ABSTRACT
Oxazolidinones are promising candidates for the treatment of Mycobacterium tuberculosis infections. We isolated linezolid-resistant strains from H37Rv (Euro-American) and HN878 (East-Asian) strains; resistance frequencies were similar in the two strains. Mutations were identified in ribosomal protein L3 (RplC) and the 23S rRNA (rrl). All mutant strains were cross resistant to sutezolid; a subset was cross resistant to chloramphenicol. Mutations in rrl led to growth impairment and decreased fitness that may limit spread in clinical settings.
KEYWORDS: tuberculosis, oxazolidinones, linezolid, resistance, fitness, ribosome, Mycobacterium, Mycobacterium tuberculosis, antibiotic resistance, mycobacteria
TEXT
The oxazolidinone class of antibiotics inhibits the formation of protein synthesis initiation complexes by binding to domain V of the 23S rRNA (1). Linezolid (LZD), the first member of the oxazolidinones approved for clinical use, has recently been investigated as a potential treatment for drug-resistant strains of Mycobacterium tuberculosis (2, 3). LZD demonstrates time-dependent kill kinetics against replicating M. tuberculosis (4), bactericidal activity against nonreplicating bacilli (5), and good efficacy in mouse models (5). However, the long-term administration of LZD is limited due to side effects that include neuropathy and anemia (2, 6). Sutezolid (SZD; previously PNU-100480) is a next-generation oxazolidinone that has improved tolerance over long-term administration and improved efficacy against M. tuberculosis in a mouse model (7). Resistance to oxazolidinones has been studied in other bacteria and is mediated via mutations in domain V of the 23S rRNA (rrl), in the ribosomal protein L3 (rplC), or by the transporter OptrA (8). We sought to further characterize the mechanisms of oxazolidinone resistance in M. tuberculosis.
We isolated resistant mutant strains (RMs) against LZD by plating late-log-phase cultures of M. tuberculosis H37Rv (ATCC 25618) (Euro-American lineage) and HN878 (East-Asian lineage) on Middlebrook 7H10 agar with 10% (vol/vol) OADC (oleic acid-albumin-dextrose-catalase) supplement (Becton Dickinson) and 8 μM (2.7 μg/ml) of LZD (5× MIC) (9). We confirmed resistance by determining MICs on solid medium or in liquid medium. Solid MICs were determined in 24-well plates on 7H10-OADC agar and were defined as the lowest concentrations that prevented growth. MIC90 was determined in Middlebrook 7H9 liquid medium with 10% (vol/vol) OADC and 0.05% (wt/vol) Tween 80 (Tw); bacterial growth was measured by the optical density at 590 nm (OD590) after 5 days, and the MIC90 was defined as the concentration at which 90% of growth was inhibited (10). Sixteen resistant strains were isolated in H37Rv at a frequency of 2.3 × 10−9 (Table 1); 12 resistant strains were isolated in HN878 at a similar frequency of 3.3 × 10−9 (Table 2). All strains were confirmed as resistant to LZD and were also cross resistant to sutezolid (SZD) (Tables 1 and 2).
TABLE 1.
Oxazolidinone-resistant mutant strains of M. tuberculosis H37Rv
| Strain | MIC on solid medium (μM) |
L3 mutation | 23S RNA mutation | |
|---|---|---|---|---|
| Linezolid | Sutezolid | |||
| H37Rv | 3.1 | 1.6 | WT | WT |
| LP-LZD-RM2 | 50 | 12.5 | WT | G2814T |
| LP-LZD-RM3 | 25 | 12.5 | WT | G2814T |
| LP-LZD-RM4 | 50 | 12.5 | WT | G2814T |
| LP-LZD-RM24 | 50 | 25 | WT | G2814T |
| LP-LZD-RM1 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM11 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM12 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM13 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM14 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM15 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM16 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM17 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM21 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM22 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM23 | 50 | 12.5 | C154R | WT |
| LP-LZD-RM25 | 50 | 12.5 | C154R | WT |
| HN878 WT | 1.6 | 0.4 | WT | WT |
TABLE 2.
