ABSTRACT
As a growing number of clinical isolates of Mycobacterium abscessus are resistant to most antibiotics, new treatment options that are effective against these drug-resistant strains are desperately needed. The majority of the linkages in the cell wall peptidoglycan of M. abscessus are synthesized by nonclassical transpeptidases, namely, the l,d-transpeptidases. Emerging evidence suggests that these enzymes represent a new molecular vulnerability in this pathogen. Recent studies have demonstrated that inhibition of these enzymes by the carbapenem class of β-lactams determines their activity against Mycobacterium tuberculosis. Here, we studied the interactions of β-lactams with two l,d-transpeptidases in M. abscessus, namely, LdtMab1 and LdtMab2, and found that both the carbapenem and cephalosporin, but not penicillin, subclasses of β-lactams inhibit these enzymes. Contrary to the commonly held belief that combination therapy with β-lactams is redundant, doripenem and cefdinir exhibit synergy against both pansusceptible M. abscessus and clinical isolates that are resistant to most antibiotics, which suggests that dual-β-lactam therapy has potential for the treatment of M. abscessus. Finally, we solved the first crystal structure of an M. abscessus l,d-transpeptidase, LdtMab2, and using substitutions of critical amino acids in the catalytic site and computational simulations, we describe the key molecular interactions between this enzyme and β-lactams, which provide an insight into the molecular basis for the relative efficacy of different β-lactams against M. abscessus.
KEYWORDS: l,d-transpeptidase; Mycobacterium abscessus; beta-lactams; carbapenem; cephalosporin
INTRODUCTION
Mycobacterium abscessus is a rapid-growing nontuberculous mycobacterium that causes a wide range of opportunistic infections in humans (1). Vulnerable populations, including individuals with chronic lung disease, such as cystic fibrosis, are prone to high rates of M. abscessus infection (2, 3). Natural and acquired resistance to many antibiotics signifies limited options for the treatment of M. abscessus infections (4, 5). Although a comprehensive survey of the current drug resistance profile of clinical isolates of M. abscessus is not available, data from 28 strains isolated from cystic fibrosis patients over the past decade show high levels of resistance to most drugs used to treat this infection clinically (6). A drug that inhibits a novel target and has efficacy against strains with resistance to currently available drugs would provide a means to meet this clinical need.
β-Lactams are a powerful class of antibiotics that are widely used to treat a broad spectrum of bacterial infections. Historically, β-lactams were thought to act by inhibiting only the d,d-transpeptidases (also known as penicillin binding proteins) (7). This enzyme class catalyzes the final step of peptidoglycan biosynthesis in bacteria and forms transpeptide linkages between the fourth and third residues of peptide side chains, designated 4→3 linkages. While the peptidoglycan of M. abscessus contains classical 4→3 linkages, the majority of the peptidoglycan linkages are between the third residues of the peptide side chains (8). These linkages also predominate the peptidoglycan of Mycobacterium tuberculosis (9–11). These unique 3→3 linkages are generated by nonclassical transpeptidases, namely, l,d-transpeptidases (12). Emerging evidence shows that M. abscessus may be more susceptible to carbapenems (13) and that the potency of this β-lactam subclass is driven by its unique ability to inhibit l,d-transpeptidases (14). Currently, cefoxitin (a cephalosporin) and imipenem (a carbapenem) are the only two β-lactam agents that are included in guidelines for the treatment of M. abscessus, and between these two drugs, imipenem exhibits higher potency against this mycobacterium (15). A comprehensive study of l,d-transpeptidase interactions with additional β-lactams would yield further insight into the utility of these drugs for the treatment of M. abscessus infections.
The genome of M. abscessus encodes orthologs of five l,d-transpeptidases present in M. tuberculosis (16). In this study, we cloned, expressed, and purified two l,d-transpeptidases from M. abscessus, namely, LdtMab1 and LdtMab2, and assessed the abilities of a wide variety of β-lactams to inhibit these enzymes. Next, we evaluated whether the β-lactams that inhibited the l,d-transpeptidases also inhibited the growth of both susceptible and drug-resistant clinical isolates of M. abscessus. Additionally, we used X-ray crystallography, computational simulation, and substitutions of critical amino acids involved in the active site to determine molecular interactions between β-lactams and LdtMab2.
RESULTS
Carbapenems and cephalosporins inhibit LdtMab1 and LdtMab2 to various extents.
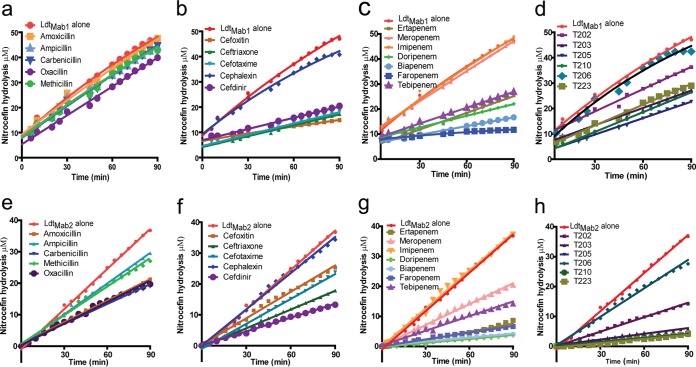
We measured the relative inhibition of LdtMab1 and LdtMab2 activities by a variety of β-lactam antibiotics. The tested β-lactams included penicillins (amoxicillin, ampicillin, carbenicillin, methicillin, and oxacillin), cephalosporins (cefdinir, cefotaxime, cefoxitin, ceftriaxone, and cephalexin), a penem (faropenem), commercially available carbapenems (biapenem, doripenem, ertapenem, imipenem, meropenem, and tebipenem), and novel experimental carbapenems (T202, T203, T205, T206, T210, and T223) (see Fig. S1 in the supplemental material) (14). Nitrocefin, a chromogenic β-lactam that is hydrolyzed by LdtMab1 and LdtMab2, was used as a substrate to monitor the activities of these enzymes in the presence of β-lactams.
Relative inhibition of LdtMab1.
None of the tested penicillins inhibited LdtMab1 (Fig. 1a). With the exception of cephalexin, which showed only minimal inhibition, all cephalosporins appeared to inhibit LdtMab1 to similar degrees (Fig. 1b). Among the carbapenems, while neither meropenem nor imipenem appeared to have an inhibitory effect, the relative order in which the others inhibited LdtMab1 was biapenem > doripenem = ertapenem > tebipenem (Fig. 1c). Similarly, experimental carbapenems inhibited LdtMab1 in the order of T205 > T210 = T203 = T223 > T202, while T206 showed no inhibition effect (Fig. 1d). Interestingly, among all the β-lactams tested, faropenem, a penem, inhibited LdtMab1 to the greatest extent (Fig. 1c).
FIG 1.
