Abstract
Secondary organic aerosol (SOA) derived from the photochemical oxidation of isoprene contributes a substantial mass fraction to atmospheric fine particulate matter (PM2.5). The formation of isoprene SOA is influenced largely by anthropogenic emissions through multiphase chemistry of its multi-generational oxidation products. Considering the abundance of isoprene SOA in the troposphere, understanding mechanisms of adverse health effects through inhalation exposure is critical to mitigating its potential impact on public health. In this study, we assessed the effects of isoprene SOA on gene expression in human airway epithelial cells (BEAS-2B) through an air-liquid interface exposure. Gene expression profiling of 84 oxidative stress and 249 inflammation-associated human genes was performed. Our results show that the expression levels of 29 genes were significantly altered upon isoprene SOA exposure under non-cytotoxic conditions (p<0.05), with the majority (22/29) of genes passing a false discovery rate (FDR) threshold of 0.3. The most significantly affected genes belong to the nuclear factor (erythroid-derived 2)-like 2 (Nrf2) transcription factor network. The Nrf2 function is confirmed through a reporter cell line. Together with detailed characterization of SOA constituents, this study reveals the impact of isoprene SOA exposure on lung responses, and highlights the importance of further understanding its potential health outcomes.
Graphical Abstract
Introduction
Airborne fine particulate matter (PM2.5) plays a key role in air quality, climate, and public health. Globally, the largest mass fraction of PM2.5 is organic, dominated by secondary organic aerosol (SOA) formed from atmospheric oxidation of volatile organic compounds (VOCs).1 Isoprene from vegetation (~600 Tg yr−1)2 is the most abundant non-methane hydrocarbon emitted into Earth’s atmosphere and has been recognized as one of the major sources of global SOA production through multiphase chemistry of its oxidation products.3, 4 The principal pathways of isoprene oxidation in the atmosphere are via reactions with atmospheric oxidants, such as hydroxyl radicals (OH), nitrate radicals (NO3) and ozone (O3).5 The OH-initiated oxidation of isoprene represents a dominant pathway during daytime, and it has been recognized as an important source of SOA, generating a broad range of reactive intermediates that can further react to yield condensable species. The formation processes of isoprene SOA are known to be greatly influenced by the presence of certain anthropogenic emissions, such as nitrogen oxides (NOx = NO + NO2) and sulfur dioxide (SO2), which determine the fate of organic peroxy radical transients (RO2), promote the acid-catalyzed reactive uptake of SOA precursors, and ultimately alter the SOA composition and yields.6, 7 Isoprene SOA is a major contributor to submicron organic aerosol mass measured in densely forested areas across the world.8 In the southeastern US, where substantial biogenic VOC emissions and anthropogenic pollutants interact in summertime, isoprene SOA has been shown to account for up to 40% of organic submicron aerosol mass.9, 10 Considering the importance of isoprene emissions as a source of PM2.5, understanding the potential health effects of isoprene SOA is crucial for addressing public health concerns stemming from community exposure to PM2.5.
A few studies have investigated the potential health effects of isoprene oxidation products by in vivo or in vitro assays. Upper airway irritation was reported in mice exposed to mixtures of isoprene and oxidant (O3 and NO2) by Wilkins et al.11 The observed effects were only partially explained by residual O3 and NO2 (known inducers of airway irritation) and the identified gaseous reaction products formaldehyde, formic acid, acetic acid, methacrolein, and methylvinyl ketone, implicating unidentified gaseous products as major contributing irritants. We have previously exposed A549 human lung cells in vitro to a gas-phase mixture of first-generation products from photooxidation of isoprene and 1,3-butadiene in the presence of NOx and reported that the irritant response as measured by interleukin-8 (IL-8) protein secretion could not be based solely on O3 generated by the systems.12 These data suggested that effects of additional products, both known and yet to be identified would be required to explain the observations. Wilkins et al.13 examined the relative humidity- and time-dependence of the irritant response of mice exposed to the ozonolysis products of isoprene by measuring respiration rate. Maximum irritation was observed for low humidity and fresh mixtures, implying that during aging reaction of highly irritating intermediates with water vapor to produce less irritating mixtures. Recent studies on isoprene photooxidation have identified hydroxyhydroperoxides (ISOPOOH),14–17 isoprene epoxydiols (IEPOX),18, 19 methacrylic acid epoxide (MAE),20 and hydroxymethyl-methyl-α-lactone (HMML)21, 22 as highly reactive gas-phase intermediates leading to SOA through multiphase chemistry or further oxidation to low-volatility condensable products. Formation of highly oxidized gaseous oligomeric hydroperoxides has also been reported from isoprene ozonolysis under low RH (relative humidity) conditions,23 which may provide a more complete explanation of cellular responses and respiratory effects.
