Abstract
Background
Pediatric oncology patients are at increased risk for invasive bacterial infection due to immunosuppression. The risk of such infection in the absence of severe neutropenia (absolute neutrophil count (ANC) ≥500/μl) is not well established and a validated prediction model for blood stream infection (BSI) risk offers clinical utility.
Methods
A six-site retrospective external validation was conducted using a previously published risk prediction model for BSI in febrile pediatric oncology patients without severe neutropenia, the Esbenshade/Vanderbilt (EsVan) model. A reduced model (EsVan2) excluding two less clinically reliable variables was also created using the initial EsVan model derivative cohort, and validated using all five external validation cohorts. One dataset was only used in sensitivity analyses due to missing some variables.
Results
From the five primary data sets, there were a total of 1197 febrile episodes and 76 episodes of bacteremia. The overall C-statistic for predicting bacteremia was 0.695 with a 0.50 calibration slope for the original model and 1.0 calibration slope when recalibration was applied to the model. The model performed better in predicting high-risk bacteremia (Gram negative or Staphylococcus aureus infection) versus BSI alone with a C-statistic of 0.801 and a calibration slope of 0.65. The EsVan2 model outperformed the EsVan model across datasets with a C-statistic 0.733 for predicting BSI and 0.841 for high-risk BSI.
Conclusions
This external validation shows that the EsVan and EsVan2 models are able to predict BSI across multiple performance sites and could assist in decision making in clinical practice once validated and implemented prospectively.
Keywords: Febrile neutropenia, pediatric oncology, risk prediction, supportive care, health services research
Introduction
Due to immune suppression from cancer and its treatment, pediatric oncology patients are at risk for bacterial blood stream infections (BSI), particularly in the setting of indwelling central venous catheters (CVC)1. There are evidence-based guidelines supporting empiric broad spectrum antibiotics in these patients for fever in the setting of an absolute neutrophil count (ANC) <500/μl1. However, for patients with fever and ANC ≥500/μl, there is little evidence-based data to guide optimal management2–5. The Esbenshade/Vanderbilt (EsVan) predictive model was created to predict the likelihood of BSI in pediatric cancer patients with ANC ≥500/μl4. However, as a model developed in a single institution, it requires external validation prior to implementation in evidence-based clinical care. Therefore, using three previously published datasets, and three newly-created datasets, we sought to externally validate the EsVan model and to evaluate its performance in order to provide evidence to support its implementation in clinical practice.
Methods
Original Model
The EsVan model was based on 932 episodes of febrile pediatric oncology subjects with an ANC ≥500/μl with the purpose of identifying clinical variables that can help predict the likelihood of BSI4. The variables which were all assessed at time of presentation that were determined to increase the risk of a BSI were tunneled external catheters, highest temperature reported or measured within 24 hours prior to presentation or within 1 hour after presentation, reported or observed chills, hypotension and increased absolute neutrophil count (ANC). Factors associated with decreased risk of BSI were exposure to chemotherapy drugs that cause fever (i.e cytarabine, anti-thymocyte globulin, and anti-GD2 antibody therapy) within 24 hours of the fever, older age and acute lymphoblastic leukemia (ALL) diagnosis. Other factors included in the model were location at time of presentation (inpatient versus outpatient), presence of upper respiratory symptoms (cough, congestion or rhinorrhea mentioned in medical record) and history of stem cell transplant. This model performed very well with a C-statistic of 0.8984. The formula of the original logistic regression prediction model is given in Supplemental Figure 3, and a web-based module can be accessed at (https://cqs.mc.vanderbilt.edu/shiny/RiskPrediction/).
Inclusion/Exclusion Criteria for Model
Episodes to be included in a dataset for external validation of the EsVan model followed the same inclusion criteria previously established for the EsVan predictive model: fever ≥38.0° for > 1 hour or ≥38.3° for any duration, in agreement with the 2010 Infectious Diseases Society of America guidelines update6, 7, central venous catheter (CVC), and ANC ≥500/μl at presentation4. Exclusion criteria included events occurring within seven days of a previous febrile episode in order to prevent the possibility of fever episodes being linked, during administration of empiric/treatment antibiotics for previous fever or an infection, or within 30 days following a stem cell transplant. Pediatric oncology subjects were defined as anyone <23 years at diagnosis with a diagnosis of cancer, Langerhans cell histiocytosis, or hemophagocytic lymphohistiocytosis.