Nitrocefin hydrolysis by LdtMab1 and LdtMab2 in the presence of β-lactams. Profiles of nitrocefin hydrolysis by LdtMab1 in the presence of penicillins (a), cephalosporins (b), commercially available carbapenems and penem (c), and experimental carbapenems (d) and nitrocefin hydrolysis by LdtMab2 in the presence of penicillins (e), cephalosporins (f), commercially available carbapenems and penem (g), and experimental carbapenems (h) are illustrated.
Relative inhibition of LdtMab2.
In contrast to LdtMab1, all of the tested penicillins exhibited moderate inhibition of LdtMab2. The greatest to least inhibition was observed in the following order: carbenicillin = oxacillin = amoxicillin > methicillin = ampicillin (Fig. 1e). Among the cephalosporins, while cephalexin failed to inhibit LdtMab2, the relative extent to which the others inhibited the enzyme was cefdinir > ceftriaxone > cefotaxime > cefoxitin (Fig. 1f). With the exception of imipenem, the carbapenems were the most potent inhibitors of LdtMab2, with the following relative inhibition profile: doripenem = biapenem > ertapenem > tebipenem > meropenem (Fig. 1g). Among the experimental carbapenems, T206 showed no inhibition, and the others inhibited the enzyme in the following descending order: T223 = T210 = T205 > T203 > T202 (Fig. 1h). Faropenem, a penem, was also a potent inhibitor of LdtMab2 (Fig. 1g). As a class, the carbapenems were the most efficient inhibitors of LdtMab2, with doripenem and biapenem showing the highest degree of inhibition.
Doripenem and cefdinir exhibit synergy against M. abscessus.
We hypothesized that the combination of a carbapenem and a cephalosporin, the two subclasses of β-lactams that inhibited LdtMab1 and LdtMab2 to the greatest extent, may exhibit synergistic activity by targeting enzymes in M. abscessus that are differentially susceptible to these β-lactam subclasses. To test this hypothesis, we selected the following carbapenems based on their relative abilities to inhibit LdtMab1 and LdtMab2 (Fig. 1a to h): doripenem, biapenem, tebipenem, ertapenem, T205, T210, and faropenem (a penem). Among the cephalosporins, we chose cefdinir, as it was the most potent inhibitor of these enzymes. Drug combinations containing a carbapenem and cefdinir, each at various concentrations, were evaluated in a checkerboard assay against a pansusceptible M. abscessus strain, ATCC 19977. The combination of doripenem and cefdinir inhibited M. abscessus growth with a fractional inhibitory concentration (FIC) index (FICI) of 0.50, an indication of synergy between these drugs against this bacterium (Table 1). The combination of an experimental carbapenem, T210, and cefdinir was also synergistic against M. abscessus, with an FICI of 0.50. The FICI of the combination of biapenem and cefdinir was 0.62, an indication of mild synergy (see Table S1 in the supplemental material).
TABLE 1.
In vitro activities of cefdinir and doripenem, alone and in combination, against five M. abscessus strainsa
| M. abscessus strain | MIC (μg/ml) against M. abscessus |
MIC (μg/ml) of cefdinir/MIC (μg/ml) of doripenem against M. abscessus (FICI) | |
|---|---|---|---|
| Cefdinir | Doripenem | ||
| ATCC 19977 | 8–16 | 4–8 | 2–4/1–2 (0.5) |
| 1N | 8–16 | 4–8 | 1–2/2–4 (0.62) |
| 2N | 8–16 | 8–16 | 1–2/4–8 (0.62) |
| 5N | 4–8 | 4–8 | 0.25–0.5/2–4 (0.56) |
| 13N | 4–8 | 4–8 | 0.25–0.5/2–4 (0.56) |
MICs of cefdinir and doripenem and FICI values for the combination of cefdinir and doripenem are shown. ATCC 19977 is a reference M. abscessus strain, and 1N, 2N, 5N, and 13N are clinical isolates.
Next, we assessed whether the combination of doripenem and cefdinir exhibited synergy against clinical isolates of M. abscessus. For this task, we chose four strains isolated from cystic fibrosis patients (6), which exhibit a range of resistance against the antibacterials currently used to treat M. abscessus infections (Table S1). Notably, strains 5N and 13N were resistant to imipenem, and strains 1N and 2N displayed intermediate resistance to this carbapenem. The FICI of the combination of doripenem and cefdinir was <1 for all four strains, indicating mild synergy between these two β-lactams against these strains (Table 1). Most interestingly, the reduction of the MIC of doripenem and cefdinir, when used as a combination, is highly significant for therapeutic applications.
Crystal structure of LdtMab2 provides insight into its inhibition by β-lactams.
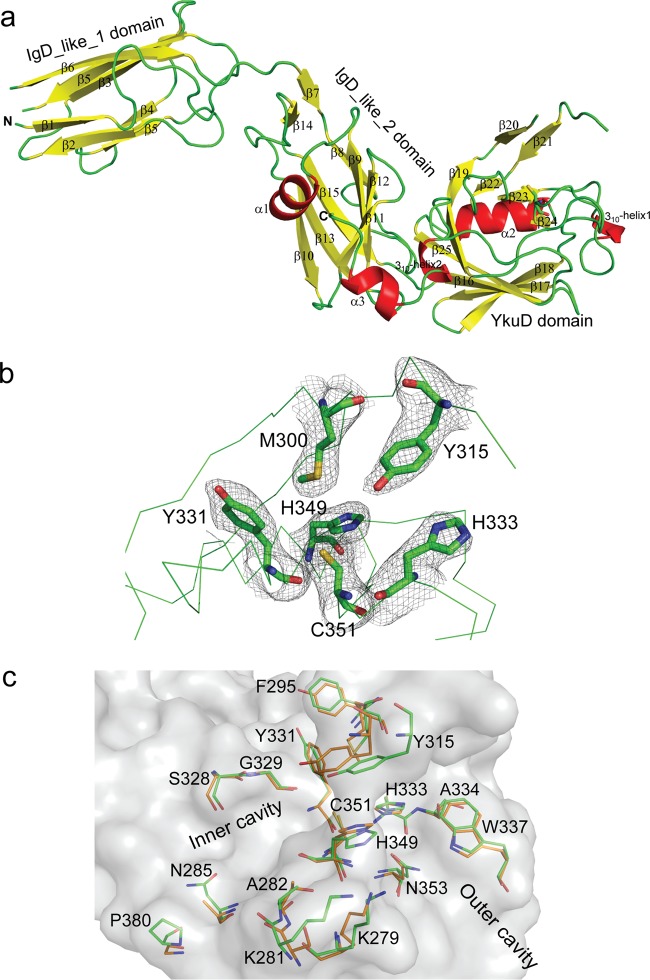
We hypothesized that the various levels of inhibition of LdtMab1 and LdtMab2 by different β-lactams could be attributed to differences in the molecular interactions of the drugs with these enzymes. As each individual β-lactam is unique in structure and chemical composition, the interactions of β-lactams with the enzymes and, consequently, the levels of enzyme inhibition are likely to differ. We determined the crystal structure of LdtMab2 and performed molecular dynamics studies with select β-lactams to gain insight into the molecular interactions between the drugs and the enzyme. LdtMab2 was selected for structural studies, as its ortholog LdtMt2 is the primary l,d-transpeptidase in M. tuberculosis (10, 17).
