Abstract
Background
Migration and travel are major drivers of the spread of infectious diseases. Geographic proximity and a common language facilitate travel and migration in Mesoamerica, which in turn could affect the spread of HIV in the region.
Methods
6,092 HIV-1 subtype B partial pol sequences sampled from unique antiretroviral treatment-naive individuals from Mexico (40.7%), Guatemala (24.4%), Honduras (19%), Panama (8.2%), Nicaragua (5.5%), Belize (1.4%), and El Salvador (0.7%) between 2011–2016 were included. Phylogenetic and genetic network analyses were performed to infer putative relationships between HIV sequences. The demographic and geographic associations with clustering were analyzed and viral migration patterns were inferred using the Slatkin-Maddison approach on 100 iterations of random subsets of equal number of sequences per location.
Results
A total of 1,685/6,088 (27.7%) of sequences linked with at least one other sequence, forming 603 putative transmission clusters (range: 2–89 individuals). Clustering individuals were significantly more likely to be younger (median age 29 vs 33 years, p<0.01) and men-who-have-sex-with-men (40.4% vs 30.3%, p<0.01). Of the 603 clusters, 30 (5%) included sequences from multiple countries with commonly observed linkages between Mexican and Honduran sequences. Eight of the 603 clusters included more than 10 individuals, including two comprised exclusively of Guatemalans (52 and 89 individuals). Phylogenetic and migration analyses suggested that the Central and Southern regions of Mexico along with Belize were major sources of HIV throughout the region (p<0.01) with genetic flow southward from Mexico to the other nations of Mesoamerica. We also found evidence of significant viral migration within Mexico.
Conclusion
International clusters were infrequent, suggesting moderate migration between HIV epidemics of the different Mesoamerican countries. Nevertheless, we observed important sources of transnational HIV spread in the region, including Southern and Central Mexico and Belize.
Keywords: Mesoamerica, Migration, Network, Clusters, HIV
1. INTRODUCTION
Migration and travel have been implicated as major factors in the spread of HIV since the earliest days of the epidemic (1, 2). The association of travel with HIV infection largely reflects the long-recognized link of travel with the risk of acquiring sexually transmitted infections (3, 4). Travel may involve changes in sexual behavior, including an increase in the number of sexual partners and contacts with sex workers (3). Migrants are also at higher risk of acquiring infections due to factors such as social isolation, poor living conditions, and increased risk taking (5). Furthermore, once infected, these same individuals are at risk of bringing the infection to susceptible populations (6). Phylodynamic analyses have demonstrated how HIV likely spread first along key transportation corridors in the Democratic Republic of Congo and West Africa before spreading out into rest of the population (7, 8). Similarly, historical evidence suggests that much of the HIV epidemic in Mesoamerica, consisting of Mexico and the Central American nations of Guatemala, Nicaragua, Honduras, Belize, El Salvador, Panama, and Costa Rica, has its origins in the United States (US) (7, 9–12). It is likely that the epidemic spread from the US to Mesoamerica as labor and economic migrants from Mesoamerica to the US returned home (9, 10, 12–14). Many of the Mesoamerican nations have also suffered tremendous sociopolitical upheaval all along the 20th century, with geopolitical forces leading to substantial shifts of populations across the region (15). However, little is known about how HIV spread throughout Mesoamerica, and the role of migration and travel in the diffusion of this epidemic in the region is still to be addressed.
The current HIV epidemic in Mesoamerica is concentrated in several high-risk groups, with HIV prevalence in these groups varying from nation to nation. Men who have sex with men (MSM) are highly affected across Mesoamerica, with prevalence as high as 20% in Panama (16). In the region, a high level of stigma is associated with HIV (17) and with homosexuality (18–20) causing marginalization of this population, which in turn leads to non-disclosure of sexual risk increased risk-taking behavior (21). As a consequence, many MSM in Mesoamerica consider themselves heterosexual, but are sexually active with both men and women (22). Sex workers are another group at high-risk for acquiring HIV infection, with HIV prevalence higher in transgender or male sex workers than female sex workers in the region(16). However, less is known about the potential transmission links between these high-risk populations and their role in the dynamics of the overall Mesoamerican epidemic.
Here we explore the phylodynamics of HIV across Mesoamerica, using a large cohort of individuals recruited over the past 5 years as part of the Mesoamerican Project to better understand regional epidemics, the viral transmission links between them, and risk groups across the region.
2. METHODS
2.1. Study Populations
Antiretroviral treatment (ART)-naive HIV-infected individuals were enrolled between October 2010 and February 2016 as part of the multicenter cross-sectional Mesoamerican Project to assess HIV transmitted drug resistance, molecular epidemiology and viral diversity across Mexico and Central America. Individuals were included at the time of diagnosis at participating clinics, or at follow-up visits prior to starting ART according to national guidelines. Every eligible person attending each participating site was offered the opportunity to participate in the study. No exclusion criteria were applied except for known previous exposure to ART. All participants gave written informed consent to participate in the study.
Socio-demographic data from participants were collected through direct application of a questionnaire at the time of sample donation. Each participant provided a single blood sample that was processed at the Centre for Research in Infectious Diseases (CIENI) of the National Institute of Respiratory Diseases (INER) in Mexico City, a WHO-designated laboratory for HIV genotyping, fulfilling procedural and infrastructure requirements for good laboratory practices and quality assurance, after a maximum of 72 hours from collection.
Participating institutions in Central America included: Belize: Ministry of Health, Belmopan; Guatemala: Roosevelt Hospital, Guatemala City (a national referral center) (23); El Salvador: Rosales National Hospital, San Salvador; Honduras: University School Hospital, Tegucigalpa; National Cardio-Pulmonary Institute, Tegucigalpa; Mario Catarino Rivas Hospital, San Pedro Sula; Atlántida Hospital, La Ceiba; South Hospital, Choluteca (five of the largest HIV clinics across the country) (24); Nicaragua: Roberto Calderón Hospital, Managua (the largest reference center in the country) (25); Panama: Gorgas Memorial Institute for Health Studies, Panama City (a national referral center) (26). In the case of Mexico, a national collaborative network was established with clinics in 12 states (27) and further expanded in 2015 to include a total of 17 states (28). The estimated sampling depth of the new diagnoses in each of the participating nations is shown in Supplemental Table S3 and Figure S5. The protocol was reviewed and approved by the Ethics Committee of the INER and by local Committees in each Central American country.
2.2. Sequencing
Partial HIVpol (HXB2 positions 2253–3554) was bulk sequenced from free plasma virus (extraction from 1 mL of plasma, QIAmp Viral RNA Kit, QIAGEN, Valencia, CA), using a previously described in-house protocol (23), with a 3730×l Genetic Analyzer instrument (Thermo Fisher, Waltham, MA). Sequences were assembled using the web-based automated sequence analysis tool RECall (University of British Columbia, Vancouver, Canada) (29).
2.3. Transmission Cluster Inference
We employed HIV-TRACE (HIV TRAnsmission Cluster Engine; www.hivtrace.org)_using the protocol described by Wertheim et al (30) to infer transmission clusters. Briefly, all partial HIV pol sequences were aligned to HXB2 reference sequence. All pairwise distances were computed and a putative linkage between two individuals was considered whenever their pol sequences were ≤0.015 substitutions/site divergent (TN93 substitution model) (31–33). When calculating pairwise genetic distance, all nucleotide ambiguities were resolved and only sequences with less than 1.5% ambiguities were retained (34). Problematic sequences, including those closely related to HXB2 and NL4-3, which could be deemed as possible contaminants, were flagged and repeated based on phylogenetic analyses performed on a monthly basis as an internal laboratory control. Multiple linkages were then combined into putative transmission clusters. Clusters comprised of only two linked nodes were identified as dyads. This approach will detect clusters of recent transmission in which the clustering viruses are genetically similar, implying a direct or indirect epidemiological connection (34).
Convergent evolution due to ART resistance pressure has the potential to confound evolutionary analyses (35). However, pure sampled population included only sequences from purportedly ART-naive individuals. Furthermore, HIV-TRACE analysis has been shown to be robust to the inclusion of codons associated with drug resistance (30, 34). Nonetheless, we explored the effect of inclusion/exclusion of sites associated with drug resistance.
