Abstract
Non-apoptotic regulated cell death is not fully characterized, particularly for ferroptosis, the iron- and ROS-dependent form of regulated cell death. A systematic approach using modulatory profiling and cell line sensitivity analysis has unraveled the association of lipid metabolism with ferroptosis and enabled the discovery of a novel specific ferroptosis inducer.
Regulated cell death, despite its negative connotation, is essential in many physiological processes, including tissue sculpting during embryogenesis, development of the immune system and destruction of damaged cells1. The dysregulation of cell death has been associated with many pathological processes, such as cancer, neurodegeneration and tissue injury. A well-characterized form of regulated cell death is apoptosis; however, alternative mechanisms such as necroptosis2, autophagy3 and ferroptosis4 have been classified but remain less understood. To gain further insight and dissect the mechanisms governing these alternative forms of cell death, Shimada et al.5 embarked on a small-molecule phenotypic screen for inducers of non-apoptotic cell death. In this issue, the authors identify components of lipid metabolism as novel regulators of ferroptosis and a small molecule, FIN56, that specifically induces ferroptosis with a mechanism that is distinct from that of previously described ferroptosis inducers.
Different forms of regulated cell death mechanisms have distinct morphological and biochemical features, although some crosstalk exists between components of these different processes. Understanding the critical components and mechanisms of cell death regulation and developing approaches to systematically study and compare the different cell death phenotypes would be highly desired. Concurrently, the identification of highly specific chemical probes for distinct cell death mechanisms offers new opportunities to dissect the diverse forms of cell death in physiological and disease contexts and may provide new therapeutic approaches for regulating cell death and survival in human disease. For example, because improper survival and resistance to apoptosis is a known hallmark of cancer development and progression, drugs targeting key components of the apoptotic pathway have been developed6. However, it is increasingly recognized that other forms of regulated cell death can be modulated in order for cancer cells to survive7,8. Thus, the identification of new targets and probes for the alternative forms of regulated cell death will enable further drug development toward a more comprehensive and effective treatment approach to induce cancer cell death.
Ferroptosis is characterized by increased levels of lipid peroxidation products and reactive oxygen species (ROS) derived from iron metabolism9. Previous studies have identified several proteins that directly and indirectly regulate these processes. Specifically, mitochondrial voltage-dependent anion channels (VDAC2 and VDAC3) and NADPH oxidase are known positive regulators of ferroptosis, while glutathione peroxidase 4 (GPX4), the cysteine/glutamate transporter SLC7A11 and heat shock protein β-1 are negative regulators. However, identifying the core regulators of ferroptosis and determining how the cell promotes ferroptosis over other cell death forms still remain major objectives. To address this, Shimada et al. screened 3,169 lethal compounds for induction of cell death independent of caspase activation (caspase activation is characteristic of apoptosis) and found 451 hits. Using a modulatory profiling approach, subsets of compounds were identified that induced three types of regulated non-apoptotic cell death: metal-ion-dependent cell death, necrostatin-1-dependent cell death and ferroptosis. Optimization of one compound using structure–activity relationships led to the identification of FIN56, a novel specific inducer of ferroptosis. FIN56 was found to induce the degradation of glutathione peroxide 4 (GPX4), a lipid repair enzyme that was previously found to negatively affect ferroptosis10. Further dissection of the FIN56 mechanism of action using a chemoproteomics and short interfering RNA (siRNA) screening approach revealed that FIN56 binds and activates squalene synthase (SQS), an enzyme that acts downstream of HMG-CoA reductase in the mevalonate pathway. Activation of SQS downstream in the pathway may suppress non-steroidogenic metabolites such as the coenzyme Q10. However, activation of SQS has no effect on the loss of GPX4 protein levels. An independent activity of FIN56 linked to the loss of GPX4 was evidenced through the activity of acetyl-CoA carboxylase (ACC), an enzyme involved in fatty acid synthesis. Therefore, the mechanism of FIN56-induced ferroptosis involves two distinct pathways in association with the mevalonate pathway and fatty acid synthesis (Fig. 1).