We have recently reported on oxidative potential and cellular responses to organic extracts of isoprene-derived SOA generated via the major pathway of reactive uptake of IEPOX and MAE onto acidified sulfate aerosol.24, 25 In the acellular dithiothreitol (DTT) assay,24 MAE-derived SOA exhibited stronger thiol reactivity than IEPOX-derived SOA. In accord with this report, extracts of MAE-derived SOA were found to be a stronger inducer of oxidative stress response genes than IEPOX-derived SOA in human bronchial epithelial cells (BEAS-2B).25 We have also exposed BEAS-2B cells to a photochemically-generated isoprene SOA mixture at an air-liquid interface in an Electrostatic Aerosol in Vitro Exposure System (EAVES) as a physiologically relevant simulation of exposure to airborne fine particles by inhalation.26–29 Our initial findings demonstrated that isoprene SOA enhances expression of IL-8 and cyclooxygenase-2 (COX-2) genes,29 which are known mediators of inflammatory processes associated with lung diseases, such as chronic obstructive pulmonary disease (COPD).30 This study expands our previous work by exposing BEAS-2B cells in the EAVES device to isoprene SOA generated in an outdoor smog chamber and profiling the expression of over 300 oxidative stress and inflammation-associated genes. Differentially expressed genes are components of known biological pathways that provide mechanistic insights into the potential health effects of isoprene SOA.
Materials and Methods
Generation and characterization of isoprene SOA
Isoprene-derived SOA was generated by photooxidation of isoprene under natural sunlight in the presence of acidified sulfate seed aerosols. Experiments were conducted in a 120-m3 outdoor Teflon-film chamber located on the roof of the Gillings School of Global Public Health, University of North Carolina at Chapel Hill. The chamber facility has been described in detail previously.28 Real-time aerosol size distributions and volume concentrations were monitored using a differential mobility analyzer (DMA; BMI model 2002) coupled to a mixing condensation particle counter (MCPC; BMI model 1710). The aerosol-sizing instrument was periodically calibrated and checked against standard polystyrene latex spheres (PSLs) (NIST). Prior to each experiment, the chamber was continuously flushed with at least five chamber-volumes of clean air. The background particle mass concentration was < 1 μg m−3. Acidified sulfate seed aerosols were atomized into the chamber (constant output atomizer, TSI 3076) from an aqueous solution containing 0.06 M magnesium sulfate (MgSO4) and 0.06 M sulfuric acid (H2SO4) to provide a preexisting aerosol surface for reactive uptake or partitioning of isoprene SOA precursors. After seed aerosols reached the target mass concentration (~100 μg m−3, assuming unit density), 200 ppbv NO (Airgas, 1% NO/N2) and 3.5 ppmv isoprene (Sigma-Aldrich, 99%) were injected into the chamber under natural sunlight to initiate the photochemical oxidation. Isoprene mixing ratios were measured by a Varian CP-3800 Gas Chromatograph equipped with a flame ionization detector (GC/FID). Ozone (O3) was measured with an ML9811 series O3 Photometer (American Ecotech, Warren, RI). NOx was measured with an ML9841 series NOx Analyzer (American Ecotech, Warren, RI). O3 and NOx meters were calibrated prior to experiments by gas-phase titration using a NIST standard NO tank and stable O3 source. A typical experimental profile of isoprene SOA generation is shown in Figure 1. Experiments were conducted under humid conditions (50–70% RH), as monitored with a calibrated dew point meter (Dew Prime I, EdgeTech, Marlborough, MA). The average chamber temperature was 27.4 °C. Control experiments with the same concentrations of acidified sulfate seed aerosols and gaseous precursors were conducted in the absence of sunlight. After maximum SOA growth (~35 μg m−3) was attained, aerosol samples were collected onto Teflon membrane filters (47 mm diameter, 1.0 μm pore size; Pall Life Science) at a flow rate of 16–18 L min−1 for 1 h for analysis of aerosol chemical composition by gas chromatography/electron ionization-quadrupole mass spectrometry (GC/EI-MS) with prior trimethylsilylation, and ultra-performance liquid-chromatography coupled to high-resolution quadrupole time-of flight mass spectrometry equipped with electrospray ionization (UPLC/ESI-HR-QTOFMS), as described in detail elsewhere.19, 20 All filters from experiments in Table 1 were stored at −20 °C in the dark until chemical analysis. Authentic standards of certain IEPOX-derived SOA tracers (2-methyltetrols, 3-methyltetrahydrofuran-3,4-diols, and IEPOX-derived organosulfates) and MAE/HMML-derived SOA tracers (2-methylglyceric acid and MAE-derived organosulfates) were synthesized in-house using procedures previously described by our group in order to quantify contributions of IEPOX- and MAE-derived SOA to the photochemically-generated isoprene SOA mass used in this study.31, 32 In quantifying the total IEPOX-derived SOA contribution to the total isoprene SOA mass, C5-alkene triols and oligomers of IEPOX were quantified by using the 2-methyltetrol standard as a surrogate. In quantifying the ISOPOOH-derived SOA contribution, the ISOPOOH-derived organosulfate (UPLC/ESI-HR-QTOFMS negative ion measured at m/z 231.01801, C5H11O8S−),17 was quantified using the IEPOX-derived organosulfate standard as a surrogate.
