SUMMARY
Circular RNAs (circRNAs) are single-stranded RNAs that are joined head to tail with largely unknown functions. Here we show that transfection of purified in vitro generated circRNA into mammalian cells led to potent induction of innate immunity genes and confers protection against viral infection. The nucleic acid sensor RIG-I is necessary to sense foreign circRNA, and RIG-I and foreign circRNA co-aggregate in cytoplasmic foci. CircRNA activation of innate immunity is independent of a 5′ triphosphate, double-stranded RNA structure, or the primary sequence of the foreign circRNA. Instead, self-nonself discrimination depends on the intron that programs the circRNA. Use of a human intron to express a foreign circRNA sequence abrogates immune activation, and mature human circRNA is associated with diverse RNA binding proteins reflecting its endogenous splicing and biogenesis. These results reveal innate immune sensing of circRNA and highlight introns—the predominant output of mammalian transcription—as arbiters of self-nonself identity.
In Brief
Chen et al. show that exogenous circRNAs trigger an immune response that can protect against viral infection. They find that cells distinguish self from nonself circRNAs based on their biogenesis. CircRNAs spliced by endogenous human spliceosomes associate with many RNA-binding proteins that mark their origin as self molecules.
INTRODUCTION
Initially discovered as pathogen genomes such as hepatitis D virus (HDV) and plant viroids (Kos et al., 1986; Sanger et al., 1976), circRNAs were recently recognized as a pervasive class of noncoding RNAs in eukaryotic cells (Hansen et al., 2013; Memczak et al., 2013; Salzman et al., 2012), generated through back splicing (Chen, 2016; Jeck and Sharpless, 2014; Wilusz, 2016). CircRNAs have been postulated to function as microRNA sponges (Hansen et al., 2013, 2011; Memczak et al., 2013), in cell-to-cell information transfer (Lasda and Parker, 2016), as templates for translation (Legnini et al., 2017; Pamudurti et al., 2017; Yang et al., 2017), or memory due to their extraordinary stability (Fischer and Leung, 2017). However, the ability of exogenous circRNA to activate immune response or inhibit viral infection has not been investigated. We show that circRNA generated from foreign introns potently stimulates immune signaling and inhibits viral infection. Additionally, cells distinguish between self-nonself circRNAs based on the introns from which they were produced, perhaps because mature human circRNAs are associated with diverse RNA binding proteins.
RESULTS
Cell-Free Production of CircRNA by Phage Intron
To generate circRNAs that we could subsequently transfect into cells, we programmed in vitro production of circRNAs with autocatalytic-splicing introns (Figure 1A). The group I intron of phage T4 thymidylate synthase (td) gene is well characterized to circularize while the exons linearly splice together (Chandry and Belfort, 1987; Ford and Ares, 1994; Perriman and Ares, 1998). When the td intron order is permuted (5′ half placed at the 3′ position and vice versa) flanking any exon sequence, the exon is circularized via two autocatalytic transesterification reactions (Ford and Ares, 1994; Puttaraju and Been, 1995). We encoded destabilized green fluorescent protein (GFP) followed by the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) as the exon. Successful circRNA production was monitored by reverse transcription and quantitative polymerase chain reaction (qRT-PCR) analysis with divergent primers (Figure 1A, brown, and Figure S1A) that span the splice junction and only amplifies product when circRNA is present. Convergent primers (Figure 1A, green, and Figure S1A) detect both the linear and circRNA. Circularization also brings the IRES upstream of the GFP sequence, allowing protein translation. Following in vitro transcription, the permuted autocatalytic-splicing introns produce a 1.5 kilobase circRNA, as expected (Figure 1B). As a control, we transcribed a linear RNA with the same sequence as the circRNA and treated both linear and circRNAs with phosphatase so there are no 5′ phosphates present in either sample. The circRNA product is resistant to digestion with the exonuclease RNase R while the linear RNA unspliced transcript and side products were depleted (Figure 1B). We thus used RNase R treatment to purify circRNA in all subsequent experiments. Direct sequencing of the circRNA splice junction confirmed a uniform splice junction, precisely at the expected autocatalytic splice site (Figure 1C). Thus, phage autocatalytic-splicing intron enables in vitro production of circRNAs that may be suitable for mammalian expression.
Figure 1. Cell-Free Production of CircRNA.
(A) CircRNA was synthesized by in vitro transcription from a permuted intron-exon template. RNase R treatment of the transcription products enriched for circRNA. Green arrows represent the convergent qRT-PCR primers to detect both linear and circRNA; brown arrows represent the divergent qRT-PCR primers to detect circRNA.
(B) Linear and circRNA were treated with RNase R and analyzed by Bioanalyzer.
(C) Sequencing of circRNA product reveals precise and uniform splice junction.
Exogenous CircRNA Stimulates Innate Immunity Gene Expression
In the course of investigating circRNA as a vehicle for ectopic gene expression, we unexpectedly uncovered its potent immune stimulatory property. When we transiently transfected purified unmodified or Cy3-labeled circRNA into HeLa cells, we observed very little GFP expression and, surprisingly, significantly more cell death in the cells containing circRNA than linear RNA encoding the same sequence (Figure S1B). This effect required intracellular introduction of circRNA via transfection; adding circRNA to the media had no effect. Transfection of in vitro transcribed linear RNA is known to induce innate immune signaling (Warren et al., 2010), but such an effect was not known for circRNAs. RNA-seq of HeLa cells transiently transfected with linear or circRNA showed that circRNA more potently stimulated the expression of genes involved in innate immunity; the top enriched Gene Ontology terms include response to cytokine, regulation of cytokine production, cellular response to virus, and NF-κB signaling (Figure 2A). 127 genes were significantly induced by either linear or circRNA transfection (induced 4-fold, p < 0.05, FDR < 0.1), and the majority of these genes were significantly more induced by circRNA than linear RNA transfection (84 observed versus 64 expected by chance alone, p = 0.00016, hypergeometric distribution; Figure 2A). CircRNA-induced genes include well-known innate immunity regulators such as retinoic-acid-inducible gene-I (RIG-I, also known as DDX58), melanoma-differentiation-associated gene 5 (MDA5, also known as IFIH1), 2′-5′ oligoadenylate synthase 1 (OAS1), OAS-like protein (OASL), and, to a lesser extent, protein kinase R (PKR).
Figure 2. Exogenous CircRNA Stimulates Innate Immunity Gene Expression and Inhibits Viral Infection.
(A) Linear and circRNA stimulate expression of specific genes. Left: scatterplot of RNA-seq result of HeLa cells transfected with linear or circRNA encoding GFP-IRES sequence. Genes induced by at least 2-fold compared to mock control are plotted, and the color scale represents the enrichment of circRNA compared to linear RNA. Genes shown in bold were validated by qRT-PCR below. Right: Gene Ontology enrichment of genes induced by circular or linear RNA. Heatmap GO enrichment p value is shown.
(B) CircRNA stimulates expression of innate immune genes. HeLa cells were mock transfected or transfected with 500 ng of linear or circRNA encoding GFP-IRES. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR. Means ± SEM are shown (n = 3), *p < 0.05, Student’s t test, throughout.
