Abstract
TCF4, a key transcription factor of Wnt signaling system, has been recently found to be essential for maintaining stem cells. However, its signaling pathway is not well elucidated. This study was to explore the functional roles and signaling pathway of TCF4 in maintaining adult stem cell properties using human corneal epithelial stem cells as a model. With immunofluorescent staining and real-time polymerase chain reaction, we observed that TCF4 was exclusively expressed in the basal layer of human limbal epithelium where corneal epithelial stem cells reside. TCF4 was found to be well colocalized with ABCG2 and p63, two recognized epithelial stem/progenitor cell markers. Using in vitro culture models of primary human corneal epithelial cells, we revealed that TCF4 mRNA and protein were upregulated by cells in exponential growth stage, and RNA interference by small interfering RNA-TCF4 (10–50 nM) transfection blocked TCF4 signaling and suppressed cell proliferation as measured by WST-1 assay. TCF4 silence was found to be accompanied by downregulated proliferation-associated factors p63 and survivin, as well as upregulated cyclin-dependent kinase inhibitor 1C (p57). By creating a wound healing model in vitro, we identified upregulation and activation of β-catenin/TCF4 with their protein translocation from cytoplasm to nuclei, as evaluated by reverse transcription-quantitative real-time polymerase chain reaction, immunostaining, and Western blotting. Upregulated p63/survivin and down-regulated p57 were further identified to be TCF4 downstream molecules that promote cell migration and proliferation in wound healing process. These findings demonstrate that transcription factor TCF4 plays an important role in determining or maintaining the phenotype and functional properties of human corneal epithelial stem cells.
Keywords: Adult stem cell, Corneal epithelium, TCF4, Survivin, Proliferation
Introduction
Adult stem cells are small subpopulations of slow-cycling undifferentiated resident cells with high proliferative capacity and self-renewal ability, as well as pluripotent potential. These cells can be differentiated into functionally mature cells to regenerate all the cell types of the tissue where they are located [1]. Adult stem cells exhibit unique characteristics, including slow-cycling or long cell-cycle time during homeostasis in vivo; small size and poor differentiation with primitive cytoplasm; high proliferative potential after wounding or placement in culture; ability for self-renewal and functional tissue regeneration [2–5]. However, the underlying mechanism by which these properties of adult stem cells are maintained has not been well elucidated.
Wingless (Wnt) has been demonstrated as a potent morphogen that promotes the expansion of stem cell population and maintains their precursor properties throughout development by activating signaling process controlling cell proliferation and body patterning [6–8]. There are several branches of Wnt-mediated signaling cascade in mammals, the most prominent is the canonical Wnt signaling pathway, which has been implicated in a diverse array of cellular functions, such as stem cell proliferation and self-renewal; stem cell activation, fate determination and differentiation; as well as aging and senescence [9, 10].
The TCF/LEF family is a group of transcription factors that bind to DNA through a high mobility group domain. They are involved in the Wnt signaling pathway, where they recruit the coactivator β-catenin to enhance elements of genes they target [11–13]. Four TCF genes, TCF1, LEF1, TCF3 and TCF4, exist in mammals. Indeed, TCF3 and TCF4 are mostly related to stem cell property maintenance, while the TCF-1 locus acts as an intestinal tumor suppressor primarily due to the production of a truncated dominant negative isoform of TCF-1, which antagonizes TCF-4 in stem cell renewal. Stabilized β-catenin acts as a transcriptional cofactor for TCF3, TCF4, TCF1, and Lef1, as well as other DNA-binding proteins. That the accumulated nuclear β-catenin binds TCF family members is a hallmark of canonical Wnt signaling pathway [14]. Many affected genes are regulated by TCF activation, such as survivin, cyclin D1, and c-Myc [15–17]. Nevertheless, little is known about the functional role and underlying mechanisms by which TCF transcription factors maintain the adult stem cells properties.
The ocular surface is an ideal region to study adult stem cell biology because of the unique spatial arrangement of stem cells and transient amplifying cells. It has been known that corneal epithelial stem cells are located in the basal layer of human corneal limbus [18–20]. The compartmentalization of the corneal epithelial stem cells within the limbus provides a valuable opportunity to study the behavior of adult stem cells [20, 21]. With microarray analysis, we have observed that transcription factor TCF4 was one of the most upregulated genes in rapidly adherent progenitor cells isolated from limbal basal epithelium by collagen IV adhesion technique, and β-catenin/TCF4 Wnt signaling is an important pathway upregulated in the stem/progenitor cell populations [22]. However, little is known yet on TCF4 molecular biology in corneal epithelial stem cells. The present study was to explore the expression, localization, activation, and functional role of TCF4 as well as its signaling molecules in maintaining human corneal epithelial stem cells using multiple in vitro models of primary human corneal epithelial cultures.
Materials and Methods
Materials and Reagents
Cell culture dishes, plates, centrifuge tubes, and other plastic ware were purchased from Becton, Dickinson and Company (Franklin Lakes, NJ, http://www.bdbiosciences.com). Nunc Lab-Tec II eight-chamber slides were from Nalge Nunc International Corp (Naperville, IL, http://www.nalgenunc.com). Dulbecco’s modified Eagle’s medium, Ham F-12, HEPES, amphotericin B, gentamicin, and 0.25% trypsin/EDTA solution were from Invitrogen-GIBCO BRL (Grand Island, NY, http://www.invitrogen.com). Fetal bovine serum (FBS) was from Hyclone (Logan, UT). Primary antibodies against TCF4 and survivin were from Santa Cruz Biotechnology (Santa Cruz, CA, http://www.scbt.com). Antibodies against β-catenin and β-actin were from Cell Signaling Technology Inc (Beverly, MA, http://www.cellsignal.com). ABCG2 mAb (clone BXP-21) was from Millipore (Billerica, MA, http://www.millipore.com). Antibodies against p57 and p63 (clone 4A4), and HRP-conjugated secondary antibodies (goat anti-mouse, goat anti-rabbit, and rabbit anti-goat) were from Thermo Fisher Scientific (Fremont, CA, http://www.thermofisher.com). Fluorescein Alexa-Fluor 488- or 594-conjugated secondary antibodies (donkey anti-goat IgG or goat anti-mouse IgG) were from Molecular Probes (Eugene, OR, http://www.invitrogen.com). Nuclear Extract kit came from Active Motif (Carlsbad, CA, http://www.activemotif.com). Ready gels, Precision Plus Protein Unstained Standards and Precision Protein Streptactin-AP Conjugate came from Bio-Rad (Hercules, CA, http://www.bio-rad.com). The BCA protein assay kit was from Pierce Chemical (Rockford, IL). Ready-To-Go you-Prime First-Stand Beads were purchased from GE Health Care (Piscataway, NJ, http://www3.gehealthcare.com). Cell Proliferation Reagent WST-1 was from Roche Applied Scence (Mannheim, Germany, https://www.roche-applied-science.com). Taqman Gene Expressin Assays and Real-time PCR Master Mix kits were from Applied Biosystems (Foster City, CA, http://www.appliedbiosystems.com). RNeasy Mini Kit and Hiperfect transfection reagent from Qiagen (Valencia, CA, http://www.qiagen.com). Antibody to γ-tubulin, bovine insulin, human transferring, sodium selenite, hydrocortision, human epidermal growth factor, cholera toxin A subunit, propidium iodide (PI), and all other reagents came from Sigma-Aldrich (St. Louis, http://www.sigmaaldrich.com).
