Abstract
PI-3-kinases have been identified as key signaling proteins involved in many basic biological processes in health and disease. Transgenic animals have been essential tools to study the underlying molecular mechanisms in this context and have therefore been widely used to elucidate the role of these factors in many different settings. More specifically, PI3Kγ, a subunit highly expressed in the hematopoietic system, has been implicated to play an important role in many inflammatory diseases as well as cancer. Here we report identification of multiple additional previously unknown mutations in the genome of a widely used PI3Kγ-deficient mouse colony. These include a STOP-mutation in the GM-CSFRα chain leading to a complete and specific deficiency in GM-CSF signaling. PI3Kγ-deficient animals consequently lacked alveolar macrophages and succumbed rapidly to influenza virus infection. Furthermore, PI3Kγ-deficient mice carried an additional mutation that affects Mucin 2 (Muc2) transcripts. This protein is strongly involved in the regulation of colorectal cancer and indeed conflicting reports have indicated that PI3Kγ-deficient animals spontaneously develop colorectal tumors. Thus, we uncover previously unknown confounding factors present in a strain of PI3Kγ-deficient mice leading to additional deficiencies in important signaling pathways with potentially wide-ranging implications for the interpretation of previous studies. By separating the mutations, we established unique Csf2ra-/- mouse model that allows study the role of cell intrinsic GM-CSF receptor signaling in vivo without confounding variables introduced by defective IL-5 receptor and IL-3 receptor signaling in mice lacking the common β chain (Csf2rb).
Keywords: GM-CSF receptor, PI3Kγ, Influenza, alveolar macrophages
Introduction
Genetically modified organisms are important and essential tools for biomedical research and have been used extensively for the discovery of many basic biological mechanisms. Mice in particular have been genetically altered many times to understand the function of genes and their involvement in physiological responses such as inflammation. Especially loss-of-function studies using knockout mice are very commonly used to shed light on the role of a given factor in a process of interest. In recent times however it is increasingly realized that many conclusions drawn from such experiments may be ultimately flawed due to the complexity of genetic differences between and even within mouse strains [1]. These differences can potentially explain many phenotypes observed in knockout mouse strains due to inadequate backcrossing after generation of these strains. Importantly, spontaneous mutations causing phenotypic differences between mouse strains were also instrumental for understanding key immunological mechanisms such as the recognition of TLR4 as the receptor for LPS [2, 3] and the role of macrophage-colony stimulating factor in myelopoiesis [4]. Furthermore, some knockout mouse strains have been found to harbor additional genetic mutations, in some cases leading to a deficiency in multiple genes, which could potentially dramatically affect the observed phenotype. An example of this would be the recently discovered caspase-11 deficiency in the caspase-1 knockout mouse [5]. Similarly, we recently found impaired IL-1α production in one out of two different strains of IL-1β-deficient mice [6], which led to an over-interpretation of the role of IL-1β in atherosclerosis and possibly other disease models. Furthermore, it was recently shown that the deficiency in migration of DCs from NLRP10 mice was due to the presence of an additional mutation in DOCK8, which indeed was also present in all animals from the C3H/HeJ strain [7]. This underlines the notion that the presence of additional genetic confounders can often explain differences between knockout mice strains, where the same gene was targeted but in different laboratories and sometimes in different genetic backgrounds. Additionally, polymorphisms in genes linked to a knockout allele may also affect observed phenotypes and lead to conflicting results [8].
Phosphoinositide 3-kinases (PI3K) are important signal transduction proteins that are involved in mediating many essential cellular responses such as cell growth, division and survival. We started out with the aim to address the role of PI3Kγ-signaling upon infection with influenza virus in vivo using well-described PI3Kγ deficient mice. Here we show that a colony of these knockouts [9] is hyper-susceptible to influenza virus infection exhibiting pronounced morbidity as well as strongly impaired survival. We further show that this phenotype was caused by additional mutations in the GM-CSF receptor alpha (GM-CSFRα, Csf2ra) chain present in this mouse strain, as opposed to the PI3Kγ deficiency. This defect led to a translational stop before the trans-membrane region and consequently a complete absence of GM-CSF receptor signaling and thus to a lack of alveolar macrophages, which are essential for survival during an influenza virus infection. By gain of function studies we demonstrated that this mutation could be corrected with a wild-type copy of the Csf2ra gene leading to a complete rescue of GM-CSF receptor signaling. Additionally, we found a homozygous mutation of the Muc2 gene, which is a potential null allele that may be involved in intestinal tumorigenesis [10, 11]. These results exemplify the risk of associating phenotypes with targeted genes in knockout mice without genetic complementation. Moreover, the data establish the first specific GM-CSFRα knockout.
Methods
Mice
C57BL/6 were obtained from Charles River (Germany). Pik3cg–/– (Pik3cgtm1Pen ) mice (Sasaki et al., 2000) were provided by J. Penninger. Pik3cg–/– (Pik3cgtm1Wym) [12] and PI3Kγ-kinase-dead (KD) (Pik3cgtm1Ehi) mice [13] were backcrossed for 20 and 15 generations to C57BL/6, respectively, and provided by E. Hirsch. Csf2rb−/− mice were provided by B. Becher, Zürich. Csf2raA302Gfs*1 (Csf2ram1Kopf) mice were initially derived from Pik3cgtm1Pen, backcrossed for an additional 3 generations to C57BL/6 and carry wild-type alleles of Pik3cg. All animals were housed in individually ventilated cages under specific pathogen–free conditions and were used for experiments at between 7 and 12 weeks of age. All animal experiments were approved by the local animal ethics committees and were performed according to local guidelines and Swiss animal protection law.
Influenza virus infection
Influenza virus strain PR8 (A/Puerto Rico/34, H1N1) was originally provided by J. Pavlovic, University of Zurich. For infections, the mice were anaesthetized and intratracheally inoculated with 50 pfu in 50 ml endotoxin-free PBS. To determine influenza virus titers in the lungs, samples were collected on various days after infection, homogenized and serially diluted with MDCK cells as described [14]. Virus-specific antibodies were detected by ELISA as described recently [15]
Cell suspension preparations
Mice were killed by intraperitoneal injection of sodium pentobarbital at a dose of 400 mg per kg body weight (Euthasol; VIRBAC Schweiz). The lungs were then flushed three times with 400 μl PBS, and BAL fluid cells were harvested by centrifugation. The lungs were digested for 45 min at 37 °C with 2 mg/ml of type IV collagenase (Worthington) and 0.02 mg/ml DNase I (Sigma) and were passed through a 70-μm cell strainer. ACK buffer was used for erythrocyte lysis.