The crystal structure of LdtMab2 (fragment ΔN41) was solved at a 2.98-Å resolution (see Table S2 in the supplemental material). Electron density was not observed for residues 42 to 55. Therefore, the structure reported corresponds to residues 56 to 406. There are six chains in the asymmetric unit of LdtMab2. We superposed five chains (chains B, C, D, E, and F) to chain A to calculate their root mean square deviation (RMSD) values along the Cα atoms from residues 56 to 406. Chain B superposed with an RMSD of 0.432 Å, chain C superposed with an RMSD of 0.48 Å, chain D superposed with an RMSD of 0.663 Å, chain E superposed with an RMSD of 0.718 Å, and chain F superposed with an RMSD of 0.969 Å. Superposition of different chains revealed maximum variation along the Cα backbone in the IgD-like domain spanning residues 56 to 144 and in the YkuD domain spanning residues 295 to 322.
We next compared the structure of LdtMab2 (Fig. 2a) with that of LdtMt2 (PDB accession number 5DU7) (14). LdtMab2 superposed to LdtMt2 along the Cα backbone atoms corresponding to residues 56 to 406 with an RMSD of 1.52 Å. IgD-like domain 1 (18) in the N terminus of LdtMab2 showed the most conformational differences compared to the corresponding domain in LdtMt2 (Fig. S2). This suggests that the N-terminal domain of LdtMab2 is flexible. In the YkuD domain of LdtMab2, the flap loop (residues 304 to 313) is disordered. The major differences between LdtMab2 and LdtMt2 occur in the catalytic domain (Fig. 2b), which belongs to the YkuD family (Pfam 03734). We superposed the YkuD domains of the two enzymes (Fig. 2c) and compared critical structural differences. While the active sites of LdtMab2 and LdtMt2 appear structurally similar, there are several differences in amino acids that may affect their ligand specificity and enzymatic activity. Notably, many residues in the inner cavity that are involved in interactions with the side chains of β-lactams (14) are not conserved. These residues are F295, Y331, N285, P380, K281, and A282.
FIG 2.
Crystal structure of LdtMab2. (a) Crystal structure of LdtMab2 detailing the tertiary structure of the enzyme. (b) Electron density map (2Fo − Fc), contoured at 1 σ, of the active site of LdtMab2. (c) Superposition of l,d-transpeptidase active sites of LdtMab2 (green) and LdtMt2 (orange).
Molecular dynamics simulations of β-lactam binding to LdtMab2.
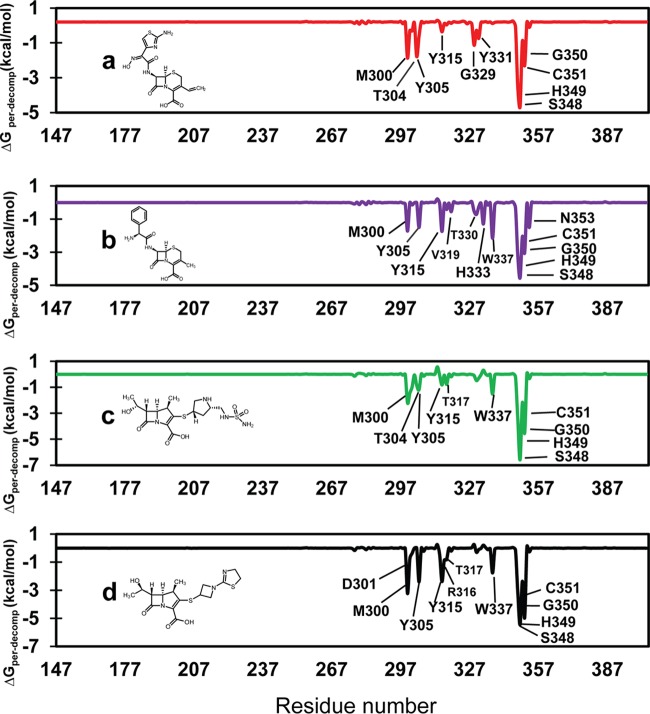
We made several unsuccessful attempts to cocrystallize LdtMab2 with β-lactams to gain insight into molecular interactions between the enzyme and this antibiotic class. As an alternate approach, we performed molecular dynamics simulations of interactions of select β-lactams with the LdtMab2 active site. Cefdinir, cephalexin, doripenem, and tebipenem were chosen for this analysis, as they represent both weak and strong inhibitors of the enzyme (Fig. 1f and g). These β-lactams were docked into the active site of the experimentally generated three-dimensional (3-D) crystal structure of LdtMab2 (see Table S3 in the supplemental material). The docked LdtMab2–β-lactam complexes were used as starting points for molecular dynamics simulations. A total of 100 ns of molecular dynamics simulations was carried out in an explicit water solvent for each LdtMab2–β-lactam pair. After the initial 30 ns, all pairs were fairly stable during the entire remaining period of the molecular dynamics simulations, with RMSD values, with respect to the average structure, of 1.14 ± 0.31, 1.49 ± 0.28, 1.38 ± 0.33, and 1.18 ± 0.35 for cefdinir, cephalexin, doripenem, and tebipenem, respectively (Fig. S3). In addition, we calculated the binding energies for the LdtMab2–β-lactam pairs using the final 20 ns of molecular dynamics simulations for a total of 2,000 snapshots (Table 2). Carbapenems consistently exhibited lower binding free energy (ΔGbind) than the cephalosporins, indicating that they bind more tightly with LdtMab2. Between the two carbapenems tested, ΔGbind values were lowest for doripenem and highest for tebipenem. Therefore, doripenem binds more tightly than tebipenem to LdtMab2. These data are concordant with the relative strengths with which doripenem and tebipenem inhibit the activity of LdtMab2, as observed in the nitrocefin hydrolysis assay (Fig. 1g). Between the two cephalosporins, the lower ΔGbind value for cefdinir indicates that it binds more tightly than cephalexin to LdtMab2, again in agreement with data from the nitrocefin hydrolysis inhibition assay, where cefdinir exhibited higher potency and cephalexin exhibited the lowest potency (Fig. 1f).
TABLE 2.
Thermodynamic parameters that describe binding interactions of LdtMab2 with β-lactamsa
| β-Lactam | EvdW (kcal · mol−1) | Eelec (kcal · mol−1) | GGB (kcal · mol−1) | Gnonpolar (kcal · mol−1) | ΔGbind (kcal · mol−1) |
|---|---|---|---|---|---|
| Cefdinir | −39.81 | −71.81 | 89.17 | −4.69 | −27.14 |
| Cephalexin | −27.80 | −72.35 | 83.24 | −5.20 | −22.11 |
| Doripenem | −43.64 | −58.95 | 69.36 | −5.15 | −38.38 |
| Tebipenem | −44.26 | −52.44 | 69.26 | −4.96 | −32.40 |
Free energy of the binding (ΔGbind) of β-lactams to LdtMab2 calculated from van der Waals interactions (EvdW), electrostatic interactions (Eelec), and polar (GGB) and nonpolar (Gnonpolar) contributions.