Donor Corneal Tissues and Primary Human Corneal Epithelial Cultures
Fresh human corneoscleral tissues (in 72 hours post-mortem), which did not meet the criteria for clinical use, from donors aged 23–64 years were obtained from the Lions Eye Bank of Texas (Houston, TX). Human tissues were handled according to the tenets of the Declaration of Helsinki. Donor corneoscleral tissues were cut through the central cornea or peripheral limbus, then frozen and sectioned using a previously described method [23, 24]. To evaluate differential gene expressions, a 9-mm trephine was used to separate central cornea from the limbal tissue, and both corneal and limbal epithelia were directly scrapped with a keratome and immediately lysed in Qiagen RLT lysis buffer before RNA extraction.
Primary human corneal epithelial cells (HCECs) were established from limbal explants using a previously described method [25, 26]. In brief, each limbal rim was cut into 12 equal pieces (approximately 2 × 2 mm size each). Two pieces with their epithelial side up were directly placed into a well of six-well culture plates, or one piece into a well of 12-well plates or eight-chamber slides and they were covered with a drop of FBS overnight. The explants were then cultured in a supplemented hormonal epidermal medium containing 5% FBS (SHEM) at 37°C under 5% CO2 and 95% humidity. The medium was renewed every 2–3 days.
RNA Interference
To explore a functional role of TCF4 in proliferation, RNA interference was performed using small interfering RNA (siRNA) according to a previous Fast-Forward Transfection method using HiperFect transfection reagent [27]. In brief, primary HCECs (6 × 104 cells per cm2) in 6- or 12-well were transfected with annealed double-stranded siRNA specific for TCF4 (siRNA-TCF4) at different concentrations (10, 25, and 50 nM), and a noncoding sequence siRNA-fluorescein (siRNA-F) was used as a negative control (also served as a visible monitor for transfection efficiency). The transfected cells were incubated for 48 hours for RNA extraction and immunofluorescent staining. For WST-1 cell proliferation assay, the mixture of siRNA and Hiperfect reagent was added in 96-well plate for 5–10 minutes incubation at room temperature, and HCECs were then seeded at 6,000 per well, which made total volume 100 µl per well, and incubated for additional 48 hours before used for WST assay.
WST-1 Cell Proliferation Assay
WST-1 assay was performed according to the manufacturer’s protocol as our previous report [28]. In brief, 10 µl of WST-1 cell proliferation agent was added to each well containing 100 µl cell culture. The cells were then incubated for 2 hours at 37°C in a 5% CO2 incubator. The plate was measured at a wavelength of 450 nm with a reference wavelength 690 nm in an Tecan Infinite M200 microplate reader (Männedorf, Switzerland, http://www.tecan.com).
In Vitro Wound Healing Model of HCECs
Primary HCECs were cultured in 6- or 12-well plates or 8-chamber slides until 90% confluent. Wound incisions were made by scraping cells in 2 mm wide area following the lines drawn on the back of the plates or slides. The cultures were carefully observed and photographed until the wounding area was completely healed in 72 hours. Cultures at different time periods of wound healing were used for RNA extraction, immunofluorescent staining, or Western blot analysis.
Immunofluorescent Staining and Laser Scanning Confocal Microscopy
Immunofluorescent staining was performed as previously described [23, 29]. Corneal frozen sections or cultured corneal epithelial cells were fixed with cold methanol (for p63, β-catenin, and TCF4) or freshly prepared 2% paraformaldehyde (for ABCG2 staining) at 4°C for 10 minutes. Cultured cells were permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) at room temperature for 10 minutes. After blocking with 10% normal goat or 20% donkey serum in PBS for 30 minutes, primary antibodies against TCF4 (1:100), β-catenin (1:50), p63 (1:300), and ABCG2 (1:100) were applied and incubated for 2 hours at room temperature. A secondary antibody, Alexa Fluor 488-conjugated donkey anti-goat IgG (1:300) or 594-conjugated goat anti-mouse IgG (1:300), was then applied and incubated in a dark chamber for 1 hour and followed by counterstaining with a DNA-binding dye PI (2 µg/ml in PBS) for 5 minutes. After washing with PBS, Antifade Gel/Mount and a coverslip were applied. Sections were examined and photographed with the laser scanning confocal microscopy (LSM 510, Zeiss, Thornwood, NY, http://www.zeiss.com).
Total RNA Extraction, Reverse Transcription and Relative Quantitative Real-Time PCR
Total RNA was isolated from tissue or cells using RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol, quantified by NanoDrop ND-1000, and stored at −80° C. As previously described [30, 31], the first strand cDNA was synthesized by reverse transcription (RT) from 1 µg of total RNA using Ready-To-Go You-Prime First-Strand Beads, and the real-time PCR was performed in the Mx3005PTM system (Stratagene) with a 20 µl reaction volume containing 5 µl of cDNA, 1 µl of TaqMan Gene Expression Assay for β-catenin (Assay ID Hs99999168_ml), TCF4 (Hs00162613_ml), survivin (Hs00153353_ml), p63 (Hs00186613_ml), p57 (Hs00175938_m1) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs99999905_ml), and 10 µl TaqMan Gene Expression Master Mix (Applied Biosystems, http://www.appliedbiosystems.com). The thermocycler parameters were 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. A nontemplate control was included to evaluate DNA contamination. The results were analyzed by the comparative threshold cycle (Ct) method and normalized by a housekeeping gene GAPDH [31, 32].
Western Blotting Assay
Western blot analysis was performed using a previously reported method [33]. Primary HCECs with wound model at different time points were collected for extraction of cytoplasmic and nuclear proteins using nuclear extract kit from Active Motif according to the manufacture’s protocol. Other cultures were lysed with RIPA buffer. Protein concentration of these extracts was measured by a Micro BCA protein assay kit. Equal amount of protein (50 µg per lane) was mixed with 6 × SDS reducing sample buffer and boiled for 5 minutes before loading. The proteins were separated on an SDS polyacrylamide gel and transferred electronically to PVDF membranes. The membranes were then blocked with 5% nonfat milk in trisbuffered saline (50 mM Tris [pH 7.5], 0.9% NaCl, and 0.1% Tween 20) for 1 hour at room temperature and incubated with primary antibodies to TCF4 (1:100), β-cantenin (1:1,000), p57 (1:500), p63 (1:1,000), or β-actin (1:2,000) overnight at 4°C The membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG (1:2,000) for 1 hour at room temperature. The signal bands were detected with enhanced chemiluminescence reagent, and the images were acquired by Kodak image station 2000R (Eastman Kodak, New Haven, CT).
