellular antiviral response against HBV (35). Similarly, activation of STING not only induces inflammatory cytokine response but also directly induces the expression of some IFN-stimulated genes that may restrict HBV replication. In order to distinguish whether STING agonist-induced antiviral effect in hepatocytes is mediated by secretion of cytokines or via direct activation of intracellular antiviral pathways, we investigated whether the antiviral response is dependent on the activity of JAK1, a key enzyme for IFN-induced tyrosine phosphorylation of STAT1/2 and induction of antiviral response. As expected, treatment of AML12HBV10 cells with either of the two JAK1 inhibitors completely abolished IFN-α suppression of HBV replication. Similarly, the two JAK1 inhibitors also efficiently inhibited cGAMP- and DMXAA-induced HBV DNA reduction in the cells (Fig. 6C and D). Our results thus imply that STING agonists inhibit HBV DNA replication in the immortalized mouse hepatocytes by induction of antiviral cytokines, such as IFNs, but not through direct induction of intracellular antiviral response.
DMXAA inhibition of HBV replication in mouse hepatocytes is STING dependent.
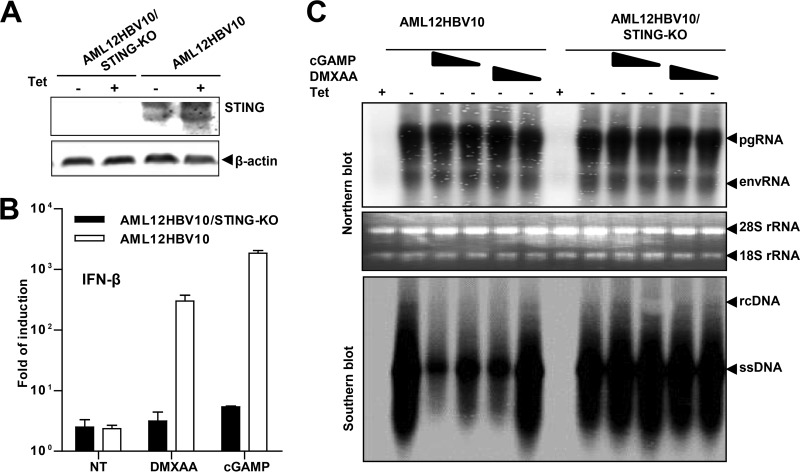
In order to ascertain that DMXAA-induced antiviral response is STING dependent and not an off-target effect, we knocked out STING in AML12HBV10 cells using CRISPR/Cas9 genome editing technology. As indicated by a Western blot assay, the AML12HBV10/STING-KO cells did not express detectable STING (Fig. 7A). As anticipated, treatment of the AML12HBV10/STING-KO cells with cGAMP or DMXAA did not induce IFN-β mRNA expression (Fig. 7B). Moreover, although treatment of parental AML12HBV10 cells with cGAMP or DMXAA suppressed HBV replication at the level of reducing the cytoplasmic core-associated DNA replication intermediates, knocking out STING completely abolished such an antiviral effect of cGAMP and DMXAA (Fig. 7C). The results thus confirmed that both cGAMP and DMXAA inhibition of HBV DNA replication in the mouse hepatocytes is STING dependent.
FIG 7.
DMXAA inhibition of HBV replication in mouse hepatocytes is STING dependent. (A) Parental AML12HBV10 cells and a cell clone selected after transfection of a plasmid expressing Cas9 and single guide RNA (sgRNA) specific to mouse STING were cultured in the absence or presence of TET for 2 days, and the expression of STING was examined by a Western blot assay. β-Actin served as a loading control. (B) Parental AML12HBV10 and derived STING knockout cell lines were mock treated or treated with 500 μM DMXAA or 10 μg/ml cGAMP for 4 h. IFN-β mRNA levels were determined by a qRT-PCR assay and are presented as the fold induction over that in untreated parental cells. Means and standard deviations are shown (n = 3). (C) The indicated cell lines were cultured in the presence of TET (+) or absence of TET (−) and mock treated or treated with 165 and 500 μM DMXAA or 3 and 10 μg/ml cGAMP. The cells were harvested 2 days after treatment. HBV mRNA levels were determined by Northern blotting assay (upper panel). pgRNA, pregenomic RNA; envRNA, viral mRNAs specifying envelope proteins. 28S and 18S rRNA served as loading controls. HBV core DNAs were determined by Southern blotting hybridization (lower panel). rcDNA, relaxed circular DNA; ssDNA, single-stranded DNA.
Activation of STING in human hepatoma cells inhibits HBV infection.