The time to most recent common ancestor (TMRCA, years) of the largest clusters (size >10 individuals) were estimated using a Markov Chain Monte Carlo (MCMC) framework as implemented in BEAST v1.8.1 (36) with BEAGLE to improve run-time (37). We used a discretized gamma distribution (GTR + Γ4) to account for among-site rate variation. The molecular clock was calibrated using sequence sampling dates under a relaxed uncorrelated lognormal molecular clock model and a gamma prior on clock rate(38). Two separate runs of 50 × 106 chain steps were performed, sub-sampling parameters every 20,000 steps. After removing 10% of burn-in and combining evolutionary parameters and trees using LogCombiner. Convergence of the chains was inspected using Tracer v1.5. The TMRCA estimates were expressed as mean and 95% highest posterior density (HPD) years before the sampling year of the most recent sequence in the cluster.
2.4. Compartmentalization of the Epidemics
We evaluated the population genetic structure of the overall Mesoamerican HIV-1 epidemic by using genetic diversity to compute the Fixation index (FST) (39) between each national epidemic in a pairwise fashion applied on 100 random subsets of 40 sequences. Briefly, the fixation index is defined as where πI is the estimate of mean pairwise intra-compartment (i.e. location) genetic distance (TN93)(31), and πD is its inter-compartment counterpart. Both quantities were computed by comparing all sequences sampled from two different countries, with the requirement that they share at least 150 aligned nucleotide positions. The null hypothesis of a random distribution of genetic variation was rejected when permutation testing, using 10,000 randomizations of sequence locations, revealed significant compartmentalization between populations from different countries. The p value of the permutation test must be ≤0.05 to establish significance.
2.5. Viral Migration
To determine the major routes of viral migration throughout Mesoamerica, we used the Slatkin-Maddison approach (40) implemented in HyPhy, where we inferred the number of migration events between the populations of each Mesoamerican nation (41). To better characterize this viral diffusion in the region, Mexico was divided into 3 regions: (1) the Northern Region (states of Baja California, Jalisco, Tamaulipas, Nuevo Leon, Sonora, Sinaloa, Colima, Chihuahua), (2) the Central Region (states of Puebla, Distrito Federal, Estado De Mexico, Morelos, Oaxaca, Michoacan, Guerrero, Tlaxcala, Hidalgo) and (3) the Southern region (states of Quintana Roo, Yucatan, Veracruz, Campeche, Tabasco, Chiapas) (Supplemental Figure 1). To accommodate for the differences in sample size (available number of sequences for each geographic location), we repeated these analyses on 100 iterations of random subsets of equal number of sequences per location (Belize, El Salvador, Honduras Guatemala, Northern Mexico, Central Mexico, Southern Mexico, Nicaragua and Panama). Given the historical relationship between the HIV epidemics in the US and across Mesoamerica (14, 42–44), we repeated these analyses after including random subsets of equal number of HIV-1 subtype B sequences collected in the United States during the same time period. HIV-1 subtype B partial pol sequences (HXB2 coordinates: 2253–3554) at least 500 nucleotides long and collected in the United States since 2011 were downloaded from the Los Alamos National Laboratory (LANL) HIV sequence database on May 23th 2017. Only 1 sequence per individual, based on classification in the LANL database, was retained. We inferred the major sources and sinks of viral migration with a one-way ANOVA approach after Bonferroni adjustment for multiple comparisons.
3. RESULTS
3.1. Mesoamerican Cohort
After quality filtering, 6,088 subtype B HIV sequences from unique individuals, with associated sociodemographic and geographic information, and sampled between 2011 and 2016 across Mesoamerica, were analyzed (Figure 1). Nearly 30% of participants were female, and 57% identified as heterosexual, with only 1.3% reporting intravenous drug use (Table 1 and Supplemental Table S1). Late presentation to clinical care was frequent, with a median CD4+ T cell count of 239 cells/mm3.
Figure 1.
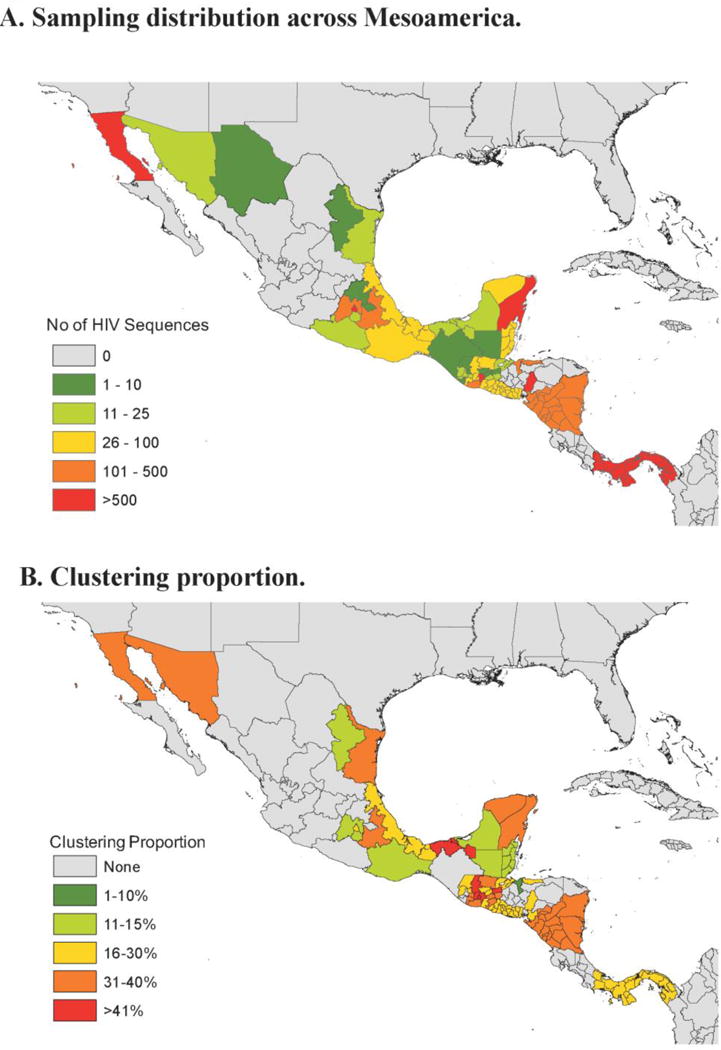
1A. Sampling distribution across Mesoamerica. The number of HIV pol sequences included in the analysis are represented. 1B. Clustering Proportion. Heat map representing the proportion of sampled HIV infected individuals within each geographic subdivision clustering with at least one other individual. Data are presented at the Country level for El Salvador, Belize, Nicaragua and Panama at the state/province level for Guatemala, Honduras, and Mexico.
Table 1.