Flow cytometry
A FACS Calibur, Canto II or LSR Fortessa (BD) was used for multi-parameter analysis, and data were analyzed with FlowJo software (TreeStar). Fluorochrome-conjugated or biotinylated monoclonal antibodies specific to mouse CD11c (N418), CD11b (M1/70), Ly-6C (HK1.4), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD4 (GK1.5), CD8α (53-6.7), Gr-1 (RB6-8C5, eBioscience), MHC class II (M5/114.15.2, eBioscience), Ly-6G (1A8, BD Biosciences) were from Biolegend unless otherwise stated. PE-conjugated peptide-MHC class I tetramers (H-2Db/NP34) with the peptide NP34 (NP366-374; ASNENMETM) from the nucleoprotein of influenza virus A/PR/8/34 were generated as described [16]). GM-CSFRα was stained with unlabeled rat anti-mouse GM-CSFRα (Clone 698423, R&D Systems) followed by detection with biotinylated mouse anti-ratIgG2a (RG7/1.30, BD Biosciences) and Streptavidin-APC (eBioscience). Prior to all staining, FcγIII/II receptors were blocked by incubation with anti-CD16/32 (2.4G2) purified from hybridoma supernatant (Swiss Federal Institute of Technology Zurich).
Bone marrow chimeras
For 4-way Pik3cg–/–/WT bone marrow chimeras, WT and Pik3cg–/– mice were lethally irradiated and reconstituted with 5–10x106 Pik3cg–/– or wild-type BM cells. For mixed bone marrow chimeras, WT and Pik3cg–/– recipient mice were lethally irradiated (9.5 Gy, using a caesium source) and reconstituted with 5–10x106 BM cells of a 1:1 mixture of CD45.1+WT:CD45.2+Pik3cg–/–. Mice were analysed 10 weeks post-reconstitution. BM chimeras were analysed 8-12 weeks post-reconstitution.
Genome sequencing
Tail genomic DNA was prepared using the Qiagen DNeasy kit according to manufacturer's instructions, and subjected to whole-genome sequencing as described [17]. Briefly, 100bp paired-end read libraries were prepared and sequenced on an Illumina HiSeq 2000, with all three sample libraries barcoded and loaded onto a single lane. Reads were aligned to the mouse reference genome (mm10) using Stampy [18] with BWA settings, and duplicate reads were identified and discarded using the Picard tool (http://broadinstitute.github.io/picard/). Variants from all three samples were joint-called by Platypus version 0.1.9 (www.well.ox.ac.uk/platypus) [19]and annotated using ANNOVAR [20] with RefSeq and dbSNP 138 annotations.
Cell culture and transfection
BM-derived DCs were differentiated in vitro in complete RPMI supplemented with 2 ng/ml GM-CSF. For retroviral transfections, mouse GM-CSFRα coding sequence was amplified by RT-PCR and cloned into pMY-IRES-GFP retroviral vector. Retrovirus-containing supernatant was produced in the Ecotropic Phoenix packaging cell line and used to infect BM cells.
Hydrodynamic gene delivery
Mouse IL-5 coding sequence was amplified by RT-PCR and cloned into pLIVE in vivo expression vector (Mirus Bio). 100 μg Endotoxin-free plasmid DNA was injected i.v. in PBS in a volume equal to 10% body weight (0.1 ml/g) within 5 s.
Statistics
Mean values, SD, SEM, and Student’s t test (unpaired) and One-way ANOVA with CI 95% were calculated using Prism software (GraphPad Software, Inc). p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****).
Results
Fatal outcome of influenza virus infection in Pik3cgtm1Pen-deficient mice
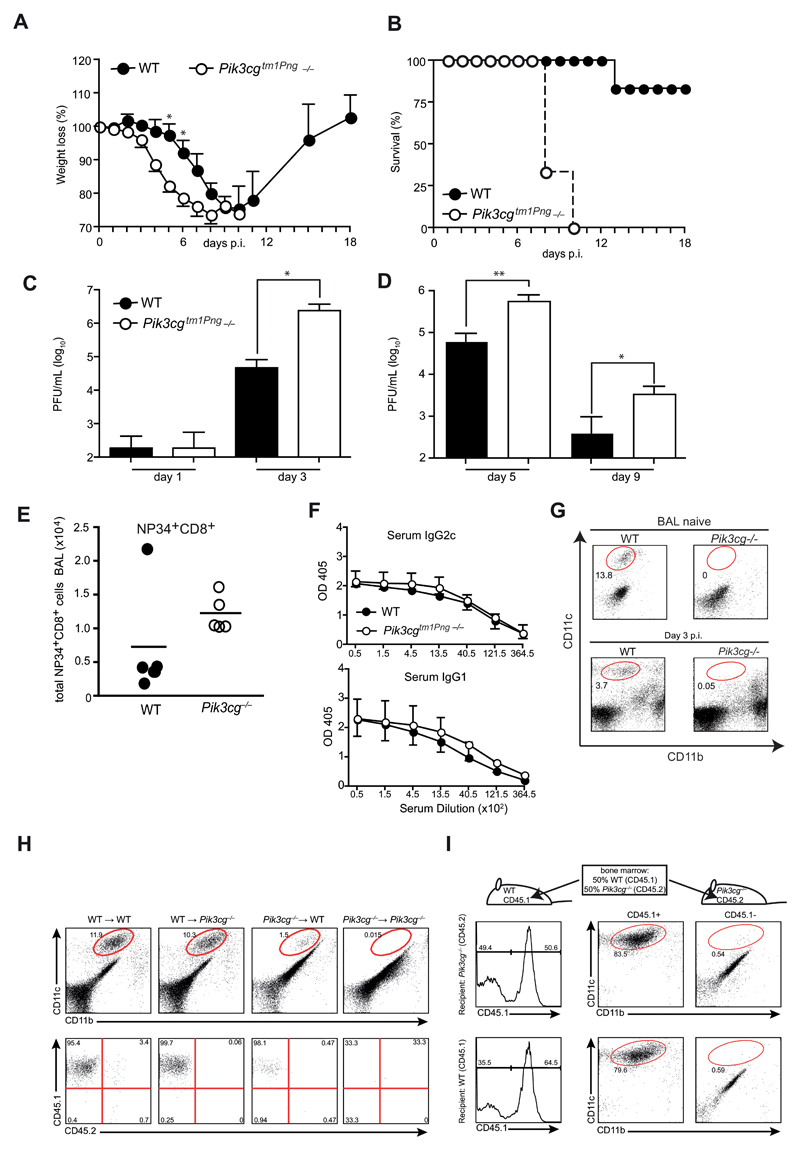