Construction of the Validation Cohort
In order to validate the EsVan model, six external datasets were obtained that met inclusion and exclusion criteria. This included three previously published datasets and three constructed specifically for the validation cohort. For those previously published, local investigators used additional medical record review to document all inclusion criteria and delete cases meeting exclusion criteria. Bartholomew et al. published a dataset of 392 episodes of pediatric non-neutropenic fever3. After applying EsVan inclusion and exclusion criteria, this resulted in a cohort of 348 episodes of which 312 episodes with complete data were used for the primary analysis with the additional episodes to be included in sensitivity analysis. Ali et al. published a dataset of 254 episodes of non-neutropenic fever and all episodes met inclusion/exclusion criteria, and so a de-identified dataset was presented for analysis2. Kelly et al. previously published a dataset of 459 episodes of pediatric non-neutropenic fever and provided a de-identified spreadsheet of the initial dataset5. Due to more limited variables available, this data set was only used for the sensitivity analysis. Three additional data sets were constructed for the external validation study. These included 209 episodes from Columbia University from 2009–2012, 193 episodes from Children’s Hospital of Philadelphia (CHOP) from 2011 – 2015, and 229 episodes from University of California San Francisco (UCSF) from 2012 – 2014. In the Columbia dataset it consisted of all non-neutropenic patients who presented to their clinic or emergency room in the outpatient setting and at UCSF it was all patients who presented to their institution with non-neutropenic fever in all settings. For the CHOP dataset it was unique as it is only one that didn’t include all subject diagnosed during a specific time frame. It instead identified all patients presenting to clinic during one specific week and the abstracted all of the events that occurred in those subjects from 2011–2015. All five primary datasets with complete data together resulted in a total cohort of 1197 episodes. When all 6 datasets were combined, including missing data for use in the primary sensitivity analysis, the total cohort was 1692 episodes. Table 1 summarizes the original EsVan dataset and the six datasets used in the external validation.
Table 1.
Characteristics of the Cohort by Treatment Group
| Characteristics | Vanderbilt | Overall1 | P value | Stanford | UCSF | CHOP | Columbia | Lebanon | Brown |
|---|---|---|---|---|---|---|---|---|---|
| Data set size | N=932 | N=1197 | N=312 | N=229 | N=193 | N=209 | N=254 | N=459 | |
| Positive blood stream infection | 9.8% (91) | 6.1% (73) | P=0.0042 | 9.3% (29) | 10.9% (25) | 3.6% (7) | 3.3% (7) | 3.1% (8) | 6.3% (29) |
| Age at time of episode3 | 3.0; 5.0; 10.0 | 3.0; 6.0; 12.0 | P=0.184 | 3.0; 6.0; 13.0 | 3.0; 7.0; 14.0; | 4.0; 6.0; 11.0 | 3.6; 5.8; 10.5 | 3.0; 5.0; 9.0 | 4.0; 6.0; 9.0 |
| Maximum temperature at presentation3 | 38.3; 38.7; 39.1 | 38.3; 38.6; 39.1 | P=0.864 | 38.3; 38.7; 39.1 | 38.3; 38.6; 39.0 | 38.5; 38.8; 39.3 | 38.3; 38.6; 39.0 | 38.2; 38.5 | 38.4; 38.8; 39.2 |
| Absolute neutrophil count3 | 1858; 3360; 5735 | 1660; 3240; 5780; | P=0.144 | 1598; 3205; 5920 | 1990; 3790; 6470 | 2020; 3631; 5727 | 1734; 3268; 5460 | 1290; 2660; 4935 | 1648; 3000; 5524 |
| Absolute monocyte count3 | 220; 460; 820 | 170; 400; 730 | P=<0.0014 | 170; 400; 730 | 170; 420; 730 | 184; 359; 590 | 182; 459; 814 | 165; 376; 757 | 138; 320; 635 |
| Type of Central Venous line | P<0.0012 | ||||||||
| Port-a-cath | 71.4% (665) | 74.6% (893) | 69.2% (216) | 38.9% (89) | 68.9% (135) | 95.2% (199) | 100% (254) | 83.9% (385) | |