Population characteristics at baseline and by clustering status
| Total | Non-clustering | Clustering | Odds Ratio | 95% Interval | Confidence | P-value | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| N | 6,088 | 4,403 (72.3) | 1,685 (27.7) | ||||
|
| |||||||
| Age in years, median (IQR) | 32 (25–41) | 33 (27–42) | 29 (24–37) | <0.0001# | |||
|
| |||||||
| Sex, n (%) | |||||||
| - Male | 4,261 (70.0) | 2,999 (68.1) | 1,449 (74.0) | 1 | |||
| - Female | 1,778 (29.2) | 1,364 (31.0) | 498 (25.4) | 0.76 | 0.67–0.85 | <0.0001 | |
| - NA | 49 (0.8) | 40 (0.9) | 12 (0.6) | 0.62 | 0.32–1.19 | 0.15 | |
|
| |||||||
| Origin, n (%) | |||||||
| - Mexico | 2,483 (40.7) | 1,799 (40.9) | 684 (40.6) | 1 | |||
| - Guatemala | 1,487 (24.4) | 898 (20.4) | 589 (35.0) | 1.73 | 1.51–1.98 | <0.0001 | |
| - Honduras | 1,157 (19.0) | 965 (21.9) | 192 (11.4) | 0.52 | 0.44–0.63 | <0.0001 | |
| - Panama | 504 (8.3) | 419 (9.5) | 85 (5.0) | 0.53 | 0.41–0.68 | <0.0001 | |
| - Nicaragua | 334 (5.5) | 216 (4.9) | 118 (7.0) | 1.44 | 1.13–1.83 | 0.0032 | |
| - Belize | 84 (1.4) | 73 (1.7) | 11 (0.7) | 0.40 | 0.21–0.75 | 0.0046 | |
| - El Salvador | 39 (0.6) | 33 (0.7) | 6 (0.4) | 0.48 | 0.20–1.15 | 0.098 | |
|
| |||||||
| Risk, n (%) | |||||||
| - HTS | 3,439 (56.5) | 2,566 (58.3) | 873 (51.8) | 1 | |||
| - MSM | 2,015 (33.1) | 1,335 (30.3) | 680 (40.4) | 1.50 | 1.33–1.69 | <0.0001 | |
| - PWID* | 85 (1.3) | 68 (1.5) | 17 (1.0) | 0.73 | 0.43–1.26 | 0.26 | |
| - Other/NA | 549 (9.0) | 434 (9.9) | 115 (6.8) | 0.78 | 0.63–0.97 | 0.026 | |
|
| |||||||
| Employment, n (%) | |||||||
| - Employed | 2,761(45.4%) | 1,941 (44.1) | 820 (48.7) | 1 | |||
| - Unemployed | 2,522 (41.4%) | 1,891 (42.9) | 631 (37.4) | 0.79 | 0.70–0.89 | 0.0001 | |
| - Student | 423 (6.9%) | 275 (6.2) | 148 (8.7) | 1.27 | 1.03–1.58 | 0.028 | |
| - Other/NA | 382 (6.3%) | 296 (6.7) | 86 (5.1) | 0.69 | 0.43–0.89 | 0.0038 | |
|
| |||||||
| Marital Status, n (%) | |||||||
| - Married | 1,009 (16.6%) | 747 (17.0) | 262 (15.5) | 1 | |||
| - Single | 3,851 (63.3%) | 2,828 (64.2) | 1,023 (60.7) | 1.03 | 0.88–1.21 | 0.70 | |
| - Domestic Partnership | 1,228 (20.2%) | 828 (18.8) | 400 (23.7) | 1.38 | 1.15–1.66 | 0.0007 | |
|
| |||||||
| CD4 (/mm3), median (IQR) | 239 (91–427) | 220 (79–413) | 279 (122–456) | <0.0001# | |||
|
| |||||||
| HIV RNA levels (Log10 copies/mL), median (IQR) | 4.7 (4.0–5.3) | 4.7 (4.0–5.3) | 4.8 (4.2–5.3) | 0.0004# | |||
|
| |||||||
| Year of sampling, median (IQR) | 2013 (2011–2014) | 2012 (2011–2014) | 2013 (2012–2015) | <0.0001# | |||
#clustering vs. non-clustering. MSM, men who have sex with men; PWID, people who inject drugs; HTS, heterosexual;
MSM/PWID were considered in the PWID category. NA, not available.
non parametric t-test
3.2. Mesoamerican Transmission Network
Inference of putative transmission links within the study cohort identified 603 clusters that included 1,685/6,088 (27.7%) individuals. Clusters ranged in size from 2 to 82 individuals, and 167 (27.7%) had 3 or more individuals (Figure 2A). Eight clusters included >10 individuals, and were considered large clusters. These results were robust to the inclusion and exclusion of site associated with drug resistance. The median age at diagnosis of clustering individuals was 29 (IQR: 24–37), which was significantly lower (p<0.001) that that of non-clustering individuals (median age of 33, IQR: 27–42). Of the 3,439 (25.4%) sequences from individuals identifying as heterosexual, 873 (25.4%) clustered with at least one other sequence, while significantly more, 680 of the 2,015 (33.7%) sequences from individuals identifying as men who have sex with men (MSM) or bisexual clustered (p<0.001). Males were significantly more likely to clusters than women (p<0.0001, Table 1). Half of the clusters were exclusively male (303/603, 50.2%). Even among the 291 (48.3%) clusters that include only self-identified heterosexuals, 51 (17.5%) did not include women. We also found 140 (23.2%) mixed clusters including both self-identified heterosexual individuals and MSM. Fourteen (2.3%) clusters included PWID, all but three of them also heterosexual men and women.
Figure 2. HIV transmission clusters.
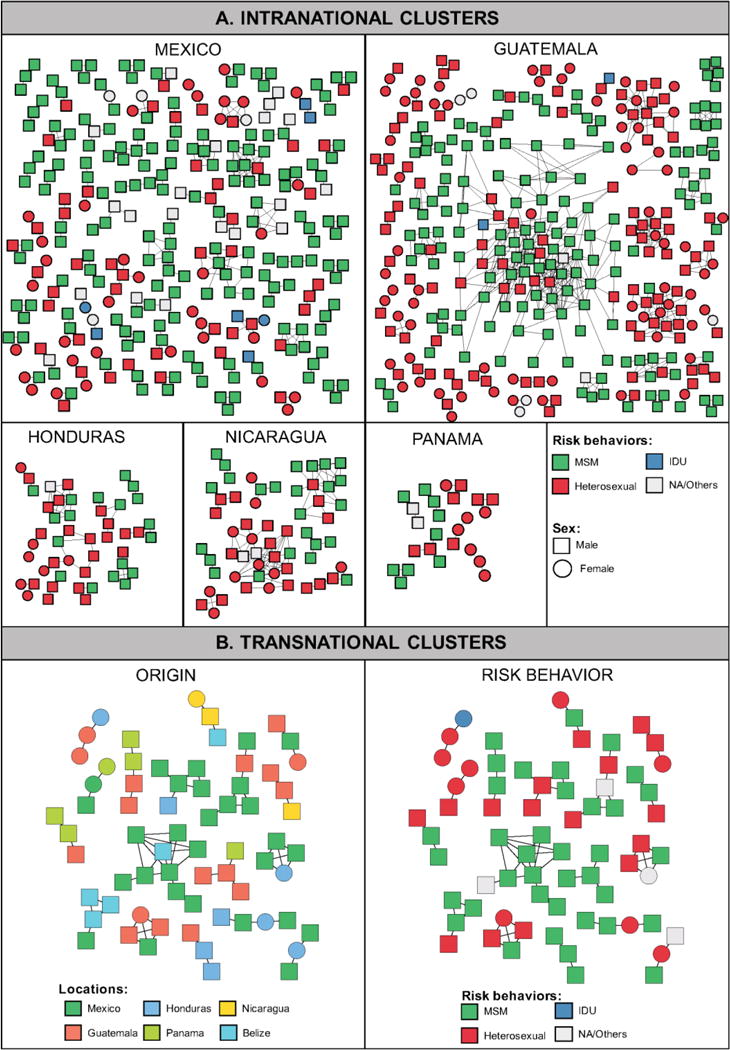
The structure and demographic make-up of inferred HIV-1 transmission clusters from the Mesoamerican cohort are illustrated. Nodes (rectangles and circles) represent connected individuals in the overall network, and putative transmission linkages are represented by edges (lines). A. Intra-national clusters. Nodes are color coded by the reported risk behavior. B. Transnational Clusters. Nodes are color coded by the country of origin (left panel) and the reported risk behavior (right panel). Only clusters with ≥3 individuals are presented. Clustering individuals from El Salvador (n=6) are not shown as they are only included in dyads.
Of the 603 clusters, 271 (44.9%) included individuals sampled in Mexico, and 182 (30.2%) included individuals sampled in Guatemala. Sequences from Guatemala (p<0.0001) and Nicaragua (p<0.0001) were significantly more likely to cluster (Table 1). Overall 573 (95%) of the clusters were comprised of individuals from only one nation. The computed FST tests revealed that the sampled epidemics in each of the Mesoamerican nations were compartmentalized (>95% positive FST, p <0.01 for each country) with respect to the overall Mesoamerican epidemic.
Nevertheless, some cross-border transmission was inferred, as 30 of the clusters (5%) included sequences from two or more countries (Figure 2B and Supplemental Table S2): 19/30 (63.3%) included Mexican sequences with commonly observed linkage between Mexico and Honduras (9/30 [30.0%] clusters).
3.3. Large Transmission Clusters
Eight of the 603 clusters included at least 10 individuals. These included four clusters with individuals exclusively from Guatemala, ranging in size from 13 to 82 individuals; two clusters with individuals exclusively from Mexico; and two with individuals exclusively from Nicaragua (Table 2; Supplemental Figure S2). The largest cluster of 82 Guatemalan individuals included almost exclusively men (81/82, 98.8%), the majority of whom reported having sex with men (64/82, 78%). The three other large clusters in Guatemala were predominantly heterosexual clusters. The Guatemalan MSM cluster included younger individuals than the heterosexual clusters (p<0.05). Individuals in all large Guatemalan clusters came mostly from Guatemala City (70/82 [85.4%], 13/21 [61.9%], 12/20 [60%], and 8/13 [61.5%] respectively). The large Mexican clusters were composed mainly of MSM (9/10 [90%] and 8/11 [73%], respectively), and from individuals from the state of Puebla (8/10 [80%] and 9/11 [82%], respectively). One of the Nicaraguan clusters was comprised mainly MSM (9/12 [75%]), whereas the other was predominantly heterosexual (15/17 [88%]). The inferred timing of the most common recent ancestor for all the large clusters varied from 1999 [95% HPD:1992–2004] to 2004 [95% HPD:2000–2008] (Table 2, Supplemental Figure S2).