To investigate the role of PI3Kγ in influenza virus infection, we used a colony of PI3Kγ-deficient (Pik3cg–/–) mice provided by the Penninger lab (Pik3cgtm1Pen -/-). Strikingly, Pik3cgtm1Pen-/- mice infected with influenza virus PR/8 showed increased weight loss and mortality (Fig. 1A,B) associated with 10-15 fold elevated virus titers in lungs from day 3 throughout the course of infection (Fig. 1C,D). Similar results were obtained with heterozygous Pik3cgtm1Pen+/- instead of pure C57BL/6 wild-type mice as controls (data not shown). To address whether the lack of PI3Kγ altered antiviral immunity, we analyzed influenza-specific T and B cell responses. The numbers of virus-specific (i.e. NP34+) and total CD8+ T cells in the bronchoalveolar lavage (BAL) of Pik3cgtm1Pen-/- mice were insignificantly increased (Fig. 1E). Furthermore, virus specific IgG2c and IgG1 antibody titers in the serum were similar in wild-type and Pik3cgtm1Pen-deficient animals (Fig. 1F) indicating intact T and B cell responses in the absence of PI3Kγ. However, a striking difference between wild-type and Pik3cgtm1Pen-/- mice could be observed in the frequency of alveolar macrophages during infection, which were completely absent in Pik3cgtm1Pen-/- animals (Fig. 1G). Notably, a deficiency in AM was already present in naïve mice and corresponded with a pronounced alveolar proteinosis (data not shown). To establish whether AM-deficiency was due to the radiosensitive (hematopoietic) or radioresistant (lung stroma, parenchyme, and epithelia) compartment in Pik3cgtm1Pen-/- mice, we generated crisscross bone marrow (BM) chimeras of wild-type and Pik3cgtm1Pen-/- mice using CD45 congenic markers to allow separation of donor and host cells. Eight weeks after adoptive transfer, wild-type (CD45.2) and Pik3cgtm1Pen-/- recipients (CD45.2) reconstituted with wild-type (CD45.1) BM showed a comparable frequency of alveolar macrophages in BAL and lung (Fig. 1H) In contrast, hematopoietic stem cells of Pik3cgtm1Pen-/- mice did not develop to AM. The few lung macrophages present in wild-type recipients were all host (CD45.1+) derived indicating that they had survived radiation. As expected, no AM were detectable in Pik3cgtm1Pen→Pik3cgtm1Pen chimeras. Thus, absence of the PI3Kγ-activity in hematopoietic cells was associated with a defect in AM. We next addressed whether PI3Kγ gene plays a cell autonomous role for AM development by reconstituting irradiated mice with bone marrow of WT and Pik3cgtm1Pen-/- mice. AM were derived only from wild-type BM (Fig. 1H) indicating a cell autonomous requirement for PI3Kγ signaling in AM development.
Figure 1. Pik3cg–/– mice are hypersusceptible to influenza virus infection.
(A and B) Pik3cgtm1Pen deficient and WT control mice were infected i.t. with 50 PFU influenza A virus PR8 and their survival (A) and weight loss (B) was monitored. (C and D) Lung viral titers were determined by plaque-assay in MDCK cells. (E and F) BAL cells from mice at day 9 p.i. were analyzed by FACS. Plots show numbers of (E) NP34-specific (left panel) and total CD8+ T cells (right panel). (F) Serum from mice at day 9 p.i. was assessed for virus-specific IgG2c and IgG1 by ELISA. (G) AM in the BAL were quantified in naïve and infected mice at d3 using flow cytometry. (H) Crisscross chimeras were generated by transfer of bone marrow (BM) from either WT or Pik3cgtm1Pen mice to lethally irradiated WT and KO recipients. FACS plots in the upper panel show CD11c and CD11b expression of BAL cells from naïve chimeric mice with gated cells indicating AM. Lower panels depict AM derived from WT (CD45.1+) versus Pik3cgtm1Pen (CD45.2) expression of gated cells. (I) Irradiated WT and Pik3cgtm1Pen recipients were reconstituted with a mixture of WT and Pik3cgtm1Pen BM in a 50:50 ratio. BAL cells from naïve mixed chimeric were analyzed by FACS. Histograms show gating strategy for CD45.1 (WT) expression, FACS plots on the right side illustrate CD11c versus CD11b expression of CD45.1-gated cells. N= 3-5 per group. All experiments have been performed at least twice.