| PICC line | 3.5% (33) | 7.8% (93) | 12.5% (39) | 18.3% (42) | 3.6% (7) | 2.4% (5) | 0% (0) | 0% (0) | |
| External tunneled catheter (CVC) | 25.1% (234) | 17.6% (211) | 18.3% (57) | 42.8% (98) | 26.4% (51) | 2.4% (5) | 0% (0) | 16.1% (74) | |
| Reports/observed chills or shaking rigors | 11.8% (110) | 7.9% (94) | P=0.0022 | 12.0% (110) | 7.0 (16) | 2.6% (5) | 3.3% (7) | 7.5%(19) | N/A7 |
| Hypotension | 1.7% (16) | 4.3% (52) | P<0.0012 | 6.7% (21) | 6.6% (15) | 3.6% (7) | 2.9% (6) | 1.2% (3) | 1.8% (7)7 |
| Drug exposure5 | 10.0% (94) | 12.0% (149) | P=0.0892 | 7.7% (24) | 26.0% (60) | 24.0% (46) | 0.5% (1) | 7.1% (18) | N/A7 |
| Upper respiratory infection symptoms6 | 45.0% (419) | 34.3% (410) | P<0.0012 | 34.9% (109) | 19.2% (44) | 27.5% (53) | 48.3% (101) | 40.6% (103) | 10.7% (49) |
| History of stem cell transplant | 7.1% (66) | 13.8% (165) | P<0.0012 | 6.4% (20) | 30.1% (61) | 38.9% (75) | 0.0% (0) | 0.4% (1) | N/A7 |
| Location at presentation | P<0.0012 | ||||||||
| Inpatient | 19.5% (182) | 30.7% (368) | 35.3% (110) | 67.7% (155) | 53.4% (103) | 0% (0) | 0% (0) | 0% (0) | |
| Outpatient | 80.5% (750) | 69.3% (829) | 64.7% (202) | 32.3% (74) | 46.6% (90) | 100% (209) | 100% (254) | 100% (459) | |
| Diagnosis | P=0.008 | ||||||||
| Acute lymphoblastic leukemia | 47.9% (446) | 42.1% (504) | 45.8% (143) | 31.0% (71) | 20.7 (40) | 50.2% (105) | 57.1% (145) | 57.1% (262) | |
| Other | 52.1% (486) | 57.9% (693) | 54.2 % (169) | 69.0% (158) | 79.3% (153) | 49.8% (104) | 42.9% (109) | 42.9% (197) |
5 primary external validation data sets excluding Brown;
Pearson test;
a; b; c represent the 25th quartile a, the median b, and the 75th quartile c for continuous variables. Numbers after percents are frequencies;
Wilcoxon test;
Exposure to cytarabine, neuroblastoma antibody therapy (Anti-GD2), or ATG within 24 hours of presentation;
Cough, rhinorrhea, and/or congestion;
Chills, Drug exposure, and history of stem call transplant information not available for this dataset and hypotension available for N=388;
Outcome
The EsVan model was designed to predict the outcome of isolated BSI and thus this outcome was maintained in the external validation, using the same criteria. A BSI was defined previously as ≥1 positive blood culture for bacteria obtained from a CVC for a recognized pathogen at the time of fever. For certain common commensals (coagulase negative staphylococci [CoNS], viridans group streptococci, diphtheroids, Micrococcus, or Bacillus species), two or more positive blood cultures were required to meet criteria for BSI8, 9. Any of these bacterial isolates recovered from a single blood culture with a corresponding negative pre-antibiotic culture drawn were classified in the analysis as non-BSI, and those where there was only one pre-antibiotic culture drawn were excluded from the analysis due to inadequate data to assess true versus false positive BSI. High-risk BSI were defined as those with a high risk of sepsis or other complications (Gram-negative organism or Staphylococcus aureus). As there were no isolated fungal BSI in the original Vanderbilt model and fungal infections are not sensitive to antibiotics, all isolated fungal infections were classified as non-BSI.
Statistical analysis
Patients’ demographics and clinical variables were summarized by study cohort. For continuous variables, the median, lower and upper quantiles were reported. The differences between the EsVan cohort and validation cohorts were assessed using Wilcoxon rank sum test. For categorical variables, the frequencies with percentages were reported, and the differences were assessed using the Pearson’s Chi-squared test.