Statistical Analysis
The Student’s t test or analysis of variance with Tukey’s post hoc testing was used for statistical comparisons. p ≤ .05 was considered statistically significant. All of these tests were performed using the GraphPad Prism 5.0 software (Graph-Pad Prism, Inc., San Diego, CA, http://www.graphpad.com).
Results
TCF4 is Exclusively Expressed by Limbal Basal Cells Where Stem Cells Reside
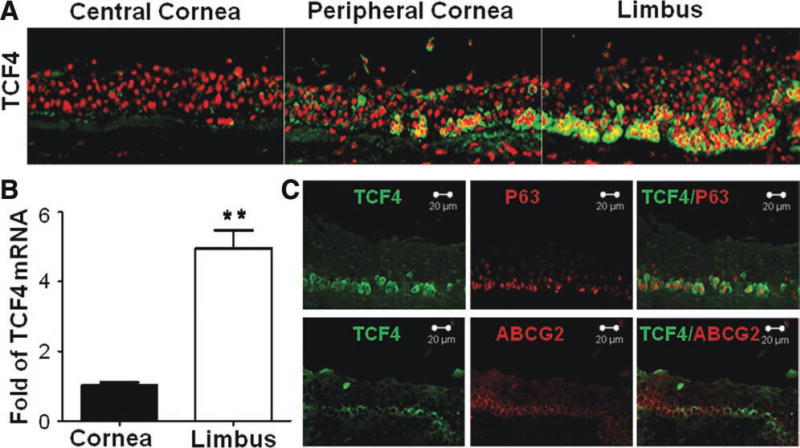
The immunofluorescent staining on corneal limbal tissue frozen sections revealed that TCF4 protein was exclusively immunolocalized at basal cells of limbal and peripheral corneal epithelia, where corneal epithelial stem cells reside. As shown in Figure 1A, TCF4 immunoreactivity was primarily located in cytoplasm and nuclei of basal cells at limbal epithelium, and the numerous TCF4-positive cells were interspersed with patches of TCF4 negative cells. TCF4-positive cells decreased and cluster-like dispersed in the basal layer of peripheral corneal epithelium. There was no TCF4 immunoreactivity detected in the suprabasal and superficial layers of limbal epithelium, nor in all layers of corneal epithelium. To verify this unique pattern of TCF4 expression in cornea and limbus, the levels of TCF4 mRNA of central corneal or limbal epithelia were evaluated by RT-quantitative real-time PCR (qPCR) with GAPDH as an internal control. The results confirmed that levels of TCF4 mRNA expression by limbal epithelium were significantly higher (4.957 ± 0.52-fold, p < .05, n = 3) than that in corneal epithelium (Fig. 1B).
Figure 1. TCF4 localization in basal layer of human limbal epithelium.
(A): Representative images showing TCF4 immunofluorescent staining (green) with propidium iodide (red) counterstaining in human central and peripheral cornea and limbus. (B): Reverse transcription-quantitative real-time polymerase chain reaction displayed TCF4 mRNA expression by corneal and limbal epithelia. Data shown as mean ± standard deviation, n = 3; **, p < .01. (C): Representative images showing double immunofluorescent staining of TCF4 (green) with p63 or ABCG2 (red).
TCF4 Protein was Colocalized with Corneal Epithelial Progenitor Markers, ABCG2 and p63 in the Basal Layer of Limbal Epithelium
ABCG2 and p63 have been accepted as stem cell-associated markers or progenitor cell markers of keratinocytes including corneal epithelial cells [34, 35]. As shown in Figure 1C, the double immunostaining of TCF4 and p63 indicated that both proteins were coimmunolocalized in limbal basal epithelium, and the most TCF4-positive cells also expressed p63. The pattern of coimmunolocalization of TCF4 and ABCG2 was different from TCF4 and p63. Although both TCF4 and ABCG2 were immunolocated within limbal basal layer, some cells expressing strong ABCG2 were weakly positive to TCF4, and other cells expressing weak ABCG2 were strongly positive to TCF4. This colocalization pattern of TCF4 with p63 and ABCG2 indicates that TCF4 may play an important role in maintaining stemness properties of corneal epithelial stem cells.
TCF4 Signaling was Related to the Growth Stage and Proliferation Capacity of HCECs
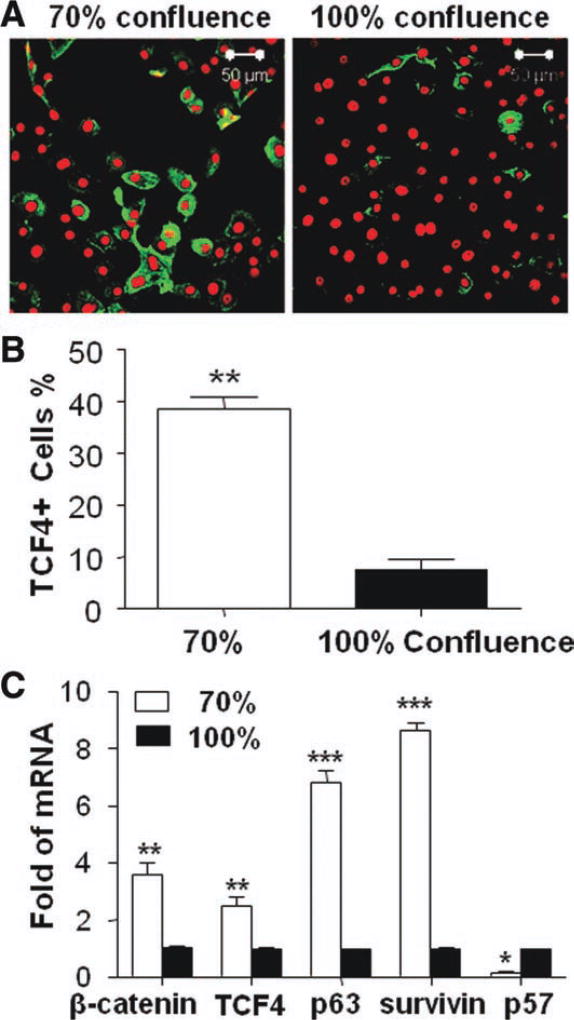
To evaluate the role of TCF4 in cell proliferation and differentiation, the primary HCECs were collected in different growing stages with 70% or 100% confluence for immunofluorescent staining and RT-qPCR. As shown in Figure 2A, TCF4 antibody was found to stain cell cytoplasm, and strong immunoreactivity was preferentially expressed by small size cells in primary HCECs. The 70% confluent culture where cells are in exponential growth contained significantly higher percentage of TCF4-positive cells (38.47 ± 2.51%, p < .05, n = 3) than 100% confluent culture (7.65 ± 1.96%, n ± 3) (Fig. 2B). RT-qPCR results shown in Figure 2C confirmed the finding by TCF4 immunofluorescent staining. TCF4 mRNA expressed significantly higher levels (2.51 ± 0.32-fold, p < .01, n = 3) by younger cells in 70% confluence than that in 100% confluent culture, which contained more quiescent and differentiated cells. Interestingly, TCF4 signaling-associated molecules were also found to be regulated in the different growth stages. β-Catenin, known as an activator of TCF4, was expressed at higher mRNA level by 70% confluent HCECs than 100% confluent cells. The mRNA expression of p63 and survivin, two proliferation-associated factors were also upregulated while the mRNA of cyclin-dependent kinase inhibitor 1C (p57), a proliferation inhibitor, was downregulated in 70% confluent HCECs, compared with 100% confluent cells (Fig. 2C).