Taken together these data indicated an essential role of PI3Kγ in AM development and that lethal outcome of influenza virus infection in Pik3cgtm1Pen-/- mice may be attributed to a deficiency in AM
Alveolar macrophage deficiency in Pik3cgtm1Pen-deficient mice is independent of PI3Kγ
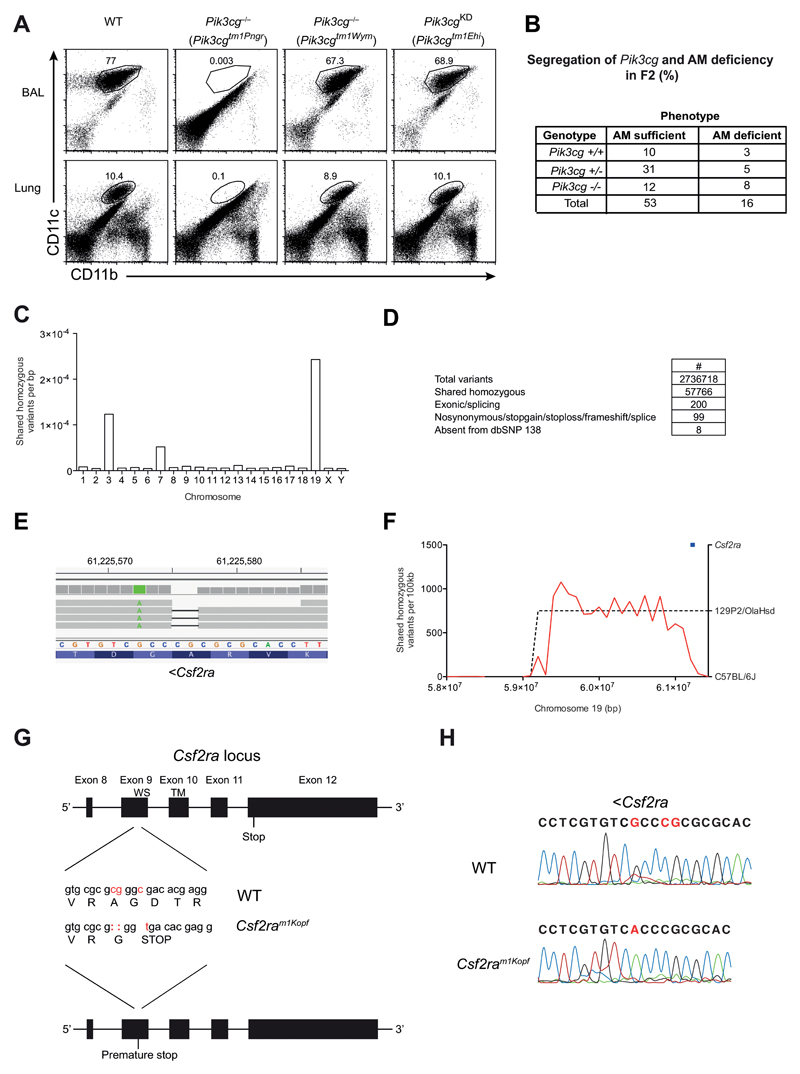
PI3Kγ has been suggested to have both kinase-dependent and independent functions [13]. To determine whether the role of PI3Kγ in AM development was dependent on the kinase function, we analysed the lungs of naive PI3Kγ-kinase dead animals (Pik3cgKD) carrying an inactivating point mutation in the kinase domain of the protein [13] for the presence of AM. The frequency of AM in these animals was comparable to wild-type mice (Fig. 2A). Surprisingly, an independently generated complete PI3Kγ knockout strain (Pik3cgtm1Wym) [12] that was included in this experiment did not show any reduction in the frequency of AM in BAL or lung (Fig. 2A) in striking contrast to the previously characterized Pik3cgtm1Pen knockouts. The lack of an AM-phenotype also corresponded to an unimpaired survival of Pik3cgtm1Wym animals to low dose (50 pfu) influenza virus infection (Fig. 2B). Overall, these results suggested that the lack of AM in Pik3cgtm1Pen animals and the concurrent mortality following low dose influenza virus infection was in fact independent from the deficiency in PI3Kγ.
Figure 2. AM deficiency is independent of PI3Kγ but caused by mutated Csf2ra gene.
(A) Flow cytometry of AMs in BAL fluid and lungs of C57BL/6, Pik3cg–/– (Pik3cgtm1Pen and Pik3cgtm1Wym) and Pik3cgKD (Pik3cgtm1Ehi) mice, gated on CD45+ cells. Numbers adjacent to outlined areas indicate percent CD11c+CD11bint AMs. (B) WT, Pik3cg–/– (Pik3cgtm1Wym) were infected i.t. with 50 PFU influenza A virus PR8 and their survival was monitored. (C) Pik3cgtm1Pen-/- mice were backcrossed once onto C57BL/6 (F1). Shown is a table containing the F2 offspring of the F1xF1 intercross with their Pik3cg genotype and AM phenotype for all analyzed mice. (D) The frequency of homozygous variants shared between three alveolar macrophage-deficient mice. (E) Filtering pipeline for variants detected by whole-genome sequencing. (F) IGV view of the homozygous Csf2ra 2bp deletion. A neighboring point mutation is indicated in green. (G) Density of shared homozygous variants (plotted as a red line) on the distal end of chromosome 19. The region of high density (indicated by a dashed line) represents a haplotype derived from the 129P2/OlaHsd background. The Csf2ra locus is indicated in blue. (H) The last 5 exons of the Csf2ra locus are depicted. The mutations in the Csf2ram1Kopf mouse strain are displayed in red and the resulting changes in the amino acid sequence are indicated (WS, WSXWS motif; TM, transmembrane domain). (I) Sanger sequencing results of the Csf2ra region encoding the mutation.
As the AM phenotype had been transmissible by bone marrow transfer, we judged a possible environmental cause for the AM phenotype such as a differing microbiota in the Pik3cgtm1Pen strain as unlikely and concluded that a genetic difference was most likely the cause for the AM phenotype. Both Pik3cg-deficient mouse strains had been shown to be complete knockouts [9, 12], even targeting the same exon. For this reason we suspected that the phenotype was likely caused by a spontaneous mutation that is present in Pik3cgtm1Pen but not Pik3cgtm1Wym mice.
To determine whether the deficiency in AM was in fact due to the presence of an additional genetic mutation, specific to the Pik3cgtm1Pen mouse strain, we crossed Pik3cgtm1Pen animals to C57BL/6 mice and then intercrossed the F1 generation (Fig. 2C). The resulting F2 mice were then screened for the presence of the AM phenotype as well as the Pik3cgtm1Pen allele. We identified individuals with homozygous wild-type alleles for Pik3cg that nonetheless lacked AM. Thus, this analysis revealed that AM-deficiency phenotype could be separated from the targeted Pik3cg locus and that the AM deficiency causing mutation was inherited in an autosomal recessive fashion, and was unlikely linked to Pik3cg (single point LOD score of 0.69 from 138 meioses) (Fig. 2C).