Interactions between β-lactams and the residues in the active site of l,d-transpeptidases affect the overall inhibition of these enzymes (14). To further understand the relevance of specific residues in the active site of LdtMab2 for binding to β-lactams, the average binding free energies of decomposition for relevant residues were measured against their interactions with cefdinir, cephalexin, doripenem, and tebipenem (Fig. 3). These β-lactams interacted with residues M300, Y305, Y315, S348, H349, G350, and C351, and their interactions exhibited a range of free energies (Table S4). Cefdinir made additional interactions with residues T304, G329, and Y331 (Fig. 3a). While T304 is part of the active-site flap, G329 and Y331 are located in the inner cavity of the active site of LdtMab2 (Fig. 2b and c). In the molecular dynamics simulations, the 100 ns snapshot shows that the 2-aminothiazole of the R2 side chain of cefdinir makes hydrogen bond interactions with these residues (Fig. S4). Unlike cefdinir, cephalexin exhibited low binding energies with active-site residues T304, G329, and Y331 but exhibited additional interactions with outer cavity residues W337 and N353, residue V319 in the active-site flap, and residue H333 in the catalytic core (Fig. 2b and c and 3b). While most of the interactions of doripenem and tebipenem with LdtMab2 were similar, the two carbapenems made critically different interactions with residues T304, Y315, and R316 (Fig. 3c and d and Table S4).
FIG 3.
Profile of binding free energies contributed by each residue to stabilize the LdtMab2–β-lactam complex. Residues with ΔGper-decomp values of ≥0.5 kcal/mol are labeled. Per-decomp refers to per residual decomposition. (a) LdtMab2-cefdinir; (b) LdtMab2-cephalexin; (c) LdtMab2-doripenem; (d) LdtMab2-tebipenem.
Role of LdtMab2 active-site residues in β-lactam ring hydrolysis.
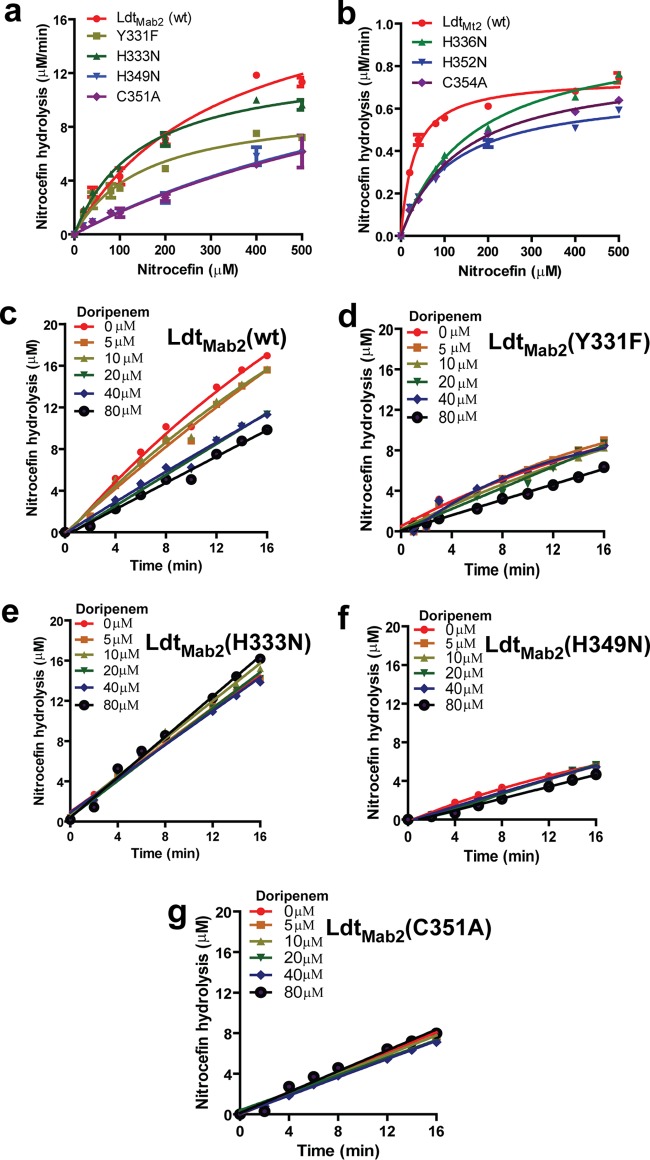
Guided by the crystal structure and molecular dynamics studies, we selected residues in the active site of LdtMab2 that make critical interactions with β-lactams to further assess their contributions to the overall enzyme activity. For this task, we generated LdtMab2 with the following amino acid substitutions: Y331F, H333N, H349N, and C351A. We observed that these mutants hydrolyzed nitrocefin at varying rates (Fig. 4a). Somewhat surprisingly, the C351A mutant hydrolyzed nitrocefin with the same Vmax as that of wild-type LdtMab2, although the mutant showed an ∼3-fold-higher Km for nitrocefin (Table 3). The H349N mutant also showed an ∼3-fold increase in the Km for nitrocefin. For the Y331F and H333N mutants, while the Vmax decreased, the Km did not change significantly. Next, we investigated whether the phenomenon of nitrocefin β-lactam ring hydrolysis in the absence of catalytically important residues also occurred with LdtMt2, the ortholog of LdtMab2 in M. tuberculosis. Based on the alignment of the amino acid sequences of these enzymes (Fig. S5), we generated similar active-site mutants in LdtMt2: H336N (corresponding to H333N in LdtMab2), H352N (corresponding to H349N in LdtMab2), and C354A (corresponding to C351A in LdtMab2). These LdtMt2 mutants hydrolyzed nitrocefin with similar Vmax values but with an increased Km (Fig. 4b).
FIG 4.
Role of l,d-transpeptidase active-site amino acid residues in β-lactam hydrolysis and inhibition by doripenem. (a) Kinetics of nitrocefin hydrolysis by LdtMab2 wild-type (wt) and mutant proteins. (b) Kinetics of nitrocefin hydrolysis by LdtMt2 wild-type and mutant proteins. (c to g) Inhibition of the nitrocefin hydrolysis activity of LdtMab2 wild-type and mutant proteins by doripenem at various concentrations.
TABLE 3.
Kinetic parameters of nitrocefin hydrolysis by LdtMab2 and LdtMt2a
| Enzyme | Mean Vmax (μM/min) ± SD | Mean Km (μM) ± SD |
|---|---|---|
| LdtMab2wt | 19.8 ± 2.03 | 328.3 ± 65.5 |
| LdtMab2Y331F | 9.7 ± 0.6 | 155.7 ± 25.8 |
| LdtMab2H333N | 12.9 ± 0.5 | 147.5 ± 15.0 |
| LdtMab2H349N | 18.8 ± 5.3 | 1,010 ± 395 |
| LdtMab2C351A | 18.6 ± 5.8 | 1,022 ± 441 |
| LdtMt2wt | 0.74 ± 0.02 | 30.0 ± 2.9 |
| LdtMt2H336N | 0.96 ± 0.03 | 163.3 ± 14.4 |
| LdtMt2H352N | 0.69 ± 0.02 | 115.7 ± 11.1 |
| LdtMt2C354A | 0.81 ± 0.02 | 140.8 ± 6.9 |
wt, wild type.