Figure 2. TCF4 expression in human corneal epithelial cells (HCECs) at different growth stages.
(A): Representative images showing TCF4 immunofluorescent staining (green) with propidium iodide (red) counterstaining in HCECs at different growth stages (70% and 100% confluence). (B): Percentages of TCF4-positive (TCF4+) cells in 70% and 100% confluent cultures. (C): Reverse transcription-quantitative real-time polymerase chain reaction displayed the expression levels (relative fold of mRNA) of β-catenin, TCF4 p63 survivin, and p57 by 70% confluent HCECs compared with 100% confluent cells. Data shown as mean ± standard deviation, n = 3; *, p < .05; **, p < .01; ***, p < .001.
Cell Proliferative Capacity is Impaired in TCF4 Silenced HCECs by siRNA Interference
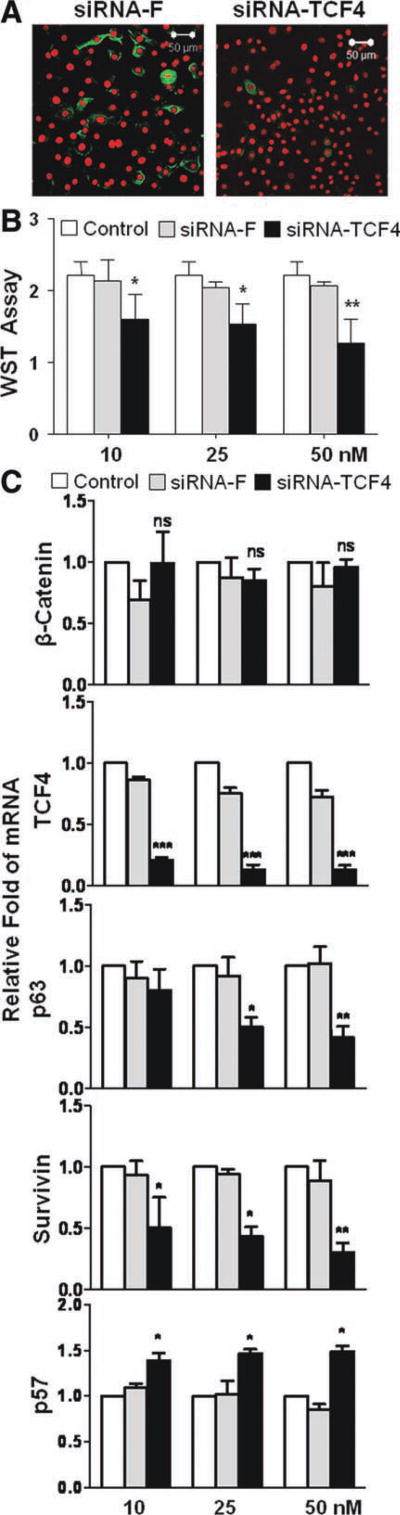
To explore a functional role of TCF4 in cell proliferation, RNA interference was performed to silence TCF4 gene by siRNA-TCF4 in HCECs. Immunofluorescent staining showed a successful knockdown of TCF4 gene by siRNA-TCF4 in 48 hours, when TCF4 protein production was almost completely suppressed in comparison of cells transfected with siRNA-F, a noncoding negative control siRNA (Fig. 3A). TCF4 mRNA expression was largely blocked by siRNA-TCF4 at all three different concentrations (10, 25, and 50 nM), when compared with untreated control or siRNA-F transfected cells (Fig. 3C). WST-1 cell proliferation assay was performed using HCECs in 96-well plates with or without transfection of siRNA-TCF4 or siRNA-F. We observed that WST proliferation index was dose-dependently (10–50 nM) suppressed by TCF4-siRNA in 48-hour growth period, when compared with untreated or siRNA-F control groups (p < .05, n = 5) (Fig. 3B).
Figure 3. Effects of TCF4 silence by siRNA interference on proliferation of human corneal epithelial cells (HCECs).
(A): Representative images showing TCF4 immunofluorescent staining (green) with propidium iodide (red) counterstaining in HCECs transfected with siRNA-TCF4 and siRNA-F. (B): WST-1 assay showing the cell proliferation index in 48 hours after siRNA-TCF4 transfection. Data shown as the mean and standard deviation of results; ns, no significance; *, p < .05; **, p < .01; ***, p < .001; n = 3, compared with siRNA-F transfection. (C): Reverse transcription-quantitative real-time polymerase chain reaction showing the effects of siRNA-TCF4 (10–50 nM) transfection on mRNA expression of β-catenin, TCF4, p63, survivin, and p57 with untreated and transfected cells with non-coding sequence siRNA-F as control, n = 5. Abbreviations: siRNA, small interfering RNA; siRNA-F, small interfering RNA-fluorescein.
To identify the molecular signaling through which TCF4 controls proliferative capacity, several potential TCF4 related factors were evaluated at mRNA levels by RT-qPCR after TCF4 knockdown (Fig. 3C). As anticipated, β-catenin, an upstream activate factor of TCF4, did not show any change at mRNA levels after TCF4 gene silenced. Interestingly, p63 and survivin were found to be downregulated significantly at mRNA levels after TCF4 knockdown (p < .05) (Fig. 3C). The extent of this decrease was dependent to the doses of siRNA-TCF4 (10–50 nM). By contrast, p57 mRNA was significantly upregulated following TCF4 silence. These findings suggest that p63 and survivin, two proliferation-associated genes, and a proliferation inhibitor p57 are potential downstream target molecules of TCF4. The downregulated p63/survivin and upregulated p57 may be the signaling pathway through which cell growth was suppressed by TCF4 gene knockdown.