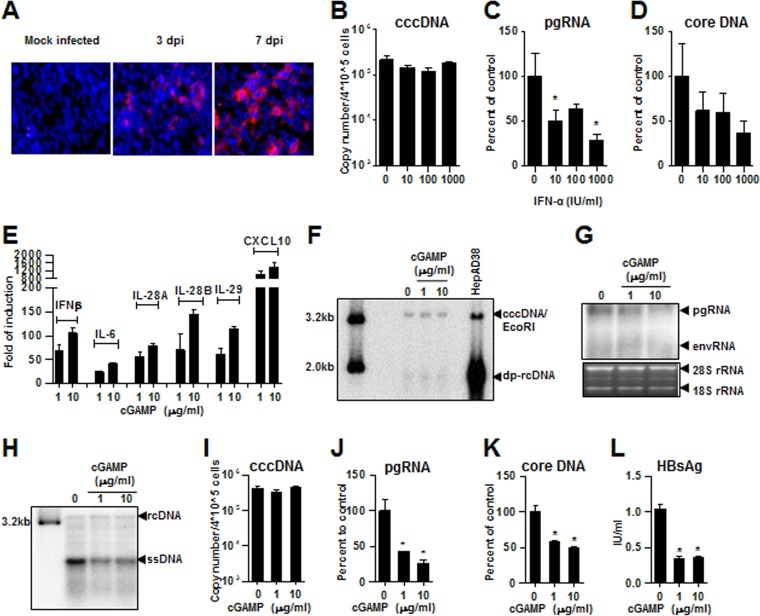
C3A is a clonal derivative of HepG2 that was selected for strong contact inhibition of growth and high albumin production (36). Because its metabolic feature is more relevant to normal hepatocytes than parental HepG2, C3A cells has been used for development of bioartificial liver devices (37). Upon constitutive expression of human NTCP via retroviral transduction, the derived cell line C3AhNTCP supports de novo HBV infection. As shown in Fig. 8A, infection of C3AhNTCP cells by HBV derived from HepAD38 cells at a multiplicity of infection (MOI) of 500 genome equivalents successfully infected C3AhNTCP cells, as indicated by immunostaining of HBcAg in the infected cells. Interestingly, HBV replication in the infected cells can be inhibited by IFN-α, which reduced viral RNA and core DNA, but not cccDNA (Fig. 8B to D). This finding is consistent with our previous report that IFN-α suppresses cccDNA transcription (38). Moreover, as observed in parental HepG2 cells, treatment of C3AhNTCP cells with cGAMP induced a robust cytokine response (Fig. 8E). Interestingly, treatment of HBV-infected C3AhNTCP starting at 48 h postinfection for 6 days did not apparently alter the amount of cccDNA but significantly inhibited HBV replication, as suggested by the dose-dependent reduction of intracellular HBV mRNA, core DNA, and secreted HBsAg (Fig. 8F to L). The results thus suggest that similar to IFN-α, the STING agonist-induced antiviral response in hepatocytes may suppress cccDNA transcription.
FIG 8.
cGAMP induces a robust cytokine response and inhibits viral replication in HBV-infected C3AhNTCP cells. (A) C3AhNTCP cells were infected with HBV at an MOI of 500 genome equivalents. The infected cells were fixed at 3 and 7 days postinfection (dpi) and HBV core protein was detected by indirect immunofluorescent staining (red). The cell nuclei were stained with DAPI (blue). (B to D) C3AhNTCP cells were infected with HBV at an MOI of 500 genome equivalents and left untreated or treated with the indicated concentrations of IFN-α for 8 days. HBV cccDNA, pgRNA, and core DNA levels were determined by qPCR assays. (E) C3AhNTCP cells were mock treated or treated with the indicated concentrations of cGAMP. The cells were harvested at 6 h posttreatment. The cytokine mRNAs were quantified by qRT-PCR assays, and findings are presented as the fold induction over mock-treated controls. (F to L) C3AhNTCP cells were infected with HBV at an MOI of 500 genome equivalents. The cells were mock treated or treated with the indicated concentrations of cGAMP at days 2, 4, and 6 postinfection and harvested at day 8 postinfection. (F) For detection of HBV cccDNA, Hirt DNA extracted from the cells was incubated at 88°C for 5 min to denature deproteinized (DP)-rcDNA into ssDNA, followed by restriction with EcoRI to convert cccDNA into unit-length double-stranded linear DNA (dslDNA; labeled cccDNA/EcoRI) and detected by Southern blotting hybridization. A 3.2-kb unit length and a 2.0-kb HBV linear DNA served as size markers. Hirt DNA extracted from HepAD38 cells cultured in the absence of TET for 10 days was used as a positive control. (G) HBV RNA levels, including pgRNA and viral mRNAs specifying envelope proteins (envRNAs), were determined by Northern blotting hybridization. 28S and 18S rRNA served as loading controls. (H) HBV DNA replication intermediates were determined by Southern blotting hybridization. rcDNA, relaxed circular DNA; ssDNA, single-stranded DNA. HBV cccDNA (I), pgRNA (J), and core DNA (K) were quantified by qPCR assays. The levels of HBsAg in culture medium at day 8 postinfection were measured by an ELISA (L). Means and standard deviations are shown (n = 4). *, P < 0.01.