(C) CircRNA transfection protects against viral infection. FACS analysis plots of HeLa cells transfected with 500 ng of Cy3-labeled linear or circRNA and exposed to VEEV-GFP virus.
(D) Percent of all HeLa cells infected with VEEV-GFP following transfection with linear or circRNA.
(E) Percent of HeLa cells that did not contain Cy3-labeled linear or circRNA infected with VEEV-GFP following transfection with linear or circRNA.
To confirm the RNA-seq results, we transfected linear or circRNA into HeLa cells, washed extensively after 24 hr to remove unincorporated RNA, and isolated total RNA for qRT-PCR. We measured the expression of a panel of innate immune genes relative to actin and also measured the levels of transfected circular and linear RNAs inside cells for normalization to account for the differences between linear and circRNA transfection efficiency and stability for all subsequent assays (STAR Methods). CircRNA potently stimulates the expression of several innate immunity genes (e.g., ~500-fold for RIG-I and 200-fold for OASL mRNA) and more potently than linear RNA of the same sequence or mock transfection control (Figures 2B, S1C, S2A, and S2B and Table S1). RIG-I induction by circRNA was also confirmed on the protein level (Figure S2C). Transfection of circRNA also induced innate immunity genes in a keratinocyte cell line (HaCaT; Figure S2D) and a mouse macrophage cell line (RAW 264.7; Figure S2E). These results suggest circRNA is an inducer of innate immunity signaling.
CircRNA Inhibits RNA Virus Infection
To test if the immunogenic response stimulated by circRNA transfection affects the ability of a virus to infect HeLa cells, we first transfected cells with linear or circRNA labeled with Cy3, waited 24 hr, and exposed cells to Venezuelan equine encephalitis virus harboring a green fluorescent protein reporter gene (VEEV-GFP, a +ssRNA virus). FACS analysis showed mock-treated cells and cells with linear RNA were infected to similar levels, while cells transfected with circRNA had a 10-fold lower VEEV-GFP infection rate (Figures 2C and 2D). Additionally in cultures transfected with circRNA, neighbor cells that did not contain transfected circRNA also had decreased infection levels, similar to cells that had circRNA (Figure 2E). This collective, non-cell-autonomous protection from virus infection is consistent with the ability of foreign circRNA to induce antiviral cytokines such as interferon-beta. Collectively, these results show that sensing of exogenous circRNA can confer functional immune protection against a viral pathogen.
RIG-I Mediates Immune Response to Foreign CircRNA
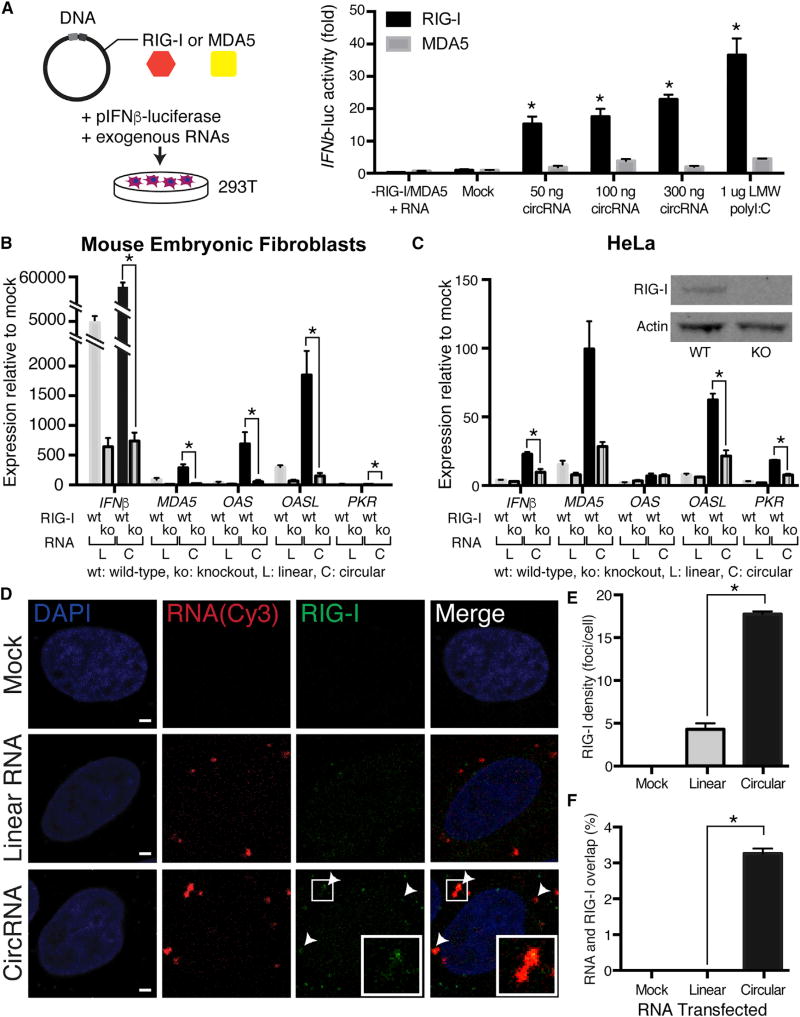
RIG-I and MDA5 are two well-known intracellular sensors of foreign RNAs in mammalian cells. RIG-I is activated by a short “panhandle” RNA structure with 5′ triphosphate (Hornung et al., 2006) while MDA5 is activated by long double-stranded RNAs (Hornung et al., 2006; Kato et al., 2008). Neither of these features is known to exist in circRNA. To investigate which cytosolic foreign nucleic acid sensor may detect exogenous circRNA, we used HEK293T cells that require exogenous RIG-I or MDA5 to be added for sensing exogenous RNAs. We found that expression of RIG-I, but not MDA5, conferred responsiveness to circRNA in HEK293T cells, as evidenced by an ~20-fold induction of IFNβ-reporter gene (Figure 3A). Quantitative comparison with low molecular weight polyI:C (range 0.2–1 kb length), which activates RIG-I strongly and MDA5 weakly, showed that accounting for molarity, circRNA is approximately 10-fold more potent than polyI:C in immune gene stimulation. In contrast, MDA5 expression conferred strong responsiveness to long dsRNA but not circRNA, suggesting distinct signaling requirements (Figures S3A–S3C).
Figure 3. RIG-I Is Necessary and Sufficient to Sense Exogenous CircRNA.
(A) RIG-I but not MDA-5 confers circRNA sensitivity in HEK293T cells. Luciferase activity of HEK293T cells transfected with plasmids encoding RIG-I or MDA5, along with mouse Ifnb promoter-firefly luciferase/Renilla luciferase vectors, and the indicated circRNA ligands or LMW polyI:C as positive control. Cells transfected with RIG-I or MDA5 only (mock) were used as a negative control. Results are presented relative to Renilla luciferase activity (*p < 0.05 compared to mock control and for RIG-I over MDA5).
(B) RIG-I is required to sense transfected circRNA in MEFs. Wild-type or RIG-I knockout (KO) MEFs were mock transfected or transfected with 500 ng of linear or circRNA. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR. Means ± SEM are shown (n = 3); *p < 0.05 throughout.