TCF4 and its Upstream β-Catenin were Activated with Nucleus Translocation in Wound Healing Model of HCECs
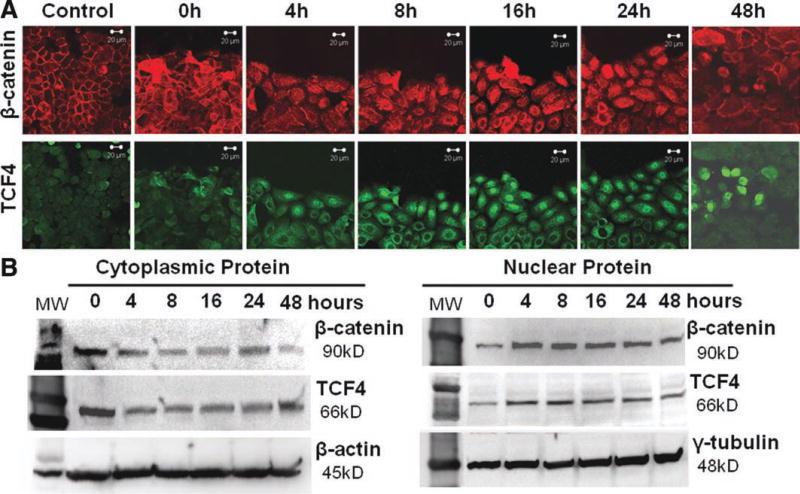
To further confirm that TCF4 is an important transcription factor determining and promoting cell proliferative capacity, we created an in vitro wound healing model in HCECs. Strikingly, we observed marked translocalization of TCF4 protein from cytoplasm to nuclei when cells migrated and proliferated during wound healing process. As shown in Figure 4A, immunofluorescent staining displayed TCF4-nuclear translocation at the wound edge as early as 4 hours after wounding, and the TCF4 nuclear-positive cells increased significantly during wounding process until wound area was completely healed in 48 hours when the TCF4 nuclear-positive cells were largely decreased. Interestingly, the similar pattern of nuclear translocation of β-catenin was observed in wounding area, especially at the wound edge during wound healing process (Fig. 4A). These results provided the evidence that TCF4 and its upstream β-catenin were activated with nucleus translocation during wound healing process in HCECs.
Figure 4. TCF4 and β-catenin activation with nucleus translocation during wound healing process of human corneal epithelial cells in vitro.
(A): Representative images of double immunofluorescent staining of TCF4 (green) and β-catenin (red) showing their nucleus translocation process at different time points after wound. (B): Western blot analysis showing changed levels of TCF4 and β-catenin in isolated cytoplasmic and nuclear proteins, indicating their nucleus translocation during wound healing process.
These findings were further substantiated by Western blot analysis, which showed quantified changes of cytoplamic and nuclear protein levels of TCF4 and β-catenin. Both TCF4 and β-catenin cytoplasmic protein decreased from 4 hours and remained the lower level until 48 hours after wound (Fig. 4B). Conversely, the nuclear protein levels of TCF4 and β-catenin increased from 4 hours and remained elevated up to 48 hours. All these results revealed that TCF4 was activated and transported into the nucleus where it bound β-catenin to activate target genes and thus promote cell migration and proliferation to repair wound in HCECs.
TCF4 Promotes Wound Healing in HCECs Through Upregulation of p63/Survivin and Downregulation of p57
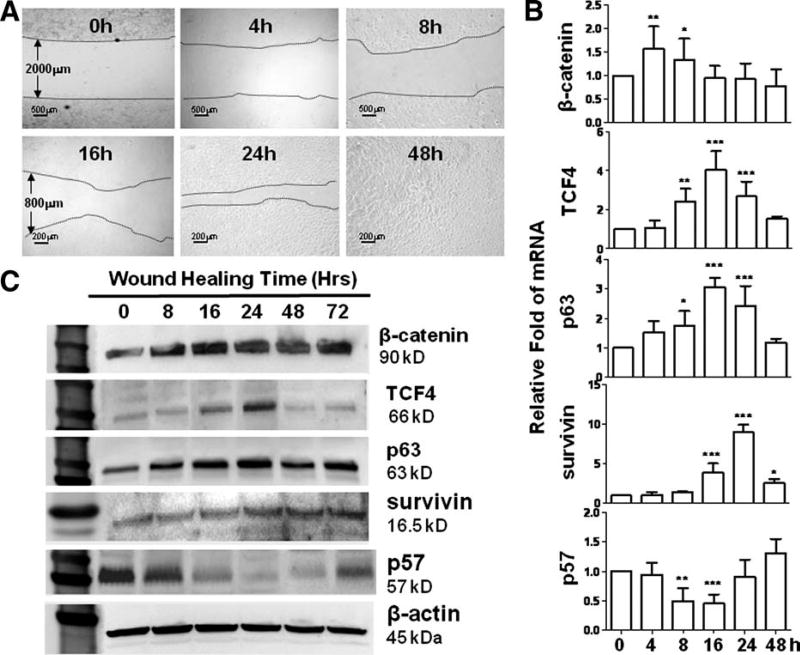
To explore the signaling pathways of TCF4 that promotes wound healing, we evaluated its potential downstream molecules regulated during wound healing process. Under our wounding condition, normal HCECs were observed to be capable of repairing a 2-mm wide wounding area within 48 hours (Fig. 5A). RT-qPCR results showed that TCF4 expression was significantly upregulated during wound healing process in HCECs (Fig. 5B). Compared with untreated control of HCECs, the mRNA levels of TCF4 was significantly increased at 8 hours, reached peak levels by fourfold high at 16 hours after wounding, and the upregulated mRNA levels lasted more than 24 hours before decreased to above-normal levels in 48 hours when the wound has been healed. The expression of β-catenin, a TCF4 activator in Wnt signaling pathway, was found to respond quickly at its mRNA levels that increased to peak at 4 hours and decreased to normal in 16 hours after wound. The upregulation of β-catenin mRNA was earlier but lasted shorter than TCF4 upregulation. The upregulation pattern of β-catenin and TCF4 indicated that Wnt signaling pathway was activated in wound healing process. Moreover, we observed the relevant regulation of proliferation-associated genes, p63, survivin, and p57. After wound, p63 mRNA increased significantly in 8 hours, reached peak by threefold at 16 hours, and then gradually reduced back to normal level within 48 hours, a pattern similar to TCF4. Survivin mRNA was increased slowly at 16 hours after wound, but strongly stimulated to peak levels by ninefold at 24 hours, and remained higher expression more than 48 hours after the wound has been healed. Interestingly, the expression of p57, a proliferation inhibitor, decreased significantly during 8–16 hours after wound. The time period of p57 mRNA downregulation was associated with upregulation of p63 and survivin genes.
Figure 5. TCF4 in wound healing model of human corneal epithelial cells (HCECs) in vitro.
(A): Representative phase images showing a 2-mm wide wound area was healed within 48 hours. (B): Reverse transcription-quantitative real-time polymerase chain reaction data showing the expression levels (relative fold of mRNA) of β catenin TCF4 p63 survivin, and p57 by HCECs at different time points after wound. Data shown as the mean and standard deviation of results, *, p < .05; **, p < .01; ***, p < .001; n = 3. (C): Western blot results showing the protein levels of β-catenin, TCF4, p63, survivin, and p57 by HCECs at different time points after wound.
This regulated pattern of β-catenin/TCF4 and proliferation-associated factors at transcriptional expression was further confirmed at protein level by Western blot analysis, which was performed using cell lysates from cultures with wound healing model up to 72 hours after wound. As shown in Figure 5C, we observed the markedly increased protein production of β-catenin, TCF4, p63, and survivin but decreased p57 protein levels during wound healing process, especially from 16 hours to 24 or 48 hours, respectively. By contrast, a house keeping protein β-actin protein was not significantly changed during these time points.