DISCUSSION
Although there has been a report suggesting that HBV pgRNA activates RIG-I to induce an IFN and proinflammatory cytokine response in hepatocytes (39), extensive studies from many other laboratories during the last decades demonstrate that HBV infection of hepatocytes, either in cultures or in vivo in humans and animals, fails to activate a detectable IFN and proinflammatory cytokine response (7, 8). In order to understand the mechanism underlying HBV evasion of PRR-mediated host innate immune response, we first examined the role of the best-studied cytoplasmic DNA sensor, cGAS, in response to HBV infection and demonstrated that HBV replication neither activates nor inhibits the cGAS-STING signal transduction pathway in a hepatocyte-derived cell line (Fig. 2). Due to tight control of the nucleocapsid uncoating at the nuclear pore complex (40), lack of HBV DNA in the cytoplasm may be responsible for the evasion of cGAS sensing. However, whether HBV DNA is capable of binding to and activating cGAS or any other DNA sensors remains to be determined.
In addition to cGAS, several other PRRs have been reported to recognize viral DNA in the cytoplasm and/or nucleus to mount a cytokine response. Besides serving as a receptor for cGAMP, STING is a molecular hub for many other DNA sensors to induce cytokine gene expression and thus a key player in host cellular response to virus infection (41). STING is highly expressed in respiratory epithelial cells, lung macrophages, and a few other cell types, but at low levels in the liver and hepatocytes, as well as in hepatoma cells (http://www.proteinatlas.org/ENSG00000184584-TMEM173/tissue). In the present study, we show that the low-level expression of STING in hepatocyte-derived cells is able to mediate a cytokine response upon activation by cGAMP and its pharmacological agonist DMXAA (Fig. 3 and 4). In contrast to a previous report that overexpression of HBV polymerase protein in hepatoma cells inhibits STING signal transduction (15), our studies in human hepatoma and immortalized mouse hepatocytes showed that HBV did not apparently inhibit STING agonist-induced cytokine response in the context of HBV replication and physiologically relevant levels of STING (Fig. 5 and data not shown). Furthermore, we showed in immortalized mouse hepatocytes and HBV-infected NTCP-expressing human hepatoma cells that activation of STING induced a robust cytokine response that restricted HBV replication, most likely by inhibition of cccDNA transcription (Fig. 6 and 8). Collectively, although STING is expressed at a low level and fails to mediate an innate immune response to HBV infection in hepatocytes, our studies suggest that STING is a potential therapeutic target for pharmacological activation of host innate, and possibly also adaptive, immune responses to cure or durably control chronic HBV infections.
Hepatocytes are the primary host cells of HBV. HBV is not a cytopathic virus and is thus able to persistently replicate in hepatocytes. However, HBV infection acquired in adult life is generally able to induce a functionally efficient, HBV-specific T-cell response that resolves the infection within 6 months. Accumulating evidence suggests that both cytopathic and noncytopathic antiviral mechanisms contribute to the clearance of HBV during acute infection (42, 43). Cytokines released by HBV-specific T cells, especially CD8+ T cells, are believed to be the main cause of the early noncytolytic clearance of HBV. Immunologically, HBV-infected hepatocytes are not only the target cells of host immune responses but also essential players in the induction of host immune responses. In particular, activation of adaptive immune response relies on viral antigens produced by HBV-infected hepatocytes. However, how and where the hepatocyte-derived HBV antigens are presented to T lymphocytes remain to be determined. Recent studies suggest that liver resident macrophages (Kupffer cells) and dendritic cells play essential roles in the priming and activation of host adaptive immune response against HBV by shaping intrahepatic lymphoid organization, antigen presentation, expression of costimulating cell surface proteins, and secretion of inflammatory cytokines (44–46).