To evaluate the performance of the EsVan model in the independent validation cohorts, model discrimination and calibration were assessed. Discrimination measures the ability of the model to differentiate between patients with and without outcome event. It is generally quantified using the area under the receive operator characteristic (ROC) curve (AUC), which is equivalent to the Harrell’s C-statistic10–12. The C-statistic of the EsVan model on the validation cohort was compared with the benchmark C statistic, which is the best possible C-statistic by refitting the model on the validation dataset. The 95% confidence intervals (CI) calculated from 300-iteration bootstraps were also reported. Calibration measures the agreement between observed outcomes and predictions. Calibration is commonly assessed graphically with calibration plot. The calibration plot has predictions on the X-axis and the outcome on the Y-axis. Perfect prediction should be on the 45° line. For binary outcomes, the plot contains only 0 and 1 values for the Y-axis. Smoothing techniques are generally used to estimate the observed probabilities of the outcome in relation to the predicted probabilities, e.g. using the loess algorithm13. The recalibration was conducted using the logistic method, in which, a logistic regression model was fitted using the linear predictors (from the original model) as the only covariates. The updated calibration slopes and intercepts were reported14, 15. The primary validation dataset (N=1197) included all dataset information from which full model variables were available. The model performance was assessed for both BSI and high risk BSI. The variables used in the original EsVan model were determined a priori. Since the EsVan model uses a large number of variables, penalized maximum likelihood estimations were used in the logistic regression to avoid overfitting. To account for the potential correlation among multiple episodes observed from same the individual, the robust covariance matrix estimates by the Huber-White method were used16, 17. More detailed methods has been described in the original article4. In the current validation study, we primarily combined all the external data sets into a master external dataset for the purposes of more precisely assessing the model performance. While, in order to give an estimate of how the model performed across institutions, individual strict validation data is given for each institution, though the estimates are generally associated with less precision due to the smaller number of events. The TRIPOD checklist for prediction model validation studies was applied18.
With the goal of external validation, the variables were re-considered. Previously, acute lymphoblastic leukemia (ALL) diagnosis was initially included in EsVan, given the hypothesis that most ALL patients were at lower risk of BSI. However, it was subsequently noted that this was dependent on phase and type of ALL therapy, with for example, subjects with relapsed ALL noted to be high-risk for BSI19. Location at presentation was also included in the original EsVan model, as it was hypothesized that subjects inpatient for chemotherapy were less likely to develop a BSI as compared to subjects in the community. However, this was dependent on the patient population and with differing admission criteria noted across centers, though not to be a reliable risk factor that could be used in a more universal model. Thus, a more parsimonious reduced EsVan2 model was created. This reduced model was then applied to the primary validation dataset and discrimination and calibration were again assessed. A nomogram for the reduced EsVan2 model is shown in Figure 1 and a web-based module can also be accessed at (https://cqs.mc.vanderbilt.edu/shiny/RiskPrediction/). As an absolute monocyte count (AMC) can be challenging for some institutions to assess quickly at presentation, a version of the reduced model (EsVan2b) was also created that eliminates AMC as a variable.
Figure 1.
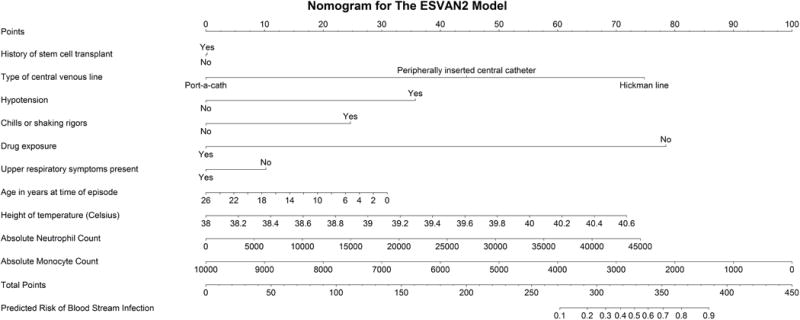
Nomogram of the reduced EsVan2 model for use to calculate the risk of a blood stream infection. Each variable value corresponds to a point value given at the top of the nomogram. The points for each variable are then added together and a predicted risk can be assessed by lining up the point total with the predicted risk given at the bottom of the nomogram.
Finally, sensitivity analyses were conducted, in which all models were applied to the larger validation dataset that included those with missing data (N=1692). For missing data, all missing variables were set first conservatively to “no” and then to “yes” (hypotension, chills, stem cell transplant, and upper respiratory symptoms) and the models were applied.
All statistical significance was considered at a two-sided 5% level. All statistical analyses were performed using R software version 3.3.120.
Results
Patient characteristics
The demographics and clinical features of the external validation cohort as well as the original EsVan derivation cohort are shown in Table 1. Statistically significant differences between the external and the EsVan derivation cohorts were noted in BSI rate (6.1 % vs 9.8%, P=.002), absolute monocyte count (400 vs 460, P<0.001), type of central line present (P<0.001), chills by history or observation (7.9% vs 11.8%, P=0.002), hypotension (4.3% vs 1.7%, P<0.001), upper respiratory symptoms (34.3% vs 45%, P<0.001), history of stem cell transplant (13.8% vs 7.1%, P<0.001), location of presentation (30.7% vs 19.5%, P<0.001), and ALL diagnosis (42.1% vs 47.9%, P=0.008). There were 17 suspected contaminants (14 CoNS and 2 Bacillus across the external data sets that were able to be reclassified as non-BSI and only one case of CoNS that was excluded due to only having one pre-antibiotic culture. There were also a total of 3 fungal isolates across the external datasets classified as non-BSI. The specific microbiologic organisms isolated at each site are provided (Supplemental Table 2).