Pik3cgtm1Pen mice are functional GM-CSFRα knockouts due to spontaneous mutations in the Csf2ra gene
To determine whether a spontaneous mutation in Pik3cgtm1Pen mice was indeed responsible for the AM–phenotype we conducted whole genome sequencing. In order to limit the amount of variation from the C57BL/6J reference strain, Pik3cgtm1Pen alveolar macrophage-deficient male mice (on a mixed 129P2/OlaHsd;C57BL/6J background) were serially outcrossed to wild-type C57BL/6 females and intercrossed, which revealed inheritance of the phenotype in a Mendelian manner (data not shown). After the third intercross, genomic DNA was prepared from three alveolar macrophage-deficient mice (wild-type at the Pik3cg locus) and subjected to whole-genome sequencing. A total of 2,736,718 unique variants were called across three samples, of which 57,766 were homozygous in all three. The majority of variants were present on chromosomes 3, 7, and 19 (Fig. 2D), suggesting that these chromosomes harbored variants from the original E14 embryonic stem cell line (129P2/OlaHsd origin) used to generate the Pik3cgtm1Pen allele [9] 200 of these variants fell within protein-coding exons or essential splice sites (i.e. within 2bp of an exon-intron boundary), 99 of these were predicted to change protein-coding sense (i.e. were nonsynonymous, stopgain, stoploss, frameshift or splice variants), and only 8 of these 99 were absent from dbSNP 138 (which includes variants from the 129P2/OlaHsd inbred strain) (Fig. 2E). Of the genes affected (Nr2c2, Tmcc1 Sugp1, Pdzd3, Nadk2, Robo1, Muc2, Csf2ra) (Table 1), Csf2ra was a prime candidate given that it encodes the GM-CSF receptor alpha (GM-CSFRα) chain, which is a subunit of the heterodimeric GM-CSF receptor. The Csf2ra variant (chr19:61225575-61225576 [mm10]) was a 2bp frameshift deletion within codon 302 (from a total of 388) in exon 9 (Fig. 2F,H). This variant was on the same allele as another at position chr19:61225572 (Fig. 2F), suggesting it had arisen on the variable 129P2/OlaHsd background. Indeed, when we examined the broader context of the variant, the 2bp Csf2ra deletion was flanked by a high density of shared homozygous variants on distal chromosome 19, where Csf2ra was the second most distal gene (Fig. 2G). Importantly, the 2bp deletion resulting in replacement of Alanine with Glycine (A302G) was directly followed by a point mutation (C ➔ T) in exon 9 that generated a premature stop signal in the extracellular part of the protein upstream of the WSXWS motif and the transmembrane domain (Fig. 2H). This mouse line (Csf2raA302Gfs*1) was designated Csf2ram1Kopf. The mutations were confirmed independently by Sanger sequencing of the genomic DNA region in mice from the colony carrying the AM-deficiency phenotype (Fig. 2I). Overall these results demonstrated that the deficiency in AM observed in Pik3cgtm1Pen was not due to absence of PI3Kγ but resulted from abrogated GM-CSF signaling as a consequence of spontaneous mutations in the Csf2ra gene causing a premature translational stop.
Notably, the homozygous Muc2 variant (Table 1) found in the Pik3cgtm1Pen mice is a 3bp deletion across a non-canonical splice site (chr7:141701485-141701487, delAAA (mm10) and therefore a potential null allele, which may promote colorectal cancer development[10, 11].
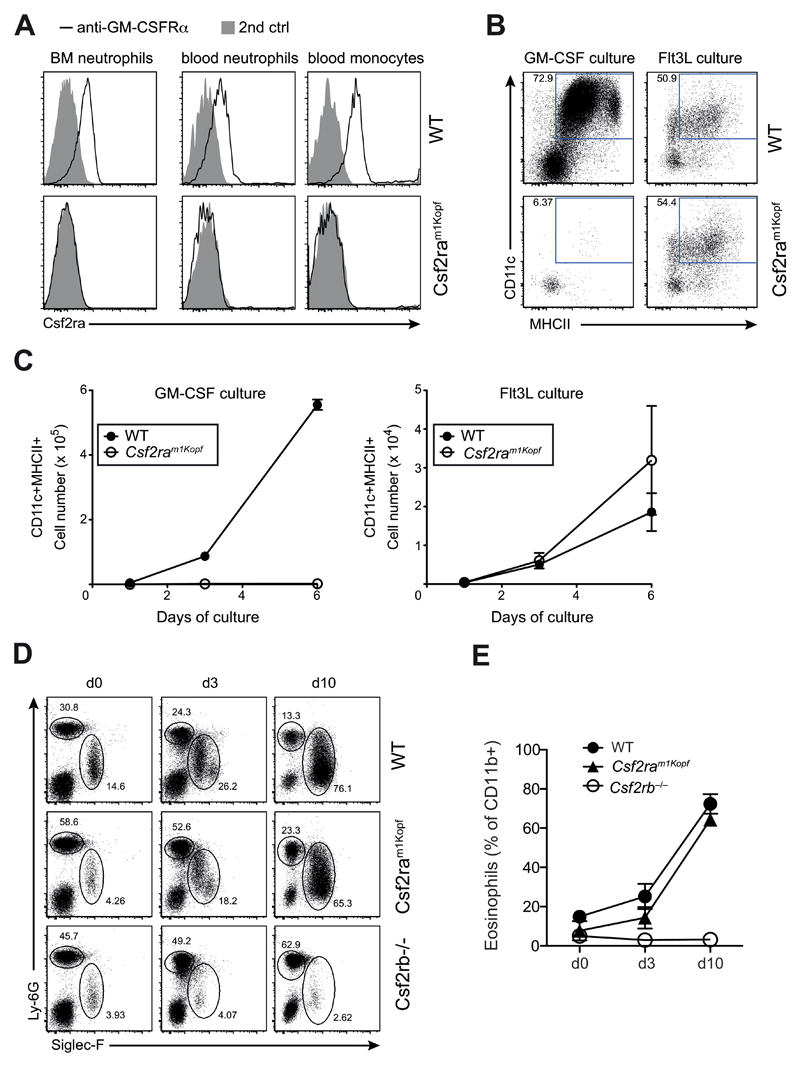
Consistent with the predicted truncation of the GM-CSFRα that consequently lacks the transmembrane domain, we did not detect membrane-bound GM-CSFRα protein on monocytes and neutrophils from Csf2ram1Kopf mice, while it was expressed on cells of wild-type mice (Fig. 3A). To demonstrate a functional consequence of the lack of GM-CSFRα we conducted an in vitro experiment using GM-CSF to differentiate dendritic cells from naïve bone marrow. In this assay differentiation of bone marrow-derived dendritic cells (BMDCs) is strictly dependent on GM-CSF. As a control we also used Flt3L to differentiate dendritic cells from bone marrow, which is also a well-established dendritic cell differentiation protocol. We found that GM-CSF-dependent dendritic cell differentiation and proliferation were completely absent in the BMDC culture derived from bone marrow of Csf2ram1Kopf mice (Fig. 3B), whereas Flt3L-mediated differentiation and expansion remained comparable to wild-type (Fig. 3C). These results confirm a GM-CSF-specific signaling defect in Csf2ram1Kopf mice. To demonstrate that signaling through the common GM-CSF receptor beta chain was still intact, we overexpressed IL-5 in vivo in wild-type, Csf2ram1Kopf, and Csf2rb–/– mice using hydrodynamic injection of an IL-5-encoding expression vector. The strong induction of IL-5 induced a pronounced blood eosinophilia in WT and Csf2ram1Kopf mice but was absent in Csf2rb–/– mice that lack the GM-CSFR common beta chain (Fig. 3D,E).