Oxazolidinone-resistant mutant strains of M. tuberculosis HN878
| Strain | MIC90 in liquid medium (μM)a |
L3 mutation | 23S RNA mutation | ||||
|---|---|---|---|---|---|---|---|
| LZD | SZD | CM | GM | KM | |||
| HN878 WT | 3 | 2 | 7 | 3 | 1 | WT | WT |
| HN-LZD-RM3 | 156 | 75 | 12 | 4 | 3 | WT | G2299T |
| HN-LZD-RM5 | 65 | 81 | WT | G2299T | |||
| HN-LZD-RM6 | WT | G2299T | |||||
| HN-LZD-RM9 | 107 | 61 | WT | G2299T | |||
| HN-LZD-RM10 | WT | G2299T | |||||
| HN-LZD-RM13 | 92 | 62 | WT | G2299T | |||
| HN-LZD-RM14 | WT | G2299T | |||||
| HN-LZD-RM1 | 60 | 31 | 412 | 3 | 2 | WT | A2689T |
| HN-LZD-RM11 | 94 | 62 | 112 | 3 | 4 | WT | G2814T |
| HN-LZD-RM2 | WT | G2814T | |||||
| HN-LZD-RM4 | WT | G2814T | |||||
| HN-LZD-RM8 | WT | G2814T | |||||
| H37Rv WT | 3.1 | 1.6 | WT | WT | |||
LZD, linezolid; SZD, sutezolid; CM, chloramphenicol; GM, gentamicin; KM, kanamycin.
We sequenced rrl and ribosomal protein L3 using primers TB-rrl-MMF1, CACACTGTTGGGTCCTGA; TB-rrl-MMF2, TGGAATCCGCTGTGAA; TB-rrl-MMF3, CAGGAGGTTGGCTTAGAA; TB-rrl-MMF4, TCGTGAACACCCTTGC; and TB-rrl-MMR1, CGCCGTAACTCTATGCA for rrl and primers TB-rplC-MMF1, TCGAGATGCGCACAC; and TB-rplC-MMR1, GGACGTCGAACAGCTC for rplC. In H37Rv, 12 strains had a C154R mutation in ribosomal protein L3 (L3C154R); four RMs had a G2814T mutation in rrl, equivalent to G2576T in Escherichia coli (Table 1). Of note, L3C154R is the dominant mutation observed in LZD-resistant clinical isolates (6, 11–13). In contrast, all of our strain HN878 resistant isolates selected in vitro had mutations in rrl; these were G2299T, A2689T, or G2814T, equivalent to G2061T, A2451T, and G2576T in E. coli, respectively (Table 2). Mutations A2689 and G2299 are located in the LZD binding site of the 23S rRNA, with A2689 being a conserved residue that is functionally important for the peptidyl-transferase activity of the ribosome (14–16). G2814 stacks on top of the active site nucleotide G2743 (equivalent to G2505 in E. coli); therefore, mutations in G2814 are likely to disrupt the LZD binding site (14). G2814T and G2299T were previously observed in M. tuberculosis LZDr strains isolated in vitro and in vivo (6, 17, 18). To the best of our knowledge, this is the first description of the A-to-T change at position 2689 encoded by rrl (rrlA2689T) being associated with LZD resistance.
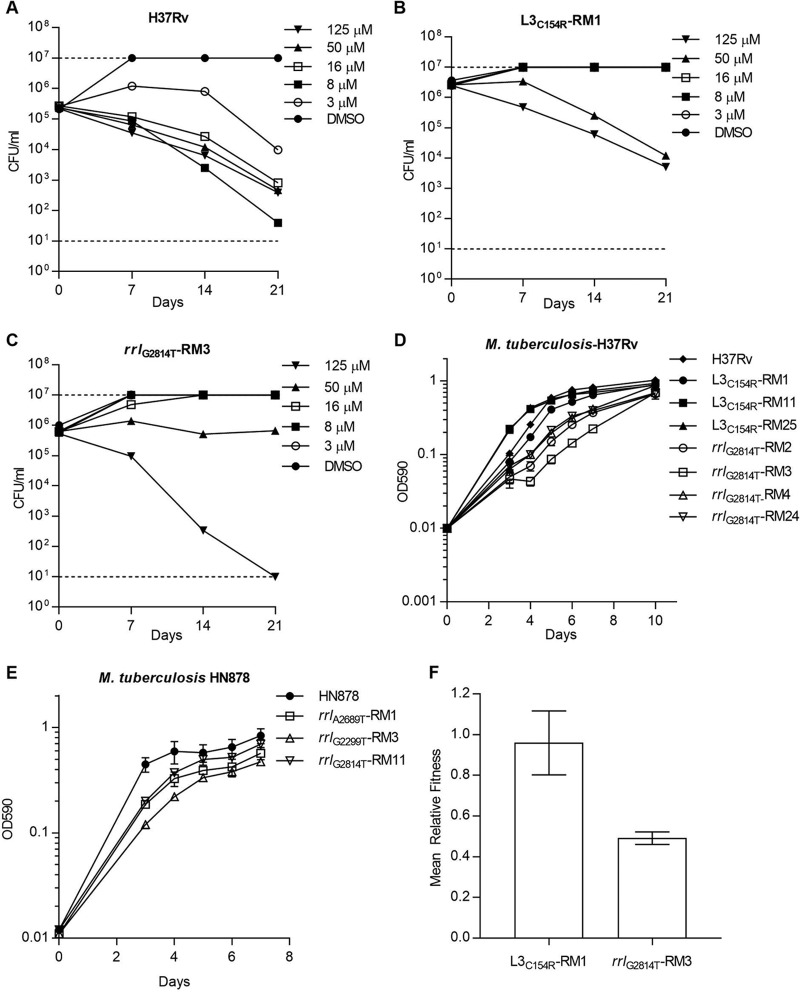
