Abstract
OBJECTIVES
The PNPLA3 rs738409 single-nucleotide polymorphism is known to promote nonalcoholic steatohepatitis (NASH), but its association with fibrosis severity and hepatocellular carcinoma (HCC) risk is less well-defined. The objectives of this study were to determine the association between PNPLA3 and liver fibrosis severity, HCC risk, and HCC prognosis among patients with liver disease.
METHODS
We performed a systematic literature review using the Medline, PubMed, Scopus, and Embase databases through May 2013 and a manual search of national meeting abstracts from 2010 to 2012. Two investigators independently extracted data on patient populations, study methods, and results using standardized forms. Pooled odds ratios (ORs), according to PNPLA3 genotype, were calculated using the DerSimonian and Laird method for a random effects model.
RESULTS
Among 24 studies, with 9,915 patients, PNPLA3 was associated with fibrosis severity (OR 1.32, 95 % confidence interval (CI) 1.20–1.45), with a consistent increased risk across liver disease etiologies. Among nine studies, with 2,937 patients, PNPLA3 was associated with increased risk of HCC in patients with cirrhosis (OR 1.40, 95 % CI 1.12–1.75). On subgroup analysis, increased risk of HCC was demonstrated in patients with NASH or alcohol-related cirrhosis (OR 1.67, 95 % CI 1.27–2.21) but not in those with other etiologies of cirrhosis (OR 1.33, 95 % CI 0.96–1.82). Three studies, with 463 patients, do not support an association between PNPLA3 and HCC prognosis but are limited by heterogeneous outcome measures. For all outcomes, most studies were conducted in homogenous Caucasian populations, and studies among racially diverse cohorts are needed.
CONCLUSIONS
PNPLA3 is associated with an increased risk of advanced fibrosis among patients with a variety of liver diseases and is an independent risk factor for HCC among patients with nonalcoholic steatohepatitis or alcohol-related cirrhosis.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the fifth most common cause of cancer and the third leading cause of cancer-related death worldwide (1). Within the United States and Europe, its increasing incidence is largely driven by the current epidemic of hepatitis C virus (HCV) cases; however, nonalcoholic fatty liver disease (NAFLD) is an increasingly common etiology for cirrhosis and HCC (1). Furthermore, the metabolic syndrome and hepatic steatosis are well-recognized cofactors in patients with other liver diseases, such as those with viral hepatitis. Underlying cirrhosis is the strongest risk factor for HCC development, and HCC is one of the leading causes of death among patients with cirrhosis.
Retrospective case-control studies have identified other risk factors for HCC, including older age, male gender, diabetes, and alcohol intake (2). In addition, family history and ethnicity are risk factors for HCC, suggesting a possible genetic component to the development of HCC. Although genome-wide association and candidate gene studies have started to explore this area, the role of genetic factors in HCC development remains poorly understood (3).
PNPLA3, also known as adiponutrin, is a member of the patatin-like phospholipase family. The rs738409 C → G single-nucleotide polymorphism (SNP), encoding the Ile 148Met variant protein of PNPLA3, is a well-described genetic determinant of hepatic steatosis (4). Several studies have established a strong link between PNPLA3 and the development of NAFLD (5). The rs738409 SNP has also recently been associated with fibrosis severity and HCC risk in patients with known liver disease. The purpose of this meta-analysis is to evaluate the association between PNPLA3 and (i) liver fibrosis severity among patients with liver disease, (ii) the development of HCC among patients with cirrhosis, and (iii) prognosis among patients with HCC.
METHODS
Literature search
We conducted a computer-assisted search with the Ovid interface to Medline, PubMed, Scopus, and Embase databases to identify relevant published articles. We searched the databases from inception through 1 May, 2013 with the following keyword combinations: “PNPLA3 or adiponutrin”. Manual searches of reference lists from applicable studies were performed to identify any studies that may have been missed by the computer-assisted search. Additional searches of Digestive Diseases Week, American Association for the Study of Liver Diseases, European Association for the Study of the Liver (EASL), International Liver Cancer Association, and American College of Gastroenterology meeting abstracts from 2010 to 2012 were performed. Finally, consultation with expert hepatologists was done to identify additional references or unpublished data. This study was conducted in accordance with the PRISMA guidelines (6).
Study selection
Two investigators (A.G.S. and H.M.) reviewed all titles of citations identified by the search strategy to generate a list of potentially relevant articles. The abstract for each potentially relevant study was then reviewed by each of the two investigators. If the applicability of a study could not be determined by title or abstract alone, the full text was reviewed. The articles were independently checked for possible inclusion and any disagreements were resolved through consensus with a third investigator (J.A.M.).
Study inclusion and exclusion criteria
Studies were included for analysis if they (i) included patients with underlying liver disease, (ii) reported stage of fibrosis, HCC incidence, and / or HCC prognosis, and (iii) stratified outcomes according to PNPLA3 genotype. We excluded studies that (i) included patients without liver disease (e.g. a cohort of obese patients), (ii) assessed outcomes in a pediatric population, (iii) assessed outcomes in post-transplant patients, or (iv) only included data regarding liver enzymes, hepatic steatosis, or inflammatory activity but not stage of fibrosis. Additional exclusion criteria included nonhuman data, lack of original data, and incomplete reports. If publications used the same cohort of patients, the manuscript with the most complete data was included.
Data extraction
Two reviewers (A.G.S. and H.M.) independently reviewed and extracted the required information from eligible studies using standardized forms. A third investigator (J.A.M.) was available to resolve any discrepancies between the two sets of extracted data. The data extraction form included the following study design items: study methodology, size and characteristics of study cohort, and inclusion and exclusion criteria. We recorded the following primary data for patients according to PNPLA3 genotype: number of patients with advanced fibrosis, number of patients who developed HCC, and tumor stage. We attempted to obtain missing data by contacting authors of included studies. One investigator (A.G.S.) assessed study quality by a modified checklist based on a modified version of the Ottawa–Newcastle scale, and discrepancies were resolved by review with an independent investigator (A.K.W.). This instrument rates observational studies based on selection of study population, comparability of study groups, and adequacy of outcome assessment (7,8).
