Abstract
This communication describes an approach for preparing monovalent semiconducting polymer dots (mPdots) with a size of 5 nmwhere each mPdot was composed of precisely a single active functional group.
Monovalent fluorescent probes with sizes less than 10 nm are desirable for in vitro and in vivo biological applications.1 Although conventional organic dyes have nanometer sizes andmonovalency, they usually suffer from low absorptivity and poor photostability. These drawbacks have limited their application in high-sensitivity imaging techniques and high-throughput assays. Semiconducting quantum dots (Qdots) have been developed as brighter and more photostable probes than conventional organic dyes, but their relatively large hydrodynamic diameter and multivalency are critical constraints for biological applications.2 Multivalency of Qdots may result in the cross-linking of surface proteins, which can activate signaling pathways and dramatically reduce receptor mobility. As a result, much effort has been devoted in the past several years to develop monovalent Qdots with smaller sizes.3
Semiconducting polymer dots (Pdots) have recently emerged as a new group of fluorescent probes which possess large absorption cross-sections, high quantum yields, and fast emission rates.4 The brightness of Pdots has been shown to be an order of magnitude higher than that of Qdots (e.g. 30 times) of comparable dimensions.4c,i Moreover, several reports5 showed that Pdots were nontoxic to cells, an important advantage over Qdots, which can be toxic if heavy metal ions leak from the inorganic core.6 Finally, Pdots with sizes comparable to typical water-soluble Qdots (~15 nm) do not blink, which is an important feature in many single-molecule experiments.
These properties of Pdots make them excellent fluorescent probes formany biological applications. For example, they recently have been used for biological detection,4l biosensing platforms,7 specific cellular4i and subcellular targeting and imaging,8 bioorthogonal labelling,4j protein detection4b and in vivo tumor targeting.9 To further optimize Pdots for biological applications, the development of small and monovalent Pdots is the next critical step. Monovalent Pdots (mPdots) have significant advantages over conventional multivalent Pdots because a single functional group is desirable for biological applications that are sensitive to protein–nanoparticle clustering and aggregation. For example, we reported a technique for counting protein copy numbers in synaptic vesicles and subcellular organelles10 using fluorescent antibodies and single-molecule counting. For these applications, it is imperative that the fluorescent labels do not have excess functional groups that may cause cross-linking and accumulate multiple antibodies per fluorescent label.
Additionally, small Pdots (<10 nm in diameter) are desirable for certain applications. For example, we have recently developed two types of small Pdots (~9 nm), a compact yellow emitting CN-PPV Pdot8 and a cross-linked Pdot.11 We found that these small Pdots were able to label subcellular features more efficiently than larger Pdots (~15 nm). For example, microtubules labelled with large Pdots tend to appear spotty in a confocal fluorescence image,4i while those same microtubules appear crisp and resolved when labelled with small Pdots.8,11 Small Pdots are also less prone than large Pdots to alter the diffusional and biological activity of the biomolecules that they label, and small Pdots can access size-restricted cellular regions, such as synapses. To meet these demands for the next generation of Pdots with monovalent functional groups and small size, this communication describes an approach based on surface attachment and washing for generating mPdots.
The strategy we developed to prepare mPdots is shown in Fig. 1. Here, we first form Pdots that have multiple polymer chains and are multivalent. We then attach the Pdots onto the surface of silica beads using Click chemistry. Once the Pdots are attached to the bead surface via a functional group (i.e. alkyne group), we wash the bead-Pdot with THF, which causes the Pdot to unfold and the entangled chains to fall apart, leaving a single chain of polymer attached to the bead. Re-introduction of aqueous solution causes the chain to re-collapse to form a single-chain Pdot, and subsequent release of the Pdot from the bead surface results in a monovalent Pdot (ESI†).
Fig. 1.
