Abstract
Bacteria adapt to adverse environmental conditions by altering gene expression patterns. Recently, a novel stress adaptation mechanism has been described that allows Escherichia coli to alter gene expression at the post-transcriptional level. The key player in this regulatory pathway is the endoribonuclease MazF, the toxin component of the toxin-antitoxin module mazEF that is triggered by various stressful conditions. In general, MazF degrades the majority of transcripts by cleaving at ACA sites, which results in the retardation of bacterial growth. Furthermore, MazF can process a small subset of mRNAs and render them leaderless by removing their ribosome binding site. MazF concomitantly modifies ribosomes, making them selective for the translation of leaderless mRNAs. In this study, we employed fluorescent reporter-systems to investigate mazEF expression during stressful conditions, and to infer consequences of the mRNA processing mediated by MazF on gene expression at the single-cell level. Our results suggest that mazEF transcription is maintained at low levels in single cells encountering adverse conditions, such as antibiotic stress or amino acid starvation. Moreover, using the grcA mRNA as a model for MazF-mediated mRNA processing, we found that MazF activation promotes heterogeneity in the grcA reporter expression, resulting in a subpopulation of cells with increased levels of GrcA reporter protein.
Keywords: Toxin-antitoxin system, mazEF module, Phenotypic heterogeneity, Gene expression, Fluorescent reporter, Flow cytometry
Introduction
Bacteria frequently experience stressful conditions in their natural habitats, such as occurrence of toxins or depletion of nutrients. To cope with continuous changes in their environment, bacteria have evolved protection programs called the bacterial stress response (Hengge, 2011). Studies on the bacterial stress response have been mostly conducted at the level of bulk populations. However, a recent genome-wide screen has shown that genes involved in the stress response display particularly high levels of variation in gene expression, which was measured in clonal populations of Escherichia coli growing in homogeneous environmental conditions (Silander et al., 2012). Many similar studies have initiated a new area of research addressing the benefits of heterogeneous gene expression for bacterial populations. Taken together, these studies provide compelling evidence that phenotypic variation in clonal bacterial populations can be beneficial in the face of environmental fluctuations by granting individual cells a higher probability to survive (Kussell & Leibler, 2005; Acar, Mettetal & Van Oudenaarden, 2008; Arnoldini et al., 2014).
In general, the bacterial stress response is characterized by a profound alteration of the transcriptional program leading to the adaptation to the given conditions (Hengge, 2011). Furthermore, bacteria can adjust to environmental constraints at the post-transcriptional level. One such mechanism has recently been described (Vesper et al., 2011): When E. coli populations encounter stress, a subset of mRNAs is processed upstream of the AUG start codon, resulting in the truncated 5′-untranslated region (UTR) and/or removal of their Shine-Dalgarno sequence (Sauert et al., 2016). In addition, ribosomes are modified and specifically translate these processed mRNAs; the 3′-end of the 16S rRNA is removed from the ribosome, thereby forming specialized stress-ribosomes that lack the anti-Shine-Dalgarno sequence (Vesper et al., 2011). Both mechanisms are mediated by the endoribonuclease MazF, the toxin component of the toxin-antitoxin (TA) system mazEF, which is a type II TA locus (Gerdes, Christensen & Løbner-Olesen, 2005; Gerdes & Maisonneuve, 2012).
TA loci are generally widespread in bacterial and archaeal genomes (Gerdes, Christensen & Løbner-Olesen, 2005; Van Melderen, 2010). Stress-induced toxin activation inhibits basic cellular processes, such as replication, translation and peptidoglycan synthesis. There are at least five different types of TA systems, which are categorized depending on the mode of action of the antitoxin (Goeders & Van Melderen, 2014). For example, the antitoxin MazE of the above mentioned TA system mazEF is a short-lived protein that neutralizes toxin expression and activity by direct binding to the toxin MazF, classifying it as a type II TA system. Besides chromosomally encoded TA systems, plasmid-encoded TA systems are likewise encoded on naturally occurring plasmids, such as the F plasmid or the R1/R100 plasmids (Gerdes, Christensen & Løbner-Olesen, 2005; Van Melderen, 2010; Ramisetty & Santhosh, 2017). These plasmid-encoded TA systems ensure plasmid maintenance in growing bacterial populations by killing those cells that do not inherit the plasmid. Even though the physiological role of plasmid-encoded TA systems is well established (by the mechanism of ‘post-segregational killing’), possible physiological roles of chromosomally encoded TA systems still remain elusive, and so far include growth modulation, persistence after exposure to antibiotics or other stress conditions, regulation of gene expression, and protection against invading plasmids and phages (Gerdes, Christensen & Løbner-Olesen, 2005; Van Melderen, 2010; Gerdes & Maisonneuve, 2012; Ramisetty & Santhosh, 2017).
Several recent studies implicated TA systems in the emergence of phenotypic heterogeneity in bacterial populations, exhibited as variation in gene expression (Maisonneuve, Castro-Camargo & Gerdes, 2013), cell size (Kasari et al., 2010) or growth rate (Klumpp & Hwa, 2014), as well as persister cell formation (Balaban et al., 2004; Maisonneuve et al., 2011). Bacterial persistence is one of the most studied functions of phenotypic heterogeneity that arises in populations of genetically identical cells independently of genetic or environmental differences. However, persister cells represent a minor fraction of a clonal population that can endure various antibiotic treatments (Balaban et al., 2004). It has been shown that MazF overproduction results in a higher number of persister cells upon treatment with different antibiotics (Maisonneuve et al., 2011).
The TA locus mazEF is transcribed into a polycistronic mRNA, where mazE encodes the short-lived antitoxin and mazF encodes for the stable toxin that cleaves single-stranded RNA regions at ACA-sequences in E. coli (Zhang et al., 2003). During periods without stress MazE forms a stable complex with MazF, thereby inhibiting the endoribonucleolytic activity of MazF (Kamada, Hanaoka & Burley, 2003). Moreover, besides MazE alone, the MazE-MazF complex can repress transcription of the operon (Marianovsky et al., 2001). When bacteria encounter stress, such as starvation, DNA damage, heat shock, and treatment with antibiotics, as for instance rifampicin, chloramphenicol and spectinomycin (Sat et al., 2001; Christensen et al., 2003; Hazan, Sat & Engelberg-Kulka, 2004), the Lon (Maisonneuve et al., 2011) and ClpAP proteases (Aizenman, Engelberg-Kulka & Glaser, 1996) degrade the antitoxin MazE, and thus enable MazF to exert its endoribonucleolytic activity.
One typical transcript identified to be processed by MazF within the 5′-UTR is the grcA mRNA (formerly called yfiD) encoding protein GrcA that has been shown to reactivate an oxidatively damaged pyruvate formate-lyase (Wagner et al., 2001). The transcript comprises two ACA sites closely upstream of the start codon, which are cleaved by MazF during stress (Vesper et al., 2011; Sauert et al., 2016). Consequently, the processed transcript is selectively translated by the MazF-modified ribosomes lacking the 3′-terminus of the 16S rRNA (Vesper et al., 2011; Sauert et al., 2016).
Here, we aimed to address the questions: (1) how transcription of the mazEF module differs between single cells in a population, and (2) how the MazF-mediated processing of distinct mRNAs affects protein synthesis at the single-cell level during stress. To this end, we followed the induction of mazEF transcription by different stressors using the chromosomally integrated transcriptional reporter Pmaz-gfp in single bacterial cells, and measured the increase in GFP fluorescence by flow cytometry. Moreover, we employed a chromosomally integrated gfp reporter fusion comprising the transcriptional and translational regulatory regions of the grcA gene. These analyses revealed that MazF-mediated processing of the grcA-gfp mRNA consequently promotes heterogeneity in the grcA reporter gene expression in clonal populations. Taken together, our results suggest that the MazF concentration is kept at low levels during adverse conditions, but can considerably shape the gene expression profile of a cell.
Material and Methods
Bacterial strains and plasmids
We used derivatives of E. coli strain K-12 MG1655 (Blattner et al., 1997; Müller et al., 2016) when addressing effects of antibiotic treatments and amino acid starvation, and derivatives of E. coli strain BW27784 containing a native copy of the mazEF operon (Khlebnikov et al., 2001) for experiments with arabinose-inducible systems. The open reading frame of the employed reporter gene EmgfpΔACA is devoid of ACA sites (Sauert, 2015; Oron-Gottesman et al., 2016), whilst the amino acid sequence corresponds to the wild-type fast-folding Emerald GFP (Tsien, 1998). The chromosomally integrated mCherry reporter (Cox III, Dunlop & Elowitz, 2010) under control of the phage λ promoter PR is used to infer constitutive gene expression. All strains and plasmids are listed in Table S1.