Table 2.
Characteristics of the large transmission clusters
| Cluster No | Cluster Size | Country - Predominant# state (No. of seq.) | Sampling Period* | Risk | Sex Male:Female | Age at diagnosis median [IQR] | tMRCA mean [95%HPD] |
|---|---|---|---|---|---|---|---|
| 1 | 82 | Guatemala - Guatemala City (70) | 2011.2–2015.7 | MSM 64/82 78% | 81:1 | 26 [23–32] | 2003 [2000–2006] |
| 2 | 20 | Guatemala - Guatemala City (12) | 2011.3–2015.7 | HTS 19/20 95% | 12:8 | 40 29–46] | 2003 [1999–2007] |
| 3 | 21 | Guatemala - Guatemala City (13) | 2011.7–2015.6 | HTS 100% | 10:11 | 37 [29–49] | 1999 [1992–2004] |
| 4 | 13 | Guatemala - Guatemala City (8) | 2011.1–2015.5 | HTS 12/13 92.3% | 10:3 | 35 [30–47] | 2004 [2000–2008] |
| 5 | 10 | Mexico - Puebla (8) | 2011.5–2014.5 | MSM 9/10 90.0% | 10:0 | 25 [21–30] | 2003 [1998–2007] |
| 6 | 11 | Mexico - Puebla (9) | 2011.7–2015.8 | MSM 8/11 72.7% | 10:1 | 22 [20–26] | 2001 [1995–2007] |
| 7 | 19 | Nicaragua - NA | 2011.9–2015.7 | HTS 15/17 (88%) | 13:6 | 37 [29–48] | 2001 [1994–2007] |
| 8 | 12 | Nicaragua - NA | 2011.6–2015.8 | MSM 9/12 75.0% | 12:0 | 25 [23–29] | 2000 [1992–2007] |
IQR: Interquartile range; tMRCA: time of the most common recent ancestor; HPD: Highest Posterior Density; NA: Not available.
For individuals belonging to each cluster;
Predominant state: accounting for > 50% of the sequences.
3.4. Migration Analysis
To better understand international viral dispersion throughout the region, we performed migration analyses as described above to infer prior and ongoing viral gene flow between different locales. Given the relatively greater size of Mexico, along with its complex geography and cultural variation, we analyzed these migration patterns after separating Mexico into three regions (North, South and Central) (Figure 3 and Supplemental Figure 3). We found that the Central (p<0.001) and Southern (p<0.05) regions of Mexico along with the nation of Belize (p<0.001) were significant sources of viral migration to the other Mesoamerican nations (Figure 3 and Supplemental Figure 3). Given the historical linkage between the HIV epidemics in Mesoamerica and in the United States, we also performed a similar analysis combining sequences collected across Mesoamerica in combination with HIV-1 Subtype B sequences collected in the United States during the same time period. Interestingly, we found significant bidirectional migration between the US and the Northern and the Central region of Mexico (p<0.001) but not the other nations of Mesoamerica (Supplemental Figure 4).
Figure 3. Matrix of HIV migration events.
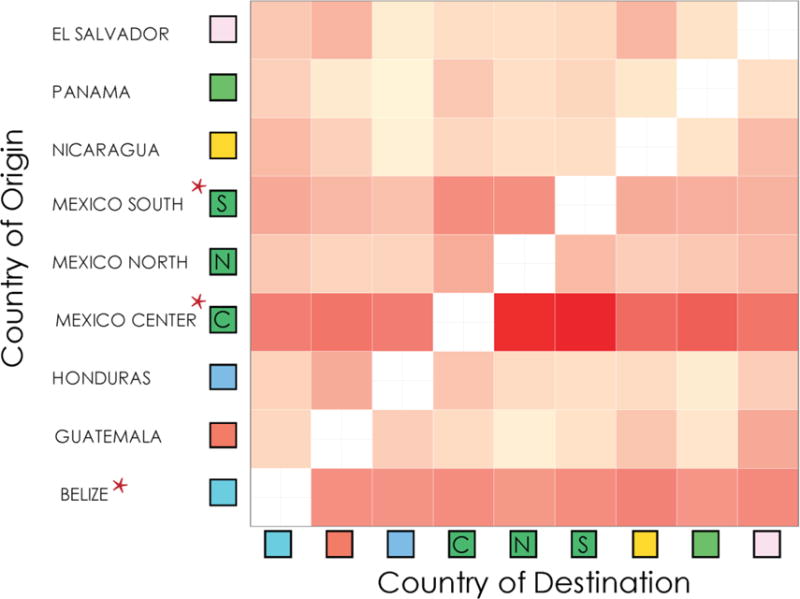
An increase in redness represents a stronger migratory signal. Red asterisk indicates significant major sources of viral migration. Viral migration patterns were inferred using the Slatkin-Maddison approach on 100 iteration of random subsets of equal number of sequences per location (see Methods for details).
4. Discussion
This large-scale analysis of data collected from across Mesoamerica for surveillance of drug resistance demonstrated similar clustering rates to published work analyzing US surveillance data (32). We extended the type of analysis by Oster et al, to examine the HIV epidemic across multiple international boundaries. This work demonstrates that socio-cultural and political boundaries still limit the spread of HIV in Mesoamerica, as the sampled epidemic was compartmentalized by nation, with small numbers of inferred transmission links between nations. However, those putative viral transmission links between individuals from different nations were likely the result of human migration or travel, facilitated by having a common language throughout the region. Such transmission links between geographically disparate populations may impact the overall epidemic by bridging susceptible populations (45), and may result in onward transmission (46).
Historically, the HIV epidemics across North and Central America was believed to have been seeded from the US and the Caribbean epidemics, beginning first in the MSM population (14, 42–44). To better characterize the role of the US epidemic in the Mesoamerican epidemic, we analyzed our data in combination with sequences from the US. The significant bidirectional migration routes from and toward the US unveiled the critical role of Mexico as a potential bridge between Northern and Southern HIV epidemics. Over time, the epidemic has spread into the heterosexual population across Mesoamerica, but particularly in Guatemala and Honduras (47). Spread of HIV between nations was then in part shaped by patterns of travel and economic migration that have occurred in the region. Eventually, after seeding at-risk populations across the region, our data and that from others (48, 49) suggest that these epidemics then evolved regionally, with limited mixing across nations, and risk groups.
To better characterize the prior and ongoing transnational spread of HIV in the region, we used phylogenetic analyses to infer putative viral migration events. Specifically, we inferred from our sample that the Central Mexico region, within which lies Mexico City the largest city in the Americas and one of the most cosmopolitan cities in the world (50), may have played an important role as a source of viral migration throughout the region. As one of the major economic engines of Mexico, the region attracts migrants not only from every region within Mexico, but also from throughout Mesoamerica (50). It has been well established that migrants are prone to more HIV risk behaviors through structural factors such as shifts in behavioral norms and social support networks (51). When migrants return to their home region, they may unknowingly infect partners, seeding local epidemics. Another group of individuals that may be contributing to viral migration are those with sexually related motivations to migrate or travel to Mexico City where they may escape from stigmatization, or be able to live a more open life than their area of origin, similar to what has been described about sexual migration to the US (52). Our data support this possibility, as 70% (21/30) of our inferred international clusters included at least one MSM, and 46 of the 94 individuals (48.9%) found in international transmission clusters were MSM. The role of MSM within the Mesoamerican epidemic may be even greater than suggested by these results. We found that 51 (17.5%) of the 291 clusters considered to be exclusively heterosexual by self-report of participants included men only. These results could be explained by other routes of transmission (i.e. unreported injection drug use) or poor sampling of the women linking these individuals, it is highly suggestive that as least a proportion of self-reported male heterosexuals are likely to have been infected through sex with men. These findings are similar to work by our group and others (53, 54). A recent study in South America also revealed limited transmission links including individuals from different countries (5.5%), with the major role of the states of São Paulo, Goiás, Rio de Janeiro, Mato Grosso, and Paraná in Brazil as potential source of international links in South America (11). While the sampling depth greatly varies across countries in both dataset, creating a potential sampling bias to conclude about their relative importance in the spread of HIV-1B in Mesoamerica or in South America and despite the great differences in the geographic distance, human mobility and spatial accessibility between South American and Central American countries, our findings (5.6% of transnational linkages) were very similar and suggest that HIV epidemics in Central and South America are predominantly driven by local transmission with limited impact of viral migrations across countries (55).