Therapeutically, activation of STING in liver-resident immune regulatory cells and hepatocytes induces a cytokine response that suppresses HBV replication. In addition, the antiviral cytokine response may also reduce viral antigen load through elimination or transcriptional suppression of cccDNA, which may attenuate the T cell exhaustion and favor the restoration of the HBV-specific T cell response (47). Moreover, activation of STING in hepatocytes, the antigen-producing cells, as well as in antigen presentation cells, may break immunotolerance against HBV and induce a functional HBV-specific adaptive immune response, particularly the cytolytic T cell response, to resolve the chronic viral infection. Encouragingly, along this line, several recent studies showed that cGAMP treatment induces an efficient antitumor immune response through activation of intratumor dendritic cells and subsequent enhancement of cross-presentation of tumor associated antigens to CD8 T lymphocytes (18, 48, 49).
In summary, our previous and current studies systematically characterized STING pathway and functional status in antigen presentation cells and hepatocytes and established a solid basis for the further development of STING agonists as immunotherapeutics for the treatment of chronic hepatitis B. The hypothesis that STING agonist therapy facilitates the induction of adaptive immune response against HBV will be investigated in future in HBV chronically infected immunocompetent animal models, such as HBV transgenic mice, adeno-associated virus vector-mediated HBV-transduced mice, or humanized mice with human immune system and liver cells (50–52).
MATERIALS AND METHODS
Cell culture.
The human hepatoma cell lines HepG2 and Huh7 and the immortalized mouse hepatocyte cell line AML12 have been described previously and cultured in DMEM/F-12 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (34, 35). C3A, a subclone of HepG2 (ATCC HB-8065), the murine macrophage cell line RAW264.7 (ATCC TIB-71), and GP2-293 cells (Clontech) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS (28, 53). HepAD38 and AML12HBV10 cell lines, which support high levels of HBV replication in a tetracycline (TET)-inducible manner, were maintained as previously described (29, 54).
Chemicals and reagents.
5,6-Dimethylxanthenone-4-acetic acid (DMXAA) was purchased from AdooQ Bioscience. IFN-α was purchased from PBL InterferonSource, and 2′3′-cGAMP was purchased from Invivogen. Janus kinase (JAK) inhibitor I and Ruxolilinib were purchased from Calbiochem and Selleckchem, respectively. Antibodies against HBV core protein and β-actin were obtained from Dako and Sigma-Aldrich, respectively. Antibodies against STING (catalog no. 13647), IRF3 (catalog no. 11904), and cGAS (catalog no. 15102) were purchased from Cell Signaling Technology. dsDNA90 was synthesized by annealing sense strand TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACATACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA to its antisense strand, with a green fluorescent dye, 6-carboxyfluorescein (FAM), labeled at the 5′ ends of both strands.
Plasmid construction.
STING cDNA was obtained from Origene cDNA clone NM_198282. Full-length STING was PCR amplified by using the STING forward primer (5′-ACACACGGATCCGCCACCATGCCCCACTCCAGCCTGCATCCA-3′) and STING reverse primer (5′-ACACACGCGGCCGCTCAAGAGAAATCC-3′). C-terminal truncated STING with deletion of amino acid 341 (aa341) to aa379 was PCR amplified by using STINGΔC forward primer (5′-ACACACGGATCCGCCACCATGCCCCACTCCAGCCTGCATCCA-3′) and STINGΔC reverse primer (5′-ACACACGCGGCCGCTCACTCTTCCTTTTCCTCCTGCCG-3′). After BamHI and NotI digestion, STING or STINGΔC cDNA was cloned into the pCX4bsr retroviral vector (Addgene) containing a blasticidin-resistant gene. Human cGAS cDNA clone was purchased from OriGene, and the coding region was subcloned into a puromycin-resistant gene expressing retroviral vector pQCXIP (Clontech) between the NotI and BamHI sites. All resulting plasmids were verified by DNA sequencing.
Package of pseudotyped retroviruses.
Pseudotyped retroviral particles were packaged in GP2-293 cells (Clontech) as previously described (53). Briefly, GP2-293 cells were cotransfected with plasmid pVSV-G that expresses vesicular stomatitis virus (VSV) glycoprotein (G) and retroviral vector expressing the desired protein in a molar ratio of 2:3. The culture media of transfected cultures were harvested every 24 h for 3 days. The pooled media were diluted at 1:1 with fresh DMEM/F-12 complete media and stored at −80°C until use.
Establishment of cell lines stably expressing cGAS and STING.
HepAD38 cells were seeded in a 12-well plate at a density of 5 × 105 cells per well and infected with medium containing pseudotyped retroviral particles expressing cGAS and STING the next day. At 2 days after infection, the cells were cultured with selection medium containing 2 μg/ml puromycin or 10 μg/ml blasticidin for 2 weeks. Antibiotic-resistant cells were pooled and expanded into cell lines. The expression of cGAS and/or STING expression of the cell lines was confirmed by Western blotting assay.