Clinical end points and statistical analysis
The primary outcomes analyzed were the odds of advanced fibrosis and the odds of developing HCC. For each individual study, an odds ratio (OR) for each outcome of interest was calculated according to PNPLA3 genotype, using both recessive (genotype GG vs. CG + CC) and dominant (genotype CC vs. CG + GG) genetic models. A pooled estimate with corresponding 95 % confidence interval (CI) was estimated using the DerSimonian and Laird method for a random effects model.
Heterogeneity was initially evaluated graphically by examination of forest plots. The χ2-test of heterogeneity was used to statistically assess the presence of significant heterogeneity and the inconsistency index (I2) was used to assess magnitude of heterogeneity (9,10). A χ2-test P value < 0.10 or I2 values > 50 % are consistent with the possibility of substantial heterogeneity. In cases of statistical heterogeneity, meta-influence analysis, in which one study is removed at a time from the model was performed to determine if there was possible undue influence of a single study (11). Publication bias was initially evaluated graphically by funnel plot analysis and then statistically by using Begg’s test (12). An asymmetric funnel plot could suggest the possibility of small studies that were not published due to unfavorable results. Subset analyses were planned for predefined variables, including (i) location of study, (ii) etiology of underlying liver disease, and (iii) definition of advanced fibrosis.
RESULTS
Literature search
The computer-assisted search yielded 406 unique titles published through 1 May, 2013. After initial review, 175 titles were potentially appropriate. Each of these abstracts was reviewed, and an additional 83 studies were excluded. Ninety-two publications underwent full-text review, of which 32 met the inclusion criteria (Figure 1). Searches of annual meeting abstracts yielded one relevant abstract, producing a total of 33 studies, with a total of 12,735 patients, for inclusion in the meta-analysis.
Figure 1.
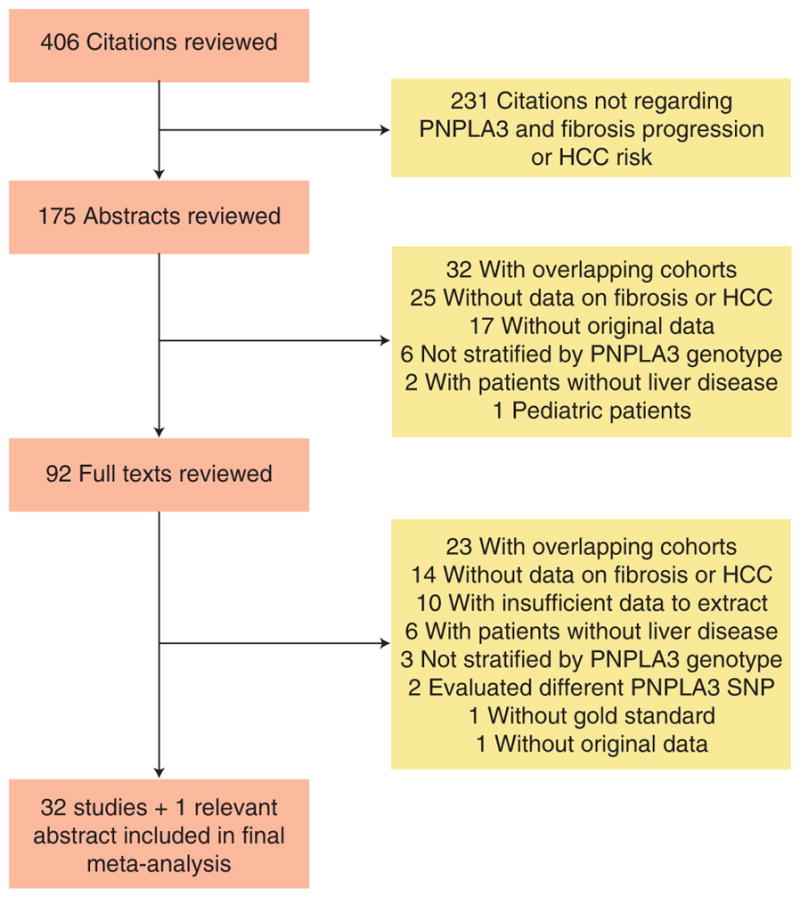
Map of literature search and selection process.
Effect of PNPLA3 rs738409 SNP on fibrosis severity
A total of 24 studies, with 9,915 patients, assessed the effect of PNPLA3 rs738409 genotype on liver fibrosis severity (13–36) (Table 1). We excluded other studies addressing the impact of PNPLA3 on fibrosis severity given the inclusion of patients without liver disease (37,38), the lack of a gold standard to assess fibrosis (39), and the use of overlapping cohorts with included publications. We also excluded a publication by Clark et al. (40) given that they assessed the rs2896019 polymorphism of PNPLA3 instead of rs738409. In brief, this SNP was associated with advanced fibrosis, defined as bridging fibrosis, under a dominant genetic model (P = 0.04) but not a recessive (P = 0.70) or additive model (P = 0.09).
Table 1.