All primers used in this study are listed in File S1, section ‘Sequences’. Briefly, the translational grcAwt reporter was constructed by fusing the fragment of the grcA gene comprising nucleotides −278 to +45 in frame to the gfp coding sequence via a linker encoding three glycine residues. The grcAATA reporter was constructed by fusion-PCR, modifying C to T in the sequence of the grcAwt reporter at positions −1 and −32 upstream of the start codon. Both grcA reporters were cloned into a modified CRIM plasmid pAH68-frt-chlor (kindly provided by T. Bergmiller; Haldimann & Wanner, 2001) via EcoRI/PstI restriction sites. The grcAwt reporter was further cloned into a low copy plasmid pZS*12-GFP (kindly provided by C. Guet) via XhoI/HindIII restriction sites. The transcriptional reporter Pmaz-gfp was cloned into a modified CRIM plasmid pAH120-frt-chlor (kindly provided by T. Bergmiller; Haldimann & Wanner, 2001), and the promoter sequence, i.e., intergenic region between relA and mazE, was flanked with XhoI/BamHI restriction sites. All constructs were inserted into neutral phage attachment sites in the E. coli genome using the CRIM system: the plasmids pAH68-frt-chlor and pAH120-frt-chlor were integrated into attHK022 and attλ, respectively (Haldimann & Wanner, 2001). After integration, the chloramphenicol resistance marker was removed by using the site-specific Flp-recombinase (Cherepanov & Wackernagel, 1995).
Growth conditions
Rich, defined media was composed of 1× M9 salts, 1 mM MgSO4, 0.1 mM CaCl2, 0.5% casamino acids (Fluka), and 10 mM maltose. Clones were first streaked from frozen glycerol stocks on LB (Lennox modification) agar to obtain single colonies. A single colony was inoculated overnight for 15–16 h, in 4 ml of media, at 37 °C with shaking at 165 rpm. Bacterial growth was monitored by measuring the optical density at 600 nm (OD600) with a spectrophotometer. The overnight cultures were diluted 1 to 1,000 into fresh media, OD600 ∼ 0.007, and grown for 2 h 15 min. The exponential cultures were then split in two or more flasks; with one flask serving as a control, while different stressors (or arabinose to induce mazF expression) were added to other flasks. After 3–4.5 h of stress, 4 ml of stressed cultures were washed with 1× PBS, resuspended in 4 ml of pre-warmed fresh media and allowed to regrowth. In general, the cultures were analyzed by flow cytometry at three time points: (t1) 2–3 h after stress induction; (t2) 5.5–7 h after stress induction, or recovered for 1.5–2.5 h after initial 3–4.5 h under stress; (t3) 21 h after stress induction, or recovered for 15.5–18 h after initial 3–4.5 h under stress.
Flow cytometry
Flow cytometers
Analysis was performed with FACS Calibur for GFP reporter strains and LSR Fortessa for mCherry-GFP systems (BD, San Jose, California, USA). Calibur is equipped with an argon laser with excitation at 488 nm. In total 100,000 or 150,000 events were acquired for each sample at low speed, with the following settings –FSC-H (forward scatter): E01, SSC-H (side scatter): 349 V, FL1-H: 813 V; log mode; primary threshold on SSC. Fortessa is equipped with lasers with excitation at 488 nm and 561 nm, and filters for detection of GFP fluorescence (BP filter 530/30 nm) and mCherry fluorescence (BP filter 610/20 nm). For each sample 50,000 events were acquired at low speed, with the following settings –FSC-H (forward scatter): 380 V, SSC-H (side scatter): 220 V, FITC-H: 430 V, PE-Texas Red-H: 590 V; log mode; compensation FITC-PE Texas Red 0.2; threshold: SSC at 200 V and FSC at 200 V.
Data analysis and visualization
Raw fcs files were exported from FlowJo (Tree Star, Inc.) into a custom R script, similar to the script published as Dataset S2 in Silander et al. (2012), with R commands for gating indicated in File S1, section ‘R commands’. Flow cytometry plots were made in FlowJo. All events were analyzed for the experiments, unless indicated otherwise. For the experiment presented in Fig. 1, in total 10,000 cells were analyzed with R script after filtering (gating) events on the basis of FSC and SSC measurements (size parameters), and visualized by using auto-gating tool on the size parameters in FlowJo.
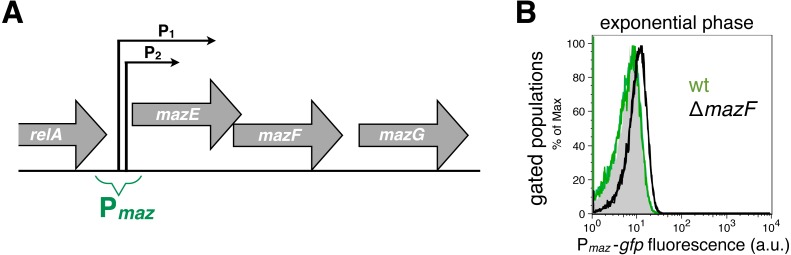
Figure 1. Strong and weak repression of the mazEF transcription.
(A) A scheme of the mazEFG operon (Keseler et al., 2013). The transcriptional reporter Pmaz-gfp includes intergenic region between the relA and mazE genes together with promoters Pmaz1 (P1) and Pmaz2 (P2), initially named P2 and P3, respectively. The promoters are located 13 nt apart, and the upstream promoter is 10 times more active than the downstream promoter (Marianovsky et al., 2001). Details of the reporter construction are given in File S1. (B) We used the reporter Pmaz-gfp to quantify weak repression of the mazEF transcription by comparing the GFP signal in the ΔmazF strain (black histogram, strain NN229) and the isogenic wild-type strain (green histogram; strain NN227), measured in the exponential phase. The GFP fluorescence level of the wild-type strain NN227 was comparable to the fluorescence of the reporterless MG1655 strain (grey shaded histogram), thus corresponding to bacterial autofluorescence (see File S1 for Source Data).
Cytometry setup and analysis for Fig. 2 was as follows. The cultures were analyzed after approximately 3 h (time point t1), 7 h (time point t2) and 23 h (time point t3). We used concentrations of stressors above their minimum inhibitory concentration (MIC), referred to as ‘harsh’ conditions (Table 1), and below MIC referred to as ‘mild’ conditions (Table 2) (Tosa & Pizer, 1971; Sulavik et al., 2001; Chaudhuri et al., 2010): ampicillin (Amp) 100 µg/ml and 1 µg/ml, chloramphenicol (Cam) 15 µg/ml and 1 µg/ml, nalidixic acid (NA) 50 µg/ml and 1 µg/ml, spectinomycin (Spec) 90 µg/ml and 1 µg/ml, and serine hydroxamate (SHX, Sigma) 100 µg/ml and 1 µg/ml, as ‘harsh’ and ‘mild’ conditions, respectively.
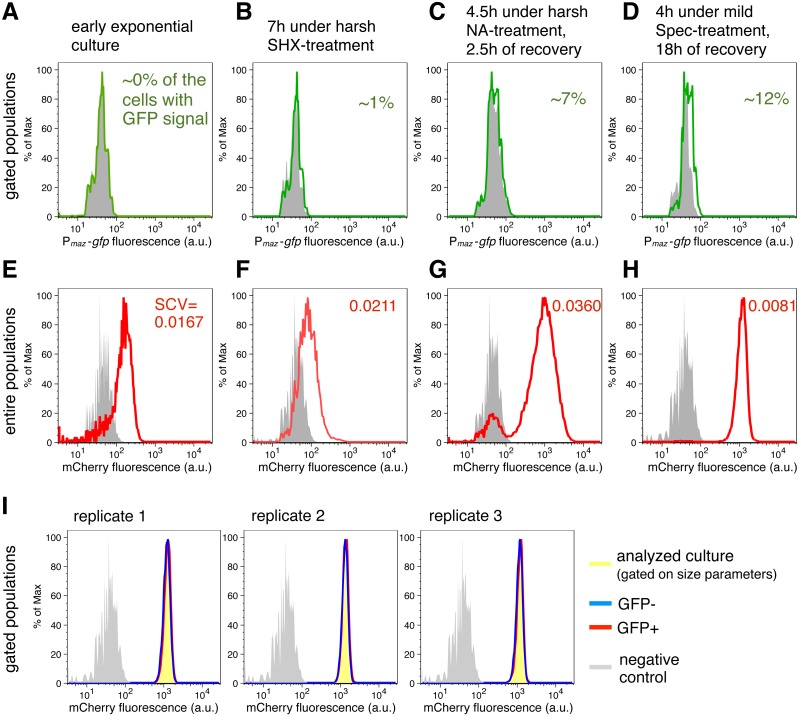
Figure 2. Transcriptional de-repression and activation of the mazEF module, and constitutive gene expression under different conditions.