We expected cysteine (C351 in LdtMab2 and the corresponding residue C354 in LdtMt2) mutants to be catalytically inactive, as this amino acid is directly involved in β-lactam ring opening in carbapenems (14). The binding of a β-lactam to an l,d-transpeptidase has been proposed to follow three steps (19): initial noncovalent binding followed by the acylation of the catalytic cysteine and the subsequent hydrolysis of the acyl complex. The rates of the final step can vary significantly among different β-lactam classes and l,d-transpeptidases. For instance, with LdtMt2 and carbapenems, this rate is very low, and stable acyl adducts are detected even after 24 h (14). LdtMab2C351A and LdtMt2C354A were able to hydrolyze nitrocefin albeit at a reduced rate (Fig. 4a and b). There is a precedent for β-lactam ring hydrolysis without the involvement of a catalytic residue. It has been speculated that a water molecule in the active site of the β-lactamase TEM-1 with a catalytic serine substitution can act as a nucleophile and open the β-lactam ring (20). Therefore, it is possible that nitrocefin reacts with water at the catalytic site. Histidine in the active site of LdtMt2 facilitates the protonation of the β-lactam ring nitrogen (14). As the H333N mutant (and LdtMt2H336N) was able to hydrolyze nitrocefin (Fig. 4a and b), the existence of an alternate proton donor is likely. A recent study suggested a role of water in the protonation of the β-lactam ring nitrogen in LdtMt2 (21).
Next, we used doripenem, a carbapenem, to probe the involvement of Y331, H333, H349, and C351 in LdtMab2, as the corresponding residues in LdtMt2 are critical for reactions with carbapenems (14). As expected, a gradual decrease in nitrocefin hydrolysis with increasing concentrations of doripenem was observed with wild-type LdtMab2 (Fig. 4c). However, the nitrocefin hydrolysis activities of the H333N, H349N, and C351A mutants were not altered by doripenem (Fig. 4d to g). This observation confirms that these residues are involved in interactions with doripenem.
DISCUSSION
In this study, we determined the relative inhibition of the M. abscessus l,d-transpeptidases LdtMab1 and LdtMab2 by different classes of β-lactams and determined the MICs of the β-lactams against M. abscessus that were the most potent inhibitors of these critical enzymes. Interestingly, we observed that combinations of β-lactams from different classes exhibit synergy against M. abscessus, which suggests broader clinical application of dual-β-lactam therapy. Finally, we determined the first crystal structure of a key M. abscessus transpeptidase, LdtMab2; performed molecular dynamics studies that predicted binding affinities of various β-lactams; and performed enzymatic assays with active-site mutants of LdtMab2 that provided an insight into the molecular interactions of this enzyme with β-lactams.
The genome of M. abscessus encodes orthologs of five l,d-transpeptidases present in M. tuberculosis (10, 16). As none of the orthologs in M. abscessus have been characterized, here, we chose to focus on LdtMab2, as its ortholog in M. tuberculosis, LdtMt2, is the dominant l,d-transpeptidase, and its loss renders M. tuberculosis susceptible to amoxicillin-clavulanate (10). LdtMab2 and LdtMt2 share 80% amino acid sequence similarity (see Fig. S5 in the supplemental material).
We hypothesized that the mechanism of β-lactam ring opening of nitrocefin by LdtMab2 would be similar to the proposed β-lactam ring opening in carbapenems and penems by LdtMt2 (14, 22). This mechanism proposed the roles of cysteine (C351 in LdtMab2) and histidine (H333 in LdtMab2) residues in β-lactam ring opening and acylation, where the cysteine acts as a nucleophile and subsequently becomes acylated by the carbonyl carbon of the β-lactam ring. Simultaneously, the histidine protonates the β-lactam ring nitrogen. However, nitrocefin hydrolysis by LdtMab2 C351A and H333N substitution mutants suggests that the opening of the β-lactam ring in nitrocefin does not follow the same mechanism and potentially involves bulk water present in the catalytic site or involves another residue. Nitrocefin is a cephalosporin, a β-lactam subclass with a chemical composition that is distinct from those of carbapenems and penems. It is likely that the interaction of cephalosporins with an l,d-transpeptidase is distinct from those of other β-lactam subclasses.
Optimization of β-lactam selection for inclusion in regimens holds significant untapped potential for the clinical treatment of M. abscessus infections. Current M. abscessus treatment guidelines include only two β-lactams: cefoxitin (a cephalosporin) and imipenem (a carbapenem) (23). However, we observed only a modest ability of these agents to inhibit LdtMab1 and LdtMab2, whereas other β-lactams were much more potent inhibitors of these enzymes (Fig. 1). In particular, faropenem (a penem), biapenem, and doripenem (both carbapenems) showed exceptional abilities to inhibit both LdtMab1 and LdtMab2. Among cephalosporins, cefdinir (an oral agent) was observed to have the greatest inhibition of LdtMab2. While some of these potent agents (for example, biapenem and faropenem) are not currently available in the United States, doripenem is FDA approved and holds promise for clinical use against M. abscessus.
Of note, several novel experimental carbapenems newly synthesized by our laboratory (14), including T205 and T223, showed inhibition of LdtMab1 and LdtMab2, respectively (Fig. 1). These promising novel agents merit further study to define their safety, efficacy, and potential for further drug development for use against M. abscessus.
Our study is the first to demonstrate that combinations of dual β-lactams exhibit synergy against M. abscessus. This synergy is likely attributed to nonredundant targets and the differential inhibition of both nonclassical (LdtMab1 and LdtMab2) and classical (d,d-transpeptidases) transpeptidases, a concept which to date has been underappreciated and underexplored in mycobacteria. An antimycobacterial regimen that contains only a single β-lactam agent may not fully inhibit mycobacterial peptidoglycan synthesis, which in turn may prevent full antimicrobial killing that occurs when this key pathway is effectively shut down. In particular, the combination of doripenem and cefdinir was synergistic and very potent against M. abscessus (Table 1), and this combination should be considered for further preclinical development for this pathogen. Importantly, dual-agent synergy was observed with both the key laboratory strain ATCC 19977 and multiple-drug-resistant clinical isolates, which shows the potential for dual-β-lactam therapy to revolutionize the design of antimycobacterial clinical regimens. Synergy of dual β-lactams against M. tuberculosis and Enterococcus faecalis was recently observed by two different groups (24, 25).