Generation of STING knockout AML12HBV10 cell line.
To knock out STING expression in AML12HBV10 cells, the cells were transfected with TMEM173 double nickase plasmid (Santa Cruz) using Lipofectamine 2000 (Life Technologies). At 2 days after transfection, the cells were reseeded at a density of 104 cells per 10-cm-diameter dish and cultured in medium containing 2 μg/ml puromycin. Approximately 2 weeks later, single cell clones were picked up and expanded into cell lines. The expression of STING in the individual cell clones was determined by a Western blot assay with a STING-specific antibody, and one cell line with undetectable STING expression was selected for further study.
Analysis of STING and cytokine expression by qRT-PCR assay.
For cytokine gene expression analysis, total cellular RNA was extracted by using TRIzol reagent (Invitrogen). cDNA was synthesized by using a SuperScript III Platinum One-Step qRT-PCR kit (Invitrogen). Real-time PCR assays were performed using a LightCycler 480 II (Roche). Primer sequence information for qRT-PCR analyses of human and mouse cytokines is provided in Table 1.
TABLE 1.
Primers for qPCR
| Genea | Species | Primer |
|
|---|---|---|---|
| Orientation | Sequence (5′–3′) | ||
| STING | Human | Sense | GAGATCTCTGCAGTGTGTGAA |
| Antisense | GCCGCAGATATCCGATGTAATA | ||
| IFN-β | Human | Sense | ACTGCCTCAAGGACAGGATG |
| Antisense | AGCCAGGAGGTTCTCAACAA | ||
| IL-29 | Human | Sense | TTCCAAGCCCACCACAAC |
| Antisense | TCCCTCACCTGGAGAAGC | ||
| IL-28A | Human | Sense | TCGCTTCTGCTGAAGGACTGCA |
| Antisense | CCTCCAGAACCTTCAGCGTCAG | ||
| IL-28B | Human | Sense | TCGCTTCTGCTGAAGGACTGCA |
| Antisense | CCTCCAGAACCTTCAGCGTCAG | ||
| TNF-α | Human | Sense | CTCTTCTGCCTGCTGCACTTTG |
| Antisense | ATGGGCTACAGGCTTGTCACTC | ||
| IL-6 | Human | Sense | AGACAGCCACTCACCTCTTCAG |
| Antisense | TTCTGCCAGTGCCTCTTTGCTG | ||
| CXCL10 | Human | Sense | GGTGAGAAGAGATGTCTGAATCC |
| Antisense | GTCCATCCTTGGAAGCACTGCA | ||
| β-Actin | Human | Sense | CACCATTGGCAATGAGCGGTTC |
| Antisense | AGGTCTTTGCGGATGTCCACGT | ||
| STING | Mice | Sense | TACTTCTGACCTGCGCCTTG |
| Antisense | CACGGTCTGTGTTTTGCTGG | ||
| IFN-β | Mice | Sense | TCCAAGAAAGGACGAACATTCG |
| Antisense | TGAGGACATCTCCCACGTCAA | ||
| IL-6 | Mice | Sense | GCCTTCTTGGGACTGATG |
| Antisense | CTGGCTTTGTCTTTCTTGTTA | ||
| TNF-α | Mice | Sense | GGCAGGTCTACTTTGGAGTCATTGC |
| Antisense | ACATTCGAGGCTCCAGTGAATTCGG | ||
| GAPDH | Mice | Sense | CATCAAGAAGGTGGTG |
| Antisense | CCTGTTGCTGTAGCC | ||
| pgRNA | HBV | Sense | GGTCCCCTAGAAGAAGAACTCCCT |
| Antisense | CATTGAGATTCCCGAGATTGAGAT | ||
| Core DNA | HBV | Sense | GGCTTTCGGAAAATTCCTATG |
| Antisense | AGCCCTACGAACCACTGAAC | ||
| cccDNA | HBV | Sense | GGGGCGCACCTCTCTTTA |
| Antisense | CCACCCAGGTAGCTAGAGTCATTAG | ||
TNF-α, tumor necrosis factor alpha; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blot assay.