(A–D) GFP fluorescence of strain NN230 harboring the Pmaz-gfp reporter is depicted in green; background GFP fluorescence of strain TB205 is depicted in grey. We measured the GFP signal in the cells gated on the size parameters and mCherry signal, to exclude filaments, dead cells and other debris that can influence the calculations. Analysis of the expression of the Pmaz-gfp reporter and the constitutive mCherry reporter upon amino acid starvation induced with serine hydroxamate, as well as upon antibiotic treatments—namely ampicillin, chloramphenicol, nalidixic acid, spectinomycin—are reported in Table 1 for ‘harsh’ conditions and in Table 2 for ‘mild’ conditions. (E–H) mCherry fluorescence of strain NN230 harboring the λPR-mCherry reporter is depicted in red; background mCherry fluorescence of strain MG1655 is depicted in grey. To infer population heterogeneity, we analyzed squared coefficient of variation (SCV; defined as squared standard deviation divided by the squared mean) in the mCherry fluorescence of the entire populations, without gating. (I) We first gated populations based on the size parameters, to minimize influence of cell size on the level of GFP fluorescence. Then, we analyzed the constitutive mCherry fluorescence in the fraction of cells without (GFP−) or with (GFP+) GFP signal of strain NN230, recovered after mild Spec-treatment. The same GFP threshold was applied in all replicates gated on approximately 20,000 cells by using FlowJo, and then two fractions per replicate, GFP- and GFP+ fraction, were analyzed in R. There is no correlation between GFP signal and mCherry signal in the analyzed replicates. For replicate 1: Spearman’s rho = 0.104, p = 5.7E − 50, N = 20,440 events; for replicate 2: rho = 0.088, p = 2.0E − 36, N = 20,546 events; for replicate 3: rho = 0.072, p = 2.5E − 25, N = 20,542 events.
Table 1. Expression of the Pmaz-gfp reporter in the presence of stressors at concentrations above the MIC levels (‘harsh’ stress conditions).
Percentage of the cells that express the transcriptional reporter Pmaz-gfp above background level, measured before, during and after experiencing stress.
| Time point | Percentage of the cells with GFP signala (gated populations) | SCV of log mCherry fluorescence (a.u.)a (entire populations) | ∼OD | ||
|---|---|---|---|---|---|
| t0 | Exponential | ∼0 | 0.0140 ± 0.001 | 0.23 | |
| t1 | 5 h of growth | Control | ∼0 | 0.0156 ± 0.002 | 2.10 |
| 3 h under stress | cAmp, 100 µg/ml | ∼0 | 0.0327 ± 0.002 | 0.07 | |
| Cam, 15 µg/ml | ∼0 | 0.0176 ± 0.002 | 0.35 | ||
| NA, 50 µg/ml | ∼0 | 0.0273 ± 0.005 | 0.68 | ||
| SHX, 100 µg/ml | 0.78 ± 0.56 | 0.0157 ± 0.001 | 0.56 | ||
| Spec, 90 µg/ml | 1.05 ± 0.35 | 0.0142 ± 0.002 | 0.42 | ||
| t2 | 9 h of growth | Control | 0.39 ± 0.07 | 0.0210 ± 0.002 | 7.16 |
| 7 h under stress | Amp, 100 µg/ml | ∼0 | 0.0265 ± 0.001 | 0.07 | |
| Cam, 15 µg/ml | 0.18 ± 0.11 | 0.0227 ± 0.004 | 0.56 | ||
| NA, 50 µg/ml | ∼0 | 0.0315 ± 0.008 | 0.77 | ||
| SHX, 100 µg/ml | b1.01 ± 0.19 | 0.0211 ± 0.004 | 3.00 | ||
| Spec, 90 µg/ml | 2.35 ± 0.46 | 0.0110 ± 0.002 | 0.62 | ||
| 4.5 h under stress, | Regrowth, Amp-treated | 0.02 ± 0.01 | 0.0300 ± 0.001 | 0.23 | |
| then 2.5 h of regrowth | Regrowth, Cam-treated | 5.41 ± 4.32 | 0.0148 ± 0.001 | 1.40 | |
| Regrowth, NA-treated | 7.01 ± 2.94 | 0.0360 ± 0.000 | 0.77 | ||
| Regrowth, SHX-treated | 0.61 ± 0.05 | 0.0165 ± 0.001 | 4.21 | ||
| Regrowth, Spec-treated | 1.31 ± 0.94 | 0.0171 ± 0.002 | 1.34 | ||
| t3 | 25 h of growth | Control | 1.13 ± 0.02 | 0.0075 ± 0.001 | 7.27 |
| 4.5 h under stress, | Regrowth, Amp-treated | 1.99 ± 0.38 | 0.0123 ± 0.001 | 3.85 | |
| then 15.5 h of regrowth | Regrowth, Cam-treated | 4.56 ± 0.38 | 0.0113 ± 0.001 | 7.81 | |
| Regrowth, NA-treated | 2.58 ± 0.75 | 0.0167 ± 0.001 | 6.71 | ||
| Regrowth, SHX-treated | 4.04 ± 0.32 | 0.0147 ± 0.004 | 6.68 | ||
| Regrowth, Spec-treated | 5.65 ± 0.92 | 0.0091 ± 0.001 | 8.62 | ||
Notes.
(mean ± standard error of the mean).
Values in bold font are the examples depicted in Fig. 2.
Amp, ampicillin; Cam, chloramphenicol; NA, nalidixic acid; SHX, serine hydroxamate; Spec, spectinomycin.
Table 2. Expression of the Pmaz-gfp reporter in the presence of stressors at concentrations below MIC levels (‘mild’ stress conditions).
Percentage of the cells that express the transcriptional reporter Pmaz-gfp above background level, measured before, during and after experiencing mild stress conditions.
| Time point | Percentage of the cells with GFP signala (gated populations) | SCV of log mCherry fluorescence (a.u.)a (entire populations) | ∼OD | ||
|---|---|---|---|---|---|
| t0 | Exponential | b∼0 | 0.0167 ± 0.002 | 0.22 | |
| t1 | 5 h of growth | Control | ∼0 | 0.0157 ± 0.000 | 1.34 |
| 3 h under stress | cAmp, 1 µg/ml | 3.13 ± 2.21 | 0.0159 ± 0.000 | 1.25 | |
| Cam, 1 µg/ml | 0.95 ± 0.34 | 0.0163 ± 0.003 | 0.64 | ||
| NA, 1 µg/ml | 2.96 ± 1.26 | 0.0113 ± 0.002 | 1.15 | ||
| SHX, 5 µg/ml | 2.36 ± 1.58 | 0.0173 ± 0.000 | 1.17 | ||
| Spec, 1 µg/ml | 2.01 ± 1.14 | 0.0125 ± 0.002 | 1.08 | ||
| t2 | 9 h of growth | Control | 1.02 ± 0.46 | 0.0211 ± 0.003 | 5.93 |
| 7 h under stress | Amp, 1 µg/ml | 1.98 ± 0.51 | 0.0162 ± 0.000 | 5.67 | |
| Cam, 1 µg/ml | 0.29 ± 0.13 | 0.0188 ± 0.000 | 3.00 | ||
| NA, 1 µg/ml | 1.22 ± 0.18 | 0.0185 ± 0.001 | 4.72 | ||
| SHX, 5 µg/ml | 1.14 ± 0.29 | 0.0173 ± 0.000 | 5.13 | ||
| Spec, 1 µg/ml | 1.65 ± 0.57 | 0.0173 ± 0.001 | 4.17 | ||
| 4 h under stress, | Regrowth, Amp-treated | 1.92 ± 0.08 | 0.0117 ± 0.001 | 5.88 | |
| then 2.5 h of regrowth | Regrowth, Cam-treated | 0.16 ± 0.06 | 0.0182 ± 0.000 | 3.48 | |
| Regrowth, NA-treated | 0.90 ± 0.04 | 0.0155 ± 0.000 | 5.34 | ||
| Regrowth, SHX-treated | 1.84 ± 0.37 | 0.0111 ± 0.001 | 7.33 | ||
| Regrowth, Spec-treated | 0.96 ± 0.18 | 0.0160 ± 0.000 | 5.37 | ||
| t3 | 25 h of growth | Control | 4.94 ± 1.77 | 0.0088 ± 0.001 | 7.83 |
| 4 h under stress, | Regrowth, Amp-treated | 3.90 ± 0.12 | 0.0072 ± 0.000 | 9.45 | |
| then 18 h of regrowth | Regrowth, Cam-treated | 2.18 ± 0.12 | 0.0118 ± 0.002 | 7.99 | |
| Regrowth, NA-treated | 11.69 ± 0.80 | 0.0094 ± 0.001 | 9.24 | ||
| Regrowth, SHX-treated | 2.95 ± 0.24 | 0.0163 ± 0.006 | 9.99 | ||
| Regrowth, Spec-treated | 12.04 ± 1.96 | 0.0081 ± 0.001 | 9.51 | ||
Notes.
(mean ± standard error of the mean).
Values in bold font are the examples depicted in Fig. 2.
Amp, ampicillin; Cam, chloramphenicol; NA, nalidixic acid; SHX, serine hydroxamate; Spec, spectinomycin.