Characteristics of studies assessing fibrosis severity
| Author and year | Number of patients | PNPLA3 genotypes | Study location | Patient population | Liver disease etiology | Fibrosis assessment | Definition of advanced fibrosis |
|---|---|---|---|---|---|---|---|
| Burlone (13) | 60 | 46% CC 32% CG 22% GG |
Europe | European | NAFLD | Liver biopsy | Bridging fibrosis |
| Dunn (14) | 417 | 57% CC 35% CG 8% GG |
United States | 81% Caucasian | Mixed cohort | Autopsy | Bridging fibrosis |
| Guichelaar (15) | 72 | 50% CC 39% CG 11% GG |
United States | Not reported | NAFLD | Liver biopsy | Portal/periportal fibrosis |
| Hotta (16) | 253 | 18% CC 42% CG 40% GG |
Japan | Japanese | NAFLD | Liver biopsy | Bridging fibrosis |
| Idilman (34) | 174 | Not available | Turkey | Turkish | NAFLD | Liver biopsy | Portal/periportal fibrosis |
| Krawczyk (17) | 899 | 54% CC 39% CG 7% GG |
Europe | European | Mixed cohort | Elastrography | Cirrhosis |
| Miyashita (18) | 235 | 31% CC 48% CG 21% GG |
Japan | Japanese | HCV | Liver biopsy | Bridging fibrosis |
| Muller (19) | 605 | Not available | Europe | Caucasian | HCV | Liver biopsy | Cirrhosis |
| Patin (20) | 1161 | 51% CC 40% CG 9% GG |
Europe | European | HCV | Liver biopsy | Bridging fibrosis |
| Payer (21) | 80 | 56% CC 39% CG 5% GG |
Europe | European | HCV-HIV | Not reported | Bridging fibrosis |
| Rembeck (35) | 359 | 60% CC 37% CG 3% GG |
Europe | European | HCV | Liver biopsy | Bridging fibrosis |
| Rotman (22) | 894 | 26% CC 45% CG 29% GG |
United States | 82% Caucasian | NAFLD | Liver biopsy | Bridging fibrosis |
| Stattermayer (23) | 478 | 56% CC 37% CG 7% GG |
Europe | European | HCV | Liver biopsy | Bridging fibrosis |
| Stattermayer (36) | 202 | Not available | Europe | European | HCV | Liver biopsy | Bridging fibrosis |
| Stickel (24) | 604 | 53% CC 38% CG 9% GG |
Europe | Caucasian | Alcohol | Liver biopsy | Cirrhosis |
| Trepo (25) | 330 | 42% CC 45% CG 13% GG |
Europe | Caucasian | Alcohol | Liver biopsy | Cirrhosis |
| Trepo (26) | 537 | 54% CC 39% CG 7% GG |
Europe | Caucasian | HCV | Liver biopsy | Bridging fibrosis |
| Valenti (29) | 819 | 52% CC 38% CG 10% GG |
Europe | Caucasian | HCV | Liver biopsy | Cirrhosis |
| Valenti (28) | 758 | 43% CC 44% CG 13% GG |
Europe | Caucasian | NAFLD | Liver biopsy | Portal/periportal fibrosis |
| Valenti (27) | 174 | 47% CC 40% CG 13% GG |
Europe | Not reported | HFE | Liver biopsy | Cirrhosis |
| Vigano (30) | 235 | 58% CC 37% CG 5% GG |
Europe | Not reported | HBV | Liver biopsy | Bridging fibrosis |
| Way (31) | 320 | 52% CC 39% CG 9% GG |
Europe | Caucasian | Alcohol | Liver biopsy | Cirrhosis |
| Zain (32) | 144 | 33% CC 44% CG 23% GG |
Malaysia | Malaysian | NAFLD | Liver biopsy | Portal/periportal fibrosis |
| Zampino (33) | 166 | 39% CC 49% CG 12% GG |
Europe | Caucasian | HCV | Liver biopsy | Bridging fibrosis |
HBV, hepatitis B virus; HCV, Hepatitis C virus; HFE, hemochromatosis; HIV, human immunodeficiency virus; NAFLD, nonalcoholic fatty liver disease.
All included articles were published in the past 5 years and most were conducted in Europe or the United States, enrolling primarily Caucasian patients, with only four being conducted elsewhere (Malaysia, Argentina, Turkey, and Japan). All studies were cross-sectional in nature, assessing the severity of fibrosis at only one point in time. Eleven studies included patients with HCV infection, seven evaluated NAFLD patients, three had patients with alcoholic cirrhosis, one included those with hemochromatosis, and two studies had mixed cohorts. We did not find any evidence of publication bias by visual inspection of the funnel plot analysis (Supplementary Figure online) or Begg’s test (P = 0.38).
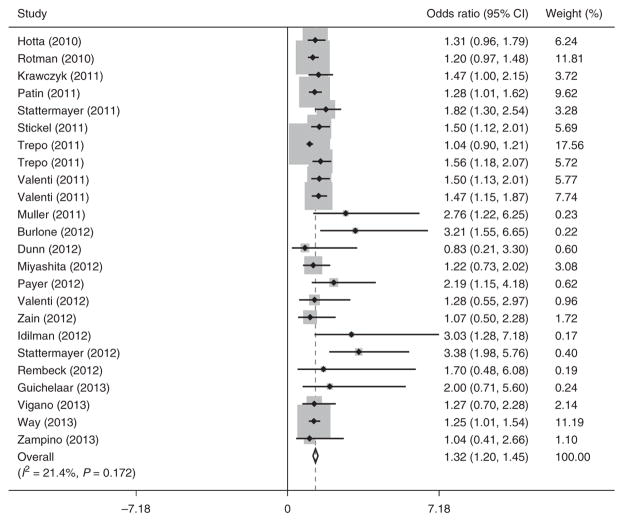
Advanced fibrosis was significantly associated with PNPLA3 polymorphism, with a similar strength of association between recessive and dominant genetic models. When assuming a recessive model, GG carriers were significantly more likely to have advanced fibrosis than did those with CG or CC genotypes (OR 1.32, 95 % CI 1.20–1.45; Figure 2). Meta-analysis with a fixed-effects approach yielded a similar association (OR 1.25, 95 % CI 1.16–1.34). A dominant model yielded a pooled OR for advanced fibrosis of 1.29 (95 % CI 1.21–1.38) for patients with CG or GG genotypes compared with those with CC genotype. On the basis of previous studies (28), a recessive model was used for subsequent analyses.
Figure 2.
Association between PNPLA3 rs738409 single-nucleotide polymorphism (SNP) and advanced fibrosis.
There was no significant heterogeneity among studies (I2 = 21.4%, P = 0.17). We performed preplanned subset analyses, according to study location and etiology of underlying liver disease, using the random-effects recessive model. The effect of PNPLA3 on fibrosis was consistent among these subsets of patients. The effect of PNPLA3 on fibrosis was similar between studies conducted in Europe or the United States and those conducted elsewhere (OR 1.35, 95 % CI 1.21–1.49 vs. OR 1.28, 95 % CI 0.95–1.60, respectively). There were also similar odds of having advanced fibrosis among patients with NAFLD or alcoholic liver disease than there were among those with other liver diseases (OR 1.23, 95 % CI 1.10–1.37 vs. OR 1.44, 95 % CI 1.28–1.61, respectively). The pooled OR for advanced fibrosis among the studies specifically evaluating HCV patients was 1.47 (95 % CI 1.27–1.67).