(C) RIG-I is required to sense transfected circRNA in HeLa cells. Wild-type or RIG-I knockout (KO) HeLa were mock transfected or transfected with 500 ng of linear or circRNA. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR. Means ± SEM are shown (n = 3). Inset: western blot of RIG-I or actin from wild-type or KO HeLa cells.
(D) CircRNA transfection induces RIG-I foci. HeLa cells were transfected with 500 ng of Cy3-labeled linear or circRNA encoding GFP-IRES and costained for RIG-I. Subcellular distribution of Cy3-labeled RNA (red), RIG-I (detected with secondary antibody conjugated with -Atto647N, pseudo-colored green), and 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclei (blue) were visualized by confocal microscopy. Scale bar, 2 µm. White arrows indicate areas of RIG-I and RNA colocalization.
(E) Quantification of the RIG-I density. Means ± SEM are shown; n ≈ 41 cells for each condition.
(F) Quantification of the overlap fraction between RIG-I and linear or circRNA. Means ± SEM are shown; n ≈ 41 cells for each condition.
Next, we investigated the requirement of RIG-I in sensing circRNA. Transfection of wild-type or RIG-I knockout (KO) MEFs (Kato et al., 2005) showed that RIG-I KO completely abrogated circRNA-induced innate immune genes (Figure 3B). RIG-I KO MEFs are still inducible by interferon stimulatory DNA, suggesting that RIG-I is needed in a proximal step of circRNA sensing (Figure S4A). Similarly, we generated HeLa cells knocked out for human RIG-I by CRISPR-Cas9 technology and found that human RIG-I KO also substantially reduced the response of innate immunity genes to circRNAs (Figures 3C and S4B). These results suggest that RIG-I is necessary and sufficient to sense foreign circRNA, at least in the cell types examined.
To probe whether RIG-I may directly sense circRNAs, we performed immunofluorescence microscopy of Cy3-labeled transfected RNA and endogenous RIG-I (Figure 3D). We found that RIG-I in cells with transfected circRNA formed more and larger intracellular foci compared to with linear RNA (p < 0.05 for each; Figures 3E, S4C, and S4D). Notably, circRNA was significantly more likely to colocalize in RIG-I foci than linear RNA (Figure 3F). The colocalized foci typically have RIG-I signal in the center surrounded by circRNA and are distributed throughout the cytoplasm. These results implicate RIG-I in the proximal sensing of circRNA. Because RIG-I is itself induced by circRNA and needed to sense circRNA, circRNA induction of RIG-I may constitute a positive feedback loop. RIG-I may directly sense circRNA, or it may be required for a proximal signal amplification step for immune gene induction.
Immune Response to CircRNA Is Not Explained by Known Ligands
How might circRNA activate RIG-I? We were able to rule out two possible features. First, the known pathogen-associated molecular pattern (PAMP) for RIG-I includes a 5′ triphosphate on RNA (Hornung et al., 2006). By definition, circRNA does not have a 5′ end, and treatment of our circRNA preparation with phosphatases that selectively cleaves 5′ phosphates did not diminish its ability to stimulate innate immune genes (Figure 4A). As positive control, phosphatase treatment of 5′ triphosphate-containing linear RNA greatly reduced its immune signaling activity (Figure S5A). These results suggest that circRNA activation of RIG-I is not due to contaminating 5′ triphosphates that are present in the circRNA preparation. Second, long double-stranded RNA (dsRNA) can activate immune signaling (typically via MDA5 rather than RIG-I). We tested the possibility that circularization may cause significant differences in secondary structure that lead to innate immune gene activation. We performed in vitro selective 2′-hydroxyl acylation and profiling (SHAPE) experiments, whereby single-stranded bases are mapped by high SHAPE reactivity (Spitale et al., 2015). Sequencing of the SHAPE libraries showed that in vitro-folded linear and circRNA, arising from the same primary sequence, have nearly identical SHAPE reactivity scores, suggesting that they have comparable secondary structures (Figure 4B). There was also no significant difference in the longest consecutive stretch of dsRNA in circular or linear RNA (38 versus 39 bases; Figure 4B). Thus, sensing of circRNA appears independent of known PAMPs of foreign RNAs.
Figure 4. CircRNA Stimulation of Innate Immune Genes Is Not Explained by Known RIG-I Ligands.
(A) Exogenous circRNA does not contain 5′ triphosphate. HeLa cells were transfected with 500 ng of circRNA or circRNA treated with phosphatase. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR. Means ± SEM are shown (n = 3).
(B) Exogenous circRNA does not contain long dsRNA duplex. In vitro selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) was conducted on linear and circRNA. SHAPE reactivity scores were calculated for each nucleotide. Black line denotes the longest consecutive dsRNA region.
(C) Transfection of circRNA made from ligating the ends of linear RNA stimulates innate immune genes. HeLa cells were mock transfected or transfected with 500 ng of linear or splint ligated circRNA encoding GFP-IRES. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR. Means ± SEM are shown (n = 3); *p < 0.05 throughout.
(D) CircRNA samples do not contain aberrant products that stimulate immune response. CircRNA was treated with RNase H in the presence of hybridizing oligonucleotide. HeLa cells were mock transfected or transfected with linear RNA, circRNA, or circRNA treated with RNase H and hybridizing oligonucleotide. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR. Means ± SEM are shown (n = 3).
We performed two additional experiments to rule out the possibility that aberrant, unknown products in the preparation of circRNA may cause immune stimulation. First, circRNA generated by in vitro splint ligation (without using the autocatalytic-splicing td intron) also induced innate immune genes (Figure 4C), suggesting that any circRNA not generated by mammalian splicing is perceived as foreign. Second, after circRNA production by in vitro transcription and purification, we used a complementary DNA oligonucleotide and RNase H to specifically cleave the circRNA. Linearization of circRNA abrogated its immune stimulatory property (Figure 4D), indicating that it is indeed the circRNA and not any other product that is responsible for the immune activation. Taken together, these experiments demonstrate that the circular properties of the RNA cause immune gene stimulation.