The regulation pattern of these proliferation-associated factors following TCF4 upregulation in wound healing process supported aforementioned RNA interference results that TCF4 gene knockdown by siRNA-TCF4 suppressed cell proliferation with downregulated p63/survivin, and upregulated p57. These findings suggest that TCF4 controls corneal epithelial cell proliferation through its downstream signaling molecules p63, survivin, and p57, the balance between upregulated p63/survivin and downregulated p57.
Discussion
TCF4 is a basic helix-turn-helix transcription factor. Mammalian TCF4 is highly expressed in the midbrain, intestine, and mammary epithelium [36–39]. In late embryonic and early neonatal gut, TCF4 was found to be present in the proliferative intervillus pockets and essential for maintenance of the progenitor compartment of gut epithelium [40, 41]. TCF4 was also found to be essential for maintaining stemness in skin epithelial stem cells [11]. However, it is still not clear through what signaling pathway TCF4 maintains these adult epithelial stem cells. Corneal epithelial stem cells have been identified to reside at the corneal limbus for more than 2 decades [18–20], but it is little known how TCF4 plays an important role in maintaining the properties of corneal epithelial stem cells. The present study revealed that TCF4 was exclusively localized in the basal layer of human limbal epithelium and well colocalized with ABCG2 and p63, two recognized epithelial stem cell/progenitor markers [34, 35]. Using different in vitro culture models of primary HCECs, we evaluated the differential expression of TCF4 by cells in exponential growth and quiescent confluent stages, investigated the TCF4 signaling and proliferative capacity in TCF4-silenced cultures, and explored TCF4 activation and downstream signaling molecules in wound healing process. All these findings identified the novel role and potential signaling pathways of TCF4 in maintaining corneal epithelial stem cells.
TCF4 Plays an Important Role in Maintaining the Phenotype of Corneal Epithelial Stem Cells
There is no report on TCF4 expression in ocular surface until our recent findings through human genome microarray analysis for human corneal epithelial progenitor populations isolated from donor limbal tissues [22], as well as through the cultured corneal epithelial progenitors in a low calcium serum-free medium CnT-20 that promotes ex vivo expansion of corneal epithelial progenitor cells via β-catenin/Tcf4/survivin signaling, a novel intrinsic pathway [42]. The present study revealed that TCF4 was uniquely expressed by the limbal basal cells and may determine the phenotype of corneal epithelial stem cells (Fig. 1). The immunofluorescent staining demonstrated that TCF4 was exclusively localized in a subpopulation of basal layers of limbal epithelium where corneal epithelial stem cells reside, but absent in the suprabasal limbal and full central corneal epithelia. TCF4 was found to be colocalized with major epithelial progenitor markers p63 and ABCG2 in limbal basal cells. Interestingly, the most TCF4 positive basal cells consistently expressed p63, while some basal cells expressing strong TCF4 were weakly positive to ABCG2, or vice versa. This differential pattern of TCF4 colocalization with p63 and ABCG2 may suggest the different functions of these proteins. TCF4 may have similar function to p63 that promotes epithelial cell proliferation [43] but may be less common to ABCG2 that protects stem cells from toxic damage as a multidrug resistant gene [35, 44]. This observation was further substantiated by RT-qPCR that message RNA expression of TCF4 was statistically higher in limbal than corneal epithelia isolated from donor tissues. These findings revealed that TCF4 expression may represent a characteristic of corneal epithelial stem cells.
Our previous studies [45, 46] have shown that the corneal epithelial cells in confluent cultures are more differentiated than younger growing cells in less confluent condition. When comparing the cells in different growth stages, the number of TCF4 immunoreactivity-positive cells was statistically greater in exponentially grown cells with 70% confluence than that in quiescent differentiated cells with 100% confluence (Fig. 2). This finding was confirmed by RT-qPCR at transcriptional levels that showed higher TCF4 expression accompanied by upregulated mRNA levels of several proliferation factors in cells with exponential growth. β-Catenin that binds and activates TCF4 [12], p63 that associates with cell proliferation [47], and survivin that is a potential target gene of β-catenin/TCF4 [48] were all found to be upregulated in 70% confluent younger cells. Interestingly, cycline-dependent kinase inhibitor p57 was conversely decreased in these younger cells, consistent with a previous report that p57 was downregulated by Wnt signaling activation [36]. Ultimately, the increased p63 and survivin with decreased p57 may contribute to the proliferative capacity of these younger cells that contained more progenitors with exponential growth.
TCF4 Activation Promotes Cell Proliferation Via Nuclear Translocation
TCF4 protein, as the pivotal transcription factor for the canonical Wnt pathway, is responsible for transactivation of cell proliferation and survival genes [49, 50]. To investigate the underlying mechanism of TCF4 signaling pathway, we created wound healing model of HCECs in vitro (Fig. 4). Strikingly, we observed increasing translocalization of TCF4 from cytoplasm into nucleus activated by wound condition. The immunofluorescent staining clearly showed the dynamic process of TCF4 nucleus translocation in cells at the area of wounding edges. The key for TCF4 activation is known to be bound by its activator β-catenin in the nucleus. With double immunofluorescent staining, we observed a similar pattern of β-catenin protein translocation from cytoplasm to nucleus in cells at wounding edges. Western blot analysis using isolated cytoplasmic and nuclear proteins gave a quantitative evidence for the nuclear translocation of both β-catenin and TCF4. These results suggest that TCF4 is mainly quiescent and localizes in cytoplasm under normal circumstance, but TCF4 would be activated through translocation into nucleus and then trigger target genes that promote cell migration and proliferation during wound healing process. Our findings are consistent with a previous report that TCF4 activation was induced by injury [51].
TCF4 Controls Cell Proliferation Through Balanced Signaling Pathways with p63, Survivin, and p57
To explore signaling pathways of TCF4 in regulating cell proliferation, RNA interference and subsequent WST proliferation assay were performed in HCEC cultures (Fig. 3). We observed that TCF4 gene knockdown sharply inhibited cell proliferation, suggesting an important role of TCF4 in maintaining proliferative capacity of human corneal epithelial stem cells. Interestingly, TCF4-silenced HCECs expressed lower p63 and survivin but higher p57, indicating that TCF4, p63, survivin, and p57 were involved in maintaining proliferative capacity of human corneal stem cells. Nuclear factor p63 has been accepted as a proliferation-related factor and a stem cell-associated or progenitor marker [34, 43, 47, 52, 53]. Survivin is a bifunctional member of the inhibitor of apoptosis gene family that counteracts cell death and controls mitotic progression [54]. It has been documented to be regulated by TCF/β-catenin in cancer cells [48, 55, 56] and stem cells [57, 58]. Survivin is also found to be a potential target gene of the β-catenin/TCF signaling axis, coupling increased cell proliferation to enhance cell survival in intestinal crypt cells [56]. As a member of Cip/Kip family, p57 specifically inactivates G1 cyclinE/CDK2 and cyclinD/CDK4/6 and involves in limbal epithelial cells proliferative inhibition [27, 59]. The consequence of TCF4 RNA interference that causes decreased p63/survivin and increased p57 may further indicate a potential signaling pathway of TCF4.