This comprehensive analysis also provided insights in to the epidemics of each of the Mesoamerican nations. Below we have organized these observations by nation.
Panama
In our sample (n=504), we found links between the Panamanian epidemic and Mexico and Guatemala. Inference of the viral migration patterns also suggested some flow of HIV from central Mexico to Panama. In Panama, the epidemic is known to be concentrated along the economic corridor of the Panama Canal, including the central districts of San Miguelito and Panama (which includes Panama City) (48). As an internationally important economic zone, an increase in migration of workers, businessmen and executives, mainly from Guatemala and Mexico to Panama for business reasons has been reported during recent years (48, 56). In addition to tourism, this could explain the inclusion of Guatemalan and Panamanian MSM in many international clusters observed in this study. Interestingly, even with this increase in immigration, the main spread of the Panamanian epidemic seems to remain national with Panama not representing a significant source of seeding of other regional epidemics. This is consistent with recently published work suggesting a recent intra-national spread of Panamanian clades with little overlap with other Central American countries (48, 49), although it is important to acknowledge that most of the Panamanian sequences included in this study were obtained from individuals residing in the central districts, which could limit detection of other possible connections.
Belize
In our sampled cohort from 2011–2016, Belize and Mexico were the predominant sources of viral gene flow in Mesoamerica. Although with only ~330,000 citizens, Belize was reported to have the highest estimated adult prevalence and incidence of HIV in Mesoamerica at 2.1% in 2007(57), but estimates have decreased to 1.4% in 2012 (58). As a more mature epidemic, this could explain why Belize was frequently inferred to be a source in our sampled epidemic. Consistent with this, in our network analysis we found several links between individuals from Belize and Mexico, Guatemala, and Nicaragua. Clusters including sequences from Mexico and Belize were mostly MSM clusters, while those including sequences from other countries were mostly heterosexual. Interestingly, no local networks were observed in our sample. Although sampling biases most surely exist as our Belize cohort is small and highly biased to include persons from the districts of Belize, Orange Walk, and Corozal, our observations could suggest an important role of sexual tourism in the observed network.
Guatemala
Although the estimated prevalence of HIV in Guatemala is low (0.7%) (58), analysis of our sequence data suggested that Guatemala was a destination for viral gene flow in the region, with evidence of migration from Honduras, Belize, El Salvador, and central Mexico. We also inferred putative transmission links between sequences from Guatemalan individuals, and those from Honduras, Nicaragua, Panama and Mexico as well. Given its proximity to Mexico, Guatemala City and Guatemalan border towns are key transit points for migrants traveling northward (59). Thus, while we did not find significant number of links between southern Mexico and Guatemala during our sample period, the links to these other regions may be explained by its role in a northward migration route to Mexico or the US (60, 61). Notably, we did identify several large transmission clusters entirely comprised of individuals sampled in Guatemala, including a large cluster of MSM, a population with an HIV prevalence 10–20 times higher than the general population in Guatemala (62). The observation of these large clusters could be partly explained by the high sampling density or the Guatemalan cohort in our study, as participating individuals were recruited in one of the largest reference hospitals in Guatemala City, which largely centralize clinical care of people on antiretroviral treatment all around the country (23). Several other large clusters were predominantly comprised of self-reported heterosexuals. While we cannot exclude unobserved intermediate of shared sources of infections (63), these clusters highlight the distinct transmission patterns in MSM and heterosexuals in Guatemala. The temporal dynamics of the large MSM cluster, with the large number of infections identified in 2014–2015, also highlights the need to increase screening and treatment in this vulnerable population to slow its growth.
Nicaragua
Although one of the poorest countries in the region, Nicaragua also has the lowest estimated adult HIV prevalence (0.3 %)(58). Civil war in Nicaragua lasted until the early 1990s, and keeping many individuals isolated (64) possibly also limiting the spread of HIV into the country. After the war, individuals began traveling to nearby nations for work. Consistent with this, we inferred links between HIV infected individuals sampled in Nicaragua and the neighboring nations of Guatemala, Belize and Honduras. We also found two large clusters comprised only of Nicaraguans. As in the case of Guatemala, these large Nicaraguan clusters could be explained by a high sampling density, as participating individuals were recruited at the largest referral center in the country, caring for approximately a third of all persons under antiretroviral treatment in Nicaragua (25). Our Bayesian phylogenetic inferences of these clusters suggested that most of these infections occurred prior to 2009. Interestingly, this cessation of growth corresponds with a massive increase in screening (from 18,000 individuals in 2006 to 106,726 in 2008) (65). Additionally, analysis of the demographic characteristics of persons included in the large clusters evidenced at least two distinct epidemics in the country: transmission among young MSM with early infection and a heterosexual epidemic involving older individuals in a more chronic stage of the infection, which may require distinct targeted prevention efforts.
Honduras
During the early 1990s, the major focus of the central American HIV epidemic (not including Mexico) was the nation of Honduras, with an estimated 60% of the region’s infections found in the country (66). It has been hypothesized that the HIV epidemic, initially most prominent in the port city of San Pedro Sula, was seeded by Caribbean sailors (67). Particularly affected have been the Garifuna population, an ethnic minority that has settled along the Atlantic coast and who have also settled Belize, Nicaragua, and Guatemala (68). Interestingly, we found putative transmission links between Hondurans and infected individuals in the nations of Nicaragua, Guatemala and Mexico. Infection among sex workers also has played a major role in the Honduran epidemic with a prevalence of 10% reported in 2007. In our sample (n=1157 sequences), nearly 40% of Honduran women had an HIV sequence that clustered with another individual, which was the highest rate across the Mesoamerican nations. Furthermore, we found that most of the females (9/19, 47.4%) involved in international clusters were sampled in Honduras, which may be reflective of the importance of sex work in viral migration in and out of Honduras.
Mexico
As the largest nation in Mesoamerica, the phylodynamics of the Mexican HIV epidemic is complex. Regional epidemics exist throughout the country, but they are also linked together. To better understand the links between the regional Mexican epidemics, we divided the country into three sections as described above. As expected, we found much closer ties between southern and central Mexico with the rest of Mesoamerica, with our migration analyses suggesting that central Mexico has been an important viral source to the epidemics in the other Mesoamerican nations. Interestingly, although economic migrants through the region are typically traveling northward through Guatemala into Mexico, our phylogenetic and migration analyses suggest that viral genetic flow is southward from Mexico to the other nations, including Guatemala. It remains unclear if this is a consequence of migrants returning home after acquiring the infection (51), or another travel related explanation.
There are several potential limitations to our analysis, related to sampling and quality of data. First, our sample represents a convenience sample of individuals receiving HIV care at centers participating in the Mesoamerican drug resistance monitoring program. However, this sample is the largest collection of sequences sampled in Mesoamerica, with broad geographic representation. In addition, sampling was not uniform across sites. To address this, we used an iterative subsampling approach with 100 replicates to thus provide a fair estimate of viral dynamics in the sampled period. Moreover, although we made every effort to identify duplicates within our database (cross-referencing similar sequences by demographic information), it is possible that some sequences represent individuals sampled more than once in different locations. Our analysis also relied on self-reported transmission risk data. Therefore, males who practiced sex with other men may have reported themselves as heterosexual given concerns about stigma or differing perceptions on what constitutes an MSM (69). Given our stringent thresholds for inferring linkages, and our manual screening to remove duplicates, we anticipate that any remaining duplicates did not significantly affect our analysis. Another limitation was the uncertainty in the duration of infection at the time of sampling. While this would not impact our network analyses, the inferred temporal dynamics of a cluster could be skewed if the entire cluster had been chronically infected for a long period of time. Inferences from our viral migration analyses are also reliant on our sample, and thus may not accurately represent the migration of virus from one region to another. For example, if the epidemic in Guatemala was seeded from US epidemic in 1980, and the Honduran epidemic was seeded from the US in 1990, when looking only at the Mesoamerican data, we may mistakenly infer that the Honduran epidemic was seeded from Guatemala. Finally, it is important to note that potential transmission linkages inferred in this study are not necessarily direct transmission partners, but may include intermediate infections separating the observed connected sequences. Nonetheless, these inferred linkages provide a reasonable proxy for understanding the viral dynamics across populations.