We determined bactericidal activity for LZD against wild-type (WT) and resistant strains. Bacterial viability was monitored over 21 d under replicating conditions using exponential-phase cultures of M. tuberculosis (5 × 105 CFU/ml) in 7H9-OADC-Tw. CFU were counted by serial dilution and plating after 3 to 4 weeks of incubation at 37°C. LZD demonstrated bactericidal activity against WT H37Rv with a minimum bactericidal concentration (MBC) equivalent to the MIC (3 μM or 1 μg/ml) (Fig. 1A). The L3C154R and rrlG2814T mutant strains were resistant to LZD bactericidal activity up to 16 μM (5.4 μg/ml) (Fig. 1B and C). At 50 μM (17 μg/ml), LZD had activity against the L3C154R strains and was bacteriostatic against the rrlG2814T strain. Full kill was achieved against all strains at 125 μM (42 μg/ml). Thus, the increased MICs of L3 and rrl mutant strains were translated into proportional increases in MBCs.
FIG 1.
Phenotypes of LZD-resistant M. tuberculosis. In vitro kill kinetics of LZD against the WT (A), L3C154R (B), and rrlG2814T (C) strains of M. tuberculosis H37Rv. Growth of mutant strains in liquid medium in an M. tuberculosis H37Rv background (D) and an M. tuberculosis HN878 background (E). Results are the means and standard deviations from three biological replicates. (F) Fitness of the H37Rv L3C154R and rrlG2814T strains relative to that of the parental WT strain as determined by in vitro coculture experiments. Results are the means and standard deviations from three biological replicates. DMSO, dimethyl sulfoxide.
Since LZD and chloramphenicol share a binding site in the 23S rRNA (14), we looked at cross-resistance. Strains carrying rrlA2689T or rrlG2814T were cross resistant to chloramphenicol, while rrlG2299T did not confer resistance (Table 2). As expected, rrl mutant strains remained susceptible to kanamycin and gentamicin, both of which bind to the 16S rRNA (Table 2). Cross-resistance to chloramphenicol is consistent with data from Staphylococcus aureus (15, 19) and Mycobacterium smegmatis (20). However, the lack of cross-resistance for rrlG2299T strains is surprising since the equivalent nucleotide in E. coli (G2061) interacts with the hydroxyl group of chloramphenicol (21). Furthermore, mutations of G2061 and A2062 are associated with chloramphenicol resistance in Thermus thermophilus (22). These results suggest that there are sufficient structural differences in the 23S rRNA of M. tuberculosis that cause chloramphenicol to bind in a different manner.
Mutations in 23S rRNA are commonly associated with growth defects in a diverse range of bacterial species, including M. smegmatis and M. tuberculosis (17, 19, 23). We conducted growth curves for representative strains in liquid medium; cultures were grown in 16-mm borosilicate tubes containing 5 ml of 7H9-OADC-Tw and incubated at 37°C with stirring at 250 rpm using an 8-mm stirrer bar. All strains with rrl mutations demonstrated impaired growth compared to WT strains (Fig. 1D and E), while strains with mutations in L3 were unimpaired (Fig. 1D).