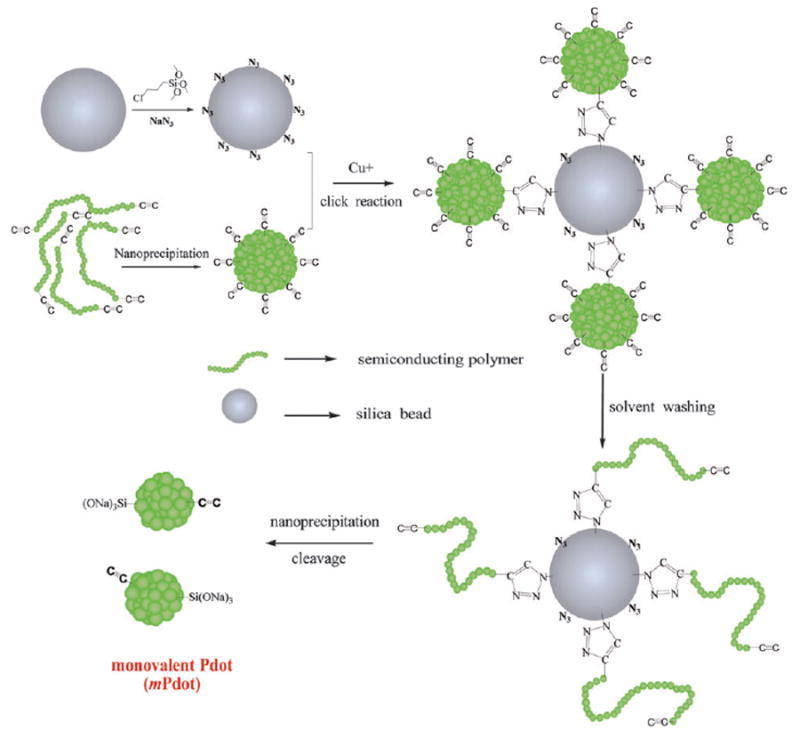
The procedure to prepare monovalent Pdots (mPdots). A silica particle with a diameter of ~200 nm was prepared and its surface modified with a layer of chloride (SiO2–Cl) via the hydrolysis and condensation of chloridetrimethoxysilane. The SiO2–Cl groups were then modified to azide to form a clickable silica nanoparticle (ESI†). Separately, regular multivalent PPV–PPA Pdots were prepared using nanoprecipitation (ESI†); these Pdots had alkyne groups so they could react with SiO2–N3 on the silica surface via click chemistry. Once the regular PPV–PPA Pdots were clicked onto the surface of the silica nanoparticle, the solvent was changed from aqueous solution to THF and the silica–polymer complex was washed with THF several times. This step removed all the polymer chains in the regular Pdot that were not covalently attached to the silica surface. The single polymer chains attached to the surface of the silica nanoparticles were then reprecipitated into small and monovalent mPdots by reintroducing the silica–polymer complex into aqueous solution from THF. Finally, the mPdots were cleaved from the silica surface and released into solution in the presence of NaOH and Triton 100. To remove NaOH and Triton 100, the mPdot solution was dialyzed overnight in water or buffer.
To implement the above strategy, we designed and synthesized a green emitting semiconducting polymer (alkyne terminated linear poly( p-phenylenevinylene) derivative containing two pendent pentaphenylene groups (PPV–PPA)). The PPV–PPA polymer had only two terminating alkyne click-functional groups (Fig. S1 and S2, ESI†). And because there was only two alkyne groups in the initial PPV–PPA polymer chain, during the preparation process one of the two alkyne groups was used to covalently bind to the silica surface, and later converted to the terminated Si(ONa)3 group after NaOH cleavage when single chain PPV–PPA was formed. Therefore, there is only one alkyne group left in the single chain PPV–PPA Pdot, resulting in an mPdot with a monovalent alkyne group.
Fig. 2 shows the size information for the silica beads, regular PPV–PPA Pdots, and PPV–PPA mPdots. As established by transmission electron microscopy (TEM), the silica bead (SiO2–N3) had a diameter of ~200 nm and the regular PPV–PPA Pdot had a 32 nm diameter (Fig. 2a and b). A representative TEM image of themPdots (Fig. 2c) shows that the mean diameter was 5.4 ± 0.5 nm (from 88 mPdots measured; Fig. 2g). Atomic force microscopy (AFM) measurements reported similar results (Fig. 2f); the mean height of mPdots was 4.5 ± 0.4 nm (from 100 mPdots imaged) (Fig. 2h). DLS showed that the hydrodynamic diameter ofmPdots was 7 nm(Fig. 2i). These three measurements are consistent because the lateral dimensions of the collapsed mPdot imaged using a TEM should be slightly larger than the height of the collapsed mPdot measured using an AFM. The hydrodynamic size is ~1–2 nm larger than TEM and AFM measurements as anticipated because of slight swelling of mPdots in aqueous solution. The molecular weight of PPV–PPA we synthesized was 86 000 g mol−1. For a single chain of PPV–PPA that is fully collapsed into a Pdot with a density of ~1.0 g cm−3, the resulting mPdot would have a diameter of ~6 nm, consistent with the results from both the TEM and AFM experiments.