In total, 50,000 events were acquired by the flow cytometer Fortessa for strain NN230. Minimally 20,000 events were processed per each sample, by filtering on the basis of FSC and SSC measurements (size parameters). The gating captured cells in the densest part of the FSC-SSC plot and therefore the gates consisted of cells with similar size and physiology due to the narrow range of FSC and SSC, respectively. Such analysis allows one to minimize the variation in fluorescence due to different cell size or cell damage, differences in stage of cell cycle, etc. Of those gated events, we only considered events that contained a mCherry signal above background level. The mCherry background level is defined as the 99th percentile of the measurements for the strain that does not contain the mCherry fluorescent reporter gene, which corresponds to autofluorescence of the MG1655 strain. This was one way how we could determine that the cells were alive or at least their membranes were not damaged during stress. For instance, cells damaged or lysed due to Amp-treatment could still be detected by flow cytometry within defined FSC and SSC parameters, but a constitutive mCherry signal would not be detected, and therefore we do not include those dead cells into further analysis.
The GFP background level is defined as the 99th percentile of measurements for the strain that does not contain the gfp fluorescent reporter gene (which corresponds to autofluorescence of the TB205 strain; Bergmiller et al., 2017), averaged over all conditions. These subsets were used for the subsequent analysis to calculate the number of cells that expressed gfp above background level. Finally, the number of GFP-positive cells was corrected with the number of cells with GFP fluorescence above background level measured in reporterless strains in the same condition, to eliminate false-positive GFP signals.
Microscopy and image analysis
An exponentially growing culture of strain NN207 was divided into two flasks, and 0.2% Ara was added to one flask to induce mazF expression. Samples were taken from untreated cultures and cultures treated for 3 h and 22 h with Ara. The cells were fixed with 1% formaldehyde in 1× PBS, incubated for 2 h in the dark at room temperature using a nutator, followed by two washing steps with 1× PBS. The samples were kept in dark at 4 °C until microscopy the following day. 1.2 µl of a fixed bacterial sample was applied on a 1.5% agarose pad using a cavity slide as previously described (Bergmiller et al., 2011). Images were acquired with Olympus IX-81 inverse widefield epifluorescence microscope using objective UPlanSApo 100×/1.4 Oil, and Hamamatsu ORCA-ER detection system. The lamp intensity was set on 100%; exposure time of 130 ms for phase contrast images; and exposure time of 200 ms (filter TexasRed U-MWG, Ex. 510–550 nm, Em. LP 590 nm) for mCherry images. Cells were analyzed with the Matlab-based package Schnitzcells (Young et al., 2011), and segmented using mCherry fluorescence images.
Statistical analysis
Statistics was done in SPSS, R and Microsoft Excel. Error bars in all graphs depict standard deviation from N replicates (independent cultures); N is indicated in every Figure Legend. We log10-transformed fluorescence data prior to analysis. The squared coefficient of variation (SCV) was used as a measure of variation in the reporter fluorescence. SCV is the squared value of coefficient of variation (CV), and CV is defined as standard deviation divided by the mean. We computed Pearson’s second skewness coefficient (median skewness) as skewness = 3*(mean − median)/standard deviation; and kurtosis type 2 in R. We used 2-tailed, heteroscedastic Student’s t-test to infer differences between two datasets for each measurement: between the fluorescence values of the grcAwt and grcAATA reporters, and between the fluorescence values of the strain harboring Pmaz-gfp reporter (NN230) and the strain without gfp reporter gene (TB205). We used One-way ANOVA with Post Hoc Test Bonferroni to infer differences in four datasets concomitantly (fluorescence measurements of strains NN200, NN211, NN208, NN221). We used non-parametric Spearman’s two-tailed test to assess correlations between mCherry and GFP signals.
Results
Monitoring repression and de-repression of the mazEF transcription using the transcriptional reporter Pmaz-gfp
Expression of the mazEF module is negatively autoregulated, and the mazEF transcription from promoters Pmaz1 and Pmaz2 is strongly repressed under non-stressful conditions by the MazE protein or the MazE-MazF complex (Marianovsky et al., 2001; Keseler et al., 2013). However, repression by the MazE antitoxin alone is reduced by approximately 40% when compared to the MazE-MazF complex, as determined in the E. coli strain MC4100 (Marianovsky et al., 2001). In contrast, stressful conditions promote degradation of the MazE antitoxin (Aizenman, Engelberg-Kulka & Glaser, 1996; Maisonneuve et al., 2011), which subsequently results in de-repression of the mazEF transcription. Thus, the state of mazEF transcription depends on the ratio between toxin T and antitoxin A within a cell: weak repression (when T ≪ A), repression (when T ≈ A), or de-repression (when T ≫ A). This complex mechanism of transcriptional autoregulation of TA systems is generally termed ‘conditional cooperativity’ (Cataudella et al., 2013; Gelens et al., 2013).
Here, we used the transcriptional reporter Pmaz-gfp to indirectly monitor the regulation of the mazEF transcription in the E. coli strain MG1655 via GFP fluorescence (Fig. 1A). As a proof-of-principle for using this fluorescent reporter to infer the conditional cooperativity mechanism, we first quantified the activity of the Pmaz promoters in the presence and absence of the MazF toxin. To this end, we calculated the difference of the mean levels of the GFP signal between the strain MG1655 ΔmazF and the isogenic wild-type strain, divided by the signal obtained with the wild-type strain (see File S1 for Source Data). Our data show that in the early exponential phase the mean GFP fluorescence level is increased by 60% in the ΔmazF strain relative to the wild-type strain (Fig. 1B), what can be attributed to the weak repression of transcription in the absence of MazF, i.e., when [MazF] ≪ [MazE].
During and after exposure to antibiotics or amino acid starvation mazEF is transcribed at low levels
Next, we aimed to quantify mazF expression levels during adverse conditions that were reported to activate MazF using the transcriptional reporter Pmaz-gfp. Concomitantly, we determined the degree of cell-to-cell heterogeneity under these conditions using the constitutive mCherry reporter (Fig. 2). We measured the GFP fluorescence by flow cytometry upon treatment with ‘mild’ (below MIC values) and ‘harsh’ (above MIC values) concentrations of ampicillin (Amp), chloramphenicol (Cam), nalidixic acid (NA), or spectinomycin (Spec), as well as upon mimicking amino acid starvation by adding serine hydroxamate (SHX) (Table 1 for ‘harsh’ and Table 2 for ‘mild’ stress). To calculate the percentage of cells that display GFP signals above background level, we first gated the populations based on the size parameters, and second, based on a constitutively expressed mCherry. Such gating allowed us to analyze live cells with similar size, as described in details in ‘Material and Methods’.
Our analysis using the transcriptional reporter Pmaz-gfp revealed that GFP fluorescence was not detectable in the exponential phase (Fig. 2A). However, during SHX-treatment (Fig. 2B) and antibiotic treatments (Tables 1 and 2) the obtained GFP signals exceeded background levels. GFP fluorescence was generally lower during ‘harsh’ than during ‘mild’ stress because the high concentrations of the stressors either killed a fraction of the stressed populations or severely inhibited growth (OD600 values are given in Tables 1 and 2). Severe growth retardation implies reduction in overall translational capacity of cells, including decreased GFP synthesis. However, increased GFP signals were measured in all recovered cultures after removal of the stressor (Fig. 2C) when translation of the reporter gfp mRNA was no longer inhibited by the stressor. The maximal GFP signal was observed during recovery after treatment with 1 µg/ml Spec (Fig. 2D), i.e., about 12% of the cells expressed gfp above background level. We also included an additional statistical test indicating significantly higher mean levels of GFP fluorescence for the strain harboring Pmaz-gfp reporter compared to the strain without gfp reporter gene (t-test, p < 0.05), during stress and in the recovery periods (all p-values in File S1). These results suggest that during antibiotic treatment and amino acid starvation, and following stress removal, de-repression of the mazEF transcription occurs at low levels. Taken together, transcription and therefore de novo production of MazF is maintained at low levels during stress.
Besides determining the GFP fluorescence, we concomitantly monitored the expression of the reporter gene λPR-mCherry to infer constitutive expression (Fig. 2E), as the λ promoter PR is specifically regulated only in the presence of the phage λ proteins (Berthoumieux et al., 2013). Given that expression of bacterial genes is regulated both through specific factors, such as transcription factors and post-transcriptional regulators, and global factors, such as the number of available RNA polymerases and ribosomes (Berthoumieux et al., 2013; Gerosa et al., 2013), the constitutive mCherry expression, which is solely affected by global factors, provides an estimate for the influence of the growth rate on gene expression in single cells (Klumpp, Zhang & Hwa, 2009; Scott et al., 2010; Klumpp & Hwa, 2014) (N Nikolic et al., 2017, unpublished data). Moreover, mCherry is a highly stable protein, thus the amount of mCherry fluorescence reflects mCherry production and dilution through cell division. As growth is impeded during adverse conditions (and mCherry is not diluted through cell division), and some stressful conditions additionally impair translation, these global factors would collectively affect the mCherry fluorescence signal.
Measurements of the entire bacterial populations, without gating, showed a high degree of variation in the constitutive mCherry expression during stressful conditions (Figs. 2F and 2G), which can signify heterogeneity in cell size, viability, and growth rate. The variation in mCherry fluorescence was highest during and after treatments with high concentrations of nalidixic acid (Fig. 2G), or ampicillin (Table 1), suggesting that cell lysis and severe growth retardation increase population heterogeneity. In contrast, the variation in mCherry fluorescence was lowest in populations experiencing longer recovery periods, i.e., in cultures initially stressed and analyzed after overnight recovery (Fig. 2H, Tables 1 and 2).