5. Conclusions
Overall, our work highlights the use of phylodynamic analyses to provide insights into the role of migration and travel in the spread of HIV. These results provide objective support for some of the epidemiologic theories regarding historical viral introductions and migrations across Mesoamerica. Continued observations and phylodynamic analyses, particularly with greater depth of sampling, may also have the potential to evaluate the impact changes in demographic trends and policies on the Mesoamerican epidemic.
Supplementary Material
Highlights.
Socio-cultural and political boundaries limit the spread of HIV across MesoAmerica
Mexico and Belize remain important of transnational HIV spread
Large transmission clusters were identified in Mexico, Nicaragua, and Guatemala.
Heterosexual and same sex HIV epidemics are compartmentalized in Guatemala
Acknowledgments
The Mesoamerican Project Group includes: Ingrid Escobar, Sabrina Navas, Rodolfo Pinzón, Leticia García, Cristina Quintana (Roosevelt Hospital, Guatemala, Guatemala); Yamitzel Saldívar, Yaxelis Mendoza (Gorgas Memorial Institute for Health Studies, Panama City); Sumaya Moreira, Bismarck Hernández (Roberto Calderón Hospital, Managua, Nicaragua); Wendy Murillo, Candy Carbajal, Leda Parham (National Autonomous University of Honduras, Tegucigalpa, Honduras); Diana Valladares, Luisa Pineda (Mario Catarino Rivas Hospital, San Pedro Sula, Honduras); Dixiana Flores, Roxana Motiño (Hospital Atlántida, La Ceiba, Honduras); Víctor Umanzor, Oneyda Méndez, Nadina Romero (Hospital del Sur, Choluteca, Honduras); Jonahi Lizama (Ministry of Health, Belmopan, Belize), and the HIVDR MexNet Group. The HIVDR MexNet Group incudes: Akio Murakami-Ogasawara, Karla A. Romero-Mora, María Gómez Palacio, Verónica S. Quiroz-Morales, Ramón Hernández-Juan, Edna Rodríguez (National Institute of Respiratory Diseases, Mexico City); María L. Méndez, David de los Santos Cebrero (CAPASITS Acapulco, Guerrero), César Rivera-Benítez (General Hospital, Mexico City), Juan Sierra-Madero, Audelia Alanis-Vega (National Institute of Medical Sciences and Nutrition, Mexico City), Luz A. González-Hernández, Jaime Andrade-Villanueva (Civil Hospital Fray Antonio Alcalde, Guadalajara, Jalisco), Jaime Álvarez-Zayas (CAPASITS Puerto Vallarta, Jalisco), Héctor Carrillo-Martínez (CAPASITS Nezahualcóyotl, Estado de México), José L. Centeno (CAPASITS Ecatepec, Estado de México), Everardo Barreto, Tanya Campos (CAPASITS Tlalnepantla, Estado de México), Jesus Oaxaca-Navarro (CAPASITS Cuernavaca, Morelos), Ricardo Aya-de la Fuente (CAPASITS Monterrey, Nuevo León), César A. Carrasco-Ayala, Lesvia M. Rivera-Abarca, Gabriela Velázquez (SEAI Oaxaca), Elizabeth Papaqui-Limón, Indiana Torres-Escobar (CAPASITS Puebla), María J. del Carmen-Ricalde, David Valenzo-Loaeza, Carlos A Barrera-Arellano (CAPASITS Cancún, Quintana Roo), Adrián Flores-Gaxiola (CAPASITS Culiacán, Sinaloa), Carlos A Avilez-Gaxiola (CAPASITS Hermosillo, Sonora), Adonay Jiménez-Jiménez (Dr. Juan Graham Casasus Hospital, Tabasco), Juan Beltrán-Saldaña (CAPASITS Tampico), Arturo Arteaga-Martínez (General Hospital, Veracruz), Elizabeth Domínguez-Ramírez (Subregional Hospital, Coatzacoalcos, Veracruz), Jorge M. de la Roca-Chiapas (Subregional Hospital, Rio Blanco, Veracruz), Miriam J. García-Collins, Hilda Basilio-Badillo (Subregional Hospital, Poza Rica, Veracruz), Dulce M. Cruz-Lavadores, Carlos R. González-Álvares (CAPASITS Mérida, Yucatán), Luis E Arias-Tlaculio (CAPASITS Valladolid, Yucatán), and Samuel Navarro-Álvarez (General Hospital Tijuana, Baja California).
Funding
This work was supported by grants from the Mexican Government (Comisión de Equidad y Género de las Legislaturas LX–LXI, Comisión de Igualdad de Género de la Legislatura LXII de la H. Cámara de Diputados de la República Mexicana) and Consejo Nacional de Ciencia y Tecnología (CONACyT SALUD-2013-01-202475), by California HIV Research Program Idea awards (ID15-SD-052), NIH-NIAID Career Development Award K01AI110181 to Joel O. Wertheim, NIH Grant AI093163 and by the department of Veterans Affairs. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
Antoine Chaillon, Santiago Avila-Ríos, Ann Dennis, Claudia García-Morales, Daniela Tapia-Trejo, Juan M. Pascale, Guillermo Porras-Cortés, Carlos J. Quant-Durán C, Ivette Lorenzana, Rita I. Meza, Elsa Y. Palou, Marvin Manzanero, Rolando A. Cedillos, Gustavo Reyes-Terán, Sanjay R. Mehta do not have any commercial or other associations that might pose a conflict of interest. Carlos R Mejia Villatoro has received supports from Merck, Gilead, Roche and Joel O. Wertheim has received grant support from Gilead Science Inc.
Authors Contributions
AC, SAR and SRM participated in the data analysis, and wrote the primary version of the manuscript; CGM and JPS participated the data analysis; JOW participated in the data analyses and revised the manuscript; AD, DTT, JMP, GPC, JQDC, IL, RIM, EYP, MM, RAC and GRT contributed to the manuscript. All authors read and approved the final manuscript.