Fig. 2.
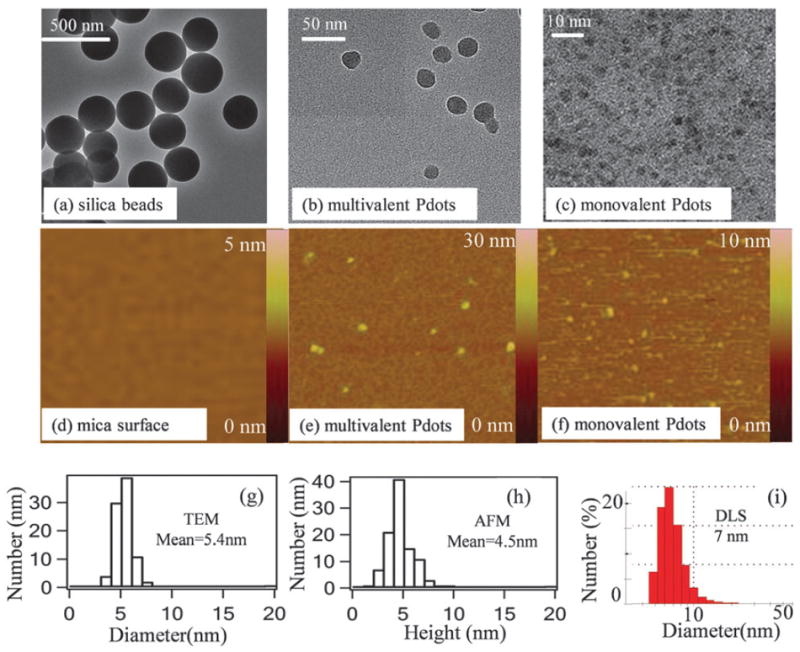
Size distribution of mPdots measured using TEM, AFM, and DLS. (a) TEMimage of silica beads showing a diameter of ~200 nm. (b) TEM image of regular PPV–PPA Pdots, which have an average diameter of 34 ± 4 nm, from measurements on 80 Pdots. (c) TEM image of PPV–PPA mPdots. (d) AFM image (1 μm × 1 μm) of the 3-aminopropyltriethoxysilane (APTEOS)-coated mica surfacewithout any nanoparticles. (e) AFM image (0.6 μm × 0.6 μm) of multivalent regular PPV–PPA Pdots. (f) AFM image (1.2 μm × 1.2 μm) of PPV–PPA mPdots. (g) Size distribution of mPdots shown in (c); the average diameter was 5.4 ± 0.5 nmfromimages of 88 mPdots. (h) Height distribution of mPdots measured using AFM; the average value was 4.5 ± 0.4 nm from measurements on 100 mPdots. (i) DLS results of mPdots in aqueous solution showing a hydrodynamic diameter of 7 nm.
Our PPV–PPA mPdot had a linear chain polymer with a Si(ONa)3 group at the end of the polymer attached to the silica surface and an alkyne group at the other end. To validate that each mPdot had only a single alkyne functional group, we followed the established approach described in the literature for confirming monovalency of nanoparticles,12 and carried out two experiments. In the first experiment, we introduced a linker with two azide groups to crosslink mPdots. Fig. 3a shows the linker (PEG7-Bis-Azide). If mPdot was monofunctional, then after crosslinking, we expect to see dumb-bell structures (Fig. 3a). If the mPdots had more than one alkyne group, then we should see aggregates of mPdots.
Fig. 3.