Figure 3. Csf2ram1Kopf mice are functional knockouts for the GM-CSFRα.
(A) Flow cytometry analysis of GM-CSFRα on neutrophils in the bone marrow (BM) or blood and monocytes in the blood of WT and Csf2ram1Kopf mice using anti-GM-CSFRα or secondary anti-ratIgG2a control. (B) In vitro differentiation of DCs from BM cells of mice as in (A) using GM-CSF or Flt3L. Expression of CD11c and MHCII on live cells is depicted. (C) Total numbers of CD11c+MHCII+ cells from in vitro DC cultures as in (B). (D) Flow cytometry of eosinophils and neutrophils in the blood after hydrodynamic injection of an IL-5 expression plasmid in WT, Csf2ram1Kopf and Csf2rb–/– mice. Numbers adjacent to outlined areas indicate percent Siglec-F+ eosinophils and Ly-6G+ neutrophils gated on CD11b+ cells. (E) Frequency of eosinophils as gated in (D).
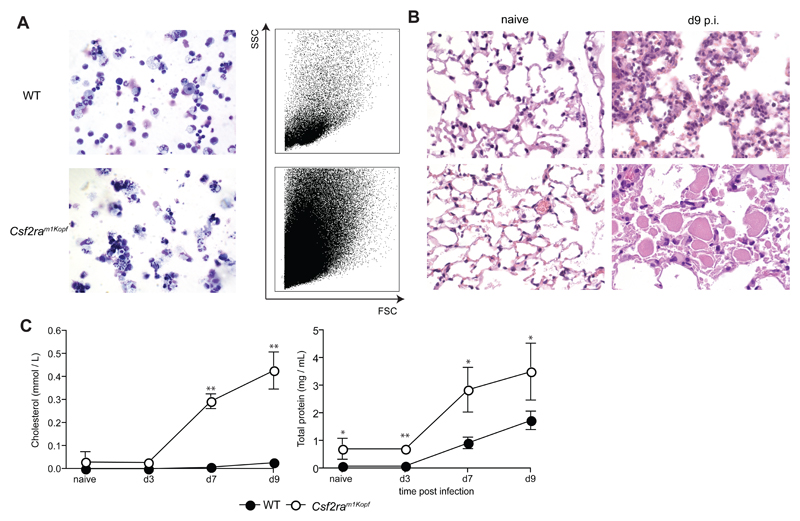
In addition, Csf2ram1Kopf mice with intact Pik3cg gene showed severe pulmonary proteinosis, accumulation of dead cells in the alveoli, and severe morbidity following influenza infection (Fig. 4 and data not shown) reminiscent of Csf2-/- and Csf2Rb-/- mice [15] providing further evidence of a complete defect of GM-CSF signaling.
Figure 4. Csf2ram1Kopf develop fatal pulmonary alveolar proteinosis during the course of influenza infection.
Groups of mice were infected with 50 PFU influenza virus PR8 before mice were sacrificed for analysis at the time points indicated. (A) BAL cells were harvested at day 9 p.i. before being analyzed by cytospins (left panel, 10x) and FACS. (B) Panels show H&E-stained histological sections of naïve and day 9-infected mice (10x). (C) BAL fluid was obtained at the time points indicated. Total protein content in BAL fluid was determined by BCA Protein assay (Thermo Scientific) according to the manufacturer’s instructions. Cholesterol levels were measured as described [15] All experiments have been performed at least twice with n = 4 per group.
Overall, these results demonstrated that Csf2ram1Kopf mice are functional knockouts for the GM-CSFRα, which selectively affects the GM-CSF receptor complex while signaling through the common beta chain is intact.
Overexpression of wild-type Csf2ra restores GM-CSF signaling in Csf2ram1Kopf mice
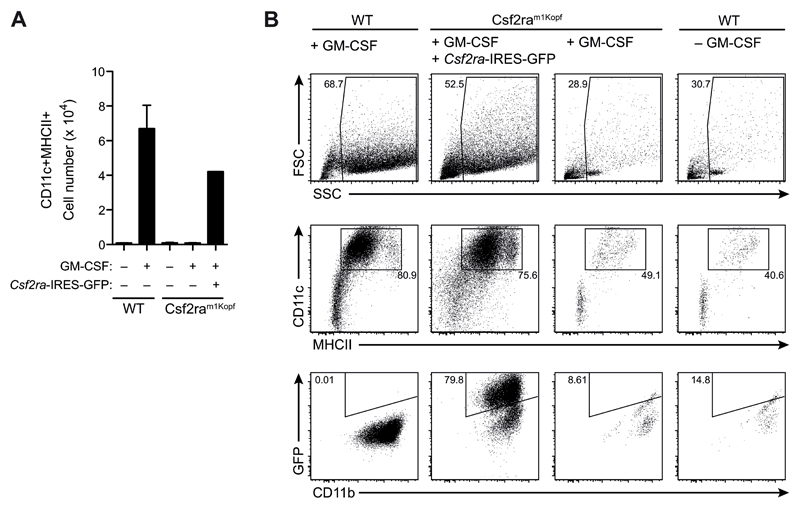
To prove that the mutation in Csf2ra was indeed responsible for the complete block in GM-CSF signaling, we infected Csf2ram1Kopf bone marrow cells with a retrovirus encoding the wild-type GM-CSFRα followed by IRES-GFP and cultured them under GM-CSF-dependent BMDC-differentiating conditions. Overexpression of wild-type GM-CSFRα restored capacity of BM cells to respond and proliferate in the presence of GM-CSF (Fig. 5A). Moreover, the differentiation into BM-derived CD11c+MHCII+ DCs was rescued in cells transfected with the wild-type GM-CSFRα, while untransfected BM cells from Csf2ram1Kopf mice were not able to generate BMDCs similar to wild-type cells that were cultured without GM-CSF (Fig. 5B).
Figure 5. Reconstitution of Csf2ram1Kopf cells with GM-CSFRα restores GM-CSF signaling.
(A) Bone marrow cells from Csf2ram1Kopf mice were infected with retrovirus encoding Csf2ra followed by IRES-GFP (Csf2ra-IRES-GFP) and stimulated in vitro with GM-CSF. Total numbers of CD11c+MHCII+ cells on d5 from the culture of Csf2ra-transfected Csf2ram1Kopf cells and untransfected WT and Csf2ram1Kopf cells. (B) Flow cytometry analysis on d8 of the GM-CSF culture of cells as in (A).