Association between PNPLA3 rs738409 SNP and HCC
Nine studies, with a total of 2,937 patients, assessed the risk of HCC according to rs738409 genotype (29,41–48) (Table 2). We excluded one study addressing the impact of PNPLA3 on HCC risk, given the inclusion of patients without liver disease (49). Six of the remaining nine studies were conducted solely in patients with underlying cirrhosis. All studies were conducted in Europe among Caucasian adult populations except for one study performed in Japan. There have been six retrospective cohort studies, two case-control studies, and one prospective cohort study — all published in the past 2 years. Two studies included only patients with HCV, two assessed patients with NAFLD, one included alcohol-induced cirrhosis patients, and four studies had mixed cohorts.
Table 2.
Characteristics of studies assessing risk of developing hepatocellular carcinoma
| Author and year | Number of patients | PNPLA3 genotypes | Study location | Patient population | Study design | Liver disease etiology | Number of patients with HCC |
|---|---|---|---|---|---|---|---|
| Corradini (41) | 221 | Not available | Europe | Caucasian | Retrospective cohort | HCV cirrhosis | 141 |
| Falletti (42) | 483 | 35% CC 45% CG 20% GG |
Europe | Caucasian | Retrospective cohort | Mixed cirrhosis cohort | 91 |
| Guyot (43) | 532 | 47% CC 39% CG 14% GG |
Europe | Caucasian | Prospective cohort | Mixed cirrhosis cohort | 159 |
| Hamza (44) | 304 | Not available | Europe | Not reported | Case control | Mixed cirrhosis cohort | 152 |
| Liu (45) | 318 | 42% CC 44% CG 14% GG |
Europe | Caucasian | Retrospective cohort | NAFLD | 33 |
| Nischalke (46) | 322 | 42% CC 44% CG 14% GG |
Europe | Caucasian | Case control | Mixed cirrhosis cohort | 161 |
| Takeuchi (47) | 107 | 7% CC 39% CG 53% GG |
Japan | Japanese | Case control | NAFLD | 50 |
| Trepo (48) | 325 | Not available | Europe | Caucasian | Retrospective cohort | Alcohol-related cirrhosis | 50 |
| Valenti (29) | 325 | 50% CC 39% CG 11% GG |
Europe | Caucasian | Retrospective cohort | HCV | 50 |
HCC, hepatocellular carcinoma; HCV, Hepatitis C virus; NAFLD, nonalcoholic fatty liver disease.
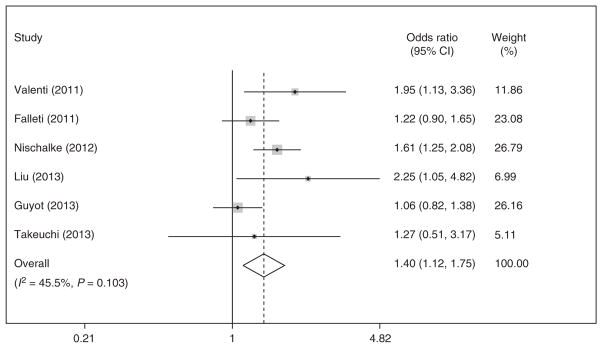
HCC risk was significantly associated with PNPLA3 polymorphism but the strength of this association depended on the genetic model. When assuming a recessive model, GG carriers were significantly more likely to have HCC than did those with CG or CC (OR 1.45, 95 % CI 1.11–1.80) (Supplementary Figure). With the recessive model, we found significant heterogeneity (I2 = 62.7 %, P = 0.006). Meta-analysis with a fixed-effects approach yielded a similar association (OR 1.33, 95 % CI 1.15–1.51). Among the six studies in which all patients had underlying cirrhosis, PNPLA3 continued to be associated with an increased risk of HCC (OR 1.58, 95% CI 1.35–1.81). Sufficient data were available from six studies to assess a pooled OR for HCC assuming a dominant model. A dominant model yielded pooled OR of 1.40 (95 % CI 1.12–1.75) for patients with CG or GG genotypes compared with those with CC genotype, without significant statistical heterogeneity (I2 = 45.5 %, P = 0.10; Figure 3).
Figure 3.
Association between PNPLA3 rs738409 single-nucleotide polymorphism (SNP) and development of hepatocellular carcinoma.
We also performed preplanned subset analyses, according to study location and etiology of underlying liver disease, using the random effects dominant model. The one study conducted outside of Europe, by Takeuchi et al. (47), had a similar point estimate for HCC risk compared with the others conducted in Europe (OR 1.45, 95 % CI 1.15–1.83). However, the effect of PNPLA3 on HCC risk appeared to be dependent on liver disease etiology. Among the subset of patients with NAFLD or alcohol abuse, PNPLA3 continued to be associated with increased odds of HCC (OR 1.67, 95 % CI 1.27–2.21). However, the association between PNPLA3 CG or GG genotype and HCC was not statistically significant among patients with other causes of cirrhosis, including HCV (OR 1.33, 95 % CI 0.96–1.82).
HCC prognosis
Three studies included extractable data about HCC prognosis according to rs738409 genotype (Table 3) (42,47,50). As each study used different surrogates of prognosis, we were not able to pool the results for the meta-analysis. Faletti and colleagues (42) assessed the effect of the PNPLA3 rs738409 on tumor biology among 94 patients with cirrhosis and HCC who had undergone liver transplantation. They found no association between the PNPLA3 SNP and Edmonson grading score or the presence of macro/ microvascular invasion (p > 0.05 for both). Takeuchi et al. (47) evaluated rs738409 among 104 patients with nonviral cirrhosis and HCC (47). Patients had a combination of liver diseases, although most had NAFLD (n = 57) or alcoholic liver disease (n = 34). The PNPLA3 SNP was associated with larger tumors (median 3.8 vs. 2.8 cm, P = 0.047), although there was no significant difference in tumor, node, metastases staging (P = 0.21). Valenti et al. (50) assessed 265 patients with cirrhosis and HCC and found a direct association between tumor size and the PNPLA3 rs738409 SNP, assuming an additive genetic model, although this did not achieve statistical significance (P = 0.06). On Cox regression, the PNPLA3 SNP alleles were associated with a nonsignificant higher risk of death (hazard ratio 1.39, 95 % CI 0.94–2.02), independent of age, gender, etiology of liver disease, and smoking status.