Importantly, a significant degree of intrinsic resistance to β-lactams in M. abscessus is conferred by a chromosomally encoded highly active class A β-lactamase, BlaMab (26). BlaMab inactivates both β-lactams and β-lactamase inhibitors with the β-lactam chemical nucleus (for example, clavulanate and sulbactam). Surprisingly, compared to penicillins and carbapenems, cephalosporins are more resistant to hydrolysis by BlaMab (26). Among β-lactamase inhibitors, BlaMab is effectively inhibited by avibactam, a non-β-lactam agent (27), and the addition of avibactam to carbapenems has shown promise in vitro against M. abscessus (6, 28). We anticipate that even more profound synergy would be observed with the addition of avibactam to a dual-β-lactam regimen. In the future, we intend to examine the potential of avibactam and other β-lactamase inhibitors to further enhance the potency of dual-β-lactam regimens.
MATERIALS AND METHODS
Bacterial strains, growth media, and drugs.
Mycobacterium abscessus ATCC 19977 was used as the primary strain. In addition, four M. abscessus clinical strains isolated from cystic fibrosis patients, labeled 1N, 2N, 5N, and 13N, were used for MIC determinations. All strains were grown in Middlebrook 7H9 broth with albumin-dextrose-catalase enrichment with constant shaking at 37°C. β-Lactams (amoxicillin, ampicillin, carbenicillin, methicillin, oxacillin, cefoxitin, ceftriaxone, cefotaxime, cephalexin, cefdinir, ertapenem, meropenem, imipenem, doripenem, biapenem, faropenem, and tebipenem) were procured from a commercial vendor (Sigma-Aldrich). The following novel experimental carbapenems synthesized by our group were also tested: T202, T203, T205, T206, T210, and T223 (14).
Cloning, expression, and purification of proteins.
The amino acid sequences of the l,d-transpeptidases of M. tuberculosis, namely, LdtMt1 and LdtMt2 (9, 10), were used to identify orthologs in M. abscessus by using a BLAST search. These respective orthologs, MAB_3165c and MAB_1530, are referred to here as LdtMab1 and LdtMab2, respectively. A putative transmembrane domain, similar to that in LdtMt2 (29), exists in LdtMab2. During cloning, this domain was excluded and only the fragment containing residues 42 to 406 (ΔN41) was included to facilitate the overexpression and purification of the soluble protein. The corresponding gene fragment from M. abscessus ATCC 19977 was cloned into the multiple-cloning site in pET28a+ with an N-terminal His6 tag that can be removed with tobacco etch virus (TEV) protease (29). Proteins were overexpressed and purified as follows. Escherichia coli BL21δε3 (catalog number C2527H; NEB Labs) cells transformed with a pET28a+ vector carrying the desired gene fragments were grown in Luria-Bertani (LB) medium at 37°C to exponential phase (culture A600 of ∼0.8), induced with 0.25 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG), and grown overnight at 16°C at 220 rpm with orbital shaking before harvesting by centrifugation. The cells were lysed by sonication in a buffer containing 50 mM Tris (pH 8.0), 400 mM sodium chloride, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 1 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) and centrifuged to separate the soluble protein. The supernatant was passed through a 5-ml HisTrap HP nickel affinity column (GE Healthcare Life Sciences) preequilibrated with wash buffer containing 50 mM Tris (pH 8), 400 mM sodium chloride, 1 mM TCEP, and 0.1 mM PMSF. After the column was washed with 25 column volumes of wash buffer, the bound protein was eluted with a gradient of 1 to 1,000 mM imidazole. The His6 tag was removed from LdtMab2 by overnight incubation with 2 mg of TEV protease at 4°C while dialyzing the protein against a buffer containing 50 mM Tris (pH 8), 150 mM sodium chloride, 0.5 mM TCEP, and 0.1 mM PMSF. This mixture was then passed over the nickel affinity column to remove the His6 tag, the remaining uncleaved fusion protein, and the His6-tagged TEV protease. Fractions containing LdtMab2 were pooled, further purified on a Superdex 200 16/60 column (GE Life Sciences), concentrated up to 30 mg/ml by using a 10,000-molecular-weight-cutoff (MWCO) Vivaspin 20 concentrator, and stored at −80°C. The LdtMab1 fragment containing residues 32 to 248 (ΔN31) was also cloned, expressed, and purified in a similar manner. The N-terminal 31 residues in LdtMab1 that encompass a putative transmembrane domain were excluded to facilitate the overexpression and purification of the soluble protein.
Site-directed mutagenesis.
Single-amino-acid substitutions of LdtMab2 (fragment ΔN41) were constructed by site-directed mutagenesis as described previously (30). Briefly, the following primers were used to generate the indicated amino acid substitutions: GTCGTACAGCGGTATTTTCGTGCACGCGGCACCG and CGGTGCCGCGTGCACGAAAATACCGCTGTACGAC for Y331F, CAGCGGTATTTACGTGAATGCGGCACCGTGGTCCG and CGGACCACGGTGCCGCATTCACGTAAATACCGCTG for H333N, GCGCACCAACACCAGCAATGGCTGCTTGAACGTCAG and CTGACGTTCAAGCAGCCATTGCTGGTGTTGGTGCGC for H349N, and CAACACCAGCCACGGCGCGTTGAACGTCAGCACGGC and GCCGTGCTGACGTTCAACGCGCCGTGGCTGGTGTTG for C351A (the sequence representing the mutagenized codon is in italic type in each sequence). For each mutagenesis procedure, two separate PCR mixtures, with a forward or reverse primer, NEB fusion polymerase, and high-fidelity buffer, were used on a pET28a+ vector carrying the wild-type sequence for LdtMab2 (fragment ΔN41) as the template. DNAs from the two reactions were purified, combined, denatured at 95°C, slowly renatured to 37°C, and digested with DpnI. This DNA was used to transform E. coli DH5α (catalog number C2987H; NEB Labs), and the plasmid harboring the desired mutation was purified. E. coli BL21δε3 (catalog number C2527H; NEB Labs) was used to overproduce proteins.
Nitrocefin hydrolysis assays.
Nitrocefin (CalBioChem) is a chromogenic β-lactam whose native and hydrolyzed forms can be monitored at 496 nm. LdtMab1 and LdtMab2, each at 10 μM, were reacted with 100 μM nitrocefin in a 100-μl reaction mixture volume at room temperature in a solution containing 25 mM HEPES-morpholineethanesulfonic acid (MES)-diethanolamine and 300 mM NaCl (pH 5.0). Nitrocefin hydrolysis was monitored by using a microplate reader (Bio-Rad). The concentration (c) of hydrolyzed nitrocefin was calculated from its absorbance (A), molar extinction coefficient (ε) (20,500) and path length (l) (0.5 cm) values by using Beer's law: A = εcl. To assess nitrocefin hydrolysis by LdtMab1 and LdtMab2 in the presence of β-lactams, these proteins were added, in separate reaction mixtures at a 10 μM final concentration, to a reaction mixture containing 100 μM nitrocefin and 200 μM various β-lactams. The amount of hydrolyzed nitrocefin was then determined spectroscopically as described previously (14). Nitrocefin in buffer alone (without enzyme) was included to correct for the low level of non-enzyme-driven hydrolysis of nitrocefin.