Cells in a well of a 24-well plate were lysed with 100 μl of NuPAGE LDS sample buffer (Thermo Fisher Scientific) supplemented with 2.5% 2-mercaptoethanol (Sigma). The cell lysate was subjected to denaturing gel electrophoresis with NuPAGE 4–12% Bis-Tris Gel and NuPAGE MOPS SDS running buffer (Thermo Fisher Scientific). Proteins were transferred from the gel onto a polyvinylidene difluoride membrane using an iBlot 2 dry blotting system (Thermo Fisher Scientific). Membranes were blocked with TBST (50 mM Tris-HCl, pH 7.6; 150 mM NaCl; 0.1% Tween 20) containing 5% nonfat milk for 1 h, followed by incubation with the desired antibody overnight at 4°C. After a washing step with TBST, the membrane was incubated with LI-COR IRDye secondary antibodies. Membranes were again washed with TBST and imaged with a LI-COR Odyssey system (LI-COR Biotechnology).
Establishment and HBV infection of C3AhNTCP cells.
Human sodium taurocholate cotransporting polypeptide (NTCP) gene coding sequence was amplified from a cDNA clone purchased from Origene. A C-terminal C9 tag was added using PCR with the primers harboring C9 tag sequence and NotI and BamHI restriction enzyme sites. The purified PCR fragments were digested with restriction enzymes NotI and BamHI and cloned into a pQCXIP vector (Clontech). VSV-G protein pseudotyped retroviruses were packaged in GP2-293 cells. The C3AhNTCP cell line stably expressing human NTCP was established by infection of C3A cell line with the pseudotyped retroviruses and selected with medium containing 2 μg/ml puromycin. Puromycin-resistant cells were expanded into the cell line and designated C3AhNTCP. For HBV infection, C3AhNTCP cells were seeded into collagen-coated 24-well plates at a density of 4 × 105 cells per well and cultured in complete DMEM containing 3% dimethyl sulfoxide (DMSO). One day later, the cells were infected with HBV prepared from HepAD38 cell culture media at an multiplicity of infection (MOI) of 500 genome equivalents per cell in DMEM containing 4% polyethylene glycol 8000 (PEG-8000). The inocula were removed 24 h later, and the infected cultures were maintained in complete DMEM containing 3% DMSO until harvesting.
Analyses of HBV DNA and RNA.
HBV core DNA extraction from HepAD38, AML12HBV10, or C3AhNTCP cells and analyses using Southern blotting hybridization and real-time PCR assays were performed as described previously (34, 54). Total cellular RNA was extracted using TRIzol reagent (Invitrogen). HBV RNA and core DNA were analyzed by Northern and Southern blot hybridization assays, respectively, with [α-32P]UTP-labeled full-length riboprobes (55). Pregenomic RNA and core DNA were quantified by quantitative PCR (qPCR) assays with the primers listed in Table 1. HBV cccDNA was extracted by a modified Hirt DNA extraction procedure (56). The Hirt DNA preparation was digested with plasmid-safe ATP-dependent DNase (Epicentre), and cccDNA was then quantified by a qPCR assay with primers listed in Table 1.
ELISA.
HBsAg in culture medium was determined with an enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's directions (Autobio).
Immunofluorescence assay.
STING translocation and IRF3 nuclear localization, as well as the expression of HBV core protein, were detected by immunofluorescence assay. Briefly, cells were fixed with phosphate-buffered saline containing 4% paraformaldehyde, followed by incubation with 0.1% Triton X-100 for 20 min. The cells were then blocked and incubated with antibodies against STING, IRF3, or HBV core protein. Bound primary antibody was visualized by using Alexa Fluor 488-conjugated secondary antibodies (Invitrogen). Cell nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole).
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health (AI113267), Arbutus Biopharma, Inc., and the Commonwealth of Pennsylvania through the Hepatitis B Foundation.