The fitness cost of resistance mutations is an important contributor to the emergence and expansion of drug-resistant strains (24, 25). To investigate the fitness cost of LZD resistance, we conducted in vitro competition experiments as described previously (25). Briefly, 100 ml of 7H9-OADC was inoculated with ∼106 CFU of the WT and mutant strains in a 450-cm2 roller bottle. Coculture experiments were grown at 37° until stationary phase (OD590, ∼1). Serial dilutions were plated onto 7H10-OADC with and without 5 μM (1.7 μg/ml) LZD at day 0 and at stationary phase. CFU were counted after 4 to 5 weeks of incubation at 37°C. The relative fitness (W) of resistant (R) compared to that of susceptible (S) strains was calculated by W = ln(RF/RI)/ln(SF/SI) (25), where RI and SI are the number of resistant and susceptible cells at day 0, and RF and SF are the number of resistant and susceptible cells at stationary phase. Experiments were performed in biological triplicate. The H37Rv rrlG2814T strain had a fitness cost compared to that of the susceptible parental strain (Fig. 1F). In contrast, the L3C154R mutant strain had no fitness cost relative to that of the parent (Fig. 1F). Relative fitness costs have been previously shown to influence the spread of resistance, with low-cost resistance phenotypes being the most prevalent within clinical populations (25). From a limited number of clinical studies, the L3C154R single nucleotide polymorphism (SNP) is more prevalent than rrl SNPs within LZD-resistant strains (6, 11, 13). Whether or not this is because of the associated fitness cost requires further investigation. Resistance defects can be overcome by compensatory mechanisms as, for example, in S. aureus where changes in the copy number of 23S rRNA can achieve a balance between fitness and resistance (19). M. tuberculosis is unique in that it contains only a single copy of 23S rRNA, so it may not have access to the same compensatory mutations. Identifying compensatory mechanisms that overcome the fitness defects associated with rrl SNPs would provide further insights.
In conclusion, we demonstrate that mutations in the 23S rRNA (rrl) and the ribosomal protein L3 (RplC) are associated with resistance to the oxazolidinones LZD and SZD. Resistance led to decreased bactericidal activity from LZD. Mutations in rrl, but not L3, had a competitive fitness cost in vitro, suggesting that their appearance may be limited in clinical settings.
ACKNOWLEDGMENTS
We thank Ian Orme of Colorado State University for M. tuberculosis HN878. We thank Rhea N. Coler and Susan L. Baldwin at IDRI for useful discussion.
This research was supported with funding from the Bill and Melinda Gates Foundation under grant OPP1024038 and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award R01AI125160.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders played no role in the study, the preparation of the article, or the decision to publish.
REFERENCES
- 1.Leach KL, Swaney SM, Colca JR, McDonald WG, Blinn JR, Thomasco LM, Gadwood RC, Shinabarger D, Xiong L, Mankin AS. 2007. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell 26:393–402. doi: 10.1016/j.molcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Migliori GB, Eker B, Richardson MD, Sotgiu G, Zellweger JP, Skrahina A, Ortmann J, Girardi E, Hoffmann H, Besozzi G, Bevilacqua N, Kirsten D, Centis R, Lange C. 2009. A retrospective TBNET assessment of linezolid safety, tolerability and efficacy in multidrug-resistant tuberculosis. Eur Respir J 34:387–393. doi: 10.1183/09031936.00009509. [DOI] [PubMed] [Google Scholar]
- 3.Chang KC, Yew WW, Tam CM, Leung CC. 2013. WHO group 5 drugs and difficult multidrug-resistant tuberculosis: a systematic review with cohort analysis and meta-analysis. Antimicrob Agents Chemother 57:4097–4104. doi: 10.1128/AAC.