CircRNA Biogenesis Dictates RNA Activation of Immune Signaling
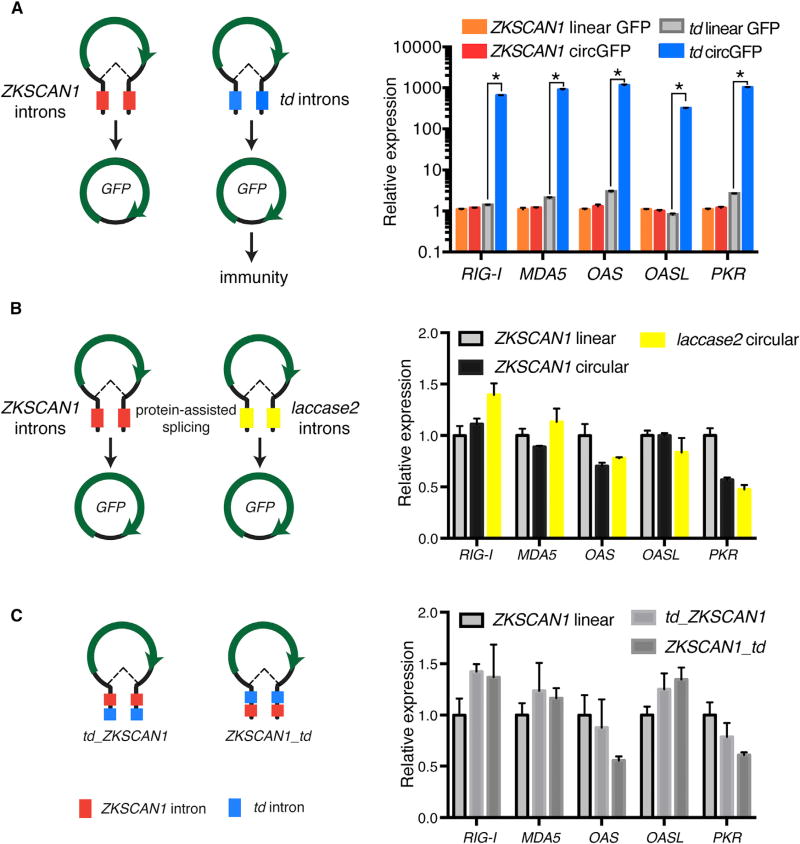
Next, we mapped the feature of circRNA required to activate innate immune gene expression. We used a phage td intron to produce linear and circRNA in vitro containing only the mCherry coding sequence—replacing the entire exonic sequence of GFP-IRES circRNA. The mCherry circRNA potently simulated innate immune genes over linear RNA and control, similar to GFP-IRES circRNA (Figures S5B–S5D). These results suggest that immune activation is based on the circularity of the RNA rather than primary sequence or ribosome binding by IRES. We reasoned that if the circRNA exon does not define “self” versus “foreign,” the only remaining feature is the intron that programs the circularization. We transfected a DNA construct that expresses the phage autocatalytic-splicing GFP-IRES circRNA and compared it to a control DNA construct that lacks the second intron half, making it only encode linear RNA. We found that DNA-programmed expression still induced robust immune gene activation, highlighting a potent mode of circRNA induction of immune genes (~600-fold over linear RNA; Figure 5A). We kept the GFP-IRES circRNA exon and replaced the phage td intron with endogenous human ZKSCAN1 introns (Liang and Wilusz, 2014), which do not have autocatalytic-splicing properties (Figures 5A and S6C). Complementary Alu repeats are present in these flanking introns, which enable human ZKSCAN1 to splice GFP-IRES into a circRNA. The linear control is the vector missing a portion of the upstream ZKSCAN1 intron so that base pairing of the 5′ and 3′ introns and circularization would not occur. The full-length ZKSCAN1 introns led to copious production of GFP-IRES circRNA (confirmed by qRT-PCR and used for normalization; Figures S6A and S6B), but completely abrogated innate immune gene induction (Figure 5A). These results suggest that splicing by a “self intron” in human cells confers self-identity to circRNAs irrespective of their exonic sequence, and distinguishes endogenous circRNAs from foreign nucleic acids.
Figure 5. Intron Sequence Determines Immune Signaling by CircRNA.
(A) HeLa cells were transfected with 500 ng of DNA plasmid programming ZKSCAN1 or td introns that produce a linear RNA or circRNA encoding GFP-IRES. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR. Means ± SEM are shown (n = 3); *p < 0.05 throughout.
(B) CircRNA produced from Drosophila laccase2 introns flanking GFP-IRES exon does not stimulate innate immune genes. HeLa cells were transfected with 500 ng of DNA plasmid that produce a linear RNA or circRNA encoding GFP-IRES directed by laccase2 introns.
(C) Co-expression of programmed protein-assisted and autocatalytic-splicing td circRNA still stimulates innate immune genes. HeLa cells were transfected with 500 ng of DNA plasmid programming ZKSCAN1 intron or sequential ZKSCAN1 and td introns encoding circRNA GFP-IRES. Relative expression of the indicated mRNA and transfected RNA are measured by qRT-PCR, to confirm both the autocatalytic-splicing and protein-assisted splicing processes occur. Means ± SEM are shown (n = 3).
We further interrogated the properties of the foreign circRNA sensing mechanism. First, transfection of a DNA construct encoding the Drosphila laccase2 introns (Kramer et al., 2015) flanking GFP-IRES exon into HeLa cells programmed circRNA production, as confirmed by qRT-PCR, but did not stimulate innate immune genes (Figure 5B). Thus, because circRNA programmed by Drosophila introns but processed by the human spliceosome is sensed as “self,” it is likely that the human splicing machinery or an associated process—rather than the sequence of human intron itself—confers self-identity to endogenously produced circRNAs. Second, encoding both td and ZKSCAN1 introns on the same RNA molecule yielded circRNA products that did not activate innate immune genes (Figure 5C). The same result is obtained with human introns either flanking phage intron or nested within phage intron. This result suggests that the “self circRNA” identity conferred by human introns is dominant over the foreign identity. Third, co-expression of ZKSCAN1 intron-programmed circRNAs and td circRNAs in the same cells still led to immune gene activation, indicating that human intron-mediated protection of immune activation operates solely in cis and lacks transitive property (Figure S6D).
Endogenous CircRNA Associated with a Diverse Set of RNA-Binding Proteins
Finally, to understand how endogenous human circRNA may be marked, we performed comprehensive identification of RNA binding proteins by mass spectrometry (ChIRP-MS) (Chu et al., 2015) to compare proteins that are associated with model circular or linear RNAs (Table S2). We transfected HeLa cells with DNA plasmids encoding RNAs with combinations of (i) ZKSCAN1 or phage td introns, which direct protein-assisted splicing or autocatalytic splicing, respectively; (ii) ZKSCAN1 endogenous exons or the GFP-IRES exon; (iii) control linear RNAs that lack one of the flanking introns required for back splicing or circRNAs with full introns (Figure 6A). The endogenous ZKSCAN1 circRNA arises from a canonical “forward splice” between exons 2 and 3 and a “back splice” from the 3′ of exon 3 back to the 5′ of exon 2 (Liang and Wilusz, 2014); the mix of forward and back splice typifies most circRNAs (Jeck and Sharpless, 2014; Jeck et al., 2013; Salzman et al., 2012; Wilusz, 2016). After DNA transfection, we performed FISH against td-intron programmed circGFP-IRES and found the circRNA to be predominantly localized to the cytoplasm. Cells were crosslinked with formaldehyde to trap endogenous complexes; we then used oligonucleotide probes to isolate ZKSCAN1 or GFP-IRES RNA products, along with their associated proteins. The oligonucleotide probes are directed against the exonic sequences only; hence, this experiment tests the consequence of introns on the subsequent fate of RNA binding proteins that associate with circRNA.
Figure 6. Splicing Mechanism Determines Self versus Non-self CircRNA.
(A) Schematic for DNA-programmed splicing of linear or circular ZKSCAN1 or td-directed RNA with endogenous ZKSCAN1 or GFP-IRES exons.
(B) Heatmap of associated proteins identified by ChIRP-MS for each of the spliced RNAs. Enlarged portion of the heatmap shows the proteins for the most enriched circZKSCAN1 RNAs generated by ZKSCAN1 introns. Proteins shown in bold are found enriched in ZKSCAN1 intron-directed circRNAs (both ZKSCAN1 and GFP-IRES exons).