REFERENCES
- 1.Ott JJ, Stevens GA, Groeger J, Wiersma ST. 2012. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2013 Mortality and Causes of Death Collaborators. 2015. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479–480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Falth M, Stindt J, Koniger C, Nassal M, Kubitz R, Sultmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate cotransporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Guo JT, Guo H. 2015. Metabolism and function of hepatitis B virus cccDNA: implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res 122:91–100. doi: 10.1016/j.antiviral.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang J, Block TM, Guo JT. 2012. The innate immune response to hepatitis B virus infection: implications for pathogenesis and therapy. Antiviral Res 96:405–413. doi: 10.1016/j.antiviral.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Wieland SF, Chisari FV. 2005. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol 79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu S, Chen J, Wu M, Chen H, Kato N, Yuan Z. 2010. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKεand DDX3. J Gen Virol 91:2080–2090. doi: 10.1099/vir.0.020552-0. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Ryu WS. 2010. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3: implications for immune evasion. PLoS Pathog 6:e1000986. doi: 10.1371/journal.ppat.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woltman AM, Op den Brouw ML, Biesta PJ, Shi CC, Janssen HL. 2011. Hepatitis B virus lacks immune activating capacity, but actively inhibits plasmacytoid dendritic cell function. PLoS One 6:e15324. doi: 10.1371/journal.pone.0015324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent IE, Zannetti C, Lucifora J, Norder H, Protzer U, Hainaut P, Zoulim F, Tommasino M, Trepo C, Hasan U, Chemin I. 2011. Hepatitis B virus impairs TLR9 expression and function in plasmacytoid dendritic cells. PLoS One 6:e26315. doi: 10.1371/journal.pone.0026315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar M, Jung SY, Hodgson AJ, Madden CR, Qin J, Slagle BL. 2011. Hepatitis B virus regulatory HBx protein binds to adaptor protein IPS-1 and inhibits the activation of beta interferon. J Virol 85:987–995. doi: 10.1128/JVI.01825-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei C, Ni C, Song T, Liu Y, Yang X, Zheng Z, Jia Y, Yuan Y, Guan K, Xu Y, Cheng X, Zhang Y, Wang Y, Wen C, Wu Q, Shi W, Zhong H. 2010. The hepatitis B virus X protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J Immunol 185:1158–1168. doi: 10.4049/jimmunol.0903874. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Li J, Chen J, Li Y, Wang W, Du X, Song W, Zhang W, Lin L, Yuan Z. 2015. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J Virol 89:2287–2300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertoletti A, Ferrari C. 2012. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut 61:1754–1764. doi: 10.1136/gutjnl-2011-301073. [DOI] [PubMed] [Google Scholar]
- 17.Thomsen MK, Nandakumar R, Stadler D, Malo A, Valls RM, Wang F, Reinert LS, Dagnaes-Hansen F, Hollensen AK, Mikkelsen JG, Protzer U, Paludan SR. 2016. Lack of immunological DNA sensing in hepatocytes facilitates hepatitis B virus infection. Hepatology 64:746–759. doi: 10.1016/j.jhep.2015.11.039. [DOI] [PubMed] [Google Scholar]
- 18.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paijo J, Doring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, Witte G, Messerle M, Hornung V, Kaever V, Kalinke U. 2016. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog 12:e1005546. doi: 10.1371/journal.ppat.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam E, Stein S, Falck-Pedersen E. 2014. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J Virol 88:974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Zhao K, Su X, Lu L, Zhao H, Zhang X, Wang Y, Wu C, Chen J, Zhou Y, Hu X, Wang Y, Lu M, Chen X, Pei R. 2017. MITA/STING and its alternative splicing isoform MRP restrict hepatitis B virus replication. PLoS One 12:e0169701. doi: 10.1371/journal.pone.0169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Hao R, Liu D, Liu X, Wu S, Guo S, Wang Y, Tien P, Guo D. 2016. Inhibition of hepatitis B virus replication by activation of the cGAS-STING pathway. J Gen Virol 97:3368–3378. doi: 10.1099/jgv.0.000647. [DOI] [PubMed] [Google Scholar]
- 26.Dansako H, Ueda Y, Okumura N, Satoh S, Sugiyama M, Mizokami M, Ikeda M, Kato N. 2016. The cyclic GMP-AMP synthetase-STING signaling pathway is required for both the innate immune response against HBV and the suppression of HBV assembly. FEBS J 283:144–156. doi: 10.1111/febs.13563. [DOI] [PubMed] [Google Scholar]