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian V, Solapure S, Iyer H, Ghosh A, Sharma S, Kaur P, Deepthi R, Subbulakshmi V, Ramya V, Ramachandran V, Balganesh M, Wright L, Melnick D, Butler SL, Sambandamurthy VK. 2014. Bactericidal activity and mechanism of action of AZD5847, a novel oxazolidinone for treatment of tuberculosis. Antimicrob Agents Chemother 58:495–502. doi: 10.1128/AAC.01903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, Sala C, Dhar N, Vocat A, Sambandamurthy VK, Sharma S, Marriner G, Balasubramanian V, Cole ST. 2014. In vitro and in vivo activities of three oxazolidinones against nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:3217–3223. doi: 10.1128/AAC.02410-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee M, Lee J, Carroll MW, Choi H, Min S, Song T, Via LE, Goldfeder LC, Kang E, Jin B, Park H, Kwak H, Kim H, Jeon H-S, Jeong I, Joh JS, Chen RY, Olivier KN, Shaw PA, Follmann D, Song SD, Lee J-K, Lee D, Kim CT, Dartois V, Park S-K, Cho S-N, Barry CE III. 2012. Linezolid for treatment of chronic extensively drug-resistant tuberculosis. N Engl J Med 367:1508–1518. doi: 10.1056/NEJMoa1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallis RS, Jakubiec W, Kumar V, Bedarida G, Silvia A, Paige D, Zhu T, Mitton-Fry M, Ladutko L, Campbell S, Miller PF. 2011. Biomarker-assisted dose selection for safety and efficacy in early development of PNU-100480 for tuberculosis. Antimicrob Agents Chemother 55:567–574. doi: 10.1128/AAC.01179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, Wang D, Wang Z, Shen Y, Li Y, Fessler AT, Wu C, Yu H, Deng X, Xia X, Shen J. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 9.Ioerger TR, O'Malley T, Liao R, Guinn KM, Hickey MJ, Mohaideen N, Murphy KC, Boshoff HIM, Mizrahi V, Rubin EJ, Sassetti CM, Barry CE, Sherman DR, Parish T, Sacchettini JC. 2013. Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS One 8(9):e75245. doi: 10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ollinger J, Bailey MA, Moraski GC, Casey A, Florio S, Alling T, Miller MJ, Parish T. 2013. A dual read-out assay to evaluate the potency of compounds active against Mycobacterium tuberculosis. PLoS One 8(4):e60531. doi: 10.1371/journal.pone.0060531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckert P, Hillemann D, Kohl TA, Kalinowski J, Richter E, Niemann S, Feuerriegel S. 2012. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother 56:2743–2745. doi: 10.1128/AAC.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makafe GG, Cao Y, Tan Y, Julius M, Liu Z, Wang C, Njire MM, Cai X, Liu T, Wang B, Pang W, Tan S, Zhang B, Yew WW, Lamichhane G, Guo J, Zhang T. 2016. Role of the Cys154Arg substitution in ribosomal protein L3 in oxazolidinone resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 60:3202–3206. doi: 10.1128/AAC.00152-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter E, Rüsch-Gerdes S, Hillemann D. 2007. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 51:1534–1536. doi: 10.1128/AAC.01113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci U S A 105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson J, Kim DF, O'Connor M, Lieberman KR, Bayfield MA, Gregory ST, Green R, Noller HF, Dahlberg AE. 2001. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci U S A 98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youngman EM, Brunelle JL, Kochaniak AB, Green R. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117:589–599. doi: 10.1016/S0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 17.Hillemann D, Rüsch-Gerdes S, Richter E. 2008. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother 52:800–801. doi: 10.1128/AAC.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother 56:603–612. doi: 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besier S, Ludwig A, Zander J, Brade V, Wichelhaus TA. 2008. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob Agents Chemother 52:1570–1572. doi: 10.1128/AAC.01098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long KS, Munck C, Andersen TM, Schaub MA, Hobbie SN, Bottger EC, Vester B. 2010. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob Agents Chemother 54:4705–4713. doi: 10.1128/AAC.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlünzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 22.Gregory ST, Carr JF, Rodriguez-Correa D, Dahlberg AE. 2005. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J Bacteriol 187:4804–4812. doi: 10.1128/JB.187.14.4804-4812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sander P, Belova L, Kidan YG, Pfister P, Mankin AS, Böttger EC. 2002. Ribosomal and non-ribosomal resistance to oxazolidinones: species-specific idiosyncrasy of ribosomal alterations. Mol Microbiol 46:1295–1304. doi: 10.1046/j.1365-2958.2002.03242.x. [DOI] [PubMed] [Google Scholar]
- 24.Cohen T, Murray M. 2004. Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nat Med 10:1117–1121. doi: 10.1038/nm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]