Crystallization of LdtMab2.
Crystals of LdtMab2 (ΔN42) were grown by using the sitting-drop vapor diffusion method at 20°C in a solution containing 13% polyethylene glycol 8000 (PEG 8000) and 110 mM sodium citrate tribasic dihydrate (pH 5.5). The crystals appeared in 4 to 5 days and grew to their final size within 3 to 4 weeks. Crystals were flash-frozen and stored in liquid N2 until use.
Data collection and structure determination.
X-ray diffraction data were collected at a wavelength of 1.03 Å at beamline 19-ID at the Advanced Photon Source (Argonne National Laboratories). The diffraction data were recorded with a Pilatus3 detector and processed with HKL3000 (31). The LdtMab2 crystal belongs to the primitive monoclinic space group P21, with 6 molecules in the asymmetric unit. The crystal structure of LdtMab2 was solved by the molecular replacement method using PHASER-MR (32) in the PHENIX suite of programs. The coordinates of the M. tuberculosis protein LdtMt2 (PDB accession number 5DU7) were used as a template. The initial density modification, model building, and refinement were performed by using AutoBuild (33), subjected to multiple rounds of crystallographic refinement with phenix.refine from the Phenix suite of programs (34), and rebuilt to fit the electron density with COOT (35). The final model of LdtMab2 has an R factor value of 20.37% and an Rfree value of 27.09% for all data between 47.08- and 2.98-Å resolutions. The crystallographic parameters and final refinement statistics are summarized in Table S2. Structure figures were prepared by using the PyMOL Molecular Graphics System, version 1.5.0.4 (Schrödinger, LLC).
Molecular docking, molecular dynamics, and analysis of binding free energy.
The crystal structure of LdtMab2 was used as the starting point for docking and molecular dynamics simulations. The initial docking of cefdinir, cephalexin, doripenem, and tebipenem with LdtMab2 was performed by using Molegro Virtual Docker (MVD) software (36). Docked results were visually inspected to ensure that an acceptable drug-enzyme interaction was present. The docked LdtMab2-inhibitor complexes were used as starting structures for the classical molecular dynamics simulations. The simulations performed here were similar to the procedures applied previously to the l,d-transpeptidase LdtMt2 of M. tuberculosis (37).
MIC and fractional inhibitory concentration.
A standard broth microdilution method (38) was used to determine the MIC of β-lactams against M. abscessus strains. Briefly, an M. abscessus culture grown to exponential phase (A600 of ∼0.5 to 0.6) was used to inoculate ∼105 bacilli into each well of microtiter culture plates containing a β-lactam in 2-fold serial dilutions with concentrations ranging from 128 to 0.5 μg/ml. The M. abscessus cell pellet size was recorded by visual inspection after 3 days of incubation at 30°C without shaking, according to CLSI guidelines (39). The MIC is the lowest concentration of the drug at which an M. abscessus cell pellet could not be observed. The checkerboard titration assay, a modification of the broth dilution assay, was used to determine the fractional inhibitory concentration (FIC) of each β-lactam in a combination, as described previously (40). Briefly, two β-lactams, each starting at a concentration equal to its MIC and serially diluted 2-fold, were added to wells in a microtiter culture plate. In this setup, all possible 2-fold dilution combinations up to 1/32 the MIC of each drug were included. An M. abscessus culture grown to exponential phase (A600 of ∼0.5 to 0.6) was used to inoculate ∼105 bacilli into the wells. The mixtures were incubated at 30°C, and cell pellet sizes were recorded after 3 days. The FIC of each drug and the FIC index (the sum of the FICs of each drug in the combination) were calculated as described previously (40). The final MIC and FIC data represent the averages of results from two biological replicates.
Accession number(s).
Coordinates and structure factors have been submitted to the PDB under accession number 5UWV.
Supplementary Material
ACKNOWLEDGMENTS
G.L. was responsible for overall study conception, study design, cloning, data interpretation, and manuscript writing; P.K. was responsible for study design, X-ray crystallography, microbiology and biochemical studies, data analysis, and manuscript preparation; V.C. was responsible for protein expression and purification and crystallization; J.R.A.S., J.L., and C.N.A. were responsible for molecular dynamics studies; F.D. was responsible for microbiology studies; S.L.G. was responsible for diffraction of crystals; S.B. was responsible for interpreting structure and enzyme activity data; K.A.C. was responsible for data interpretation and manuscript preparation; and S.L. and J.S.F. were responsible for the design and the synthesis of the evolved carbapenems.
This work was supported by National Institutes of Health awards DP2OD008459 and R21AI121805 and by Cystic Fibrosis Foundation award LAMICH16GO.
Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank CNPQ Brazilian Agency for financial support (process no. 407096/2016-7) and CHPC from South Africa and HiPerGator Computational Center at the University of Florida for computational resources.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00866-17.
REFERENCES
- 1.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 2.Olivier KN, Weber DJ, Wallace RJ Jr, Faiz AR, Lee JH, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, Edwards LJ, Chakraborti S, Knowles MR, Nontuberculous Mycobacteria in Cystic Fibrosis Study Group. 2003. Nontuberculous mycobacteria. I. Multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med 167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
- 3.Leung JM, Olivier KN. 2013. Nontuberculous mycobacteria: the changing epidemiology and treatment challenges in cystic fibrosis. Curr Opin Pulm Med 19:662–669. doi: 10.1097/MCP.0b013e328365ab33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 5.Benwill JL, Wallace RJ Jr. 2014. Mycobacterium abscessus: challenges in diagnosis and treatment. Curr Opin Infect Dis 27:506–510. doi: 10.1097/QCO.0000000000000104. [DOI] [PubMed] [Google Scholar]
- 6.Kaushik A, Gupta C, Fisher S, Story-Roller E, Galanis C, Parrish N, Lamichhane G. 16 February 2017. Combinations of avibactam and carbapenems exhibit enhanced potencies against drug-resistant Mycobacterium abscessus. Future Microbiol doi: 10.2217/fmb-2016-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh C, Wencewicz TA. 2016. Antibiotics: challenges, mechanisms, opportunities, p 37–68. ASM Press, Washington, DC. [Google Scholar]