Table 3.
Characteristics of studies assessing hepatocellular carcinoma prognosis
| Author and year | Number of patients | Study location | Patient population | Liver disease etiology | Prognosis surrogate | Outcome |
|---|---|---|---|---|---|---|
| Falletti (42) | 94 | Europe | Caucasian | Mixed cohort | Edmonson grade and vascular invasion | P > 0.05 |
| Takeuchi (47) | 104 | Japan | Japanese | Non-viral | Tumor, node, metastases staging | P=0.21 |
| Valenti (50) | 265 | Europe | Not reported | Mixed cohort | Tumor size | P=0.06 |
Quality assessment
The quality assessment for included studies is described in Table 4. All studies adequately ascertained PNPLA3 exposure and outcomes (of advanced fibrosis or HCC development), through medical records. Although all studies assessing HCC outcomes had representative cohorts, several studies assessing fibrosis outcomes had convenience samples that may not be representative of larger populations. For example, most studies assessing fibrosis in HCV patients only included those who were undergoing pretreatment biopsy, which may underrepresent patients with morbid obesity or other significant medical comorbidities. Similarly, most studies only included patients undergoing biopsy for clinical purposes, thereby excluding any patients with cirrhosis and overt hepatic decompensation. There is only one study by Dunn et al. (14) that attempts to assess this association from a population basis, using autopsy data (14). Furthermore, we found that several studies also failed to control for important confounders, such as age, gender, and alcohol intake for presence of significant fibrosis or presence of cirrhosis, age, and gender for development of HCC.
Table 4.
Quality assessment of studies
| Author and year | Cases representative | Controls from same population | Ascertainment of PNPLA3 | Controls and cases comparablea | Outcome assessment | Statistical analysis |
|---|---|---|---|---|---|---|
| Studies assessing fibrosis severity | ||||||
| Burlone (13) | No | Yes | Yes | Yes | Yes | Yes |
| Dunn (14) | Yes | Yes | Yes | No | Yes | Yes |
| Guichelaar (15) | No | Yes | Yes | No | Yes | Yes |
| Hotta (16) | Yes | Yes | Yes | Yes | Yes | Yes |
| Idilman (34) | No | Yes | Yes | Yes | Yes | Yes |
| Krawczyk (17) | Yes | Yes | Yes | Yes | Yes | Yes |
| Miyashita (18) | No | Yes | Yes | Yes | Yes | Yes |
| Muller (19) | No | Yes | Yes | Yes | Yes | Yes |
| Patin (20) | No | Yes | Yes | No | Yes | Yes |
| Payer (21) | No | Yes | Yes | No | Yes | Yes |
| Rembeck (35) | No | Yes | Yes | No | Yes | Yes |
| Rotman (22) | Yes | Yes | Yes | Yes | Yes | Yes |
| Stattermayer (23) | No | Yes | Yes | No | Yes | Yes |
| Stattermayer (36) | No | Yes | Yes | No | Yes | Yes |
| Stickel (24) | Yes | Yes | Yes | No | Yes | Yes |
| Trepo (25) | Yes | Yes | Yes | Yes | Yes | Yes |
| Trepo (26) | No | Yes | Yes | Yes | Yes | Yes |
| Valenti (29) | Yes | Yes | Yes | Yes | Yes | Yes |
| Valenti (28) | No | Yes | Yes | No | Yes | Yes |
| Valenti (27) | No | Yes | Yes | Yes | Yes | Yes |
| Vigano (30) | No | Yes | Yes | Yes | Yes | Yes |
| Way (31) | No | Yes | Yes | No | Yes | Yes |
| Zain (32) | No | Yes | Yes | No | Yes | Yes |
| Zampino (33) | Yes | Yes | Yes | No | Yes | Yes |
| Studies assessing development of HCC | ||||||
| Corradini (41) | Yes | Yes | Yes | Yes | Yes | Yes |
| Falletti (42) | Yes | Yes | Yes | Yes | Yes | Yes |
| Guyot (43) | Yes | Yes | Yes | Yes | Yes | Yes |
| Hamza (44) | Yes | Yes | Yes | Yes | Yes | Yes |
| Liu (45) | Yes | Yes | Yes | Partial | Yes | Yes |
| Nischalke (46) | Yes | Yes | Yes | Yes | Yes | Yes |
| Takeuchi (47) | Yes | Yes | Yes | No | Yes | Yes |
| Trepo (48) | Yes | Yes | Yes | Yes | Yes | Yes |
| Valenti (29) | Yes | Yes | Yes | Partial | Yes | Yes |
HCC, hepatocellular carcinoma.
Confounders of interest for the presence of significant fibrosis were age and gender. Confounders of interest for risk of HCC were age and the presence of cirrhosis.
DISCUSSION
Our meta-analysis highlights the point that PNPLA3’s role is not limited to the development of hepatic steatosis and NAFLD, as it also appears to effect subsequent progression to advanced fibrosis and the development of HCC. We demonstrated that PNPLA3 rs738409 is associated with fibrosis severity among patients with underlying liver disease and that it is an independent risk factor for HCC among those with cirrhosis. We did not find any evidence for an association between PNPLA3 genotype and HCC prognosis; however, larger studies with a wide distribution of tumor stages are still needed.
Although its exact biological function is debated, PNPLA3 is highly expressed on the surface of lipid droplets of hepatocytes and adipose tissue (51). The rs738409 variant appears to interfere with lipoprotein export as well as to favor lipogenic activity over lipase activity, leading to hepatic fat accumulation (51–53). We found that PNPLA3 was associated with an increased risk of significant fibrosis, independent of liver disease etiology. Patients with steatosis, either due to NAFLD or alcohol abuse, had the same increased odds of having significant fibrosis as did those with other causes of liver disease, including HCV infection. However, it should be noted that most included studies assessed either NAFLD or HCV infection, with only one study assessing HBV infection and another assessing patients with hemochromatosis. Further studies assessing the association between PNPLA3 and the presence of advanced fibrosis in other liver disease etiologies are still needed.