Primary validation of EsVan model
When the original EsVan model was applied to the primary external validation cohort, it showed reasonable discrimination, with a C-statistic of 0.695 (95% CI 0.633–0.762) (Figure 2a), with the benchmark C-statistic of this model refitted on the validation cohort of 0.759. The model calibrated well, particularly for those with low predicted risk of developing BSI (Figure 2b). After recalibration, the model achieved an intercept of 0 and slope of 1 (Figure 2c). In predicting the high-risk BSI, the model performed better with C-statistic of 0.801 (95% CI 0.693–0.894) (Supplemental Figure 1a) and the calibration plot is shown in Supplemental Figure 1b. The reduced model (EsVan2), with the removal of location and ALL diagnosis, had a C-statistic of 0.895 in the initial model derivation cohort and this updated regression model is given in Supplemental Figure 4. When the reduced EsVan2 model was applied to the validation cohorts it showed a C-statistic of 0.733 (95%CI 0.672–0.792) (Figure 3a), with the benchmark C-statistic of this model refitted on the validation cohort of 0.755. The calibration and recalibration are shown in Figure 3b and 3c. The EsVan2 model also performed better when applied to high-risk BSI with a C-statistic of 0.841 (95% CI 0.781–0.899) (Supplemental Figure 1c). The calibration is shown in Supplemental Figure 1d. When AMC was also removed from EsVan2, the C-statistic for the EsVan2b model on the external validation cohort fell to 0.731 for BSI and 0.832 for high-risk BSI. Figure 4 using the external datasets provides a distribution for the predicted probability from the model for each episode of non-high-risk BSI and high-risk bacteremia. This shows that 85.1% of the cases were low risk (<10% predicted risk) with the BSI rate in that group being 4.3% with only 27.9% of the BSI infections in that group being high-risk. There is also an increase in actual BSI rates in those with a predicted risk over 40% with an actual BSI rate of 35% in these subjects.
Figure 2.
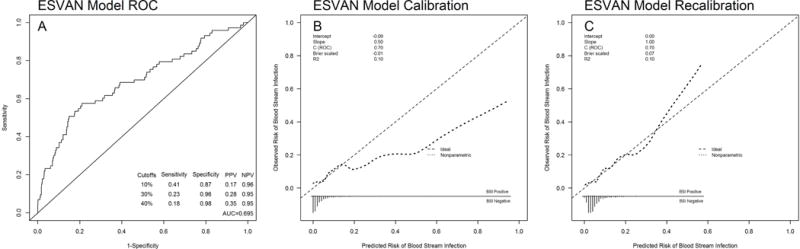
A. Receiver operating curve for the EsVan model when applied to 5 external datasets. Sensitivity, specificity, positive predictive value and negative predictive value given at cut points of 10%,30%, and 40%. B. Calibration of the EsVan model when applied to 5 external datasets. C. Calibration of the EsVan model once recalibrated.
Figure 3.
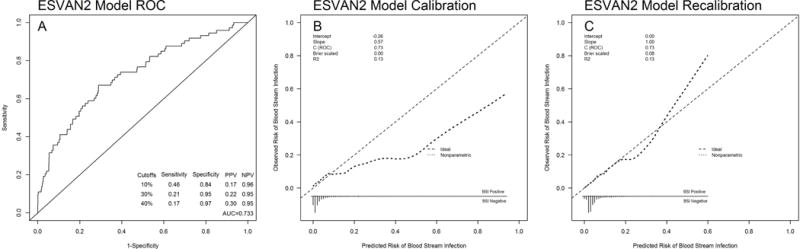
A. Receiver operating curve for the EsVan2 model when applied to 5 external datasets. Sensitivity, specificity, positive predictive value and negative predictive value given at cut points of 10%,30%, and 40%. B. Calibration of the EsVan2 model when applied to 5 external datasets. B. Calibration of the EsVan2 model once recalibrated.
Figure 4.