Interestingly, we found no correlation between the GFP and mCherry signals in the analyzed populations after mild Spec-treatment (gated only based on size parameters, Fig. 2I), and the same trend is also found in other populations initially stressed and analyzed after overnight recovery (see File S1 for Source Data). If we assume that constitutive mCherry fluorescence is an indicator of growth (Leveau & Lindow, 2001; Klumpp, Zhang & Hwa, 2009; Scott et al., 2010; Klumpp & Hwa, 2014) (N Nikolic et al., 2017, unpublished data), and GFP fluorescence is an indicator of the mazEF transcription, this lack of correlation could imply that de-repression and activation of the mazEF transcription do not depend on the cellular growth rate. This result is in line with previous population-based studies that have shown that the expression of an unregulated gene is growth-rate dependent, whereas the expression of a negatively autoregulated gene is a growth-rate independent process (Klumpp, Zhang & Hwa, 2009; Scott et al., 2010).
MazF-mediated mRNA processing monitored by gfp reporter gene fusions
The increasing evidence that TA systems might introduce phenotypic heterogeneity in genetically identical populations (Gerdes & Maisonneuve, 2012; Klumpp & Hwa, 2014) prompted us to study the impact of the MazF-mediated processing on the translation of the respective mRNAs (Vesper et al., 2011; Sauert et al., 2016). To this end, we selected the grcA transcript, which has been shown to be cleaved by MazF at two ACA sites closely upstream of the start codon (Vesper et al., 2011; Sauert et al., 2016). The toxin MazF can thus modify the grcA mRNA by keeping a short leader sequence or making the mRNA leaderless, which allows its selective translation by stress-ribosomes (Vesper et al., 2011). Hence, the grcA mRNA represents a bona fide model to investigate the alteration of gene expression by MazF-mediated mRNA processing.
To follow the grcA expression pattern at the single-cell level, we constructed a fluorescent reporter gene comprising the 5′-UTR and the first 15 codons of grcA fused in frame to the gfp coding sequence, under control of the native PgrcA promoters (Fig. 3A). This wild-type reporter grcAwt was used as a proxy for the impact of MazF-mediated mRNA processing on gene expression. As a control, we generated the grcAATA fluorescent reporter by substituting the ACA sequence motifs by ATA, to prevent MazF-mediated processing. In both reporters, we used an Emerald gfp version that does not contain ACA sites in the open reading frame (EmgfpΔACA) (Sauert, 2015; Oron-Gottesman et al., 2016), while the amino acid sequence remains the same as in the wild-type Emerald GFP (Tsien, 1998).
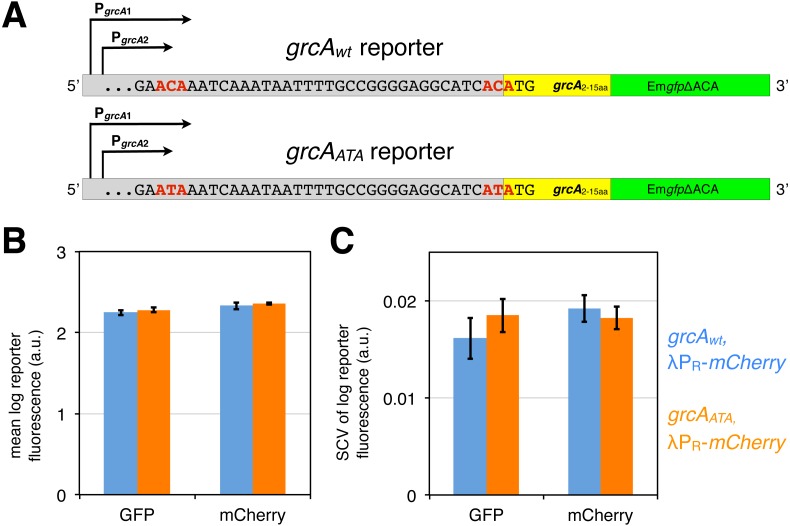
Figure 3. grcA reporters constructed to infer MazF-mediated mRNA processing.
(A) The grcAwt reporter comprises 278 nt upstream of the start codon of grcA, as well as the first 15 codons fused in frame to EmgfpΔACA, which is a gfp variant devoid of ACA sites in the open reading frame (Sauert, 2015; Oron-Gottesman et al., 2016). The grcAATA reporter is a modified grcAwt reporter harboring two nucleotide substitutions (C to T) at the positions previously determined to be processed by MazF (Vesper et al., 2011). (B) GFP and mCherry signals do not significantly differ between the grcAwt and the grcAATA reporter measured in the exponential phase. Strains NN207 (depicted in blue) and NN214 (depicted in orange) harbor the grcAwt and the grcAATA reporter, respectively, together with the constitutive λPR-mCherry reporter. (C) The differences are neither significant for the variation in GFP and mCherry fluorescence. Variation is defined as the squared coefficient of variation, SCV. We analyzed N = 5 independent replicates.
First, we confirmed by flow cytometry that both, the grcAwt and the grcAATA reporters display similar fluorescence signals during exponential growth without stressors, which would indicate that there are no significant differences in translation of the reporter mRNAs. Indeed, we observed no significant difference in the mean GFP fluorescence level between the grcAwt and the grcAATA reporter (Fig. 3B, Fig. S1). Likewise, there was no difference in the variation in GFP fluorescence, measured as squared coefficient of variation (SCV, Fig. 3C). Alongside, we measured constitutive mCherry expression from the reporter λPR-mCherry. Correspondingly, we observed no significant differences in the mean level and variation in the mCherry fluorescence in strains harboring the different grcA reporters (Figs. 3B and 3C).
Heterogeneity in the grcAwt reporter fluorescence during recovery from mazF expression
Next, we aimed to investigate differences in fluorescence between the grcAwt and grcAATA reporters in conditions that promote MazF activation. Because our fluorescent system is based on the GFP translation and subsequent fluorescence, and most of the known inducers of MazF activation are translational inhibitors (Sat et al., 2001; Christensen et al., 2003; Hazan, Sat & Engelberg-Kulka, 2004), we opted for an experimental setup in which mazF expression is tightly controlled by a synthetic promoter. Thus, we employed a chromosomally integrated system to express mazF under transcriptional control of the arabinose-inducible promoter PBAD in order to investigate direct effects of MazF activation on translation of reporter mRNAs (sequence PBAD-mazF taken from the plasmid-based system pBAD-mazF (Amitai, Yassin & Engelberg-Kulka, 2004) (N Nikolic et al., 2017, unpublished data)). In this setup, mazF expression was ectopically induced at low levels (Fig. S2), as our results suggested that MazF is transcribed at low levels during antibiotic treatments or amino acid starvation (Tables 1 and 2).
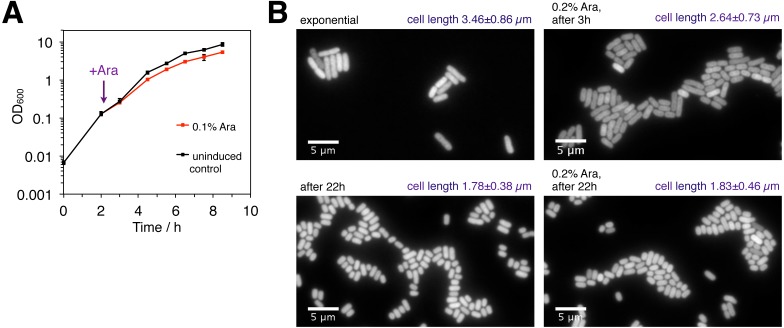
The analysis was performed using derivatives of the E. coli strain BW27784 (Khlebnikov et al., 2001). Strain BW27784 is suitable for arabinose-inducible expression systems such as PBAD-mazF because it constitutively takes up L-arabinose but is devoid of L-arabinose metabolism (Fig. S2). mazF expression was ectopically induced by addition of 0.1% arabinose (Ara) to exponentially growing cultures. It is important to note that bacterial growth did not cease under these conditions. This low level of ectopic mazF expression led to a 20% reduction in the bacterial growth rate and a decrease in the final optical density (OD600) by 36% (Fig. 4A). In contrast to previous observations that ectopic toxin overexpression causes cell elongation (Kasari et al., 2010), the mild ectopic mazF expression used here did not result in the formation of filamentous cells (Fig. 4B; all values in File S1).
Figure 4. Effect of the mild MazF stress on bacterial growth and cell length.