Overall, we here describe the discovery of a mouse line carrying spontaneous mutations in the Csf2ra gene that resulted in a premature translational stop and truncation of the GM-CSFRα chain. As a consequence, GM-CSF signaling is completely abrogated resulting in the failure of AM to develop. This mouse strain could be useful to study GM-CSF signaling without affecting the signaling of other cytokines that share the common beta chain.
Discussion
GM-CSF is strictly required for development of alveolar macrophages and in humans and mice [21–24]. We have recently established that Csf2-/- and Csf2Rb-/- mice develop lung failure and succumb following low dose influenza virus infection due the absence of alveolar macrophages [15]. In this study we found that Pik3cgtm1Pen mice succumb to a low dose (i.e. 50 pfu) of pulmonary influenza virus infection with apparently unimpaired T and B cell responses. This is in contrast to Pik3cgtm1Wym and Pik3cgKD (Pik3cgtm1Ehi) mice that exhibit increased morbidity only after high dose infection (i.e. >200 pfu) due to a defect in lung DC mediated CD8+ T cell responses. Reconciling these conflicting data, we show that severe morbidity to influenza infection of Pik3cgtm1Pen mice was due to a deficiency in alveolar macrophages caused by an additional mutation in the Csf2ra locus in Pik3cgtm1Pen mice that results in a complete block in GM-CSF signaling. By reconstituting with a wild-type copy of Csf2ra, we show that GM-CSF responsiveness can be restored, thus proving that it is indeed this mutation, which is responsible for the observed AM deficiency in Csf2ram1Kopf mice. The reduced anti-viral CD8+ T cell response observed in infected Pik3cgtm1Wym and Pik3cgKD (Pik3cgtm1Ehi) mice [25] is blurred in Pik3cgtm1Pen mice probably because of increased pulmonary inflammation and proteinosis in the absence of alveolar macrophages already prior infection.
The findings presented above are significant as they for the first time report a mouse strain deficient in the GM-CSF receptor alpha chain due to a spontaneous mutation similar to those found in humans [26] [21]. To date, studies addressing the role of GM-CSF receptor in PAP have strongly relied on tools targeting either GM-CSF or the receptor beta chain [27], and most recently Csf2rb-/- mice have been used to demonstrate the therapeutic potential of transfer of pulmonary macrophages to treat PAP [28]. However, due to the fact that Csf2rb is also used in IL-3 signaling [29], and is necessary for signaling to IL-5 [27], this is not a clean system to study the role of GM-CSF. The future use of the mouse presented in this paper will allow a much more precise investigation of the role of the GM-CSF receptor signaling in both AM development in general as well as provide a better model to test therapeutic interventions for PAP in humans. In addition, in some diseases such as for example pulmonary allergic inflammation both GM-CSF and IL-5 have been implicated to play an important role in disease development, regulating dendritic cell activation [30] and eosinophil migration respectively [31]. Here a GM-CSF specific receptor knockout will allow to dissect the roles of this cytokine in this context more precisely as it leaves IL-5 signaling completely intact.
Furthermore, GM-CSF has also been implicated to play a role in various inflammatory diseases and is currently a target in different clinical trials [32]. The cells responding to this key cytokine have however not been characterized in depth, and these studies have been hampered by the lack of a tool providing GM-CSF receptor deficiency while leaving IL-3 and IL-5 signaling intact. Thus, in this case also Csf2ra-/- mice will allow a more stringent experimental set up to study the role of GM-CSF signaling in development of autoimmunity. Naturally, there is still a need for cell type specific cytokine receptor knockout animals to dissect the roles of GM-CSF in these settings.
Besides the Csf2ra mutation in Pik3cgtm1Pen mice, we also found a potential null allele of Muc2 in Pik3cgtm1Pen mice. Mice lacking the Mucin Muc2 due to a targeted mutation develop colorectal cancer [11]. Similarly, Pik3cgtm1Pen mice were originally also reported to develop intestinal adenoma and carcinoma [10], which was not seen in other lines of Pik3cg-deficient mice [12], [10](Erratum in Nature 2003). Thus, it is tempting to speculate that the Muc2 gene variant in Pik3cgtm1Pen mice contributed to colorectal cancer development. The discovery of spontaneous additional mutation(s) in a commonly used knockout mouse strain presented in this paper highlights a phenomenon that is increasingly appreciated as hampering progress in biomedical research. The general assumption that the targeted genetic allele in a knockout mouse is the only genetic difference does not hold in many cases as additional genetic changes have been found to cause phenotypes previously associated with the targeted allele [5]. Our results therefore further underline the necessity to be cautious when interpreting data obtained from knockout animals and suggest that gain of function studies would often be essential to validate the interpretation of experiments based on targeted genetic loss of function.
In conclusion, our study identifies to our knowledge the first GM-CSF receptor alpha knockout mouse, which will serve as a useful mouse model to study therapeutic interventions in hereditary PAP as well as the role of GM-CSF signaling in autoimmunity and other inflammatory diseases. The discovery of this Csf2ra mutation further highlights the necessity to warrant caution when using genetically modified animals and thus serves as a general warning for scientists to be aware of factors confounding interpretation of experimental data.
Supplementary Material
Summary Sentence.
Identification of a spontaneous Csf2ra null mutation in PI3Kγ-/- mice allowed generation of unique Csf2ra-/- mice essential for study of cell intrinsic GM-CSF receptor signaling
Acknowledgements
This project was supported by grants from Swiss National Science Foundation (SNF 310030-163443/1) and ETH Zürich (ETH-34-13-1). OS was supported by the Wellcome Trust (100083/Z/12/Z). We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z and MRC Hub grant G0900747 91070) for the generation of sequencing data.
Abbreviations
- AM
Alveolar macrophage
- BAL
Bronchoalveolar lavage
- BM
Bone marrow
- GM-CSF
Granulocyte-Macrophage Colony Stimulating Factor
- IL
Interleukin
- LPS
Lipopolysaccharide
- PAP
Pulmonary alveolar proteinosis
- PI3Kγ
Phosphoinositide – 3 – kinase γ
- TLR
Toll-like receptor
Footnotes
Author contributions: S.P.N., C.S., A.K.H. and O.M.S. performed experiments, S.P.N., C.S. A.K.H., O.M.S. and M.K. designed experiments, J.P. and E.H. provided mice, and S.P.N, C.S. and M.K wrote the manuscript.