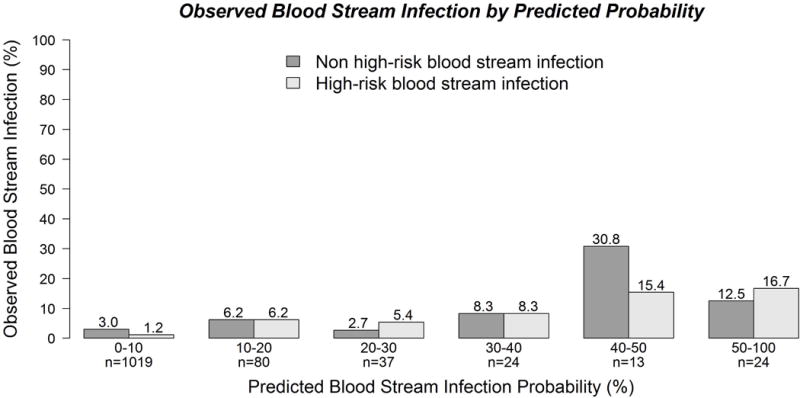
Distribution of the observed blood stream infections in the 5 external datasets by the predicted probability as calculated by the EsVan model.
Sensitivity analysis
When the EsVan model was applied to the secondary data set with all missing values set conservatively to “no”, it showed a C-statistic for BSI of 0.678 and high-risk BSI of 0.781. For the updated reduced model, EsVan2, the C-statistic was 0.711 for BSI and 0.800 for high risk BSI. When the EsVan model was applied to the secondary data set with all missing values set to “yes”, it showed a C-statistic for BSI of 0.672 and high-risk BSI of 0.759. For the updated reduced model, EsVan2, the C-statistic was 0.690 for BSI and 0.757 for high risk BSI. As results from these very conservative approaches were quite similar, further multiple imputation methods on the missing values were not attempted (Supplemental Figure 2).
Strict application of the model to individual datasets
The EsVan model when applied to strictly to each dataset showed the following c-statistics: UCSF (0.705), Stanford (0.738), Columbia (0.725), Lebanon (0.483), CHOP (0.475), Brown (0.657). When the EsVan2 model was applied it showed improved discrimination: UCSF (0.725), Stanford (0.743), Columbia (0.748), Lebanon (0.629). CHOP (0.545), and Brown (0.677).
Discussion
The EsVan model is the first to attempt to predict BSI in febrile pediatric oncology subjects without severe neutropenia, and the current study externally validates this model by showing that in applying it to new datasets it was still able to show good discrimination in predicting BSI (C-statistic 0.695) with improved discrimination for high-risk BSI (C-statistic 0.801). Compared with the C-statistic from the initial model derivation cohort, there is a small drop in C-statistic in this validation. This may due to the differences in case-mix between this validation cohort and initial derivation cohort. This validation cohort has significantly lower BSI incidence, and is different than the initial derivation cohort in several patient characteristics. It is worth noting, however, when we refit the model on the validation cohort, the benchmark C-statistic rose from nearly 0.70 to 0.76, indicating that the EsVan model performed reasonably to the best possible model that one can achieve from this particular validation cohort.
Overall the EsVan model also showed good calibration in this validation cohort, though the upper tail of the curve was not calibrated perfectly in that it over predicts the risk of BSI in those with a risk over 40% (actual BSI rate is 35% in these subjects). It however was quite good for patients with a predicted BSI risk under 10% which was 85% of the cohort 4.
It has been suggested that a model when applied to a new dataset should be recalibrated so that the calibration and predictions become more accurate in the new population, while preserving discrimination15, 21. When the EsVan model was recalibrated on this validation cohort, it performance improved, achieved calibration intercept of 0 and slope of 1 (Figure 2b), further demonstrating usefulness of the model. For other institutions to use this model, the performance and prediction accuracy could potentially be improved by updating the model with local institutional data, though the EsVan model did show relatively good calibration and so could be directly adapted and used in a clinical setting22.
The EsVan2 model was created to assess if reduction of the EsVan model, removing two variables thought to be less reliable across larger multi-institutional populations, would improve the predictive ability across external datasets, while maintaining its performance on the initial Vanderbilt dataset. The EsVan2 model had a similar C-statistic to EsVan model when applied to the Vanderbilt dataset and indeed performed better when applied to the external datasets. As some institutions cannot get an AMC back in a timely fashion to incorporate into a predictive model, a version of the EsVan2 model was created that removed AMC. This model EsVan2b decreased the predictive ability only slightly.
Since each of the individual datasets were small compared to the initial EsVan dataset, this analysis combined the external data into one large cohort. The model performed better in datasets that had higher BSI rates (UCSF/Stanford) than in those with low BSI rates (CHOP/Lebanon). It is not clear why the dataset performed much worse in the CHOP dataset although it was the smallest, only contained a small portion of patients treated during given time period at CHOP, and had a surprisingly low BSI rate of 3% with only one BSI being high-risk despite containing patients with high rate of stem cell transplant and external CVCs. This is likely due to random case mix in a small dataset but could be related to unknown factors such as infection control practices.