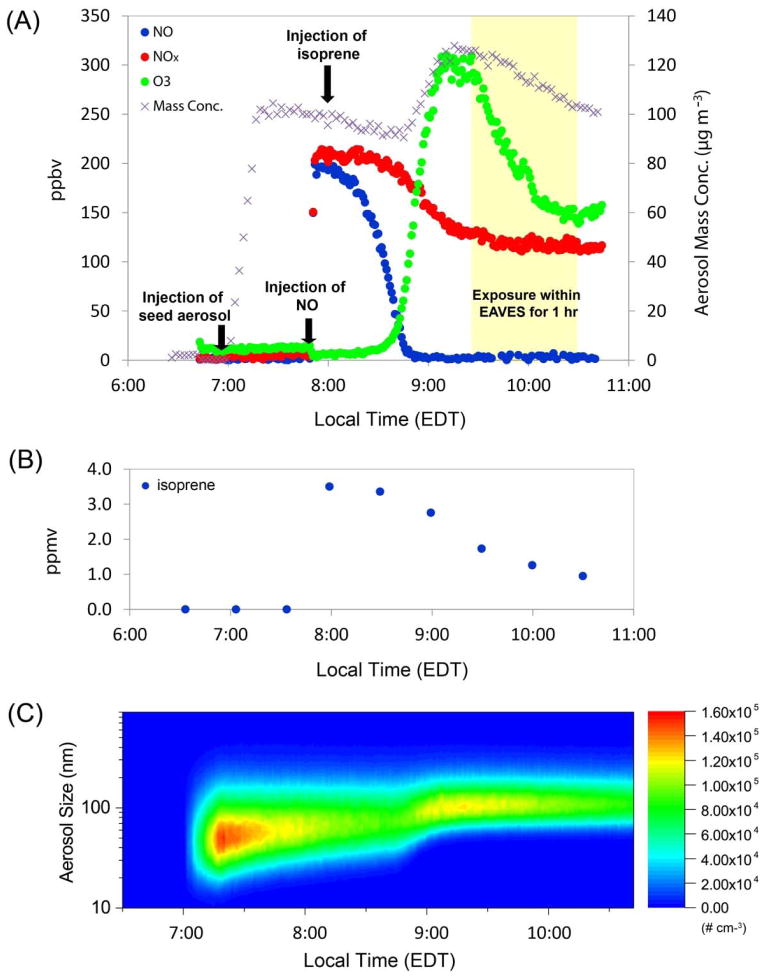
Figure 1.
Experimental profile of the photochemical oxidation experiment of isoprene. (A) Measurements of NO, NOx, O3 and aerosol mass concentration during the time course of experiment; (B) Decay of isoprene; (C) Shift of aerosol size distribution to greater values following the photochemical oxidation of isoprene, consistent with condensational SOA growth.
Table 1.
Summary of control chamber experiments for cell exposures within EAVES, with + and − representing presence or absence, respectively, of given parameters in each experiment.