Wound healing is a good model to study cell migration and proliferation. Our wounding condition with HCEC cultures successfully activated TCF4 signaling, as evidenced by nuclear translocation of both β-catenin and TCF4, and also stimulated their expression and production at mRNA and protein levels (Fig. 5). HCECs expressed the increased p63 and survivin but decreased p57 following TCF4 upregulation and activation during wound healing process. This is consistent to the regulatory changes of TCF4 and downstream molecules in the cells with 70% confluence; but it is opposite to the pattern observed by aforementioned RNA interference experiments, which revealed the decreased p63 and survivin but increased p57 following TCF4 knockdown. These findings indicate that TCF4 controls cell proliferative capacity through the balanced signaling pathways between p63/survivin and p57.
Conclusion
We have explored a unique pattern of TCF4 expression that is exclusively localized in the basal layer of human limbal epithelium and colocalized with limbal stem cell-associated markers ABCG2 and p63. TCF4 was found to be expressed higher in younger HCECs in exponential growth stage. TCF4 upregulation and activation with nuclear translocation were identified to maintain cell proliferation and promote wound healing of HCECs through downstream molecules p63, survivin, and p57. Taken together, all findings presented in this study demonstrated that transcription factor TCF4 may determine the adult stem cell phenotype and maintain the functional properties of human corneal epithelial stem cells.
Acknowledgments
We thank the Lions Eye Bank of Texas for their great support in providing human corneoscleral tissues. This study was supported by DOD CDMRP FY06 PR064719 (DQL), NIH Grant EY11915 (SCP), National Natural Science Foundation of China (30901634), Fight for Sight (YQ), Research to Prevent Blindness, Oshman Foundation, William Stamps Farish Fund.
Footnotes
Author contributions: R.L.: conception and design, provision of study material, collection and/or assembly of data, data analysis and interpretation, and manuscript writing; Y.Q.: provision of study material, collection and/or assembly of data, data analysis and interpretation, and manuscript writing; J.G.: conception and design, financial support, manuscript writing, and final approval of manuscript; L.Z. and Z.S.: provision of study material or patients and collection and/or assembly of data; S.C.P.: conception and design, financial support, and manuscript writing; and D.-Q.L.: conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Diaz-Flores L, Jr, Madrid JF, Gutierrez R, et al. Adult stem and transit-amplifying cell location. Histol Histopathol. 2006;21:995–1027. doi: 10.14670/HH-21.995. [DOI] [PubMed] [Google Scholar]
- 2.Blau HM, Brazelton TR, Weimann JM. The evolving concept of a stem cell: Entity or function? Cell. 2001;105:829–841. doi: 10.1016/s0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- 3.Watt FM, Hogan BL. Out of Eden: Stem cells and their niches. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 4.Lavker RM, Sun TT. Epidermal stem cells: Properties, markers, and location. Proc Natl Acad Sci USA. 2000;97:13473–13475. doi: 10.1073/pnas.250380097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotsarelis G, Kaur P, Dhouailly D, et al. Epithelial stem cells in the skin: Definition, markers, localization and functions. Exp Dermatol. 1999;8:80–88. doi: 10.1111/j.1600-0625.1999.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 6.Polesskaya A, Seale P, Rudnicki MA. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 7.Pinto D, Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 8.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Beildeck ME, Gelmann EP, Byers SW. Cross-regulation of signaling pathways: An example of nuclear hormone receptors and the canonical Wnt pathway. Exp Cell Res. 2010;316:1763–1772. doi: 10.1016/j.yexcr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen H, Merrill BJ, Polak L, et al. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poy F, Lepourcelet M, Shivdasani RA, et al. Structure of a human Tcf4-beta-catenin complex. Nat Struct Biol. 2001;8:1053–1057. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- 13.Phillips BT, Kimble J. A new look at TCF and beta-catenin through the lens of a divergent C. elegans Wnt pathway. Dev Cell. 2009;17:27–34. doi: 10.1016/j.devcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham TA, Ferkey DM, Mao F, et al. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- 15.Camac KS, Thompson FM, Cummins AG. Activation of beta-catenin in the stem cell region of crypts during growth of the small intestine in infant rats. Dig Dis Sci. 2007;52:1242–1246. doi: 10.1007/s10620-006-9200-7. [DOI] [PubMed] [Google Scholar]
- 16.Takayama S, Rogatsky I, Schwarcz LE, et al. The glucocorticoid receptor represses cyclin D1 by targeting the Tcf-beta-catenin complex. J Biol Chem. 2006;281:17856–17863. doi: 10.1074/jbc.M602290200. [DOI] [PubMed] [Google Scholar]
- 17.Yoo JH, Lee HJ, Kang K, et al. Lignans inhibit cell growth via regulation of Wnt/beta-catenin signaling. Food Chem Toxicol. 2010;48:2247–2252. doi: 10.1016/j.fct.2010.05.056. [DOI] [PubMed] [Google Scholar]
- 18.Schermer A, Galvin S, Sun T-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotsarelis G, Cheng SZ, Dong G, et al. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 20.Tseng SC. Concept and application of limbal stem cells. Eye. 1989;3(2):141–157. doi: 10.1038/eye.1989.22. [DOI] [PubMed] [Google Scholar]
- 21.Dua HS, Azuara-Blanco A. Limbal stem cells of the corneal epithelium. Surv Ophthalmol. 2000;44:415–425. doi: 10.1016/s0039-6257(00)00109-0. [DOI] [PubMed] [Google Scholar]
- 22.Bian F, Liu W, Yoon KC, et al. Molecular signatures and biological pathway profiles of human corneal epithelial progenitor cells. Int J Biochem Cell Biol. 2010;42:1142–1153. doi: 10.1016/j.biocel.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, de Paiva CS, Luo L, et al. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004;22:355–366. doi: 10.1634/stemcells.22-3-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian F, Qi H, Ma P, et al. An immunoprotective privilege of corneal epithelial stem cells against Th17 inflammatory stress by producing glial cell-derived neurotrophic factor. Stem Cells. 2010;28:2172–2181. doi: 10.1002/stem.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HS, Jun SX, de Paiva CS, et al. Phenotypic characterization of human corneal epithelial cells expanded ex vivo from limbal explant and single cell cultures. Exp Eye Res. 2004;79:41–49. doi: 10.1016/j.exer.2004.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Paiva CS, Pflugfelder SC, Li D-Q. Cell size correlates with phenotype and proliferative capacity in human corneal epithelial cells. Stem Cells. 2006;24:368–375. doi: 10.1634/stemcells.2005-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, Li D-Q, Tong L, et al. Targeted inhibition of p57 and p15 blocks transforming growth factor beta-inhibited proliferation of primary cultured human limbal epithelial cells. Mol Vis. 2006;12:983–994. [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng X, de Paiva CS, Rao K, et al. Evaluation of the transforming growth factor-beta activity in normal and dry eye human tears by CCL-185 cell bioassay. Cornea. 2010;29:1048–1054. doi: 10.1097/ICO.0b013e3181cf98ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Paiva CS, Chen Z, Corrales RM, et al. ABCG2 transporter identifies a population of clonogenic human limbal epithelial cells. Stem Cells. 2005;23:63–73. doi: 10.1634/stemcells.2004-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo L, Li D-Q, Doshi A, et al. Experimental dry eye stimulates production of inflammatory cytokines and MMP-9 and activates MAPK signaling pathways on the ocular surface. Invest Ophthalmol Vis Sci. 2004;45:4293–4301. doi: 10.1167/iovs.03-1145. [DOI] [PubMed] [Google Scholar]