(C) Proteomic analysis of linear and circRNA ribonucleoprotein complexes. Venn diagrams show enriched proteins detected by ChIRP-MS of ZKSCAN1 intron-directed linear or circular ZKSCAN1 or GFP-IRES exons. Gene ontology enrichment for the proteins detected in both linear and circular ZKSCAN1 or GFP-IRES samples.
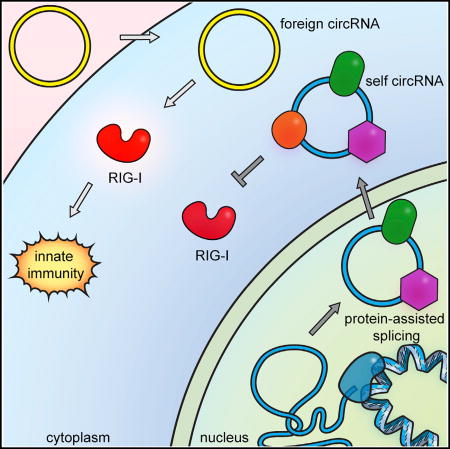
(D) Model of self versus non-self discrimination of mammalian circRNAs.
Following mass spectrometry to identify the proteins, we defined enrichment as containing ten or more spectral counts with 4-fold or greater than the RNase-treated negative control. We found that both circular and linear ZKSCAN1 RNAs are associated with a large and common set of RNA binding proteins (145 of 192 RBPs [75%]) involved in RNA splicing, ribonucleocomplex assembly, and nucleic acid transport (lane 1 versus lane 2 in Figure 6B, and Table S3). The shared RBPs include multiple components of the spliceosome (e.g., U1, U2, and U4/U6/U5 tri-snRNP subunits), the exon junction complex that is deposited as splice junctions (EIF4A3, MGN2, RNPS1), and factors that mediate nuclear export of RNAs (DDX39B, THOC4, XPO5). In addition, we found a set of 36 proteins that are at least 3-fold more enriched on circZKSCAN1 than linear ZKSCAN1 RNA (Figure 6C and Table S3). These include MOV10, a RNA helicase required for replication of HDV circRNA genome (Haussecker et al., 2008) and RNA interference (Tomari et al., 2004), components of the nonsense-mediated decay pathway (UPF1, UPF2), RNA N6-methyladenosine reader proteins (YTHDF2, YTHDF3) (Dominissini et al., 2012), and the antiviral protein OASL.
To identify proteins associated with RNAs recognized as “self,” we compared ZKSCAN1 intron-directed linear and circRNAs. Many of the proteins common to the ZKSCAN1 introns expressing endogenous ZKSCAN1 exons were also detected in the ChIRP-MS results from ZKSCAN1 introns expressing a foreign GFP-IRES exon (lane 1 versus lane 3 in Figures 6B and 6C, and Table S3). This demonstrates that the introns direct the placement of RBPs onto the circular and linear RNAs. The proteins unique to circGFP-IRES generated by ZKSCAN1-directed circularization also overlap with the circZKSCAN1, whereas the circGFP-IRES generated by td-directed circularization have very few proteins associated (lane 3 versus lane 5 in Figure 6C, and Table S3). We did not detect RIG-I associated with the autocatalytically spliced circRNA, perhaps suggesting that the interaction is transient and was not captured by the ChIRP-MS.
Thus, a diverse and distinctive set of RBPs associate with endogenously produced human circRNA. Recent identification of m6A-marked endogenous human circular RNAs is fully consistent with our observation (Molinie et al., 2016; Yang et al., 2017). These results demonstrate that introns program the protein cargos of circular RNAs, and also refine the set of RNA binding proteins that can mark the circRNA as “self.”
DISCUSSION
CircRNAs are prevalent in mammalian cells, but much is still unknown about their function, biosynthesis, degradation, and associated cellular machinery. Endogenous messenger RNAs possess features on their ends—the 5′ cap and 3′ polyA tail— that identify them as products of RNA polymerase II. Do circular RNAs escape immune surveillance? Our results suggest a system of self-nonself discrimination for circRNAs, based on the intron that programs the circularization (Figure 6D). This strategy is elegant and general: cells may sense diverse foreign circRNAs, such as HDV and viroids, irrespective of their primary sequence, while not responding to numerous self circRNAs. Another potential advantage of using introns to distinguish self versus foreign circRNAs is that the large size of human introns (spanning multiple kilobases on average) makes it difficult for compact viral genomes to simply incorporate introns in order to mask foreign circRNAs. We speculate that it may be the expansiveness of human “junk DNA” in introns that confers its utility for self-nonself discrimination and thereby defines humanness.
CircRNAs generated by human intron splicing are endowed with numerous RBPs and potentially other features that mark their origin. How exogenous circRNA activates RIG-I, and how one or more factors deposited by human introns prevent RIG-I activation are key questions for future studies. Our discovery of a large family of proteins associated with human-spliced circRNAs opens the door for systematic analyses. The recognition that circRNAs deemed foreign can activate immune signaling raises the question of circRNA involvement in autoimmune diseases. Conversely, exogenous introduction of circRNAs may boost immune activation for therapeutic purposes.