| # | Experiment | isoprene | NO | Seed | Sunlight | SOA formed |
|---|---|---|---|---|---|---|
| 1 | Clean air control | − | − | − | − | − |
| 2 | Seed only control | − | − | + | − | − |
| 3 | Dark control | + | + | + | − | − |
|
| ||||||
| Photochemical run | + | + | + | + | + | |
Cell culture
BEAS-2B cells were cultured in keratinocyte growth medium (KGM-Gold™, Lonza), containing serum-free keratinocyte basal medium (KBM) supplemented with bovine pituitary extract (0.4%, w/v), human epidermal growth factor (0.1 %), insulin (0.1%), hydrocortisone (0.1%), and GA-1000 (0.1% gentamicin, 0.1% amphotericin B). Cells were grown at 37°C under an atmosphere containing 5% CO2 in a humidified incubator. Prior to experiments, cells were subcultured on collagen-coated Millicell (Human VitroCol®, Advanced BioMatrix) cell culture inserts (12 mm diameter, hydrophilic polytetrafluoroethylene (PTFE), 0.4 μm pore size, 0.69 cm2 filter area, EMD Millipore) at a density of 5×104 cells per insert for 24 h. Cell culture inserts were housed in 12-well plates with growth medium (KGM) supplied. At the time of exposure when cells reached 60–70% confluence, KGM was completely removed from the apical and basolateral cell surfaces. Cells were washed twice with phosphate buffered saline (PBS, Sigma) and placed in fresh media deprived of growth factor (KBM).
Direct air-liquid interface exposure to isoprene SOA
Air-liquid interface exposures were conducted using the Electrostatic Aerosol in Vitro Exposure System (EAVES)26 for direct deposition of isoprene SOA on cultured BEAS-2B cells. The characteristics and applications of the EAVES device have been described in detail previously.26, 27 Briefly, isoprene SOA entrained in an air stream was charged by corona discharge, and directly deposited onto the cells at the air-liquid interface by an electric field applied above the cells. The EAVES device was housed in an incubator maintained at 37 °C, and air from the chamber passed over the cells at a flow rate of 1 L min−1. The temperature and the humidity of the air stream were the same as the chamber conditions. At the time of exposure, the cells grown on permeable Millicell cell culture inserts (n=4) were transferred to a sterile titanium dish containing 1.5 mL of fresh KBM. Cell culture inserts were placed above the medium to maintain cell viability and allow exposures to be performed at an air-liquid interface. Cells were exposed to chamber aerosol using the EAVES device for 1 hour. A set of unexposed controls was maintained in the incubator. Following exposure in the EAVES, cells culture inserts were transferred to new 12-well plates containing 1 mL fresh KBM per well underneath of the cell culture inserts. The basal extracellular medium was collected after incubation for 9 h, based on prior time course experiments monitoring induction of oxidative stress and expression of inflammation-associated genes (COX-2 and IL-8) in BEAS-2B cells in response to isoprene SOA exposure through filter resuspension published in Arashiro et al.29 Total RNA was harvested using 200 μL of Trizol (Life Technologies). Extracellular medium and the isolated RNA samples were stored at −20 °C and −80 °C, respectively, until further analysis.
Cytotoxicity analysis
The release of lactate dehydrogenase (LDH) within the extracellular medium of each sample well was measured, and compared to that of unexposed controls maintained in the incubator to ensure that toxicity of exposure would not interfere with gene expression analysis. LDH levels were assessed using the LDH cytotoxicity detection kit (Takara) according to the manufacturer’s protocol. Briefly, 50 μL of each sample was mixed with 50 μL of the assay mixture containing the catalyst and dye solution (iodotetrazolium chloride (INT) and sodium lactate), and incubated at room temperature in the dark for 30 min. The reaction was stopped by adding 50 μL of HCl (1N) to each well. The absorbance of samples was measured at 492 nm, against a reference wavelength of 620 nm. Low and high controls, which measure the levels of spontaneous LDH release from untreated cells and the maximum LDH activity that can be released from 100% killing of cells (treatment with 1% Triton X-100), were determined to calculate the percentage of induced cytotoxicity by isoprene SOA exposure.