(A) Exponentially growing cultures of strain NN204 were divided into two flasks: 0.1% Ara was added to the first flask to ectopically induce mazF expression from the chromosomally integrated PBAD-mazF system, and the second flask served as a control. The maximum bacterial growth rate was measured as the slope of ln-transformed OD600 data measured at t1 = 3 h and t2 = 4.5 h, and the final OD600 was measured at t = 8.5 h. Here depicted are average values of N = 3 independent replicates; error bars present standard deviation. (B) An exponentially growing culture harboring the λPR-mCherry reporter and the chromosomally integrated system PBAD-mazF (strain NN207) was split in two flasks; 0.2% Ara was added to one flask, and the second flask served as the uninduced control. The samples were taken and cells were fixed after 3 h and 22 h. mCherry images were acquired with an epifluorescence microscope to infer variation in the cell length, indicated as mean ± standard deviation.
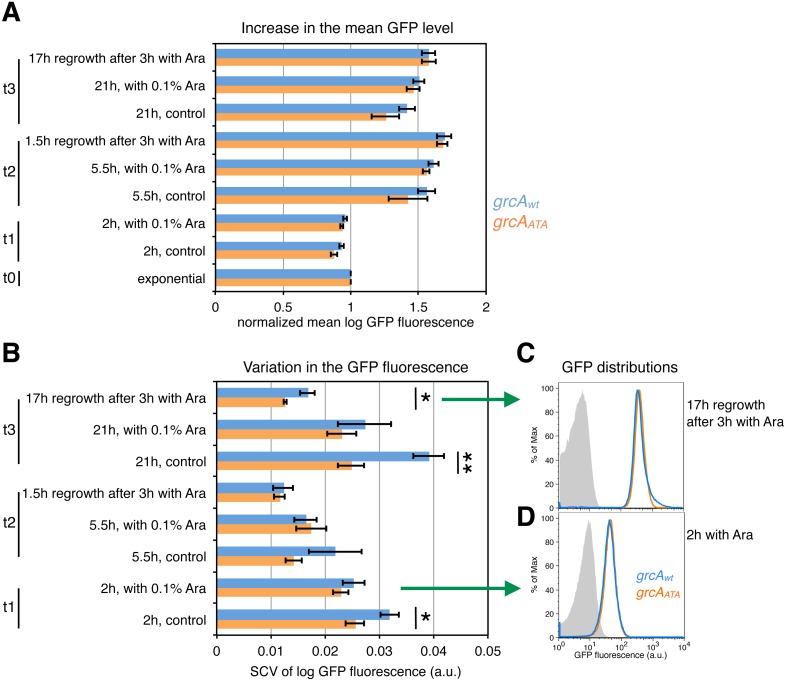
After arabinose induction of mazF expression and during the recovery period after the removal of the inducer, we analyzed the increase in the mean level of GFP fluorescence encoded by the grcAwt and the grcAATA reporters. We calculated normalized mean GFP fluorescence as the mean GFP level of each sample divided by the mean GFP level of the respective culture measured in the exponential phase. Despite the normalized mean levels of GFP fluorescence were similar between the grcA reporters (Fig. 5A), there were significant differences in variation in the GFP fluorescence (Fig. 5B). Interestingly, variation in the grcAwt reporter fluorescence was higher than the grcAATA reporter fluorescence 17 h after recovery from 3 h of ectopic mazF expression. Likewise, we observed increased variation when cultures experienced nutrient deprivation during the overnight growth (stationary phase), without addition of the inducer (‘control’, time point t3 in Fig. 5B). Moreover, variation in the grcAwt reporter fluorescence after overnight growth was significantly higher in the wild-type strain MG1655 containing native mazEF operon compared to the ΔmazF background (Fig. S3). These results suggest that conditions that promote MazF activation increase heterogeneity in the translation of the reporter mRNA that is processed by MazF, compared to the reporter mRNA that is not affected by MazF cleavage.
Figure 5. Effect of ectopic mazF expression on translation of the grcAwt and grcAATA reporters.
Fluorescence was measured at different time points: during exponential phase before mazF expression (t0), and 2 h (t1), 5.5 h (t2), and 21 h (t3) without or with addition of 0.1% Ara to ectopically induce mazF expression. In addition, we analyzed cultures that were washed 3 h after arabinose induction and recovered for 1.5 or 17 h in fresh media, respectively. (A) For analysis of the mean level of GFP fluorescence, the mean of each replicate was divided by the mean GFP fluorescence of the respective culture in the exponential phase, and referred to as ‘normalized mean’. The normalized mean level of log10-transformed GFP fluorescence was not significantly different between strain NN206 harboring the grcAwt reporter (blue) and strain NN212 harboring the grcAATA reporter (orange). (B) Variation in the GFP fluorescence (SCV) was significantly higher in the grcAwt reporter than in the grcAATA reporter in the overnight recovery phase after 3 h of stress caused by ectopic mazF expression (time point t3), and throughout the growth curve without added Ara. Significant p-values of t-tests are depicted (*p < 0.05, **p < 0.01). All p-values are listed in File S1. (C) The GFP fluorescence distribution is skewed towards higher values for grcAwt reporter in the overnight recovery phase after 3 h of ectopic mazF expression (blue). The histogram of the grcAATA reporter is depicted in orange, and the reporterless strain NN204 is depicted in grey as the background level of GFP fluorescence, i.e., bacterial autofluorescence. The average value of skewness (kurtosis) is 0.023 (26.50) for the grcAwt reporter and −0.227 (42.37) for the grcAATA reporter, measured in N = 3 replicates. The differences between the reporters are significant (p = 0.003 for skewness, p = 0.00098 for kurtosis), and all skewness and kurtosis values are reported in File S1. Here, the histograms of one replicate per strain are plotted. (D) For comparison, the GFP fluorescence distributions are not significantly different between grcAwt and grcAATA reporters after 2 h of ectopic mazF expression.
To further understand variable gfp expression within clonal populations during ectopic mazF expression, we employed two additional measures of variation, namely skewness and kurtosis, which signify the degree of asymmetry and the “tailedness” of GFP fluorescence distributions. Quantitative analyses of skewness and kurtosis showed statistically significant differences (p < 0.01) between the grcA reporters only for the fluorescence measurements 17 h after recovery from 3 h of ectopic mazF expression (see File S1 for Source Data). Interestingly, the corresponding histograms of GFP fluorescence revealed that the grcAwt reporter GFP distributions had a right tail (Fig. 5C), i.e., the distributions were skewed toward higher values (for comparison see Fig. 5D). Specifically, GFP distributions of the grcAwt reporter contained more highly fluorescent cells as compared to the grcAATA GFP distributions. This result is indicative of the generation of a small subpopulation of cells with an increased level of reporter gene expression as a consequence of the MazF-mediated processing of the grcAwt mRNA. It has been previously established that stress response promotes functional ribosome heterogeneity (Byrgazov, Vesper & Moll, 2013; Starosta et al., 2014), and that during MazF-induced stress the total ribosome pool is comprised of canonical ribosomes as well as stress-ribosomes that are able to translate MazF-processed mRNAs (Vesper et al., 2011; Sauert et al., 2016). Thus, one plausible explanation is that during ectopic mazF expression both, the un-processed and processed forms of the grcAwt reporter mRNA can be translated by canonical and stress-ribosomes, respectively, whereas the grcAATA reporter mRNA can solely be translated by canonical ribosomes that comprise one part of the total ribosome pool. Given that MazF processing affects more than 300 mRNA species in E. coli by trimming their 5′-UTRs various lengths (Sauert et al., 2016), it is feasible that processed mRNAs maintain different translational regulatory elements that influence their translation in a specific manner. To generally investigate translational heterogeneity of other MazF-processed mRNAs, a reporter system similar to the grcA reporter could be used. However, the reporter sequence has to be carefully optimized and adapted to each candidate mRNA, since we already observed differences in the reporter fluorescence between the wild-type and the ΔACA reporter prior to MazF stress for another MazF-processed mRNA, namely the rpsU mRNA (Fig. S4).
Discussion
In this study we investigated how de-repression and transcriptional activation of the mazEF module differ between cells within a clonal population during and after antibiotic treatment, and nutritional stress. In all our experiments, we employed fluorescent reporters that are highly stable systems, and thus do not provide information on current physiological state (e.g., transient bursts in mazEF transcription) but rather describe cumulative changes in reporter gene expression between two flow cytometry measurements. The transcriptional reporter Pmaz-gfp was not expressed during exponential growth, indicating strong repression of the mazEF transcription in conditions without stress (Marianovsky et al., 2001; Christensen et al., 2003). During stressful conditions and in the recovery phase upon removal of the stressor, we measured an increase in the GFP fluorescence of bacterial populations indicating de-repression of the mazEF transcription. Interestingly, the reporter Pmaz-gfp was not expressed above background level in all analyzed cells within a population, which suggests that the mazEF module is transcribed at a low level during adverse conditions. A recent analysis of available transcriptome data has shown non-significant or minor changes in the mazEF mRNA level in response to stressful conditions that have not been addressed in this study, namely, treatment with tetracycline and rifampicin, starvation in 48 h-cultures, and heat shock (Muthuramalingam, White & Bourne, 2016). Another study measuring fluorescence of plasmid-based transcriptional reporter systems for TA modules indicated that the mazEF module is significantly induced by isoleucine starvation, osmotic stress, and phosphate starvation, albeit not continuously during the entire stress periods (Shan et al., 2017).