STAR*METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Microbe strains
-
METHOD DETAILS
Plasmids
RNA synthesis and purification
Reverse transcription and real time PCR analysis (qRT-PCR)
-
CELL LINES AND MAINTENANCE
Cell Culture and Transient Transfection
RNA extraction, selection, library preparation, and sequencing
Western blot analysis
VEEV-GFP infection
Luciferase activity assay
Construction and validation of RIG-I−/− in HeLa cells
Immunofluorescence
ChIRP-mass spectrometry
Flow cytometry
-
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA expression analysis
Image analysis
DATA AND SOFTWARE AVAILABILITY
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-RIG-I | Cell Signaling Technology | 3743S; RRID: AB_2269233 |
| Goat polyclonal IgG anti-actin | Santa Cruz Biotechnology | SC-1616; RRID: AB_630836 |
| IRDye 800CW Goat anti-rabbit IgG | LI-COR | 926–32211; RRID: AB_621843 |
| IRDye 680CW Donkey anti-goat IgG | LI-COR | 926–68074; RRID: AB_10956736 |
| Secondary antibody goat anti-rabbit -Atto647N | Sigma-Aldrich | 50185-1ML-F |
| Bacterial and Virus Strains | ||
| TC83 vaccine strain VEEV-GFP | Dr. Jan Carette | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cy3-UTP | GE Healthcare | PA53026 |
| FastAP | ThermoFisher Scientific | EF0652 |
| RNase R | Epicenter | RNR07250 |
| T4 DNA ligase | New England Biolabs | M0202M |
| RNase H | New England Biolabs | M0297L |
| DNase I | Ambion | AM2222 |
| T4 PNK | New England Biolabs | M0201S |
| Ribolock RNase Inhibitor | Life Technologies | EO0384 |
| T4 RNA ligase 1, high concentration | New England Biolabs | M0437M |
| RecJf | New England Biolabs | M0264L |
| Deadenylase | New England Biolabs | M0331S |
| T7 Endonuclease I | New England Biolabs | M0302L |
| TRIzol | Invitrogen | 15596018 |
| Brilliant II SYBR Green qRT-PCR Master Mix | Agilent | 600825 |
| Low molecular weight poly(I:C) | Invivogen | tlrl-picw |
| High molecular weight poly(I:C) | Invivogen | vac-pic |
| Interferon stimulatory DNA | Invivogen | tlrl-isdn |
| Lipofectamine 2000 | Invitrogen | 11668-019 |
| Antibody dilution reagent | ThermoFisher Scientific | 00-3218 |
| DAPI | Vector Labs | H-1200 |
| Annexin V-FITC | BD PharMingen | 556420 |
| Critical Commercial Assays | ||
| mMessage mMachine T7 transcription kit | Ambion | AM1344 |
| MEGAscript T7 transcription kit | Ambion | AM1334 |
| RiboMinus Eukaryote Kit for RNA-Seq | Invitrogen | A1083708 |
| LIVE/DEAD Fixable Blue Dead Cell Stain Kit | ThermoFisher | L23105 |
| Dual-Luciferase Reporter Assay | Promega | E1910 |
| Deposited Data | ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE81345 |
| ChIRP-MS data | This paper | Table S1 |
| Experimental Models: Cell Lines | ||
| HeLa | ATCC | CCL-2 |
| HEK293T | ATCC | CRL-3216 |
| HaCaT | ATCC | PCS-200-010 |
| RAW 264.7 | ATCC | TIB-71 |
| Wild-type MEF | Kato et al., 2005 | N/A |
| RIG-I KO MEF | Kato et al., 2005 | N/A |
| RIG-I KO HeLa | This paper | N/A |
| Oligonucleotides | ||
| Primers | This paper | Table S2 |
| ChIRP probes | This paper | Table S3 |
| Recombinant DNA | ||
| Plasmid: autocatalytic-splicing linear GFP-IRES | This paper | N/A |
| Plasmid: autocatalytic-splicing circular GFP-IRES | This paper | N/A |
| Plasmid: autocatalytic-splicing linear mCherry | This paper | N/A |
| Plasmid: autocatalytic-splicing circular mCherry | This paper | N/A |
| Plasmid: ZKSCAN1-directed linear GFP-IRES | This paper | N/A |
| Plasmid: ZKSCAN1-directed circular GFP-IRES | This paper | N/A |
| Plasmid: laccase2-directed linear GFP-IRES | This paper | N/A |
| Plasmid: laccase2-directed circular GFP-IRES | This paper | N/A |
| Plasmid: ZKSCAN1-directed linear exon2_intron_exon3 | Liang and Wilusz, 2014 | N/A |
| Plasmid: ZKSCAN1-directed circular exon2_intron_exon3 | Liang and Wilusz, 2014 | N/A |
| Plasmid: MLM3636 | Addgene | 43860 |
| Plasmid: p3s-Cas9HC | Addgene | 43945 |
| Software and Algorithms | ||
| DESeq2 | Love et al., 2014 | N/A |
| Metascape | Tripathi et al., 2015; http://metascape.org | N/A |
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Dr. Howard Y. Chang, at Stanford University (howchang@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Wild-type and RIG-I KO MEFs were provided by Dr. Shizuo Akira and Dr. Takashi Satoh (Osaka University). MEFs were grown in Dulbecco’s modified Eagle’s medium (DMEM, 11995-073) supplemented with 100 units/ml penicillin-streptomycin (GIBCO, 15140-163), 10% (v/v) fetal bovine serum and 50 µM 2-Mercaptoethanol (Sigma-Aldrich, 516732). Human HeLa (cervical adenocarcinoma, ATCC CCL-2), human HEK293T (embryonic kidney, ATCC CRL-3216), human HaCaT (immortalized keratinocyte, ATCC PCS-200-010) and mouse RAW 264.7 (macrophage, ATCC TIB-71) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, 11995-073) supplemented with 100 units/ml penicillin-streptomycin (GIBCO, 15140-163) and 10% (v/v) fetal bovine serum (Invitrogen, 12676-011). Cell growth was maintained at 37°C in a 5% CO2 atmosphere.
Microbe strains
TOP10 E. coli competent cells were obtained from ThermoFisher Scientific. Cells were stored at −80°C and grown in LB medium at 37° C.
METHOD DETAILS
Plasmids
The plasmid containing the permuted intron-exon from the td gene of T4 bacteriophage was provided by Dr. Manuel Ares, Jr. (University of California, Santa Cruz). Plasmids expressing ZKSCAN1 and laccase2 were provided by Dr. Jeremy E Wilusz. ZKSCAN1 100–2232 plasmid was used for circRNA production and ZKSCAN1 450–2232 plasmid for linear RNA production (Liang and Wilusz, 2014). Gibson assembly (NEB) was used to construct the autocatalytic-splicing and ZKSCAN1 plasmids containing the GFP-IRES or mCherry exons.
RNA synthesis and purification
Unmodified RNA was synthesized by in vitro transcription using mMessage mMachine T7 transcription kit (Ambion, AM1344) following the manufacturer’s instructions. Cy3-labeled RNA was synthesized by in vitro transcription using MEGAscript T7 transcription kit (Ambion, AM1334) and adding Cy3-UTP (GE Healthcare, PA53026) in a 1:25 ratio with the transcription kit’s CTP. Transcribed RNA was purified by RNeasy Mini column (QIAGEN), treated with FastAP (ThermoFisher Scientific, EF0652) following the manufacturer’s instructions, and purified again by RNeasy Mini column. CircRNA was purified by treatment with RNase R (Epicenter, RNR07250) following the manufacturer’s instructions. Splint ligation circRNA was generated by treatment of in vitro transcribed linear RNA and DNA splint ATATGACGCAATATTAAACGGTAGACCCAAGAAAACATCTACTGAG with T4 DNA ligase (Moore, 1999; Moore and Sharp, 1992) (New England Bio, Inc., M0202M), and circRNA was isolated following RNase R treatment. For the RNase H assay, circRNA was hybridized with DNA complementary to the splice junction (ATTAAACGGTAGACCCAAGAAAAC) and treated with RNase H (New England Biolabs Inc., M0297L) following the manufacturer’s instructions. Quality of the RNA was assessed by agarose gel or Bioanalyzer (Agilent).
Reverse transcription and real time PCR analysis (qRT-PCR)
Total RNA was isolated from cells using TRIzol (Invitrogen, 15596018) and an RNeasy Mini kit (QIAGEN) following the manufacturer’s instructions. qRT-PCR analysis was performed in triplicate using Brilliant II SYBR Green qRT-PCR Master Mix (Agilent, 600825) and a LightCycler 480 (Roche). Primers used found in Table S1. mRNA levels were normalized to actin, GAPDH, or HPRT values. Relative expression of indicated mRNA genes for circRNA transfection are normalized by level of transfected RNA and plotted as the fold change to the expression level of cells with mock or linear RNA transfection. For the luciferase assays, results are presented relative to renilla luciferase activity.