Gene expression profiling
Isolated total RNA was further purified using Direct-zol RNA MiniPrep (Zymo Research) and stored in nuclease free water as previously described.25 The quality and quantity of RNA were assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies) and the NanoDrop 2000c spectrophotometer (Thermo Scientific). Gene expression analysis was performed according to the manufacturer’s instructions using 50–100 ng of total purified RNA with two pathway-focused panels: (1) nCounter® GX Human Inflammation Kit comprised of 249 human genes (NanoString), and (2) Human Oxidative Stress Plus RT2 Profiler PCR Array (Qiagen, 96-well format, catalog #: PAHS-065Y) comprised of 84 oxidative stress-associated genes. Four replicates from the same treatment were tested for the gene expression analysis, which was a pool of cells from two different experiments to take into account both biological and technical replicates. The data for the inflammation and the oxidative stress pathways were analyzed separately for data normalization to account for their technical differences. Analysis and normalization of the NanoString raw data was conducted using nSolver Analysis Software v2.5 (NanoString Technologies). Raw counts were normalized to a panel of 6 housekeeping genes: clathrin heavy chain (CLTC), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), beta-glucuronidase (GUSB), hypoxanthine phosphoribosyltransferase 1 (HPRT1), phosphoglycerate kinase 1 (PGK1), and tubulin beta class I (TUBB). Human Oxidative Stress Plus RT2 Profiler PCR Array data were analyzed using RT2 Profiler PCR Array Data Analysis software, version 3.5. The housekeeping gene beta-actin (ACTB) was used for normalization. Then, the differential expression of individual genes between the exposure and control groups was calculated as fold changes (FC) using the comparative cycle threshold (2 −ΔΔCT) method.33 Results with FC cutoffs ≥ 1.5 and p < 0.05 were considered significant.34 The false discovery rate (FDR)-adjusted p-value was calculated based on the Benjamini-Hochberg method.35
Data analysis
Pathway-based analysis was performed for significantly altered genes using the ConsensusPathDB database,36–38 which is a database system for the integration of human gene functional interactions to provide biological pathway information for a gene set of interest. Over-representation analysis was used to interpret the function of altered genes. The significance (p-value) of the observed overlap between the significantly altered gene set and the members of predefined pathways was calculated based on the hypergeometric distribution. Criteria of a minimum overlap of 2 genes between the input gene list and the pathway set at a p-value cutoff of 0.01 were applied.39 Pathway annotation was performed to provide biological pathway information for each gene set. The gene-gene interaction networks were constructed and visualized using the GeneMANIA Cytoscape app (version 3.4.1) to predict the putative function of altered genes.40, 41
Measurement of the nuclear factor (erythroid-derived 2)-like 2 (Nrf2) activity
The function of Nrf2 was measured through a reporter cell line to support the pathway analysis results given that the transcription factor is activated post-transcriptionally and/or post-translationally often without changing its overall expression level.42, 43 BEAS-2B cells were transduced with a lentiviral vector containing tandem repeats of the human Nrf2 consensus DNA-binding site linked to the firefly luciferase gene and a constitutively expressed green fluorescent protein (GFP) reporter (a kind gift from Dr. Steve Simmons, National Health and Environmental Effects Research Laboratory, US EPA, Durham, NC) at a multiplicity of infection (MOI) of 20. Stably transduced BEAS-2B Nrf2-luciferase reporter cells were plated in white 96-well plates (Corning Inc, Corning, NY) at 6.0×105 cells/well in KGM and cultured overnight at 37°C. Cells were then exposed to photochemically generated isoprene SOA constituents in KGM (0.1 mg/mL and 0.01 mg/mL), dark controls in KGM (0.1 mg/mL and 0.01 mg/mL), or KGM alone (media controls) for 24 h. The detailed procedure for filter resuspension of isoprene SOA constituents could be found in Lin et al.25 Following challenge, cell culture medium was aspirated, cells were lysed, and cell lysates were analyzed for GFP expression and luciferase activity on a BMG CLARIOstar microplate reader (BMG Labtech, Ortenberg, Germany). Luciferase activity was normalized to GFP expression and reported as percent Nrf2 activation observed in cells exposed to cell culture medium alone.
Results
Characterization of exposure conditions
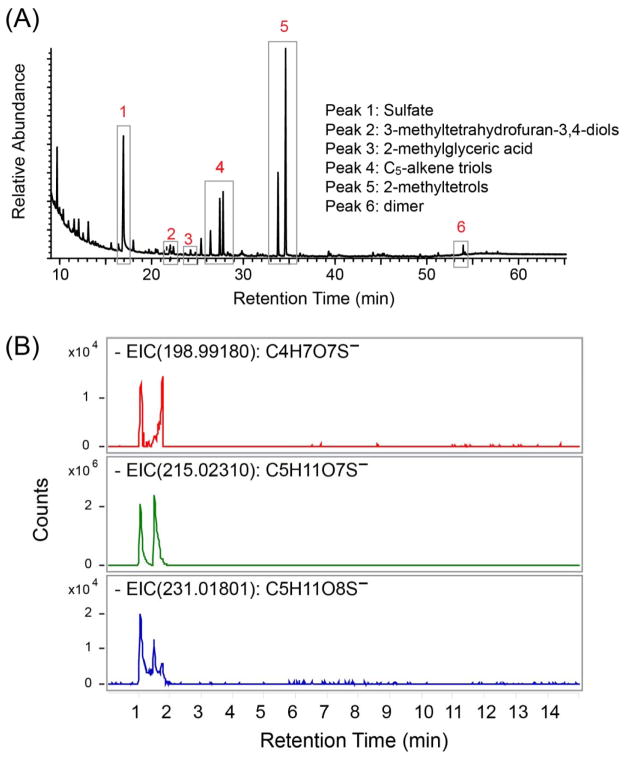