Furthermore, the exact mechanism behind this association remains poorly understood. PNPLA3 does not appear to exert any direct association on HCV, as there is no known effect on viral load or treatment response (54). It is possible that this association is simply related to confounders, such as increased fibrosis from concomitant alcohol abuse or metabolic syndrome, among patients with PNPLA3 GG genotype. Supporting this hypothesis, Müller and colleagues (19) found that PNPLA3’s effect on fibrosis was restricted to those who had significant alcohol abuse. HCV-positive patients with significant alcohol use had 4.8-odds (95 % CI 1.39–6.38) of having cirrhosis, compared with only 1.65-odds (95 % CI 0.54–5.03) in abstainers with HCV infection (19). Unfortunately, few other studies report rates of alcohol abuse and / or metabolic syndrome among the PNPLA3 genotypes to replicate this analysis.
Only a small fraction of patients with HCV or NAFLD progress to cirrhosis and / or HCC (55). Studies using clinical risk factors alone to predict fibrosis severity and / or HCC development have been limited by modest accuracy, and inclusion of genetic risk factors may enhance risk stratification (56). It is possible that risk factors, including genetic factors such as PNPLA3, may be different according to liver disease etiology. For example, we found that PNPLA3 was associated with increased HCC risk in patients with nonalcoholic steatohepatitis or alcohol-related cirrhosis but not in patients with HCV-related cirrhosis. This finding may simply be related to smaller sample sizes in subset analyses; however, it may suggest differential mechanisms of hepatocarcinogenesis according to liver disease etiology. Identification of high-risk patients could better define a subgroup of patients, in whom liver function should be closely monitored and at whom interventions should be targeted. For example, a study from Europe suggests that weight loss in obese individuals may mitigate the increased HCC risk associated with PNPLA3 (ref. 49), although large confirmatory studies assessing the impact of weight loss on both fibrosis and HCC risk are still necessary. Similarly, Burlone et al. (13) suggested that liver biopsies could be targeted to a subset of patients with NAFLD, based on PNPLA3 genotype and liver stiffness measurement (13). Finally, accurate assessment of HCC risk among patients with cirrhosis may allow a targeted application of HCC surveillance programs (57,58).
We found that most studies to date were conducted in homogenous Caucasian populations from Europe or the United States. Studies among racially and ethnically diverse cohorts, including both Hispanics and African Americans, are needed. Previous studies have demonstrated that Hispanics and Asian Indians have higher rates of NAFLD compared with Caucasians, who in turn have higher rates when compared with African Americans (59,60). Although racial/ ethnic differences in NAFLD and HCC incidence could be attributed to the frequency of PNPLA3 variants, it is alternatively possible that race and / or ethnicity could moderate the risk of these outcomes associated with the PNPLA3 variant (4).
High quality studies, including retrospective and prospective cohort studies, have established an association between PNPLA3 and HCC risk. However, all studies to date assessing fibrosis severity according to PNPLA3 genotype have been cross-sectional in nature. Trepo et al. (26) estimated a fibrosis progression rate by using the ratio of fibrosis stage at diagnosis and estimated duration of disease. When dichotomizing patients by the median progression rate, PNPLA3 was associated with fibrosis progression using a recessive model (OR 2.65, 95 % CI 1.28–5.51). However, longitudinal studies, preferably with an assessment of fibrosis at several time points, are still needed. Furthermore, higher quality studies with more representative sampling of at-risk patients are still needed.
In summary, the PNPLA3 rs738409 C → G SNP is associated with a significantly increased risk of both advanced fibrosis and HCC among patients with liver disease. In the age of personalized medicine using genetic determinants, PNPLA3 may help define a subgroup of patients toward whom future interventions, such as weight loss techniques and / or HCC surveillance, should be directed. An improved understanding of the biological role of PNPLA3, as well as its interactions with other genes and proteins, could potentially lead to the development of therapeutic targets to reduce the risk of fibrosis progression and / or HCC susceptibility.
Supplementary Material
Study Highlights.
WHAT IS CURRENT KNOWLEDGE
Hepatocellular carcinoma (HCC) is one of the leading causes of death among patients with cirrhosis.
Nonalcoholic fatty liver disease is an increasingly common etiology for cirrhosis and HCC.
PNPLA3 rs738409 is associated with the development of nonalcoholic fatty liver disease.
WHAT IS NEW HERE
PNPLA3 is associated with an increased risk of advanced fibrosis in both nonalcoholic fatty liver disease and hepatitis C patients.
PNPLA3 is associated with an increased risk of HCC in patients with nonalcoholic steatohepatitis (NASH) or alcohol-related cirrhosis.
There is insufficient evidence to support an association between PNPLA3 and HCC prognosis but further studies in large cohorts are needed.
Acknowledgments
Financial support: This work was conducted with support from UTSTAR, NIH / NCATS grant number KL2 TR000453, NIH / NCATS grant number UL1-TR000451, and the American College of Gastroenterology Junior Faculty Development Award was awarded to Amit G. Singal. Akbar K. Waljee’s work is funded by a VA HSR & D CDA-2 Career Development Award 1IK2HX000775-01.
Footnotes
CONFLICT OF INTEREST
Guarantor of the article: Amit G. Singal, MD, MS.
Specific author contributions: Amit G. Singal was involved in study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, and study supervision. Hema Manjunath was involved in acquisition of data and critical revision of the manuscript for important intellectual content. Adam C. Yopp was involved in critical revision of the manuscript for important intellectual content. Jorge A. Marrero was involved in critical revision of the manuscript for important intellectual content. Purva Gopal was involved in critical revision of the manuscript for important intellectual content. Akbar K. Waljee was involved in analysis and interpretation of data and critical revision of the manuscript for important intellectual content.