- 31.Yoon KC, de Paiva CS, Qi H, et al. Expression of Th-1 chemokines and chemokine receptors on the ocular surface of C57BL/6 mice: Effects of desiccating stress. Invest Ophthalmol Vis Sci. 2007;48:2561–2569. doi: 10.1167/iovs.07-0002. [DOI] [PubMed] [Google Scholar]
- 32.de Paiva CS, Corrales RM, Villarreal AL, et al. Corticosteroid and doxycycline suppress MMP-9 and inflammatory cytokine expression, Mapk activation in the corneal epithelium in experimental dry eye. Exp Eye Res. 2006;83:526–535. doi: 10.1016/j.exer.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Li D-Q, Luo L, Chen Z, et al. JNK and ERK MAP kinases mediate induction of IL-1beta, TNF-alpha and IL-8 following hyperosmolar stress in human limbal epithelial cells. Exp Eye Res. 2006;82:588–596. doi: 10.1016/j.exer.2005.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellegrini G, Dellambra E, Golisano O, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarkadi B, Ozvegy-Laczka C, Nemet K, et al. ABCG2—A transporter for all seasons. FEBS Lett. 2004;567:116–120. doi: 10.1016/j.febslet.2004.03.123. [DOI] [PubMed] [Google Scholar]
- 36.Castelo-Branco G, Wagner J, Rodriguez FJ, et al. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc Natl Acad Sci USA. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans PM, Chen X, Zhang W, et al. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol Cell Biol. 2010;30:372–381. doi: 10.1128/MCB.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theodosiou NA, Tabin CJ. Wnt signaling during development of the gastrointestinal tract. Dev Biol. 2003;259:258–271. doi: 10.1016/s0012-1606(03)00185-4. [DOI] [PubMed] [Google Scholar]
- 39.van Es JH, Jay P, Gregorieff A, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 40.Van der Flier LG, Sabates-Bellver J, Oving I, et al. The intestinal Wnt/TCF signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 41.Liao Y, Lonnerdal B. Beta-catenin/TCF4 transactivates miR-30e during intestinal cell differentiation. Cell Mol Life Sci. 2010;67:2969–2978. doi: 10.1007/s00018-010-0366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu R, Bian F, Zhang X, et al. The beta-catenin/Tcf4/survivin signaling maintains a less differentiated phenotype and high proliferative capacity of human corneal epithelial progenitor cells. Int J Biochem Cell Biol. 2011;43:751–759. doi: 10.1016/j.biocel.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 44.Ejendal KF, Hrycyna CA. Multidrug resistance and cancer: The role of the human ABC transporter ABCG2. Curr Protein Pept Sci. 2002;3:503–511. doi: 10.2174/1389203023380521. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z, Evans WH, Pflugfelder SC, et al. Gap junction protein connexin 43 serves as a negative marker for a stem cell-containing population of human limbal epithelial cells. Stem Cells. 2006;24:1265–1273. doi: 10.1634/stemcells.2005-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi H, Shine HD, Li DQ, et al. Glial cell-derived neurotrophic factor gene delivery enhances survival of human corneal epithelium in culture and the overexpression of GDNF in bioengineered constructs. Exp Eye Res. 2008;87:580–586. doi: 10.1016/j.exer.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chilosi M, Poletti V, Murer B, et al. Abnormal re-epithelialization and lung remodeling in idiopathic pulmonary fibrosis: The role of deltaN-p63. Lab Invest. 2002;82:1335–1345. doi: 10.1097/01.lab.0000032380.82232.67. [DOI] [PubMed] [Google Scholar]
- 48.Zhu H, Zhang G, Wang Y, et al. Inhibition of ErbB2 by Herceptin reduces survivin expression via the ErbB2-beta-catenin/TCF4-survivin pathway in ErbB2-overexpressed breast cancer cells. Cancer Sci. 2010;101:1156–1162. doi: 10.1111/j.1349-7006.2010.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong CF, Chou YT, Lin YS, et al. MAD2B, a novel TCF4-binding protein, modulates TCF4-mediated epithelial-mesenchymal transdifferentiation. J Biol Chem. 2009;284:19613–19622. doi: 10.1074/jbc.M109.005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang YS, Shih HM. Daxx positively modulates beta-catenin/TCF4-mediated transcriptional potential. Biochem Biophys Res Commun. 2009;386:762–768. doi: 10.1016/j.bbrc.2009.06.126. [DOI] [PubMed] [Google Scholar]
- 51.Chakraborty PK, Lee WK, Molitor M, et al. Cadmium induces Wnt signaling to upregulate proliferation and survival genes in sub-confluent kidney proximal tubule cells. Mol Cancer. 2010;9:102–117. doi: 10.1186/1476-4598-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di IE, Barbaro V, Ruzza A, et al. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc Natl Acad Sci USA. 2005;102:9523–9528. doi: 10.1073/pnas.0503437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keyes WM, Pecoraro M, Aranda V, et al. DeltaNp63alpha is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8:164–176. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez DA, Tapia JC, Fernandez JG, et al. Caveolin-1-mediated suppression of cyclooxygenase-2 via a beta-catenin-Tcf/Lef-dependent transcriptional mechanism reduced prostaglandin E2 production and survivin expression. Mol Biol Cell. 2009;20:2297–2310. doi: 10.1091/mbc.E08-09-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim PJ, Plescia J, Clevers H, et al. Survivin and molecular pathogenesis of colorectal cancer. Lancet. 2003;362:205–209. doi: 10.1016/S0140-6736(03)13910-4. [DOI] [PubMed] [Google Scholar]
- 57.Li F, Cheng Q, Ling X, et al. Generation of a novel transgenic mouse model for bioluminescent monitoring of survivin gene activity in vivo at various pathophysiological processes: Survivin expression overlaps with stem cell markers. Am J Pathol. 2010;176:1629–1638. doi: 10.2353/ajpath.2010.090414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoon K, Lim YS, Yu SB, et al. The expression of survivin and its related genes in adipocyte-derived stem cell by demethylation. Korean J Anesthesiol. 2010;58:383–390. doi: 10.4097/kjae.2010.58.4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barbaro V, Testa A, Di IE, et al. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J Cell Biol. 2007;177:1037–1049. doi: 10.1083/jcb.200703003. [DOI] [PMC free article] [PubMed] [Google Scholar]