CELL LINES AND MAINTENANCE
Cell Culture and Transient Transfection
1 ug of low molecular weight poly(I:C) (Invivogen, tlrl-picw), high molecular weight poly(I:C) (Invivogen, vac-pic), or interferon stimulatory DNA (Invivogen, tlrl-isdn) were transfected using Lipofectamine 2000 (Invitrogen, 11668-019). Briefly, cells were transfected at 70 to 80% confluence using Lipofectamine 2000. The nucleic acids and Lipofectamine 2000 were diluted, mixed in Opti-MEM (Invitrogen, 31985-088), and incubated for 5 min at room temperature (RT). Following, the nucleic acids and Lipofectamine 2000 were mixed together, incubated for 15 min at RT and then the nucleic acids-Lipofectamine 2000 complexes were applied to the monolayer cultures. 500 ng of linear or circRNA was transfected into one well of a 24-well plate.
RNA extraction, selection, library preparation, and sequencing
Total RNA was isolated using TRIzol reagent (Invitrogen), according to manufacturer’s instructions. RNA was re-suspended in ultrapure water and treated with DNase I (Ambion, AM2222) for 30 min at 37°C and subjected to RNA clean up with RNeasy Midi Kit (QIAGEN), according to manufacturer’s instructions. RNA was eluted in ultrapure water. Total RNA was subjected to ribosomal RNA depletion with RiboMinus, and larger RNAs (> 200nt) were enriched by selective binding to RNeasy Mini columns in the following ratio: 100 µL RNA in water to 350 µL RLT to 120 µL of 100% EtOH. The RNeasy manual was followed for washes and elution.
Library preparation for high-throughput sequencing was performed following the library cloning method described in Flynn et al. (Flynn et al., 2015). The following notes are minor deviations for RNA-seq: Ribo-depleted RNA was fragmented to ~100 nt by incubation for 45 s at 95°C with Zinc chloride buffer (10 mM ZnCl2,10mM Tris-HCl, pH 7.0). The reaction was stopped with 0.2M EDTA and immediately placed on ice. The fragmented RNA was recovered using the RNEasy Mini Kit (QIAGEN). Fragmented RNA was treated with 10 Units of T4 PNK (New England Biolabs, Inc., M0201S), 1 Unit of FastAP (ThermoFisher Scientific) in 1x PNK buffer with 40 Units of Ribolock RNase Inhibitor (Life Technologies, EO0384) for 30 min at 37°C, to repair the 3′ of the RNA. For the ligation of the 3′ end adaptor, 0.8 µM of 3′ end biotin blocked preadenylated adaptor, 6.6 Units of T4 RNA ligase 1, high concentration (New England Biolabs, Inc., M0437M), in 1x RNA ligase buffer with 5 mM DTT and 50% PEG8000, were added, and the reaction was incubated at RT for 3 hr. To remove excess linker, 30 Units of RecJf (New England Biolabs, Inc., M0264L) and 20 Units of 5′ Deadenylase (New England Biolabs, Inc., M0331S) in 1x RecJ buffer with Ribolock RNase Inhibitor were added to the reaction, and incubated for 1 hr at 37°C. The reaction was cleaned with RNA Clean & Concentrator columns (Zymo Research, R1016).
Reverse transcription, biotin capture of ligated fragments, cDNA circularization, library amplification, and PAGE purification were carried out exactly as written in the previously published method (Flynn et al., 2015). One microliter of each sample was used for quantification with Kapa Library quantification (Kapa Biosystems) and then sent for sequencing on the Illumina MiSeq for 1 × 50-bp cycle run.
Western blot analysis
HeLa cells were collected and lysed 24 hr after transfection to extract total proteins. RIPA buffer (150 mM sodium chloride, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris, pH 8.0) was used to lyse the cells. Proteins were fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, blocked in phosphate-buffer saline containing 3% (wt/vol) nonfat milk for 1 hr at RT, and then incubated overnight at 4°C with the indicated primary antibody. Rabbit polyclonal antibody was used to detect RIG-I (Cell Signaling Technology, 3743S) and goat polyclonal IgG antibody was used to detect actin (Santa Cruz Biotechnology, SC-1616). IRDye 800CW Goat anti-rabbit IgG (LI-COR, 926–32211) or IRDye 680CW Donkey anti-goat IgG (LI-COR, 926–68074) secondary antibodies were used according to the manufacturer’s instructions. Western blot detection and quantification was done using an Odyssey infrared imaging system (LI-COR).
VEEV-GFP infection
Transient transfection of linear or circRNA into HeLa cells as described above was performed 24h prior to infection with TC83 vaccine strain VEEV-GFP at MOI of 3.72. Samples were stained with LIVE/DEAD Fixable Blue Dead Cell Stain Kit, for UV excitation (Thermo-Fisher, L23105), 1:500 in PBS +1% FBS for 15 min. Cells were fixed with 4% paraformaldehyde in PBS for 10 min. Cells were processed and analyzed on a special order FACS Aria II (BD Biosciences).
Luciferase activity assay
Luciferase activity was determined by Dual-Luciferase Reporter Assay system (Promega, E1910) following the manufacturer’s instructions. Briefly, cultured cells were lysed in the Passive Lysis Buffer. Firefly and renilla luciferase activity in the supernatant was measured with SpectraMax i3x (Molecular Devices) and analyzed with Softmax pro 6.4 (Molecular Devices).
Construction and validation of RIG-I−/− in HeLa cells
CRISPR target sequence of RIG-I was chosen using casoffinder (http://www.rgenome.net/cas-offinder/) to have less than 4 mismatches for NGG PAM sequence. Self-complementary oligonucleotides (acaccgGATTTCTGCTGTTCATACACg and aaaacGTGTATGAACAGCAGAAATCcg, ordered from Elim Biopharm) targeting a portion of DDX58 (GATTTCTGCTGTTCATACAC) were annealed, and ligated into BsmbI-digested MLM3636 (Addgene, 43860) to create a plasmid that would transcribe the guide RNA. 650 ng of MLM3636 containing the DDX58 target sequence and 650 ng of p3s-Cas9HC (Addgene, 43945) were transiently transfected into 24-well plates of HeLa as described above. After 48 hr, colonies were plated in 96-well with a density of 0.3 cells/well for single colony isolation. To validate RIG-I knockout clones, genomic DNA was isolated during sub-culture using DNeasy Blood &Tissue Kit (QIAGEN). DDX58 locus containing the target sequence was amplified by PCR (Forward: GGCTGTTGGCATGCTACTTA, Reverse: CCTTAGCACAGAGCCTGACA, and nested Forward: TCAGCTCAGTGGTATTAGAAGAGC, nested Reverse: TGTGCCACGTAAACATCAAA) followed by T7 Endonuclease I (New England Biolabs Inc., M0302L) digestion. Clones with appropriate T7 Endonuclease I activity were sequenced by MiSeq (Illumina), after amplifying by PCR (Forward: GGGGAAAGTTGTCTTTTTTGC, Reverse: TACCTACCCATGTCTTTCAAAGTATT) followed by True-seq adaptor ligation. Clones that did not contain an in-frame deletion or wild-type sequence in the MiSeq sequencing were probed by western blot to assess for the presence of RIG-I protein.