One limitation of the study is the variables were collected through retrospective chart review, and were limited by the quality of data in the primary records. However this is outweighed by validation of this model across different academic centers with diverse patient populations, antimicrobial and admission practices.
Incorporation of this model
Since the EsVan2 model showed the best overall discrimination and calibration it is the model that will be moved forward in prospective trials. Since the majority of patient (85%) had a risk under 10% it is group that potentially could have initial empiric antibiotics withheld. Supporting this is the published data from Stanford, included in the validation cohort, that well-appearing febrile pediatric oncology patients without severe neutropenia do well without the administration of empiric antibiotics3. For those at high-risk (predicted risk over 40%), the original model may actually over predict their risk slightly which is likely represents some BSI (particularly anaerobic) are culture-negative, as well as those with significant bacterial or viral illnesses without bacteremia, however if an actual risk of a BSI is over 35% it is likely warranted that these subjects be treated more aggressively with antibiotics likely to cover the BSI’s isolated.
It must be emphasized that evidence-based risk prediction models should never be used to replace clinical judgment, but should be used to as an aid to guide management. The practice of limiting antibiotics because of the model is not recommended for ill-appearing subjects with any concern for severe sepsis, however inclusion of these variables is important to help the precision of the model and help to identify subjects a high-risk that an individual may not initially identify that way. Similar caution would also be recommended for other high risk groups such as subjects under one year of age, those unreliable to follow-up, those with rapidly falling ANC or status post stem cell transplant. It should be noted the original model and this validation is specifically for BSI and other foci of infection should be treated independently of the BSI risk. However, with these important caveats, with the external validation of the EsVan predictive model, and improvement noted with the EsVan2 predictive model, the EsVan2 model shows promise that it may be implemented to help guide management for the prediction of BSI in febrile pediatric oncology patients without severe neutropenia. A prospective implementation trial is therefore planned.
Incorporation of the EsVan2 into clinical practice if shown to be effective prospectively has the potential for greatly impacting antibiotic stewardship in pediatric oncology patients. For institutions that currently give empiric antibiotics to all, in those determined to be at low-risk for BSI, empiric antibiotics could be potentially be withheld and in institutions that don’t routinely give empiric antibiotics to these subjects, the model could potentially help to identify subjects at high-risk for BSI where empiric treatment would be warranted. Further prospective implementation will help to clarify these questions.
Supplementary Material
Acknowledgments
Grant sponsor: NCRR/NIH; Grant number: CA090625 and KL2TR000446; NCI/NIH 2P30CA068485-19
Footnotes
- Conceptualization: A. Esbenshade, D. Friedman
- Methodology: A. Esbenshade, Z. Zhao, K.Moons
- Software: A. Esbenshade, Z. Zhao
- Validation: A. Esbenshade, Z. Zhao
- Formal analysis: A. Esbenshade, Z. Zhao, Y. Shyr
- Investigation: A. Esbenshade, C. Aftandilian, R. Saab, R.. Wattier, M.Beauchemin, T. Miller, J. Wilkes, M. Kelly, A. Fernbach, M. Jeng, C. Schwartz, C. Dvorak, M. Sulis
- Resources: A. Esbenshade, C. Aftandilian, R. Saab, R.. Wattier, M.Beauchemin, T. Miller, J. Wilkes, M. Kelly, A. Fernbach, M. Jeng, C. Schwartz, C. Dvorak, M. Sulis
- Data curation: A. Esbenshade, Z. Zhao
- Writing original draft: A. Esbenshade, D. Friedman
- Writing – review and editing: A. Esbenshade, Z. Zhao, C. Aftandilian, R. Saab, R. Wattier, M. Beauchemin, T. Miller, J. Wilkes, M. Kelly, A. Fernbach, M Jeng, C. Schwartz, C. Dvorak, Y. Shyr, K. Moons, M. Sulis, D. Friedman
- Visualization: A. Esbenshade, Z. Zhao
- Supervision: D. Friedman, Y. Shyr, C. Schwartz, C. Dvorak, M. Sulis
- Project administration: A. Esbenshade
- Funding acquisition: A. Esbenshade, D. Friedman
Disclosures: This study has not been previously presented or published. The authors have no conflict of interests with respect to this work.