We also found no direct link between the fluorescence of the Pmaz-gfp reporter and constitutive mCherry fluorescence that served as an indicator of growth. mazEF expression is negatively autoregulated at the transcriptional level (Marianovsky et al., 2001; Keseler et al., 2013), and it has been previously suggested that expression of negatively autoregulated genes does not depend on the bacterial growth rate (Klumpp, Zhang & Hwa, 2009; Scott et al., 2010). Additionally, stress response mechanisms and activation of other TA systems (Gerdes & Maisonneuve, 2012; Kasari et al., 2013) might have additional roles in the regulation of mazEF expression.
Furthermore, it has been previously shown that de-repression of the mazEF transcription occurs when the ratio of toxin to antitoxin is in favor of the MazF toxin (Aizenman, Engelberg-Kulka & Glaser, 1996; Maisonneuve et al., 2011; Cataudella et al., 2013; Gelens et al., 2013). Hence, the reporter Pmaz-gfp could be employed to assess the level of temporarily unbound MazF that exerts its endoribonucleolytic activity. Here we report on a slight increase in the GFP fluorescence of the Pmaz-gfp reporter, which might also indicate increased levels of active MazF during stressful conditions and upon stress relief. Since an increased GFP signal implies elevated levels of mazEF transcription, the concentration of free MazF possibly increases only transiently before de novo production of both MazE and MazF. Thus, plasmid-based systems frequently employed to ectopically induce mazF overexpression (Vazquez-Laslop, Lee & Neyfakh, 2006) might generate levels of MazF activation that are considerably higher than herein measured during antibiotic treatments and nutritional stress.
Our data also highlight the impact of MazF-mediated mRNA processing on reporter gene expression. Interestingly, despite the population displays comparable mean fluorescence levels, the ectopic induction of mazF expression resulted in a higher variation in the grcAwt reporter fluorescence when compared to the variation in the grcAATA reporter fluorescence. This higher variation can be attributed to increased GFP signal in a subpopulation of cells, indicating the increased grcAwt reporter gene expression in those cells. This result suggests that the ability to translate both, the canonical and processed forms of grcAwt reporter mRNAs by canonical and stress-ribosomes, respectively (Vesper et al., 2011; Byrgazov, Vesper & Moll, 2013), can lead to enhanced reporter protein synthesis in a subpopulation of cells.
A recent study has identified 330 MazF-processed, polysome-associated mRNAs that harbor 5′-UTRs of various lengths (Sauert et al., 2016). For instance, one form of the MazF-processed grcA transcript comprises 2 nucleotides upstream of the start codon, whereas the MazF-processed rpoS transcript comprises 108 nucleotides upstream of the start codon. In the case of the rpoS transcript, MazF-mediated cleavage removes only a part of the translational regulatory elements (Keseler et al., 2013), thereby possibly affecting rpoS expression dynamics in the conditions that promote MazF activation. Thus, it is feasible that MazF processing promotes a variety of changes in translational properties of the rendered mRNAs.
Taken together, our results suggest that MazF activation has a subtle effect in stressed bacterial populations during antibiotic treatment or amino acid starvation, which may further cross-activate other TA systems (Kasari et al., 2013; Wessner et al., 2015) or stress response mechanisms (Wang & Wood, 2011), collectively affecting bacterial gene expression and growth. A two-tier manifestation of cell-to-cell heterogeneity potentially occurs during the MazF-mediated stress response: first, the activation of the mazEF transcription is generally maintained at low levels, and the overall amount of MazF may be variable between single cells. Second, it is conceivable that mRNA processing by MazF fosters the variation in gene expression. Hence, we hypothesize that the heterogeneous MazF-mediated processing of mRNAs and rRNAs, and the modification of ribosomes to be selective towards translation of a small subset of MazF-processed mRNAs, could produce a variety of gene expression programs within isogenic populations. The variation in translation of MazF-processed mRNAs could depend on the specific regulatory regions that remained parts of the 5′-UTRs after MazF cleavage. In conclusion, populations may exhibit a substantial level of variation in gene expression during stressful conditions that trigger MazF and during resumption of growth upon stress relief.
Supplemental Information
Figure S1. Fluorescence of the grcA reporters integrated in strains MG1655 and the isogenic mazF deletion mutant; Figure S2. The chromosomally integrated PBAD expression system is inducible; Figure S3. Increased variation in the grcAwt reporter fluorescence after overnight growth; Figure S4. rpsU reporters constructed to infer MazF-mediated mRNA processing.
Datasets: source data, statistical tests, sequences, primers, R commands.
Acknowledgments
The authors are grateful to Konstantin Byrgazov, Tobias Bergmiller and Petra Pusic for valuable discussions. We thank Tobias Bergmiller, Călin C. Guet and Karin Schnetz for sharing strains and plasmids, and Olin Silander for R scripts. We acknowledge the MFPL BioOptics Facility—Light Microscopy, and FACS Facility together with Thomas Sauer. NN is grateful to Călin C. Guet for his support.
Funding Statement
This project was funded by the Lise Meitner grant M1697 of the Austrian Science Fund (FWF) to Nela Nikolic. Also, Nela Nikolic was supported by the Swiss National Science Foundation (SNF) fellowship for prospective researchers 145706 for the first year of the project. Zrinka Didara was supported by the Erasmus+ program of the European Commission. Isabella Moll was funded by the Austrian Science Fund (FWF) grant P22249 and FWF Special Research Program RNA-REG F43 (subproject F4316). Publication costs are covered by the FWF Open Access Policy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Nela Nikolic, Email: nelanik@gmail.com.
Isabella Moll, Email: isabella.moll@univie.ac.at.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Nela Nikolic conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper, interpreted the data.
Zrinka Didara performed the experiments, reviewed drafts of the paper.
Isabella Moll conceived and designed the experiments, wrote the paper, reviewed drafts of the paper, interpreted the data.
Data Availability
The following information was supplied regarding data availability:
All raw flow cytometry files were deposited to the FlowRepository (http://flowrepository.org) with assigned Repository IDs: FR-FCM-ZY93 (data for Fig. 1), FR-FCM-ZY9Y (Table 1), FR-FCM-ZY92 (Table 2), FR-FCM-ZY94 (Fig. 2I), FR-FCM-ZY95 (Fig. 3), FR-FCM-ZY96 (Figs. 5 and S2), FR-FCM-ZY98 (Fig. S1), FR-FCM-ZY99 (Fig. S3), and FR-FCM-ZY97 (Fig. S4).