We exposed BEAS-2B cells to the mixtures of isoprene SOA generated from the photochemical oxidation of isoprene in the presence of acidified sulfate seed aerosols under natural sunlight, representing the downwind-urban atmospheric scenario in which isoprene is oxidized primarily under low-NOx conditions and subsequent SOA formation is influenced by mixing with anthropogenic emissions. Typical experimental profiles of real-time aerosol size distribution, levels of gaseous precursors and reaction products (isoprene, NOx, and O3) are shown in Figure 1. On initiation of the photochemical experiment, acidified sulfate seed aerosol was atomized into the chamber using a constant output atomizer. Once the seed aerosol reached a target level of ~ 100 μg m−3 and had stabilized for 30 min, 200 ppbv of NO and 3.5 ppmv of isoprene were injected into the chamber. Figure 1A shows that isoprene SOA formation commenced as the NO level approached zero through reaction with isoprene-derived peroxy radicals (RO2), forming NO2 and O3 as secondary products. Figure 1B shows the rapid decay of isoprene precursor during the time course of experiment. This observation is consistent with previous work by Kroll et al.44 that suggested RO2 + NO reactions suppressed SOA formation from isoprene due to the production of volatile products that do not yield condensable species. SOA growth resulted in a mass concentration increase of ~ 35 μg m−3. The mode of the aerosol size distribution shifted from ~70 nm at the beginning of the experiment to ~ 150 nm after injection of gaseous precursors, indicating growth from condensation of photochemical oxidation products to form SOA (Figure 1C). BEAS-2B cells were exposed to isoprene SOA for 1 h within the EAVES device, when the SOA growth reached the maximum. The 1 h exposure time was optimal for cell viability within the EAVES device based on prior studies.26, 27, 29 Isoprene SOA dose to the cells was estimated to be 58 ng cm−2 based on the deposition efficiency characterized by de Bruijne et al.26 using 198 nm fluorescent-labeled polystyrene latex (PSL) sphere particles. This size of PSL sphere particles is close to the mode of isoprene SOA-coated sulfate aerosols observed in the current study. Simultaneously with exposure, Teflon filters were collected for chemical analyses of particle-phase SOA constituents (Figure 2). The results show that isoprene SOA formation under the experimental conditions occurred predominantly via the channel of the reactive uptake of IEPOX onto preexisting acidic sulfate seed aerosols. By using authentic standards synthesized in-house, high concentrations of IEPOX-SOA tracers, including 3-methyltetrahydrofuran-3,4-diols, C5-alkene triols, 2-methyltetrols, dimers, and the IEPOX-derived organosulfates (m/z 215.0231, C5H11O7S−), were measured that contributed to ~80% of the observed SOA mass. MAE/HMML-SOA tracers of isoprene oxidation via the high-NOx channel, including 2-methylglyceric acid and its organosulfate derivative (m/z 198.9918, C4H7O7S−) were also observed and contributed ~1.4% of the total isoprene SOA mass. A smaller contribution (~0.8%) was measured for the ISOPOOH-derived SOA tracer (m/z 231.01801, C5H11O8S−) via the low-NOx, non-IEPOX route.17 These results are consistent with recent kinetics studies and field measurements showing efficient reactive uptake of IEPOX and a large IEPOX contribution to ambient SOA.45–47
Figure 2.
Chemical characterization of chamber-generated isoprene SOA: (A) GC/EI-MS total ion chromatogram (TIC) and (B) UPLC/(−)ESI-HR-QTOFMS extracted ion chromatograms (EICs) at m/z 198.99180, 215.02310, and 231.01801 corresponding to the MAE-, IEPOX-, and ISOPOOH-derived organosulfates, respectively.
Cytotoxicity measurements
A series of control experiments were conducted to ensure that the observed cellular responses are attributable to isoprene SOA (Table 1). Relative to cells maintained in the cell culture incubator, LDH assays indicate no significant cytotoxicity (p > 0.05) to the BEAS-2B cells in the EAVES device exposed to clean air, acidified sulfate seed aerosol only, or to dark controls exposed to acidified sulfate seed aerosol and gaseous precursors simultaneously without sunlight. LDH release in the photochemical experiments was slightly enhanced compared to incubator controls (LDH fold increase= 1.19; p< 0.05), but the overall cell death was estimated to be only 3% based. Our prior work has also demonstrated that the cellular responses observed from EAVES were negligible to gases,28 and mostly attributable to the particle-phase products.27, 29 These results confirm that the exposure conditions were not significantly toxic, and thus were appropriate for investigation of exposure-induced early genomic response.