References
- 1.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pépin J, Posada D, Peeters M, Pybus OG, Lemey P. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346(6205):56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson MM, Najera R. Travel and the introduction of human immunodeficiency virus type 1 non-B subtype genetic forms into Western countries. Clin Infect Dis. 2001;32(12):1732–7. doi: 10.1086/320764. Epub 2001/05/22. [DOI] [PubMed] [Google Scholar]
- 3.Hawkes S, Hart GJ, Johnson AM, Shergold C, Ross E, Herbert KM, Mortimer P, Parry JV, Mabey D. Risk behaviour and HIV prevalence in international travellers. Aids. 1994;8(2):247–52. doi: 10.1097/00002030-199402000-00013. Epub 1994/02/01. [DOI] [PubMed] [Google Scholar]
- 4.Rogstad KE. Sex, sun, sea, and STIs: sexually transmitted infections acquired on holiday. BMJ. 2004;329(7459):214–7. doi: 10.1136/bmj.329.7459.214. Epub 2004/07/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mabey D, Mayaud P. Sexually transmitted diseases in mobile populations. Genitourin Med. 1997;73(1):18–22. doi: 10.1136/sti.73.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamiru M, Hailemariam D, Mitike G, Haidar J. HIV-Related Sexual Behaviors among Migrants and Non-migrants in Rural Ethiopia: Role of Rural to Urban Migration in HIV Transmission. Int J Biomed Sci. 2011;7(4):295–303. [PMC free article] [PubMed] [Google Scholar]
- 7.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346(6205):56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.UNAIDS, editor. UNAIDS. Population Mobility and AIDS: Technical Update. Geneva: 2001. [Google Scholar]
- 9.Junqueira DM, Almeida SE. HIV-1 subtype B: Traces of a pandemic. Virology. 2016;495:173–84. doi: 10.1016/j.virol.2016.05.003. Epub 2016/05/27. [DOI] [PubMed] [Google Scholar]
- 10.Cabello M, Junqueira DM, Bello G. Dissemination of nonpandemic Caribbean HIV-1 subtype B clades in Latin America. Aids. 2015;29(4):483–92. doi: 10.1097/qad.0000000000000552. Epub 2015/01/30. [DOI] [PubMed] [Google Scholar]
- 11.Junqueira DM, de Medeiros RM, Graf T, Almeida SE. Short-Term Dynamic and Local Epidemiological Trends in the South American HIV-1B Epidemic. PLoS ONE. 2016;11(6):e0156712. doi: 10.1371/journal.pone.0156712. Epub 2016/06/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junqueira DM, de Medeiros RM, Matte MC, Araujo LA, Chies JA, Ashton-Prolla P, Almeida SE. Reviewing the history of HIV-1: spread of subtype B in the Americas. PLoS ONE. 2011;6(11):e27489. doi: 10.1371/journal.pone.0027489. Epub 2011/12/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mir D, Cabello M, Romero H, Bello G. Phylodynamics of major HIV-1 subtype B pandemic clades circulating in Latin America. Aids. 2015;29(14):1863–9. doi: 10.1097/qad.0000000000000770. Epub 2015/09/16. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci U S A. 2007;104(47):18566–70. doi: 10.1073/pnas.0705329104. Epub 2007/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czaika M, de Haas H. The Globalization of Migration: Has the World Become More Migratory? International Migration Review. 2014;48(2):283–323. doi: 10.1111/imre.12095. [DOI] [Google Scholar]
- 16.UNAIDS. Global AIDS response progress reporting 2014. 2014 [Google Scholar]
- 17.Valle A, Trevino AC, Zambrano FF, Urriola KE, Sanchez LA, Elizondo JE. Perceived HIV-Associated Stigma among HIV-Seropositive Men: Psychometric Study of HIV Stigma Scale. Frontiers in public health. 2015;3:171. doi: 10.3389/fpubh.2015.00171. Epub 2015/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaux E, Leon M. Homophobic Attitudes and Associated Factors Among Adolescents: A Comparison of Six Latin American Countries. Journal of homosexuality. 2016;63(9):1253–76. doi: 10.1080/00918369.2016.1151697. Epub 2016/02/11. [DOI] [PubMed] [Google Scholar]
- 19.Orozco-Nunez E, Alcalde-Rabanal JE, Ruiz-Larios JA, Sucilla-Perez H, Garcia-Cerde R. [Discrimination and homophobia associated to the human immunodeficiency virus epidemic] Salud publica de Mexico. 2015;57(Suppl 2):s190–6. Epub 2015/11/07. [PubMed] [Google Scholar]
- 20.Verduzco IL. Barriers to Sexual Expression and Safe Sex Among Mexican Gay Men: A Qualitative Approach. American journal of men’s health. 2016;10(4):270–84. doi: 10.1177/1557988314561490. Epub 2014/12/17. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz Hernandez L, Garcia Torres MI. [Internalized oppression and high-risk sexual practices among homosexual and bisexual males, Mexico] Revista de saude publica. 2005;39(6):956–64. doi: 10.1590/s0034-89102005000600014. Epub 2005/12/13. doi: /S0034-89102005000600014. [DOI] [PubMed] [Google Scholar]
- 22.Caceres CF. HIV among gay and other men who have sex with men in Latin America and the Caribbean: a hidden epidemic? AIDS. 2002;16(Suppl 3):S23–33. doi: 10.1097/00002030-200212003-00005. [DOI] [PubMed] [Google Scholar]
- 23.Avila-Rios S, Garcia-Morales C, Garrido-Rodriguez D, Tapia-Trejo D, Giron-Callejas AC, Mendizabal-Burastero R, Escobar-Urias IY, Garcia-Gonzalez BL, Navas-Castillo S, Pinzon-Meza R, Mejia-Villatoro CR, Reyes-Teran G. HIV-1 drug resistance surveillance in antiretroviral treatment-naive individuals from a reference hospital in Guatemala, 2010–2013. AIDS Res Hum Retroviruses. 2015;31(4):401–11. doi: 10.1089/aid.2014.0057. Epub 2014/10/28. [DOI] [PubMed] [Google Scholar]
- 24.Avila-Rios S, Garcia-Morales C, Tapia-Trejo D, Meza RI, Nunez SM, Parham L, Flores NA, Valladares D, Pineda LM, Flores D, Motino R, Umanzor V, Carbajal C, Murillo W, Lorenzana I, Palou EY, Reyes-Teran G. HIV Drug Resistance Surveillance in Honduras after a Decade of Widespread Antiretroviral Therapy. PLoS ONE. 2015;10(11):e0142604. doi: 10.1371/journal.pone.0142604. Epub 2015/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avila-Rios S, Garcia-Morales C, Matias-Florentino M, Tapia-Trejo D, Hernandez-Alvarez BF, Moreira-Lopez SE, Quant-Duran CJ, Porras-Cortes G, Reyes-Teran G. HIV Drug Resistance in Antiretroviral Treatment-Naive Individuals in the Largest Public Hospital in Nicaragua, 2011–2015. PLoS One. 2016;11(10):e0164156. doi: 10.1371/journal.pone.0164156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendoza Y, Bello G, Castillo Mewa J, Martinez AA, Gonzalez C, Garcia-Morales C, Avila-Rios S, Reyes-Teran G, Pascale JM. Molecular epidemiology of HIV-1 in Panama: origin of non-B subtypes in samples collected from 2007 to 2013. PLoS ONE. 2014;9(1):e85153. doi: 10.1371/journal.pone.0085153. Epub 2014/01/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avila-Rios S, Garcia-Morales C, Garrido-Rodriguez D, Ormsby CE, Hernandez-Juan R, Andrade-Villanueva J, Gonzalez-Hernandez LA, Torres-Escobar I, Navarro-Alvarez S, Reyes-Teran G. National prevalence and trends of HIV transmitted drug resistance in Mexico. PLoS ONE. 2011;6(11):e27812. doi: 10.1371/journal.pone.0027812. Epub 2011/11/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avila-Rios S, Garcia-Morales C, Matias-Florentino M, Romero-Mora KA, Tapia-Trejo D, Quiroz-Morales VS, Reyes-Gopar H, Ji H, Sandstrom P, Casillas-Rodriguez J, Sierra-Madero J, Leon-Juarez EA, Valenzuela-Lara M, Magis-Rodriguez C, Uribe-Zuniga P, Reyes-Teran G, Group HM Pretreatment HIV-drug resistance in Mexico and its impact on the effectiveness of first-line antiretroviral therapy: a nationally representative 2015 WHO survey. Lancet HIV. 2016;3(12):e579–e91. doi: 10.1016/S2352-3018(16)30119-9. [DOI] [PubMed] [Google Scholar]
- 29.Woods CK, Brumme CJ, Liu TF, Chui CK, Chu AL, Wynhoven B, Hall TA, Trevino C, Shafer RW, Harrigan PR. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. Journal of clinical microbiology. 2012;50(6):1936–42. doi: 10.1128/JCM.06689-11. Epub 2012/03/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wertheim JO, Leigh Brown AJ, Hepler NL, Mehta SR, Richman DD, Smith DM, Kosakovsky Pond SL. The global transmission network of HIV-1. The Journal of infectious diseases. 2014;209(2):304–13. doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular biology and evolution. 1993;10(3):512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 32.Oster AM, Wertheim JO, Hernandez AL, Bañez Ocfemia MC, Saduvala N, Irene Hall H. Using Molecular HIV Surveillance Data to Understand Transmission between Subpopulations in the United States. J Acquir Immune Defic Syndr. 2015 doi: 10.1097/QAI.0000000000000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little SJ, Kosakovsky Pond SL, Anderson CM, Young JA, Wertheim JO, Mehta SR, May S, Smith DM. Using HIV networks to inform real time prevention interventions. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0098443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wertheim JO, Kosakovsky Pond SL, Forgione LA, Mehta SR, Murrell B, Shah S, Smith DM, Scheffler K, Torian LV. Social and Genetic Networks of HIV-1 Transmission in New York City. PLoS Pathog. 2017;13(1):e1006000. doi: 10.1371/journal.ppat.1006000. Epub 2017/01/10. following competing interests: JOW is a paid consultant for the Centers for Disease Control and Prevention. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheeler WH, Ziebell RA, Zabina H, Pieniazek D, Prejean J, Bodnar UR, Mahle KC, Heneine W, Johnson JA, Hall HI. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S.-2006. Aids. 2010;24(8):1203–12. doi: 10.1097/QAD.0b013e3283388742. Epub 2010/04/17. [DOI] [PubMed] [Google Scholar]