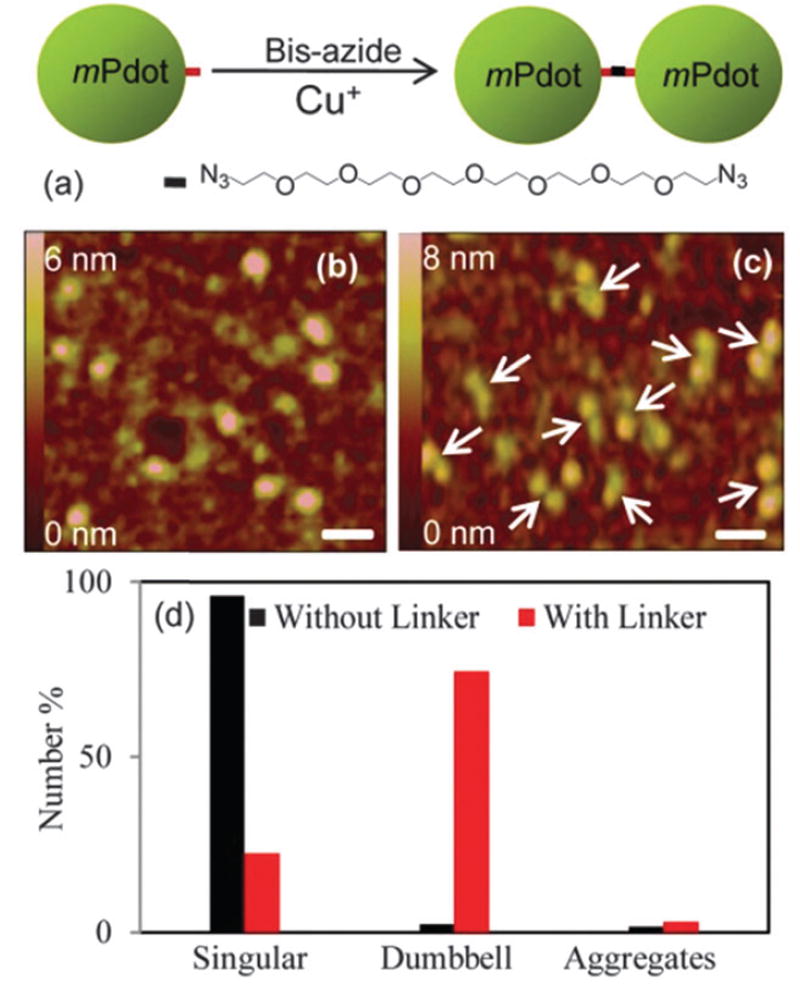
Schematic depiction and AFM images showing the formation of dumb-bell structures from crosslinking clickable mPdots with an alkyne group by using α,ω-bis-azide octa(ethylene glycol) (PEG7-Bis-Azide). (a) Chemical structure of PEG7-Bis-Azide and the schematic showing the reaction that forms the mPdot dumb-bell features observed in AFM. (b) AFM image showing individual mPdots in the presence of copper sulfate and L-sodium ascorbate but without PEG7-Bis-Azide needed for the click reaction. (c) AFM images of mPdot dumb-bell structures formed in the presence of PEG7-Bis-Azide, copper sulfate, and L-sodiumascorbate; the linker :mPdot ratio used was 1 : 2. The white arrows point to the dumb-bells. Under the same conditions, regular polyvalent Pdots formed aggregates. Scale bars in (b) and (c) represent 20 nm. (d) A plot showing the populations of mPdots (singular, dumb-bell, or aggregates) observed in the AFM images in the absence and presence of linkers. About 100 Pdots were counted for calculating the percentages.
Indeed, regular PPV–PPA Pdots aggregated after the addition of linkers in the presence of freshly prepared copper sulfate (0.5 mM) and L-sodium ascorbate (0.2 mM) needed to initiate the click reaction. Before the addition of a linker, there was no aggregation, which indicated that the aggregation of regular Pdots was only caused by the cross-linking of Pdots triggered by multiple click reactions among the Pdots with multiple alkyne groups. In contrast, when mPdots were used for the same experiment, we observed dumb-bell features (Fig. 3c). As a control, Fig. 3b shows a typical image of mPdots when no linker was added but in the presence of 0.5 mM copper sulfate. Fig. 3d displays the populations (singular, dumb-bell, or aggregates) of mPdots we observed in the absence and presence of a linker: ~96% of mPdots was singular when no linker was present (Fig. 3b), but when a linker was added, only ~23% of mPdots remained singular and ~75% of mPdots formed dumb-bell structures (Fig. 3c). It should be noted that the percentage of dumb-bell structures (~75%) formed by mPdots is similar to that of dumb-bell structures formed by monofunctional gold12b,c or silver12a nanoparticles measured by TEM as reported in the literature.12 We do not expect 100% of mPdots (or other monovalent nanoparticles) to form dumb-bell structures because of reaction kinetics and yield as well as the stoichiometry of the linker to mPdots as we will discuss in more detail below. Importantly, we observed almost no Pdot aggregates, thus confirming the absence of multivalent Pdots.