Characterization of altered gene expression
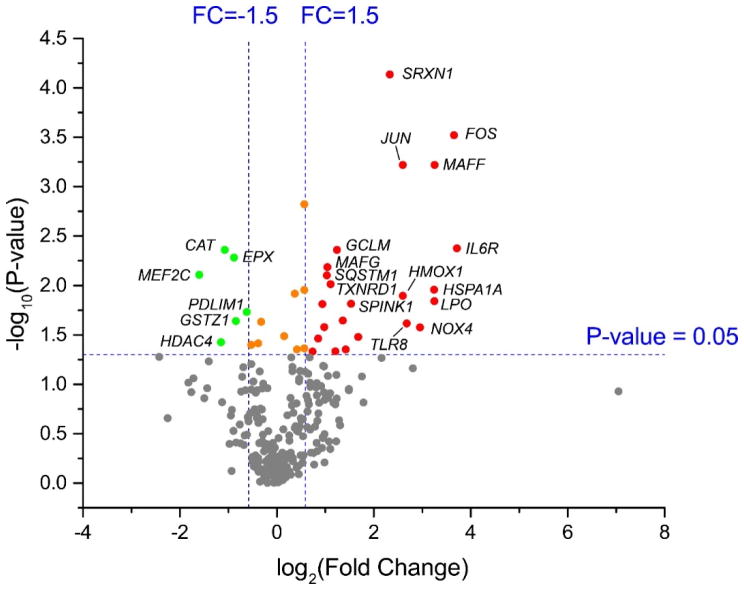
To identify differential expression of genes in lung cells associated with exposure to isoprene SOA, we extracted mRNA from dark controls and photochemical experiments. The dark controls, which take into account the effects of seed aerosol and gaseous precursors, were selected as baseline controls to normalize response in photochemical experiments. Previous studies in our laboratory demonstrated that the mass loading of sulfate seed aerosol used for the present study did not induce significant inflammatory gene expression in BEAS-2B cells over 1 hr exposure time in the EAVES.29 BEAS-2B cells exposed to isoprene SOA showed differential expression of 29 genes (Figure 3), where 23 displayed increased expression and 6 displayed decreased expression. Table 2 shows the list of genes with significant changes (fold change cut-off = ±1.5, p < 0.05). Thirteen genes associated inflammatory response were derived from the NanoString platform, and 16 genes involved in oxidative stress were from the RT2 Profiler. After adjusting for multiple comparisons, 22 genes passed a FDR threshold of 0.3,48 and 4 genes passed a stringent FDR threshold of 0.05 (Figure S2). The 22 differentially expressed genes in the gene sets exposed to isoprene SOA with FDR < 0.3 were analyzed for enrichment within biological pathways.48 The WikiPathways49-based analyses revealed that the oxidative stress (p = 7.58×10−12) and Nrf2 pathways (p = 5.77×10−11) are the most significantly enriched. Specifically, 6 of 16 genes (37.5 %) represented in the RT2 Profiler platform, and 2 of 6 genes (33.3 %) represented in the NanoString platform for the Nrf2 pathway. The BioCarta50 database also identified the pathway for oxidative stress induced gene expression via nuclear factor erythroid 2-like 2 (NRF2) (p = 1.12×10−10) to be significantly enriched, with 1 of 16 genes (6.3%) represented in the RT2 Profiler platform, and 4 of 6 genes (66.7%) represented in the NanoString platform.
Figure 3.
Volcano plot of differentially expressed genes in BEAS-2B cells upon exposure to isoprene SOA. Dots in red indicate the significantly upregulated genes (23 genes). Dots in green indicate the significantly downregulated genes (6 genes). Dots in gray represent genes that did not significantly change in expression. A full list of differentially expressed genes can be found in Table 2.
Table 2.