Immunofluorescence
HeLa cells were grown on 8-well Nunc® Lab-Tek® Chamber Slide system (Sigma-Aldrich, C7182-1CS) and transient transfection of Cy3-RNA was performed as described above. At 80%–90% confluency, cells were fixed with 1% formaldehyde (Sigma-Aldrich) for 10 min at RT. The formaldehyde fixed slide was rinsed in PBS and permeabilized in PBS 0.5% Triton X-100 for 10 min at RT. Then, the slide was blocked with antibody dilution reagent (ThermoFisher Scientific, 00–3218) for 1 hr at RT. Rabbit polyclonal primary antibody anti-RIG-I (Cell Signaling Technology, 3743S) was diluted in antibody dilution reagent 1:500, and incubated overnight at 4°C. After washing with PBS containing 0.05% Tween-20 for 3 times, 10 min each, slides were incubated with secondary antibodies goat antirabbit -Atto647N (Sigma-Aldrich, 50185-1ML-F), for 45 min at RT. The slides were washed with PBS containing 0.05% Tween-20 for 3 times 10 min each, mounted using Vectashield with DAPI (Vector Labs, H-1200) and imaged with Leica SP8.
ChIRP-mass spectrometry
ChIRP-mass spectrometry was conducted as previously described in Chu et al.(Chu et al., 2015). Probes used are found in Table S2. Spectral counts ten and above with four or more times enrichment over the RNase control were used. All samples were filtered for proteins found in the CRAPome(Mellacheruvu et al., 2013) and normalized to the signal of CRAPome proteins to account for difference in instrument running conditions. Mass spectrometry was conducted at the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry. GO term enrichment was calculated with Metascape (Tripathi et al., 2015; http://metascape.org).
Flow cytometry
Cells were stained with Annexin V-FITC (BD PharMingen, 556420) following the manufacturer’s instructions. Cells were processed and analyzed on a special order FACS Aria II (BD Biosciences).
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA expression analysis
Libraries were separated by barcode, matching reads were collapsed and barcodes removed. For all libraries, single-end RNA-Seq reads were initially mapped against human ribosomal RNA. Reads aligning to the rRNA were discarded, and remaining reads were mapped to the human (hg19 assembly) genome using STAR (Dobin et al., 2013). Reads for each transcript were extracted using HTSeq (Anders et al., 2015). Differential gene expression was calculated with DESeq2 (Love et al., 2014). GO term enrichment was calculated with Metascape (Tripathi et al., 2015; http://metascape.org).
Image analysis
Images were taken under Leica SP8 and the colocalization analysis was performed in Volocity software (PerkinElmer). In brief, the signal intensity and area from both Cy3 and RIG-I were measured in the Volocity Measurement function, and the intersection of the two channels was used to defined overlapping area. Overlapping fraction was calculated with following equation: Overlapping fraction = interest area / total area from two channels. RIG-I foci density was calculated with the following equation: number of RIG-I foci detected / number of cells. ~45 cells with independent replicates were measured and p value was calculated using Student’s t test.
DATA AND SOFTWARE AVAILABILITY
The accession number for the sequencing data reported in this paper is Gene Expression Omnibus database: GSE81345.
Supplementary Material
Highlights.
Exogenous circRNA potently stimulates immune signaling, and circRNA co-aggregate with RIG-I
Transfected cells with exogenous circRNA have increased protection against viral infection
Self-nonself discrimination depends on the intron that programs the circRNA
Mature human circRNA associated with diverse RNA-binding proteins
Acknowledgments
We thank Dr. Shizuo Akira and Dr. Takashi Satoh at Osaka University for providing wild-type and RIG-I−/− MEFs, and Dr. Jan Carette and Dr. Jennifer Lumb at Stanford University for providing VEEV-GFP virus. We thank Byron Lee for technical help and members of our labs for discussion. We thank Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry for mass spectrometry. This work was supported by Dean’s Fellowship and National Institutes of Health Postgraduate Training Program 2T32AR00742231 (Y.G.C.), James Hudson Brown-Alexander Brown Coxe Fellowship (M.V.K.), Swedish Research Council International Postdoctoral Fellowship VR-2016-06794 (X.C.), Damon Runyon Cancer Research Foundation 2090-11 and National Institutes of Health Postgraduate Training Program 2T32AR00742231 (P.J.B.), Rita Allen Foundation Scholar (J.E.W.), Howard Hughes Medical Institute (A.I.), National Institutes of Health (R00-GM104166 and R35-GM119735 to J.E.W.; R41AI120269 to A.I.; P50-HG007735 and R01-HG004361 to H.Y.C.), and Parker Institute for Cancer Immunotherapy (H.Y.C.). Stanford University has filed a patent based on this work, and Y.G.C. and H.Y.C. are named inventors.
Footnotes
Supplemental Information includes six figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2017.05.022.
AUTHOR CONTRIBUTIONS
Conceptualization: Y.G.C. and H.Y.C.; Methodology: Y.G.C. and H.Y.C.; Investigation: Y.G.C. (all except for with HEK293T and MEFs), M.V.K., S.A., and A.I. (293T and MEFs), X.C. (microscopy), and J.E.W. (ChIRP-MS design); Analysis: Y.G.C. (all except for RNA-seq data) and P.J.B. (RNA-seq data); Resources: J.E.W. (ZKSCAN1 and laccase2 plasmids); Writing: Y.G.C. and H.Y.C.
References
- Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandry PS, Belfort M. Activation of a cryptic 5′ splice site in the upstream exon of the phage T4 td transcript: exon context, missplicing, and mRNA deletion in a fidelity mutant. Genes Dev. 1987;1:1028–1037. doi: 10.1101/gad.1.9.1028. [DOI] [PubMed] [Google Scholar]
- Chen L-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. advance online publication. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR:ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A–seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Fischer JW, Leung AKL. CircRNAs: a regulator of cellular stress. Crit. Rev. Biochem. Mol. Biol. 2017;52:220–233. doi: 10.1080/10409238.2016.1276882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Martin L, Spitale RC, Do BT, Sagan SM, Zarnegar B, Qu K, Khavari PA, Quake SR, Sarnow P, Chang HY. Dissecting noncoding and pathogen RNA-protein interactomes. RNA. 2015;21:135–143. doi: 10.1261/rna.047803.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Ares M., Jr Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc. Natl. Acad. Sci. USA. 1994;91:3117–3121. doi: 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat. Struct. Mol. Biol. 2008;15:714–721. doi: 10.1038/nsmb.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS ONE. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, et al. m(6)ALAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods. 2016;13:692–698. doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. Joining RNA molecules with T4 DNA ligase. Methods Mol. Biol. 1999;118:11–19. doi: 10.1385/1-59259-676-2:11. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M., Jr Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4:1047–1054. doi: 10.1017/s135583829898061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju M, Been MD. Generation of nuclease resistant circular RNA decoys for HIV-Tat and HIV-Rev by autocatalytic splicing. Nucleic Acids Symp. Ser. 1995;33:49–51. [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J-W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. Meta-and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. Circular RNAs: Unexpected outputs of many protein-coding genes. [Published online August 29, 2016];RNA Biol. 2016 doi: 10.1080/15476286.2016.1227905. http://dx.doi.org/10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.