References
- Acar, Mettetal & Van Oudenaarden (2008).Acar M, Mettetal JT, Van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nature Genetics. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- Aizenman, Engelberg-Kulka & Glaser (1996).Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine-3′5′-bispyrophosphate: a model for programmed bacterial cell death. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai, Yassin & Engelberg-Kulka (2004).Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. Journal of Bacteriology. 2004;186:8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoldini et al. (2014).Arnoldini M, Vizcarra IA, Peña Miller R, Stocker N, Diard M, Vogel V, Beardmore RE, Hardt W-D, Ackermann M. Bistable expression of virulence genes in Salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLOS Biology. 2014;12(8):e1001928. doi: 10.1371/journal.pbio.1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban et al. (2004).Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bergmiller et al. (2017).Bergmiller T, Andersson AMC, Tomasek K, Balleza E, Kiviet DJ, Hauschild R, Tkačik G, Guet CC. Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science. 2017;356(6335):311–315. doi: 10.1126/science.aaf4762. [DOI] [PubMed] [Google Scholar]
- Bergmiller et al. (2011).Bergmiller T, Pena-Miller R, Boehm A, Ackermann M. Single-cell time-lapse analysis of depletion of the universally conserved essential protein YgjD. BMC Microbiology. 2011;11:118. doi: 10.1186/1471-2180-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoumieux et al. (2013).Berthoumieux S, De Jong H, Baptist G, Pinel C, Ranquet C, Ropers D, Geiselmann J. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Molecular Systems Biology. 2013;9 doi: 10.1038/msb.2012.70. Article 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner et al. (1997).Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Byrgazov, Vesper & Moll (2013).Byrgazov K, Vesper O, Moll I. Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Current Opinion in Microbiology. 2013;16(2):133–139. doi: 10.1016/j.mib.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataudella et al. (2013).Cataudella I, Sneppen K, Gerdes K, Mitarai N. Conditional cooperativity of toxin–antitoxin regulation can mediate bistability between growth and dormancy. PLOS Computational Biology. 2013;9(8):e1003174. doi: 10.1371/journal.pcbi.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri et al. (2010).Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLOS ONE. 2010;5(1):e8801. doi: 10.1371/journal.pone.0008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov & Wackernagel (1995).Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- Christensen et al. (2003).Christensen SK, Pedersen K, Hensen FG, Gerdes K. Toxin–antitoxin loci as stress- response elements: ChpAK/MazF and ChpBK cleave translated mRNAs and are counteracted by tmRNA. Journal of Molecular Biology. 2003;332:809–819. doi: 10.1016/S0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- Cox III, Dunlop & Elowitz (2010).Cox III RS, Dunlop MJ, Elowitz MB. A synthetic three-color scaffold for monitoring genetic regulation and noise. Journal of Biological Engineering. 2010;4 doi: 10.1186/1754-1611-4-10. Article 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelens et al. (2013).Gelens L, Hill L, Vandervelde A, Danckaert J, Loris R. A general model for toxin-antitoxin module dynamics can explain persister cell formation in E. coli. PLOS Computational Biology. 2013;9(8):e1003190. doi: 10.1371/journal.pcbi.1003190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, Christensen & Løbner-Olesen (2005).Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nature Reviews. Microbiology. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gerdes & Maisonneuve (2012).Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annual Review of Microbiology. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- Gerosa et al. (2013).Gerosa L, Kochanowski K, Heinemann M, Sauer U. Dissecting specific and global transcriptional regulation of bacterial gene expression. Molecular Systems Biology. 2013;9 doi: 10.1038/msb.2013.14. Article 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders & Van Melderen (2014).Goeders N, Van Melderen L. Toxin-antitoxin systems as multilevel interaction systems. Toxins. 2014;6:304–324. doi: 10.3390/toxins6010304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann & Wanner (2001).Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. Journal of Bacteriology. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan, Sat & Engelberg-Kulka (2004).Hazan R, Sat B, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. Journal of Bacteriology. 2004;186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge (2011).Hengge R. The general stress response in gram-negative bacteria. In: Storz G, Hengge R, editors. Bacterial stress responses. Academic Press; Washington, D.C.: 2011. pp. 251–289. [Google Scholar]
- Kamada, Hanaoka & Burley (2003).Kamada K, Hanaoka F, Burley SK. Crystal structure of the MazE/MazF complex: molecular bases of antidote–toxin recognition. Molecular Cell. 2003;11:875–884. doi: 10.1016/S1097-2765(03)00097-2. [DOI] [PubMed] [Google Scholar]
- Kasari et al. (2010).Kasari V, Kurg K, Margus T, Tenson T, Kaldalu N. The Escherichia coli mqsR and ygiT genes encode a new toxin-antitoxin pair. Journal of Bacteriology. 2010;192:2908–2919. doi: 10.1128/JB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasari et al. (2013).Kasari V, Mets T, Tenson T, Kaldalu N. Transcriptional cross-activation between toxin-antitoxin systems of Escherichia coli. BMC Microbiology. 2013;13:45. doi: 10.1186/1471-2180-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler et al. (2013).Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muñiz Rascado L, Ong Q, Paley S, Schröder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Research. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlebnikov et al. (2001).Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 2001;147:3241–3247. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- Klumpp & Hwa (2014).Klumpp S, Hwa T. Bacterial growth: global effects on gene expression, growth feedback and proteome partition. Current Opinion in Biotechnology. 2014;28:96–102. doi: 10.1016/j.copbio.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp, Zhang & Hwa (2009).Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell & Leibler (2005).Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- Leveau & Lindow (2001).Leveau JH, Lindow SE. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. Journal of Bacteriology. 2001;183:6752–6762. doi: 10.1128/JB.183.23.6752-6762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve, Castro-Camargo & Gerdes (2013).Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154(5):1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- Maisonneuve et al. (2011).Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marianovsky et al. (2001).Marianovsky I, Aizenman E, Engelberg-Kulka H, Glaser G. The regulation of the Escherichia coli mazEF promoter involves an unusual alternating palindrome. Journal of Biological Chemistry. 2001;278:5975–5984. doi: 10.1074/jbc.M008832200. [DOI] [PubMed] [Google Scholar]
- Müller et al. (2016).Müller C, Sokol L, Vesper O, Sauert M, Moll I. Insights into the stress response triggered by kasugamycin in Escherichia coli. Antibiotics. 2016;5(2) doi: 10.3390/antibiotics5020019. Article 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuramalingam, White & Bourne (2016).Muthuramalingam M, White JC, Bourne CR. Toxin-antitoxin modules are pliable switches activated by multiple protease pathways. Toxins. 2016;8:e214. doi: 10.3390/toxins8070214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron-Gottesman et al. (2016).Oron-Gottesman A, Sauert M, Moll I, Engelberg-Kulka H. A stress-induced bias in the reading of the genetic code in Escherichia coli. mBio. 2016;7(6):e01855–16. doi: 10.1128/mBio.01855-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramisetty & Santhosh (2017).Ramisetty BCM, Santhosh RS. Endoribonuclease type II toxin–antitoxin systems: functional or selfish? Microbiology. 2017 doi: 10.1099/mic.0.000487. Epub ahead of print Jul 21 2017. [DOI] [PubMed] [Google Scholar]
- Sat et al. (2001).Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger the mazEF lethality. Journal of Bacteriology. 2001;183:2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauert (2015).Sauert M. PhD Dissertation. 2015. Selective translation of leaderless mRNAs by specialized ribosomes upon MazF-mediated stress response in Escherichia coli. [Google Scholar]
- Sauert et al. (2016).Sauert M, Wolfinger MT, Vesper O, Müller C, Byrgazov K, Moll I. The MazF-regulon: a toolbox for the post-transcriptional stress response in Escherichia coli. Nucleic Acids Research. 2016;44(14):6660–6675. doi: 10.1093/nar/gkw115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott et al. (2010).Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- Shan et al. (2017).Shan Y, Gandt AB, Rowe SE, Deisinger JP, Conlon BP, Lewis K. ATP-dependent persister formation in Escherichia coli. mBio. 2017;8(1):e02267–16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander et al. (2012).Silander OK, Nikolic N, Zaslaver A, Bren A, Kikoin I, Alon U, Ackermann M. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. PLOS Genetics. 2012;8:e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta et al. (2014).Starosta AL, Lassak J, Jung K, Wilson DN. The bacterial translation stress response. FEMS Microbiology Reviews. 2014;38(6):1172–1201. doi: 10.1111/1574-6976.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulavik et al. (2001).Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, DiDomenico B, Shaw KJ, Miller GH, Hare R, Shimer G. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrobial Agents and Chemotherapy. 2001;45:1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosa & Pizer (1971).Tosa T, Pizer LI. Effect of serine hydroxamate on the growth of Escherichia coli. Journal of Bacteriology. 1971;106:966–971. doi: 10.1128/jb.106.3.966-971.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien (1998).Tsien RY. The green fluorescent protein. Annual Review of Biochemistry. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Van Melderen (2010).Van Melderen L. Toxin-antitoxin systems: why so many, what for? Current Opinion in Microbiology. 2010;13(6):781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop, Lee & Neyfakh (2006).Vazquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. Journal of Bacteriology. 2006;188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper et al. (2011).Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner et al. (2001).Wagner AF, Schultz S, Bomke J, Pils T, Lehmann WD, Knappe J. YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochemical and Biophysical Research Communications. 2001;285(2):456–462. doi: 10.1006/bbrc.2001.5186. [DOI] [PubMed] [Google Scholar]
- Wang & Wood (2011).Wang X, Wood TK. Toxin–antitoxin systems influence biofilm and persister cell formation and the general stress response. Applied and Environmental Microbiology. 2011;77:5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessner et al. (2015).Wessner F, Lacoux C, Goeders N, d’Herouel AF, Matos R, Serror P, Van Melderen L, Repoila F. Regulatory crosstalk between type I and type II toxin–antitoxin systems in the human pathogen Enterococcus faecalis. RNA Biology. 2015;12(10):1099–1108. doi: 10.1080/15476286.2015.1084465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young et al. (2011).Young JW, Locke JC, Altinok A, Rosenfeld N, Bacarian T, Swain PS, Mjolsness E, Elowitz MB. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nature Protocols. 2011;7(1):80–88. doi: 10.1038/nprot.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2003).Zhang Y, Zhang J, Hoeflich KP, Ikura M, Quing G, Inouye M. MazF cleaves cellular mRNA specifically at ACA to block protein synthesis in Escherichia coli. Molecular Cell. 2003;12:913–923. doi: 10.1016/S1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Fluorescence of the grcA reporters integrated in strains MG1655 and the isogenic mazF deletion mutant; Figure S2. The chromosomally integrated PBAD expression system is inducible; Figure S3. Increased variation in the grcAwt reporter fluorescence after overnight growth; Figure S4. rpsU reporters constructed to infer MazF-mediated mRNA processing.
Datasets: source data, statistical tests, sequences, primers, R commands.
Data Availability Statement
The following information was supplied regarding data availability:
All raw flow cytometry files were deposited to the FlowRepository (http://flowrepository.org) with assigned Repository IDs: FR-FCM-ZY93 (data for Fig. 1), FR-FCM-ZY9Y (Table 1), FR-FCM-ZY92 (Table 2), FR-FCM-ZY94 (Fig. 2I), FR-FCM-ZY95 (Fig. 3), FR-FCM-ZY96 (Figs. 5 and S2), FR-FCM-ZY98 (Fig. S1), FR-FCM-ZY99 (Fig. S3), and FR-FCM-ZY97 (Fig. S4).