References
- 1.Lehrnbecher T, Phillips R, Alexander S, et al. Guideline for the management of fever and neutropenia in children with cancer and/or undergoing hematopoietic stem-cell transplantation. J Clin Oncol. 2012;30:4427–4438. doi: 10.1200/JCO.2012.42.7161. [DOI] [PubMed] [Google Scholar]
- 2.Ali BA, Hirmas N, Tamim H, et al. Approach to Non-Neutropenic Fever in Pediatric Oncology Patients-A Single Institution Study. Pediatr Blood Cancer. 2015;62:2167–2171. doi: 10.1002/pbc.25660. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomew F, Aftandilian C, Andrews J, Gutierrez K, Luna-Fineman S, Jeng M. Evaluation of febrile, nonneutropenic pediatric oncology patients with central venous catheters who are not given empiric antibiotics. J Pediatr. 2015;166:157–162. doi: 10.1016/j.jpeds.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Esbenshade AJ, Pentima MC, Zhao Z, et al. Development and validation of a prediction model for diagnosing blood stream infections in febrile, non-neutropenic children with cancer. Pediatr Blood Cancer. 2014 doi: 10.1002/pbc.25275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly MJ, Vivier PM, Panken TM, Schwartz CL. Bacteremia in febrile nonneutropenic pediatric oncology patients. Pediatr Blood Cancer. 2010;54:83–87. doi: 10.1002/pbc.22264. [DOI] [PubMed] [Google Scholar]
- 6.Hughes WT, Armstrong D, Bodey GP, et al. 2002 guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin Infect Dis. 2002;34:730–751. doi: 10.1086/339215. [DOI] [PubMed] [Google Scholar]
- 7.Freifeld AG, Bow EJ, Sepkowitz KA, et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the infectious diseases society of america. Clin Infect Dis. 2011;52:e56–93. doi: 10.1093/cid/cir073. [DOI] [PubMed] [Google Scholar]
- 8.Mermel LA, Allon M, Bouza E, et al. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;49:1–45. doi: 10.1086/599376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Grady NP, Alexander M, Dellinger EP, et al. Guidelines for the prevention of intravascular catheter-related infections. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2002;51:1–29. [PubMed] [Google Scholar]
- 10.Harrell FE, Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 11.Harrell FE, Jr, Lee KL, Califf RM, Pryor DB, Rosati RA. Regression modelling strategies for improved prognostic prediction. Stat Med. 1984;3:143–152. doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- 12.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Steyerberg E. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. Springer; New York: 2008. [Google Scholar]
- 14.Janssen KJ, Vergouwe Y, Kalkman CJ, Grobbee DE, Moons KG. A simple method to adjust clinical prediction models to local circumstances. Can J Anaesth. 2009;56:194–201. doi: 10.1007/s12630-009-9041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steyerberg EW, Borsboom GJ, van Houwelingen HC, Eijkemans MJ, Habbema JD. Validation and updating of predictive logistic regression models: a study on sample size and shrinkage. Stat Med. 2004;23:2567–2586. doi: 10.1002/sim.1844. [DOI] [PubMed] [Google Scholar]
- 16.Huber PJ. The behavior of maximum likelihood estimates under nonstandard conditions. Berkeley, CA: University of California Press; 1967. [Google Scholar]
- 17.White H. A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica. 1980;48:817–830. [Google Scholar]
- 18.Moons KG, Altman DG, Reitsma JB, Collins GS, Transparent Reporting of a Multivariate Prediction Model for Individual Prognosis or Development I New Guideline for the Reporting of Studies Developing, Validating, or Updating a Multivariable Clinical Prediction Model: The TRIPOD Statement. Adv Anat Pathol. 2015;22:303–305. doi: 10.1097/PAP.0000000000000072. [DOI] [PubMed] [Google Scholar]
- 19.Castagnola E, Fontana V, Caviglia I, et al. A prospective study on the epidemiology of febrile episodes during chemotherapy-induced neutropenia in children with cancer or after hemopoietic stem cell transplantation. Clin Infect Dis. 2007;45:1296–1304. doi: 10.1086/522533. [DOI] [PubMed] [Google Scholar]
- 20.Team Rc. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 21.Van Houwelingen HC, Thorogood J. Construction, validation and updating of a prognostic model for kidney graft survival. Stat Med. 1995;14:1999–2008. doi: 10.1002/sim.4780141806. [DOI] [PubMed] [Google Scholar]
- 22.Moons KG, Altman DG, Vergouwe Y, Royston P. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ. 2009;338:b606. doi: 10.1136/bmj.b606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