- 8.Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, Arthur M, Mainardi JL. 2011. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by l,d-transpeptidases. J Bacteriol 193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J Bacteriol 190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. 2010. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med 16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Arora K, Lloyd JR, Lee IY, Nair V, Fischer E, Boshoff HI, Barry CE III. 2012. Meropenem inhibits D,D-carboxypeptidase activity in Mycobacterium tuberculosis. Mol Microbiol 86:367–381. doi: 10.1111/j.1365-2958.2012.08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mainardi JL, Fourgeaud M, Hugonnet JE, Dubost L, Brouard JP, Ouazzani J, Rice LB, Gutmann L, Arthur M. 2005. A novel peptidoglycan cross-linking enzyme for a β-lactam-resistant transpeptidation pathway. J Biol Chem 280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 13.Kaushik A, Makkar N, Pandey P, Parrish N, Singh U, Lamichhane G. 2015. Carbapenems and rifampin exhibit synergy against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob Agents Chemother 59:6561–6567. doi: 10.1128/AAC.01158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar P, Kaushik A, Lloyd EP, Li SG, Mattoo R, Ammerman NC, Bell DT, Perryman AL, Zandi TA, Ekins S, Ginell SL, Townsend CA, Freundlich JS, Lamichhane G. 2017. Non-classical transpeptidases yield insight into new antibacterials. Nat Chem Biol 13:54–61. doi: 10.1038/nchembio.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre AL, Dubee V, Cortes M, Dorchene D, Arthur M, Mainardi JL. 2016. Bactericidal and intracellular activity of β-lactams against Mycobacterium abscessus. J Antimicrob Chemother 71:1556–1563. doi: 10.1093/jac/dkw022. [DOI] [PubMed] [Google Scholar]
- 16.Mattoo R, Lloyd EP, Kaushik A, Kumar P, Brunelle JL, Townsend C, Lamichhane G. 2017. LdtMav2, a non-classical transpeptidase and susceptibility of Mycobacterium avium to carbapenems. Future Microbiol 12:595–607. doi: 10.2217/fmb-2016-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoonmaker MK, Bishai WR, Lamichhane G. 2014. Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to β-lactams. J Bacteriol 196:1394–1402. doi: 10.1128/JB.01396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lecoq L, Bougault C, Hugonnet JE, Veckerle C, Pessey O, Arthur M, Simorre JP. 2013. Dynamics induced by β-lactam antibiotics in the active site of Bacillus subtilis L,D-transpeptidase. Structure 20:850–861. doi: 10.1016/j.str.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Stec B, Holtz KM, Wojciechowski CL, Kantrowitz ER. 2005. Structure of the wild-type TEM-1 β-lactamase at 1.55 A and the mutant enzyme Ser70Ala at 2.1 A suggest the mode of noncovalent catalysis for the mutant enzyme. Acta Crystallogr D Biol Crystallogr 61:1072–1079. doi: 10.1107/S0907444905014356. [DOI] [PubMed] [Google Scholar]
- 21.Fakhar Z, Govender T, Maguire GEM, Lamichhane G, Walker RC, Kruger HG, Honarparvar B. 2017. Differential flap dynamics in L,D-transpeptidase2 from Mycobacterium tuberculosis revealed by molecular dynamics. Mol Biosyst 13:1223–1234. doi: 10.1039/C7MB00110J. [DOI] [PubMed] [Google Scholar]
- 22.Kim HS, Kim J, Im HN, Yoon JY, An DR, Yoon HJ, Kim JY, Min HK, Kim SJ, Lee JY, Han BW, Suh SW. 2013. Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains. Acta Crystallogr D Biol Crystallogr 69:420–431. doi: 10.1107/S0907444912048998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavollay M, Dubee V, Heym B, Herrmann JL, Gaillard JL, Gutmann L, Arthur M, Mainardi JL. 2014. In vitro activity of cefoxitin and imipenem against Mycobacterium abscessus complex. Clin Microbiol Infect 20:O297–O300. doi: 10.1111/1469-0691.12405. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalo X, Drobniewski F. 2013. Is there a place for β-lactams in the treatment of multidrug-resistant/extensively drug-resistant tuberculosis? Synergy between meropenem and amoxicillin/clavulanate. J Antimicrob Chemother 68:366–369. doi: 10.1093/jac/dks395. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Hidalgo N, Almirante B, Gavalda J, Gurgui M, Pena C, de Alarcon A, Ruiz J, Vilacosta I, Montejo M, Vallejo N, Lopez-Medrano F, Plata A, Lopez J, Hidalgo-Tenorio C, Galvez J, Saez C, Lomas JM, Falcone M, de la Torre J, Martinez-Lacasa X, Pahissa A. 2013. Ampicillin plus ceftriaxone is as effective as ampicillin plus gentamicin for treating Enterococcus faecalis infective endocarditis. Clin Infect Dis 56:1261–1268. doi: 10.1093/cid/cit052. [DOI] [PubMed] [Google Scholar]
- 26.Soroka D, Dubee V, Soulier-Escrihuela O, Cuinet G, Hugonnet JE, Gutmann L, Mainardi JL, Arthur M. 2014. Characterization of broad-spectrum Mycobacterium abscessus class A β-lactamase. J Antimicrob Chemother 69:691–696. doi: 10.1093/jac/dkt410. [DOI] [PubMed] [Google Scholar]
- 27.Soroka D, Ourghanlian C, Compain F, Fichini M, Dubee V, Mainardi JL, Hugonnet JE, Arthur M. 30 December 2016. Inhibition of β-lactamases of mycobacteria by avibactam and clavulanate. J Antimicrob Chemother doi: 10.1093/jac/dkw546. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre AL, Le Moigne V, Bernut A, Veckerle C, Compain F, Herrmann JL, Kremer L, Arthur M, Mainardi JL. 2017. Inhibition of the β-lactamase BlaMab by avibactam improves the in vitro and in vivo efficacy of imipenem against Mycobacterium abscessus. Antimicrob Agents Chemother 61:e02440-16. doi: 10.1128/AAC.02440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erdemli SB, Gupta R, Bishai WR, Lamichhane G, Amzel LM, Bianchet MA. 2012. Targeting the cell wall of Mycobacterium tuberculosis: structure and mechanism of L,D-transpeptidase 2. Structure 20:2103–2115. doi: 10.1016/j.str.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brammer Basta LA, Ghosh A, Pan Y, Jakoncic J, Lloyd EP, Townsend CA, Lamichhane G, Bianchet MA. 2015. Loss of a functionally and structurally distinct L,D-transpeptidase, LdtMt5, compromises cell wall integrity in Mycobacterium tuberculosis. J Biol Chem 290:25670–25685. doi: 10.1074/jbc.M115.660753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Thomsen R, Christensen MH. 2006. MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 37.Silva JR, Govender T, Maguire GE, Kruger HG, Lameira J, Roitberg AE, Alves CN. 2015. Simulating the inhibition reaction of Mycobacterium tuberculosis L,D-transpeptidase 2 by carbapenems. Chem Commun (Camb) 51:12560–12562. doi: 10.1039/C5CC03202D. [DOI] [PubMed] [Google Scholar]
- 38.Gavan TL, Town MA. 1970. A microdilution method for antibiotic susceptibility testing: an evaluation. Am J Clin Pathol 53:880–885. doi: 10.1093/ajcp/53.6.880. [DOI] [PubMed] [Google Scholar]
- 39.Desmond E. 2011. Susceptibility testing of mycobacteria, nocardiae and other aerobic actinomycetes. M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 40.Hsieh MH, Yu CM, Yu VL, Chow JW. 1993. Synergy assessed by checkerboard. A critical analysis. Diagn Microbiol Infect Dis 16:343–349. doi: 10.1016/0732-8893(93)90087-N. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.