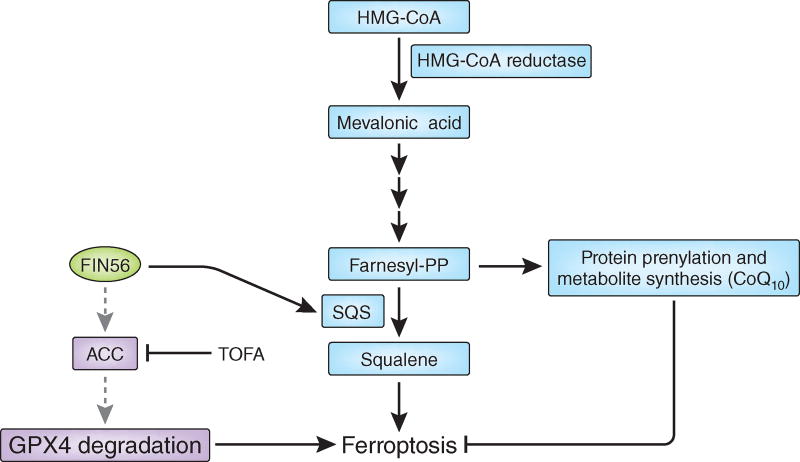
Figure 1.
Induction of ferroptosis by FIN56. FIN56 promotes ferroptosis by two distinct mechanisms: (i) negative regulation of GPX4 protein levels and (ii) activation of squalene synthase (SQS), which acts downstream of HMG-CoA reductase in the mevalonate pathway. In one mechanism (purple boxes), FIN56 promotes degradation of GPX4 in a process that requires acetyl-CoA carboxylase (ACC) activity. Inhibition of ACC by TOFA suppresses the FIN56-mediated degradation of GPX4, yet the link between FIN56, ACC and GPX4 degradation is not clear (gray dashed arrows). In the second mechanism (blue boxes), FIN56 binds and activates SQS, the enzyme that converts farnesyl pyrophosphate (FPP) to squalene, which ultimately reduces the pool of FPP available for protein prenylation and metabolite synthesis, leading to coenzyme Q10 (CoQ10) depletion, for example (black arrows). Inhibition of SQS increases the pool of available FPP and its derived products, suppressing ferroptosis. GPX4, glutathione peroxidase 4; TOFA, 5-(tetradecyloxy)-2-furoic acid; HMG-CoA, 3-hydroxy-3-methylglutaryl-coenzyme A.
The work of Shimada et al.5 exemplifies a systematic approach that allows the dissection of novel cell death phenotypes and the identification of chemical probes, enabling further characterization of the overall mechanisms of ferroptosis. Indeed, the identification of FIN56 and the linking of ferroptosis to lipid metabolism provide new avenues for investigating ferroptosis and its distinct role with respect to other forms of regulated cell death. It will be important, however, to follow up on this work by characterizing the precise mechanisms by which ACC and fatty acid synthesis regulate GXP4 protein levels and how FIN56 regulates this pathway. Moreover, it will be interesting to learn how FIN56 binds and activates SQS so as to enable further chemical optimization of FIN56 for improved selectivity and potency. Elucidating these mechanisms will allow better understanding of ferroptosis and its potential role in biological processes.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Fuchs Y, Steller H. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Degterev A, et al. Nat. Chem. Bio. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Levine B. Cell Death Diff. 2015;22:367–376. doi: 10.1038/cdd.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon SJ, et al. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada K, et al. Nat. Chem. Biol. 2016;12:497–503. doi: 10.1038/nchembio.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Souers AJ, et al. Nat. Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 7.Okada H, Mak TW. Nat. Rev. Cancer. 2004;4:592–603. doi: 10.1038/nrc1412. [DOI] [PubMed] [Google Scholar]
- 8.Vanden Berghe T, et al. Nat. Rev. Mol. Cell Biol. 2014;15:135–147. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 9.Yang WS, Stockwell BR. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang WS, et al. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]