Potential competing interests: The content is solely the responsibility of the authors and does not necessarily represent the official views of UT-STAR, the University of Texas Southwestern Medical Center and its affiliated academic and health care centers, the National Center for Advancing Translational Sciences, the Veterans Affairs, or the National Institutes of Health. The authors declare no conflict of interest.
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/ajg
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Fattovich G, Stroffolini T, Zagni I, et al. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Nishida N, Kudo M. Recent advancements in comprehensive genetic analyses for human hepatocellular carcinoma. Oncology. 2013;84(Suppl 1):93–7. doi: 10.1159/000345897. [DOI] [PubMed] [Google Scholar]
- 4.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148 M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–94. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 6.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–9. W64. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 7.Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 3rd Symposium on Systematic Reviews: Beyond the Basics 2000. [Google Scholar]
- 8.Herzog R, Alvarez-Pasquin MJ, Diaz C, et al. Are healthcare workers’ intentions to vaccinate related to their knowledge, beliefs and attitudes? A systematic review. BMC Public Health. 2013;13:154. doi: 10.1186/1471-2458-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 10.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copas JB, Shi JQ. A sensitivity analysis for publication bias in systematic reviews. Stat Methods Med Res. 2001;10:251–65. doi: 10.1177/096228020101000402. [DOI] [PubMed] [Google Scholar]
- 12.Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Stat Med. 2001;20:641–54. doi: 10.1002/sim.698. [DOI] [PubMed] [Google Scholar]
- 13.Burlone M, Rossini A, Magri A, et al. A composite score including BMI, liver stiffness, and rs738409 PNPLA3 genotype might spare liver biopsies to most NAFLD patients maintaining 95% diagnostic accuracy. Hepatology. 2012;56:885A. [Google Scholar]
- 14.Dunn W, Zeng Z, O’Neil M, et al. The interaction of rs738409, obesity, and alcohol: a population-based autopsy study. Am J Gastroenterol. 2012;107:1668–74. doi: 10.1038/ajg.2012.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guichelaar M, Gawrieh S, Olivier M, et al. Interactions of allelic variance of PNPLA3 with non genetic factors in predicting NASH and non-hepatic complications of severe obesity. Obesity. 2013;21:1935–41. doi: 10.1002/oby.20327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotta K, Yoneda M, Hyogo H, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:172. doi: 10.1186/1471-2350-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krawczyk M, Grunhage F, Zimmer V, et al. Variant adiponutrin (PNPLA3) represents a common fibrosis risk gene: non-invasive elastography-based study in chronic liver disease. J Hepatol. 2011;55:299–306. doi: 10.1016/j.jhep.2010.10.042. [DOI] [PubMed] [Google Scholar]
- 18.Miyashita M, Ito T, Sakaki M, et al. Genetic polymorphism in cyclooxygenase-2 promoter affects hepatic inflammation and fibrosis in patients with chronic hepatitis C. J Viral Hepat. 2012;19:608–14. doi: 10.1111/j.1365-2893.2011.01580.x. [DOI] [PubMed] [Google Scholar]
- 19.Müller T, Buch S, Berg T, et al. Distinct, alcohol-modulated effects of PNPLA3 genotype on progression of chronic hepatitis C. J Hepatol. 2011;55:732–3. doi: 10.1016/j.jhep.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 20.Patin E, Kutalik Z, Guergnon J, et al. Genome-wide association study identifies variants associated with progression of liver fibrosis from HCV infection. Gastroenterology. 2012;143:1244–1252. e2. doi: 10.1053/j.gastro.2012.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payer B, Reiberger T, Stift J, et al. Influence of PNPLA3 SNP on hepatic steatosis and fibrosis progression in HIV-HCV coinfected patients. J Hepatol. 2012;56:S356. [Google Scholar]
- 22.Rotman Y, Koh C, Zmuda JM, et al. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stattermayer A, Rutter K, Hofer H, et al. PNPLA3 polymorphism is patients with chronic hepatitis C—impact on steatosis, fibrosis, and treatment response. J Hepatol. 2011;54:S194. [Google Scholar]
- 24.Stickel F, Buch S, Lau K, et al. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology. 2011;53:86–95. doi: 10.1002/hep.24017. [DOI] [PubMed] [Google Scholar]
- 25.Trepo E, Gustot T, Degre D, et al. Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J Hepatol. 2011;55:906–12. doi: 10.1016/j.jhep.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 26.Trepo E, Pradat P, Potthoff A, et al. Impact of patatin-like phospholipase-3 (rs738409 C > G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology. 2011;54:60–9. doi: 10.1002/hep.24350. [DOI] [PubMed] [Google Scholar]
- 27.Valenti L, Maggioni P, Piperno A, et al. Patatin-like phospholipase domain containing-3 gene I148M polymorphism, steatosis, and liver damage in hereditary hemochromatosis. World J Gastroenterol. 2012;18:2813–20. doi: 10.3748/wjg.v18.i22.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valenti L, Nobili V, Al-Serri A, et al. The APOC3 T-455C and C-482T promoter region polymorphisms are not associated with the severity of liver damage independently of PNPLA3 I148M genotype in patients with nonalcoholic fatty liver. J Hepatol. 2011;55:1409–14. doi: 10.1016/j.jhep.2011.03.035. [DOI] [PubMed] [Google Scholar]
- 29.Valenti L, Rumi M, Galmozzi E, et al. Patatin-like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 2011;53:791–9. doi: 10.1002/hep.24123. [DOI] [PubMed] [Google Scholar]
- 30.Vigano M, Valenti L, Lampertico P, et al. PNPLA3 I148M affects liver steatosis in patients with chronic hepatitis B. Hepatology. 2013;58:1245–52. doi: 10.1002/hep.26445. [DOI] [PubMed] [Google Scholar]
- 31.Way M, McQuillin A, Gurling H, et al. The PNPLA3 I148M mutation significantly increases the risk of developing alcohol-related cirrhosis in alcohol dependent individuals. J Hepatol. 2013;58:S563. [Google Scholar]