- 36.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25(11):1370–6. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5) doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132(2):583–9. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123(3):603–13. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21(5):676–9. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 42.Bastos FI, Caceres C, Galvao J, Veras MA, Castilho EA. AIDS in Latin America: assessing the current status of the epidemic and the ongoing response. Int J Epidemiol. 2008;37(4):729–37. doi: 10.1093/ije/dyn127. Epub 2008/07/26. [DOI] [PubMed] [Google Scholar]
- 43.Nadai Y, Eyzaguirre LM, Sill A, Cleghorn F, Nolte C, Charurat M, Collado-Chastel S, Jack N, Bartholomew C, Pape JW, Figueroa P, Blattner WA, Carr JK. HIV-1 epidemic in the Caribbean is dominated by subtype B. PLoS ONE. 2009;4(3):e4814. doi: 10.1371/journal.pone.0004814. Epub 2009/03/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagan I, Holguin A. Reconstructing the timing and dispersion routes of HIV-1 subtype B epidemics in the Caribbean and Central America: a phylogenetic story. PLoS ONE. 2013;8(7):e69218. doi: 10.1371/journal.pone.0069218. Epub 2013/07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volz E, Frost SD, Rothenberg R, Meyers LA. Epidemiological bridging by injection drug use drives an early HIV epidemic. Epidemics. 2011;2(3):155–64. doi: 10.1016/j.epidem.2010.06.003. Epub 2011/03/01. doi: S1755-4365(10)00048-4 [pii] 10.1016/j.epidem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Wertheim JO, Oster AM, Hernandez AL, Saduvala N, Banez Ocfemia MC, Hall HI. The International Dimension of the U.S. HIV Transmission Network and Onward Transmission of HIV Recently Imported into the United States. AIDS Res Hum Retroviruses. 2016;32(10–11):1046–53. doi: 10.1089/aid.2015.0272. Epub 2016/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calleja JM, Walker N, Cuchi P, Lazzari S, Ghys PD, Zacarias F. Status of the HIV/AIDS epidemic and methods to monitor it in the Latin America and Caribbean region. Aids. 2002;16(Suppl 3):S3–12. doi: 10.1097/00002030-200212003-00002. Epub 2003/04/11. [DOI] [PubMed] [Google Scholar]
- 48.Mendoza Y, Martinez AA, Castillo Mewa J, Gonzalez C, Garcia-Morales C, Avila-Rios S, Reyes-Teran G, Armien B, Pascale JM, Bello G. Human immunodeficiency virus type 1 (HIV-1) subtype B epidemic in Panama is mainly driven by dissemination of country-specific clades. PLoS ONE. 2014;9(4):e95360. doi: 10.1371/journal.pone.0095360. Epub 2014/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murillo W, Veras N, Prosperi M, de Rivera IL, Paz-Bailey G, Morales-Miranda S, Juarez SI, Yang C, DeVos J, Marin JP, Mild M, Albert J, Salemi M. A single early introduction of HIV-1 subtype B into Central America accounts for most current cases. J Virol. 2013;87(13):7463–70. doi: 10.1128/JVI.01602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobs AJ. The world’s cities : contrasting regional, national, and global perspectives. 1st. New York: Routledge; 2013. p. xix, 397. [Google Scholar]
- 51.Ruiz Y, Guilamo-Ramos V, McCarthy K, Munoz-Laboy MA, de Lourdes Rosas Lopez M. Exploring migratory dynamics on HIV transmission: the case of Mexicans in New York City and Puebla, Mexico. Am J Public Health. 2014;104(6):1036–44. doi: 10.2105/AJPH.2013.301770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carillo H, Fontdevila J, Brown J, Gomez W. Risk Across Borders: Sexual Contexts and HIV Prevention Challenges among Mexican Gay and Bisexual Immigrant Men. 2008 Available from: http://caps.ucsf.edu/uploads/projects/Trayectos/monograph/EnglishFinal.pdf.
- 53.Hue S, Brown AE, Ragonnet-Cronin M, Lycett SJ, Dunn DT, Fearnhill E, Dolling DI, Pozniak A, Pillay D, Delpech VC, Leigh Brown AJ, Resistance UKCoHD, the Collaborative Hiv A-HIVDRN Phylogenetic analyses reveal HIV-1 infections between men misclassified as heterosexual transmissions. AIDS (London, England) 2014;28(13):1967–75. doi: 10.1097/QAD.0000000000000383. [DOI] [PubMed] [Google Scholar]
- 54.Esbjornsson J, Mild M, Audelin A, Fonager J, Skar H, Bruun Jorgensen L, Liitsola K, Bjorkman P, Bratt G, Gisslen M, Sonnerborg A, Nielsen C, Medstrand P, Albert J. HIV-1 transmission between MSM and heterosexuals, and increasing proportions of circulating recombinant forms in the Nordic Countries. Virus Evol. 2016;2(1):vew010. doi: 10.1093/ve/vew010. Epub 2016/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mir D, Cabello M, Romero H, Bello G. Phylodynamics of major HIV-1 subtype B pandemic clades circulating in Latin America. AIDS (London, England) 2015;29(14):1863–9. doi: 10.1097/QAD.0000000000000770. [DOI] [PubMed] [Google Scholar]
- 56.Organización Internacional para las Migraciones (OIM) Perfil Migratorio de Guatemala 2012. Organizatión Internacional para las Migraciones (OIM), 2012 06/2013. Report No. [Google Scholar]
- 57.UNAIDS. Report on the Global AIDS Epidemic. Joint United Nations Programme on HIV/AIDS (UNAIDS) 2008. 2008 [Google Scholar]
- 58.UNAIDS. UNAIDS Global Report 2013. UNAIDS; 2013. [Google Scholar]
- 59.Infante C, Aggleton P, Pridmore P. Forms and determinants of migration and HIV/AIDS-related stigma on the Mexican-Guatemalan border. Qual Health Res. 2009;19(12):1656–68. doi: 10.1177/1049732309353909. [DOI] [PubMed] [Google Scholar]
- 60.MSF. Guatemala: Confronting a growing AIDS Crisis. 2016 Dec 6; [Google Scholar]
- 61.Flannery NP. What do you need to know about central American migration. Forbes [Internet] 2016 2014 December 6. [Google Scholar]
- 62.The Global Fund to Fight AIDS, Tuberculosis, and Malaria. 2013 2013 March 13. Report No. [Google Scholar]
- 63.Romero-Severson E, Skar H, Bulla I, Albert J, Leitner T. Timing and order of transmission events is not directly reflected in a pathogen phylogeny. Mol Biol Evol. 2014;31(9):2472–82. doi: 10.1093/molbev/msu179. Epub 2014/05/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Low N, Egger M, Gorter A, Sandiford P, Gonzalez A, Pauw J, Ferrie J, Smith GD. AIDS in Nicaragua: epidemiological, political, and sociocultural perspectives. Int J Health Serv. 1993;23(4):685–702. doi: 10.2190/1P6N-BPDW-M7BM-P2DR. PubMed PMID: 8276529. [DOI] [PubMed] [Google Scholar]
- 65.Informe año 5. Proyecto Fondo Mundial: Nicaragua, compromiso y acción ante el SIDA, Tuberculosis y Malaria. [Year 5 report. Managua: 2009. (Global Fund Project: Nicaragua, commitment and action in response to AIDS, tuberculosis and malaria]). [Google Scholar]
- 66.Smallman S, editor. War and HIV in Latin America. International & Global Studies Faculty Publications and Presentations; 2009. p. 6-12-2009. [Google Scholar]
- 67.Gandhi AD, Pettifor A, Barrington C, Marshall SW, Behets F, Guardado ME, Farach N, Ardon E, Paz-Bailey G. Migration, Multiple Sexual Partnerships, and Sexual Concurrency in the Garifuna Population of Honduras. AIDS Behav. 2015;19(9):1559–70. doi: 10.1007/s10461-015-1139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalichman HF. Critical Population Spotlight: HIV and the Garifuna of Honduras–Comment on Gandhi et al. “Migration, Multiple Sexual Partnerships, and Sexual Concurrency in the Garifuna Population of Honduras”. AIDS Behav. 2015;19(9):1571–3. doi: 10.1007/s10461-015-1149-0. [DOI] [PubMed] [Google Scholar]
- 69.Kendall T, Herrera C, Caballero M, Campero L. HIV prevention and men who have sex with women and men in Mexico: findings from a qualitative study with HIV-positive men. Culture, health & sexuality. 2007;9(5):459–72. doi: 10.1080/13691050601183629. Epub 2007/08/10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