As additional control, we varied the amount of linker that we added. For the result shown in Fig. 3c where most mPdots were in dumb-bell form, the stoichiometry or the molar ratio of the linker to mPdots was ~1 : 2. When we lowered the molar ratio of the linker to mPdots to 1 : 10, or increased themolar ratio of the linker to mPdots to 10 : 1, the majority of mPdots appeared as a single isolated Pdot (Fig. S3, ESI†), similar to the image shown in Fig. 3b. For a linker to mPdot ratio of 1 : 10, there was simply insufficient linker to form dumb-bell structures, leaving mPdots as isolated nanoparticles. For a linker :mPdot ratio of 10 : 1, we also observed individual isolated mPdots because the reaction between the free linker and mPdot was much faster than between the mPdot-linker with another mPdot; this difference in reaction kinetics resulted in mPdots all attached to a single linker, which prevented the formation of dumb-bell structures. This set of experiments confirmed that each mPdot had only a single functional group.
Next we converted the alkyne functional group on the mPdot to a carboxylic acid group. The motivation for this experiment was two-fold. First, carboxylic acid is one of the most common functional groups used for bioconjugation. We have demonstrated its utility in our past experiments for covalently linking a wide range of biomolecules and dyes to Pdots and transforming them into other functional groups.4i,j,7 Therefore, this experiment acted as a gateway for changing the alkyne group to other functional groups or for attachment to biomolecules. Second, we had previously demonstrated that Cu2+ could crosslink Pdots with carboxyl groups because Cu2+ is complexed by the carboxyl groups.13 Monovalent carboxyl mPdots should also form dumb-bell structures in the presence of Cu2+ while polyvalent Pdots would form aggregates as we had previously demonstrated (Fig. 4a).13 This experiment served as another validation that mPdots were monovalent.
Fig. 4.

Generation of carboxyl-terminated mPdots and the formation of carboxyl mPdot dumb-bells in the presence of Cu2+. (a) Schematic showing the transformation of alkyne to carboxylic mPdots and then cross-linking mPdot-COOH to form dumb-bell structures. (b) AFM image of regular polyvalent carboxyl Pdots, formed from alkyne Pdots using azide-acid, in the presence of 0.5 mM Cu2+ in HEPES buffer (pH = 7.4); only aggregates were observed. (c) AFM image of mPdots with the carboxyl group in HEPES buffer (pH = 7.4) but without Cu2+. (d) AFM image of mPdots with the carboxyl group in the presence of 10 mM Cu2+ in HEPES buffer (pH = 7.4). The white arrows point to the dumb-bell structures. Scale bars in (b), (c), and (d) represent 100 nm, 25 nm, and 20 nm, respectively. (e) A plot showing the populations of regular multivalent Pdots and mPdots (singular, dumb-bell, or aggregates) in the AFM images in the absence and presence of Cu2+. About 100 Pdots were counted for calculating the percentages.
To form mPdot-COOH, we used azide-PEG-COOH to react with mPdot-alkyne via click cycloaddition in the presence of copper sulfate (0.5 mM) and L-sodium ascorbate (0.2 mM). Regular multivalent Pdots had a high density of alkyne groups on the surface and quickly aggregated in the presence of copper sulfate when the alkyne groups were transformed to COOH groups (Fig. 4b). The solution turned cloudy. Under the same conditions, mPdots did not aggregate and the solution remained clear. We then dialyzed out the remaining copper(I)/(II) in the mPdot solution and added a more concentrated Cu2+ solution at 10 mM to induce mPdots to form dumb-bell structures. Fig. 4d shows the result, in which dumb-bell features can be clearly discerned. No aggregates of mPdots were detected, even at 10 mM of Cu2+; we note for regular Pdots with COOH groups that only ~5 μM of Cu2+ was sufficient to induce significant Pdot aggregation.13 Fig. 4e shows the populations of mPdots (singular, dumb-bell, or aggregates) in the AFM images: ~94% of multivalent Pdots was observed to form aggregates in the presence of Cu2+, but for mPdots in the presence of Cu2+, they formed dumb-bell structures instead (60%). Some mPdots remained singular (38%), likely because Cu2+ isnot as strong a cross linker and the stability of two Pdots linked by a single Cu2+ ion is rather low and prone to disruption. In fact, Cu2+ attached to a single carboxyl group is not stable and two carboxyl groups are required to form a stable complex, which also explains why having more Cu2+ than mPdots in solution did not prevent formation of dumb-bell structures. From these experiments, the strong contrast in the behavior of regular multivalent Pdots and mPdots is evident.