- 32.Zain SM, Mohamed R, Mahadeva S, et al. A multi-ethnic study of a PNPLA3 gene variant and its association with disease severity in non-alcoholic fatty liver disease. Hum Genet. 2012;131:1145–52. doi: 10.1007/s00439-012-1141-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zampino R, Coppola N, Cirillo G, et al. Abdominal fat interacts with PNPLA3 I148M, but not with the APOC3 variant in the pathogenesis of liver steatosis in chronic hepatitis C. J Viral Hepatitis. 2013;20:517–23. doi: 10.1111/jvh.12053. [DOI] [PubMed] [Google Scholar]
- 34.Idilman R, Keskin O, Karatayli SC, et al. The impact of genetic variability of patatin-like phospholipase domain-containing 3 gene on histological progression of non-alcoholic fatty liver disease: A crosssectional and longitudinal follow-up study. Hepatology. 2012;56:884A–5A. [Google Scholar]
- 35.Rembeck K, Maglio C, Lagging M, et al. PNPLA 3 I148M genetic variant associates with insulin resistance and baseline viral load in HCV genotype 2 but not in genotype 3 infection. BMC Med Genet. 2012;13:82. doi: 10.1186/1471-2350-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stattermayer AF, Rutter K, Beinhardt S, et al. Association of the IL28B genotype with insulin resistance in patients with chronic hepatitis C. J Hepatol. 2012;57:492–8. doi: 10.1016/j.jhep.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 37.Francque SM, Verrijken A, Mertens I, et al. Noninvasive assessment of non-alcoholic fatty liver disease in obese or overweight patients. Clin Gastroenterol Hepatol. 2012;10:1162–8. doi: 10.1016/j.cgh.2012.06.019. quiz e87. [DOI] [PubMed] [Google Scholar]
- 38.Verrijken A, Beckers S, Francque S, et al. A gene variant of PNPLA3, but not of APOC3, is associated with histological parameters of NAFLD in an obese population. Obesity. 2013;21:2138–45. doi: 10.1002/oby.20366. [DOI] [PubMed] [Google Scholar]
- 39.Tian C, Stokowski RP, Kershenobich D, et al. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21–3. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 40.Clark PJ, Thompson AJ, Zhu Q, et al. The association of genetic variants with hepatic steatosis in patients with genotype 1 chronic hepatitis C infection. Dig Dis Sci. 2012;57:2213–21. doi: 10.1007/s10620-012-2171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corradini SG, Burza MA, Molinaro A, et al. Patatin-like phospholipase domain containing 3 sequence variant and hepatocellular carcinoma. Hepatology. 2011;53:1776. doi: 10.1002/hep.24244. author reply 1777. [DOI] [PubMed] [Google Scholar]
- 42.Falleti E, Fabris C, Cmet S, et al. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 2011;31:1137–43. doi: 10.1111/j.1478-3231.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- 43.Guyot E, Sutton A, Rufat P, et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J Hepatol. 2013;58:312–8. doi: 10.1016/j.jhep.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 44.Hamza S, Petit J, Masson D, et al. PNPLA3 rs738409 GG homozygote status is associated with increased risk of hepatocellular carcinoma in cirrhotic patients. J Hepatol. 2012;56:S281. [Google Scholar]
- 45.Liu Y, Patman G, Lethart J, et al. Carriage of PNPLA3 I148M is associated wtih an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2013;58:S516. doi: 10.1016/j.jhep.2014.02.030. [DOI] [PubMed] [Google Scholar]
- 46.Nischalke HD, Berger C, Luda C, et al. The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS ONE. 2011;6:e27087. doi: 10.1371/journal.pone.0027087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeuchi Y, Ikeda F, Moritou Y, et al. The impact of patatin-like phospholipase domain-containing protein 3 polymorphism on hepatocellular carcinoma prognosis. J Gastroenterol. 2013;48:405–12. doi: 10.1007/s00535-012-0647-3. [DOI] [PubMed] [Google Scholar]
- 48.Trepo E, Guyot E, Ganne-Carrie N, et al. PNPLA3 (rs738409 C > G) is a common risk variant associated with hepatocellular carcinoma in alcoholic cirrhosis. Hepatology. 2012;55:1307–8. doi: 10.1002/hep.25518. [DOI] [PubMed] [Google Scholar]
- 49.Burza MA, Pirazzi C, Maglio C, et al. PNPLA3 I148M (rs738409) genetic variant is associated with hepatocellular carcinoma in obese individuals. Dig Liver Dis. 2012;44:1037–41. doi: 10.1016/j.dld.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Valenti L, Motta B, Soardo G, et al. I148M PNPLA3 polymorphism is associated with clinical features in patients wtih hepatocellular carcinoma. J Hepatol. 2012;56:S129. [Google Scholar]
- 51.He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–15. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–93. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valenti L, Dongiovanni P, Ginanni Corradini S, et al. PNPLA3 I148M variant and hepatocellular carcinoma: A common genetic variant for a rare disease. Dig Liver Dis. 2013;45:619–24. doi: 10.1016/j.dld.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Valenti L, Aghemo A, Stattermayer AF, et al. Implications of PNPLA3 polymorphism in chronic hepatitis C patients receiving peginterferon plus ribavirin. Aliment Pharmacol Ther. 2013;35:1434–42. doi: 10.1111/j.1365-2036.2012.05109.x. [DOI] [PubMed] [Google Scholar]
- 55.Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–32. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- 56.Singal AG, Mukherjee A, Elmunzer BJ, et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol. 2013;108:1723–30. doi: 10.1038/ajg.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singal AG, Tiro JA, Gupta S. Improving hepatocellular carcinoma screening: applying lessons from colorectal cancer screening. Clin Gastroenterol Hepatol. 2013;11:472–7. doi: 10.1016/j.cgh.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singal AG, Yopp A, CSS, et al. Utilization of hepatocellular carcinoma surveillance among American patients: a systematic review. J Gen Intern Med. 2012;27:861–7. doi: 10.1007/s11606-011-1952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–95. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 60.Weston SR, Leyden W, Murphy R, et al. Racial and ethnic distribution of nonalcoholic fatty liver in persons with newly diagnosed chronic liver disease. Hepatology. 2005;41:372–9. doi: 10.1002/hep.20554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.