- 27.Cui X, Clark DN, Liu K, Xu XD, Guo JT, Hu J. 2016. Viral DNA-dependent induction of innate immune response to hepatitis B virus in immortalized mouse hepatocytes. J Virol 90:486–496. doi: 10.1128/JVI.01263-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo F, Han Y, Zhao X, Wang J, Liu F, Xu C, Wei L, Jiang JD, Block TM, Guo JT, Chang J. 2015. STING agonists induce an innate antiviral immune response against hepatitis B virus. Antimicrob Agents Chemother 59:1273–1281. doi: 10.1128/AAC.04321-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 41:1715–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. 2013. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu C, Guo H, Pan XB, Mao R, Yu W, Xu X, Wei L, Chang J, Block TM, Guo JT. 2010. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J Virol 84:9332–9340. doi: 10.1128/JVI.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo H, Jiang D, Ma D, Chang J, Dougherty AM, Cuconati A, Block TM, Guo JT. 2009. Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells. J Virol 83:847–858. doi: 10.1128/JVI.02008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sussman NL, Chong MG, Koussayer T, He DE, Shang TA, Whisennand HH, Kelly JH. 1992. Reversal of fulminant hepatic failure using an extracorporeal liver assist device. Hepatology 16:60–65. doi: 10.1002/hep.1840160112. [DOI] [PubMed] [Google Scholar]
- 37.Mavri-Damelin D, Damelin LH, Eaton S, Rees M, Selden C, Hodgson HJ. 2008. Cells for bioartificial liver devices: the human hepatoma-derived cell line C3A produces urea but does not detoxify ammonia. Biotechnol Bioeng 99:644–651. doi: 10.1002/bit.21599. [DOI] [PubMed] [Google Scholar]
- 38.Liu F, Campagna M, Qi Y, Zhao X, Guo F, Xu C, Li S, Li W, Block TM, Chang J, Guo JT. 2013. Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS Pathog 9:e1003613. doi: 10.1371/journal.ppat.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, Watanabe T, Iijima S, Sakurai Y, Watashi K, Tsutsumi S, Sato Y, Akita H, Wakita T, Rice CM, Harashima H, Kohara M, Tanaka Y, Takaoka A. 2015. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 42:123–132. doi: 10.1016/j.immuni.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Gallucci L, Kann M. 2017. Nuclear import of hepatitis B virus capsids and genome. Viruses 9:E21. doi: 10.3390/v9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog 8:e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Summers J, Jilbert AR, Yang W, Aldrich CE, Saputelli J, Litwin S, Toll E, Mason WS. 2003. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc Natl Acad Sci U S A 100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 44.Dienstag JL. 2009. Benefits and risks of nucleoside analog therapy for hepatitis B. Hepatology 49:S112–S121. doi: 10.1002/hep.22920. [DOI] [PubMed] [Google Scholar]
- 45.Publicover J, Gaggar A, Nishimura S, Van Horn CM, Goodsell A, Muench MO, Reinhardt RL, van Rooijen N, Wakil AE, Peters M, Cyster JG, Erle DJ, Rosenthal P, Cooper S, Baron JL. 2013. Age-dependent hepatic lymphoid organization directs successful immunity to hepatitis B. J Clin Invest 123:3728–3739. doi: 10.1172/JCI68182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Publicover J, Goodsell A, Nishimura S, Vilarinho S, Wang ZE, Avanesyan L, Spolski R, Leonard WJ, Cooper S, Baron JL. 2011. IL-21 is pivotal in determining age-dependent effectiveness of immune responses in a mouse model of human hepatitis B. J Clin Invest 121:1154–1162. doi: 10.1172/JCI44198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang L, Zhao Q, Wu S, Cheng J, Chang J, Guo JT. 2017. The current status and future directions of hepatitis B antiviral drug discovery. Expert Opin Drug Discov 12:5–15. doi: 10.1080/17460441.2017.1255195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corrales L, McWhirter SM, Dubensky TW Jr, Gajewski TF. 2016. The host STING pathway at the interface of cancer and immunity. J Clin Invest 126:2404–2411. doi: 10.1172/JCI86892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. 2017. Innate immune signaling and regulation in cancer immunotherapy. Cell Res 27:96–108. doi: 10.1038/cr.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang D, Liu L, Zhu D, Peng H, Su L, Fu YX, Zhang L. 2014. A mouse model for HBV immunotolerance and immunotherapy. Cell Mol Immunol 11:71–78. doi: 10.1038/cmi.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bility MT, Cheng L, Zhang Z, Luan Y, Li F, Chi L, Zhang L, Tu Z, Gao Y, Fu Y, Niu J, Wang F, Su L. 2014. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog 10:e1004032. doi: 10.1371/journal.ppat.1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guidotti LG, Matzke B, Schaller H, Chisari FV. 1995. High-level hepatitis B virus replication in transgenic mice. J Virol 69:6158–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao X, Guo F, Comunale MA, Mehta A, Sehgal M, Jain P, Cuconati A, Lin H, Block TM, Chang J, Guo JT. 2015. Inhibition of endoplasmic reticulum-resident glucosidases impairs severe acute respiratory syndrome coronavirus and human coronavirus NL63 spike protein-mediated entry by altering the glycan processing of angiotensin I-converting enzyme 2. Antimicrob Agents Chemother 59:206–216. doi: 10.1128/AAC.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, Guo JT. 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 87:6931–6942. doi: 10.1128/JVI.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang P, Liu F, Guo F, Zhao Q, Chang J, Guo JT. 2016. Characterization of novel hepadnaviral RNA species accumulated in hepatoma cells treated with viral DNA polymerase inhibitors. Antiviral Res 131:40–48. doi: 10.1016/j.antiviral.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo H, Mao R, Block TM, Guo JT. 2010. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J Virol 84:387–396. doi: 10.1128/JVI.01921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]