Conflict of interest disclosure:
The authors declare no conflict of interest
References
- 1.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi ST, Lariviere L, Leveque G, Clermont S, Moore KJ, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) The Journal of experimental medicine. 1999;189:615–25. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–4. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 5.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–21. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 6.Freigang S, Ampenberger F, Weiss A, Kanneganti TD, Iwakura Y, Hersberger M, Kopf M. Fatty acid-induced mitochondrial uncoupling elicits inflammasome-independent IL-1alpha and sterile vascular inflammation in atherosclerosis. Nature immunology. 2013;14:1045–53. doi: 10.1038/ni.2704. [DOI] [PubMed] [Google Scholar]
- 7.Krishnaswamy JK, Singh A, Gowthaman U, Wu R, Gorrepati P, Sales Nascimento M, Gallman A, Liu D, Rhebergen AM, Calabro S, Xu L, et al. Coincidental loss of DOCK8 function in NLRP10-deficient and C3H/HeJ mice results in defective dendritic cell migration. Proc Natl Acad Sci U S A. 2015;112:3056–61. doi: 10.1073/pnas.1501554112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuvolone M, Kana V, Hutter G, Sakata D, Mortin-Toth SM, Russo G, Danska JS, Aguzzi A. SIRPalpha polymorphisms, but not the prion protein, control phagocytosis of apoptotic cells. The Journal of experimental medicine. 2013;210:2539–52. doi: 10.1084/jem.20131274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, Bolon B, Wakeham A, Itie A, Bouchard D, Kozieradzki I, Joza N, et al. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–6. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki T, Irie-Sasaki J, Horie Y, Bachmaier K, Fata JE, Li M, Suzuki A, Bouchard D, Ho A, Redston M, Gallinger S, et al. Colorectal carcinomas in mice lacking the catalytic subunit of PI(3)Kgamma. Nature. 2000;406:897–902. doi: 10.1038/35022585. [DOI] [PubMed] [Google Scholar]
- 11.Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, Kucherlapati R, Lipkin M, Yang K, Augenlicht L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–9. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–53. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 13.Patrucco E, Notte A, Barberis L, Selvetella G, Maffei A, Brancaccio M, Marengo S, Russo G, Azzolino O, Rybalkin SD, Silengo L, et al. PI3Kgamma modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell. 2004;118:375–87. doi: 10.1016/j.cell.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Bachmann MF, Ecabert B, Kopf M. Influenza virus: a novel method to assess viral and neutralizing antibody titers in vitro. J Immunol Methods. 1999;225:105–11. doi: 10.1016/s0022-1759(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 15.Schneider C, Nobs SP, Heer AK, Kurrer M, Klinke G, van Rooijen N, Vogel J, Kopf M. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS pathogens. 2014;10:e1004053. doi: 10.1371/journal.ppat.1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–6. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 17.Bull KR, Rimmer AJ, Siggs OM, Miosge LA, Roots CM, Enders A, Bertram EM, Crockford TL, Whittle B, Potter PK, Simon MM, et al. Unlocking the bottleneck in forward genetics using whole-genome sequencing and identity by descent to isolate causative mutations. PLoS genetics. 2013;9:e1003219. doi: 10.1371/journal.pgen.1003219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SR, Consortium W. G. S. Wilkie AO, McVean G, Lunter G. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nature genetics. 2014;46:912–8. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki T, Sakagami T, Rubin BK, Nogee LM, Wood RE, Zimmerman SL, Smolarek T, Dishop MK, Wert SE, Whitsett JA, Grabowski G, et al. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. The Journal of experimental medicine. 2008;205:2703–10. doi: 10.1084/jem.20080990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. The Journal of experimental medicine. 2013;210:1977–92. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nature immunology. 2014;15:1026–37. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- 24.Stanley E, Lieschke GJ, Grail D, Metcalf D, Hodgson G, Gall JA, Maher DW, Cebon J, Sinickas V, Dunn AR. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5592–6. doi: 10.1073/pnas.91.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobs S, Schneider C, Heer A, Huotari J, Helenius A, Kopf M. PI3Kγ Is Critical for Dendritic Cell-mediated CD8+ T Cell Priming and Viral Clearance During Influenza Virus Infection. PLOS pathogens. 2016;12:e1005508. doi: 10.1371/journal.ppat.1005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Moczygemba M, Doan ML, Elidemir O, Fan LL, Cheung SW, Lei JT, Moore JP, Tavana G, Lewis LR, Zhu Y, Muzny DM, et al. Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRalpha gene in the X chromosome pseudoautosomal region 1. The Journal of experimental medicine. 2008;205:2711–6. doi: 10.1084/jem.20080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishinakamura R, Nakayama N, Hirabayashi Y, Inoue T, Aud D, McNeil T, Azuma S, Yoshida S, Toyoda Y, Arai K, et al. Mice deficient for the IL-3/GM-CSF/IL-5 beta c receptor exhibit lung pathology and impaired immune response, while beta IL3 receptor-deficient mice are normal. Immunity. 1995;2:211–22. doi: 10.1016/1074-7613(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T, Arumugam P, Sakagami T, Lachmann N, Chalk C, Sallese A, Abe S, Trapnell C, Carey B, Moritz T, Malik P, et al. Pulmonary macrophage transplantation therapy. Nature. 2014 doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitamura T, Sato N, Arai K, Miyajima A. Expression cloning of the human IL-3 receptor cDNA reveals a shared beta subunit for the human IL-3 and GM-CSF receptors. Cell. 1991;66:1165–74. doi: 10.1016/0092-8674(91)90039-2. [DOI] [PubMed] [Google Scholar]
- 30.Willart MA, Deswarte K, Pouliot P, Braun H, Beyaert R, Lambrecht BN, Hammad H. Interleukin-1alpha controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. The Journal of experimental medicine. 2012;209:1505–17. doi: 10.1084/jem.20112691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamelmann E, Gelfand EW. Role of IL-5 in the development of allergen-induced airway hyperresponsiveness. Int Arch Allergy Immunol. 1999;120:8–16. doi: 10.1159/000024215. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton JA, Achuthan A. Colony stimulating factors and myeloid cell biology in health and disease. Trends Immunol. 2013;34:81–9. doi: 10.1016/j.it.2012.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.