Finally, we investigated the stability of mPdots in different buffer solutions. Fig. S4 (ESI†) shows the normalized fluorescence intensity of mPdots in different buffers (including TRIS, TBE, PBS and HEPES buffer) as a function of time. The result indicated that the fluorescence of the mPdots buffer solution did not change over a period of three days. Because any aggregation of Pdots would result in a decrease in the measured fluorescence intensity due to self-quenching,14 this result confirmed that the Pdots are well dispersed and stable over this period, which are suitable for biological applications.
In conclusion, we have developed a method for preparing monovalent and very small Pdots. We carried out two experiments to show that mPdots only had a single functional group. When the mPdots were crosslinked, they formed dumb-bell structures as seen using an AFM. We generated clickable mPdots as well as mPdots with a single carboxyl group, which could be used for covalent attachment of a broad range of biological molecules. The importance of having monovalency has been illustrated in the literature for other nanoparticles. For example, monovalent nanoparticles have been shown to label glutamate receptors at neuronal synapses without activating EphA3 tyrosine kinase3b while polyvalent nanoparticles result in activation. Monovalent nanoparticles have also been shown to offer improved quantification in tumor targeting and imaging,3a and better performance in the tracking of individual proteins in live cells.15 The development of the very small and monovalent mPdots, coupled with the high brightness of Pdots as we have previously demonstrated,4c,i is expected to advance their adoption as useful fluorescent probes in biomedical applications.
Supplementary Material
Footnotes
Electronic supplementary information (ESI) available: Materials, characterization and Pdots preparation. See DOI: 10.1039/c4cc01689k
Notes and references
- 1.(a) Hilderbrand SA, Kelly KA, Weissleder R, Tung C-H. Bioconjugate Chem. 2005;16:1275–1281. doi: 10.1021/bc0501799. [DOI] [PubMed] [Google Scholar]; (b) Shao F, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2487–2491. doi: 10.1021/bc800417b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Shi W, Dolai S, Rizk S, Hussain A, Tariq H, Averick S, L’Amoreaux W, ElIdrissi A, Banerjee P, Raja K. Org Lett. 2007;9:5461–5464. doi: 10.1021/ol702370m. [DOI] [PubMed] [Google Scholar]; (d) Wu S, Zhu C, Zhang C, Yu Z, He W, He Y, Li Y, Wang J, Guo Z. Inorg Chem. 2011;50:11847–11849. doi: 10.1021/ic201506y. [DOI] [PubMed] [Google Scholar]
- 2.Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, Webb WW. Science. 2003;300:1434–1436. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- 3.(a) Liu HY, Gao X. Bioconjugate Chem. 2011;22:510–517. doi: 10.1021/bc200004z. [DOI] [PubMed] [Google Scholar]; (b) Howarth M, Liu W, Puthenveetil S, Zheng Y, Marshall LF, Schmidt MM, Wittrup KD, Bawendi MG, Ting AY. Nat Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Zhang X, Yu J, Rong Y, Ye F, Chiu DT, Uvdal K. Chem Sci. 2013;4:2143–2151. doi: 10.1039/C3SC50222H. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ye F, Smith PB, Wu C, Chiu DT. Macromol Rapid Commun. 2013;34:785–790. doi: 10.1002/marc.201200809. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wu C, Chiu DT. Angew Chem Int Ed. 2013;52:3086–3109. doi: 10.1002/anie.201205133. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Sun W, Yu J, Deng R, Rong Y, Fujimoto B, Wu C, Zhang H, Chiu DT. Angew Chem Int Ed. 2013;52:11294–11297. doi: 10.1002/anie.201304822. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Rong Y, Wu C, Yu J, Zhang X, Ye F, Zeigler M, Gallina ME, Wu I-C, Zhang Y, Chan Y-H. ACS Nano. 2013;7:376–384. doi: 10.1021/nn304376z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Zhu C, Liu L, Yang Q, Lv F, Wang S. Chem Rev. 2012;112:4687–4735. doi: 10.1021/cr200263w. [DOI] [PubMed] [Google Scholar]; (g) Li K, Liu B. J Mater Chem. 2012;22:1257–1264. [Google Scholar]; (h) Das S, Powe AM, Baker GA, Valle B, El-Zahab B, Sintim HO, Lowry M, Fakayode SO, McCarroll ME, Patonay G, Li M, Strongin RM, Geng ML, Warner IM. Anal Chem. 2012;84:597–625. doi: 10.1021/ac202904n. [DOI] [PubMed] [Google Scholar]; (i) Wu C, Schneider T, Zeigler M, Yu J, Schiro PG, Burnham DR, McNeill JD, Chiu DT. J Am Chem Soc. 2010;132:15410–15417. doi: 10.1021/ja107196s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Wu C, Jin Y, Schneider T, Burnham DR, Smith PB, Chiu DT. Angew Chem Int Ed. 2010;49:9436–9440. doi: 10.1002/anie.201004260. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Pecher J, Mecking S. Chem Rev. 2010;110:6260–6279. doi: 10.1021/cr100132y. [DOI] [PubMed] [Google Scholar]; (l) Wu C, Bull B, Szymanski C, Christensen K, McNeill J. ACS Nano. 2008;2:2415–2423. doi: 10.1021/nn800590n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Fernando LP, Kandel PK, Yu J, McNeill J, Ackroyd PC, Christensen KA. Biomacromolecules. 2010;11:2675–2682. doi: 10.1021/bm1007103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rahim NAA, McDaniel W, Bardon K, Srinivasan S, Vickerman V, So PTC, Moon JH. Adv Mater. 2009;21:3492–3496. [Google Scholar]; (c) Pu KY, Li K, Shi JB, Liu B. Chem Mater. 2009;21:3816–3822. [Google Scholar]
- 6.Hardman R. Environ Health Perspect. 2006;114:165–172. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Ye F, Wu C, Jin Y, Chan Y-H, Zhang X, Chiu DT. J Am Chem Soc. 2011;133:8146–8149. doi: 10.1021/ja202945g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chan Y-H, Wu C, Ye F, Jin Y, Smith PB, Chiu DT. Anal Chem. 2011;83:1448–1455. doi: 10.1021/ac103140x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye F, Wu C, Jin Y, Wang M, Chan Y-H, Yu J, Sun W, Hayden S, Chiu DT. Chem Commun. 2012;48:1778–1780. doi: 10.1039/c2cc16486h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Hansen SJ, Hou Q, Yu J, Zeigler M, Jin Y, Burnham DR, McNeill JD, Olson JM, Chiu DT. Angew Chem Int Ed. 2010;50:3430–3434. doi: 10.1002/anie.201007461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Mutch SA, Kensel-Hammes P, Gadd JC, Fujimoto BS, Allen RW, Schiro PG, Lorenz RM, Kuyper CL, Kuo JS, Bajjalieh SM, Chiu DT. J Neurosci. 2011;26:1461–1470. doi: 10.1523/JNEUROSCI.3805-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mutch SA, Fujimoto BS, Kuyper CL, Kuo JS, Bajjalieh SM, Chiu DT. Biophys J. 2007;92:2926–2943. doi: 10.1529/biophysj.106.101428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Wu C, Zhang X, Ye F, Gallina ME, Rong Y, Wu IC, Sun W, Chan Y-H, Chiu DT. Adv Mater. 2012;24:3498–3504. doi: 10.1002/adma.201201245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) VIckova B, Moskovits M, Pavel I, Siskova K, Sladkova M, Slouf M. Chem Phys Lett. 2008;455:131–134. [Google Scholar]; (b) Liu X, Worden JG, Dai Q, Zou J, Wang J, Huo Q. Small. 2006;2:1126–1129. doi: 10.1002/smll.200600162. [DOI] [PubMed] [Google Scholar]; (c) Worden JG, Dai Q, Shaffer AW, Huo Q. Chem Mater. 2004;16:3746–3755. [Google Scholar]
- 13.Chan Y, Jin Y, WU C, Chiu DT. Chem Commun. 2010;47:2820–2822. doi: 10.1039/c0cc04929h. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y, Ye F, Wu C, Chan Y-H, Chiu DT. Chem Commun. 2012;48:3161–3163. doi: 10.1039/c2cc17703j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke S, Pinaud F, Beutel O, You C, Piehler J, Dahan M. Nano Lett. 2010;10:2147–2154. doi: 10.1021/nl100825n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