List of differentially expressed oxidative stress and inflammation-associated genes in BEAS-2B cells upon exposure to isoprene SOA within the EAVES device. Fold changes in expression were determined relative to the dark controls.
| Gene Name | Fold Change | p value | FDR-adjusted p value* | Panel |
|---|---|---|---|---|
| AREG | 2.67 | 0.0443 | 0.4518 | NanoString |
| BCL6 | 1.97 | 0.0263 | 0.4518 | NanoString |
| CD55 | 3.22 | 0.0330 | 0.4518 | NanoString |
| FOS | 12.59 | 0.0003 | 0.0498 | NanoString |
| HDAC4 | −2.22 | 0.0375 | 0.4518 | NanoString |
| IL23R | 2.57 | 0.0226 | 0.4518 | NanoString |
| IL6R | 13.11 | 0.0042 | 0.2092 | NanoString |
| JUN | 6.04 | 0.0006 | 0.0498 | NanoString |
| MAFF | 9.53 | 0.0006 | 0.0498 | NanoString |
| MAFG | 2.06 | 0.0065 | 0.2698 | NanoString |
| MAFK | 1.92 | 0.0154 | 0.4261 | NanoString |
| MEF2C | −3.03 | 0.0078 | 0.2775 | NanoString |
| TLR8 | 6.40 | 0.0241 | 0.4518 | NanoString |
|
| ||||
| BAG2 | 1.67 | 0.0466 | 0.2059 | RT2 Profiler |
| CAT | −2.11 | 0.0044 | 0.1095 | RT2 Profiler |
| EPX | −1.84 | 0.0052 | 0.1095 | RT2 Profiler |
| GCLC | 2.31 | 0.0462 | 0.2059 | RT2 Profiler |
| GCLM | 2.36 | 0.0044 | 0.1095 | RT2 Profiler |
| GLA | 1.80 | 0.0343 | 0.1918 | RT2 Profiler |
| GSTZ1 | −1.80 | 0.0228 | 0.1473 | RT2 Profiler |
| HMOX1 | 6.05 | 0.0127 | 0.1165 | RT2 Profiler |
| HSPA1A | 9.47 | 0.0110 | 0.1162 | RT2 Profiler |
| LPO | 9.51 | 0.0143 | 0.1165 | RT2 Profiler |
| NOX4 | 7.73 | 0.0264 | 0.1585 | RT2 Profiler |
| PDLIM1 | −1.54 | 0.0185 | 0.1297 | RT2 Profiler |
| SPINK1 | 2.88 | 0.0153 | 0.1165 | RT2 Profiler |
| SQSTM1 | 2.04 | 0.0079 | 0.1162 | RT2 Profiler |
| SRXN1 | 5.02 | 0.0001 | 0.0061 | RT2 Profiler |
| TXNRD1 | 2.16 | 0.0097 | 0.1162 | RT2 Profiler |
Benjamini-Hochberg (BH) Method
Discussion
Exposure to fine particulate air pollution is associated with increased cardiovascular and pulmonary morbidity and mortality.51 Generation of reactive oxygen species (ROS) in the respiratory system leading to oxidative stress and inflammation are considered to be strongly correlated with PM2.5-induced health effects.52
The chemical composition of isoprene SOA to which cells were exposed in this work is formed predominantly through the low-NOx channel because of the high VOC/NOx ratio used for the experiments. Although no additional OH source was added to the chamber, the photolysis of nitrous acid (HONO) formed at the chamber walls in the early morning and photolysis of ozone in the presence of water vapor yield OH radicals. Under such NOx-limited conditions, the majority of peroxy radicals (RO2) from OH-initiated oxidation react with hydroperoxyl radicals (HO2) to yield hydroperoxides (ROOH) and a wide array of multifunctional oxidation products that can contribute to SOA formation. The chamber-generated SOA has a molecular composition similar to ambient PM2.5 observed during the summertime in the southeastern U.S.17, 46, 47
Previous reports based on the acellular DTT assay demonstrated stronger oxidizing potential for total isoprene SOA than for SOA derived from the IEPOX, which is a major isoprene photooxidation product.24 Organic peroxides could be a major contributor to this difference, in light of the observation that isoprene-derived hydroperoxide standards induced the strongest responses of the assayed isoprene SOA components.24 In addition, recent analysis of organic hydroperoxides in photochemically-generated isoprene SOA mixture by Jiang et al.53 also reveals a direct correlation of organic hydroperoxide contents with DTT consumption. Exposure of cells to extractable SOA constituents from total isoprene SOA and IEPOX-derived SOA also indicates more potent induction of oxidative stress response genes by isoprene SOA extracts.25, 29 These observations highlight the potential significance of hydroperoxide constituents in total isoprene SOA, since they would not have been present in the aerosols generated from pure IEPOX precursors. Although ISOPOOH- and MAE-derived SOA constituents account for only a small fraction of isoprene SOA produced under chamber conditions in this study (<5% based on SOA tracers shown in Figure 2), they may be the major reactive components responsible for the induced cellular responses.
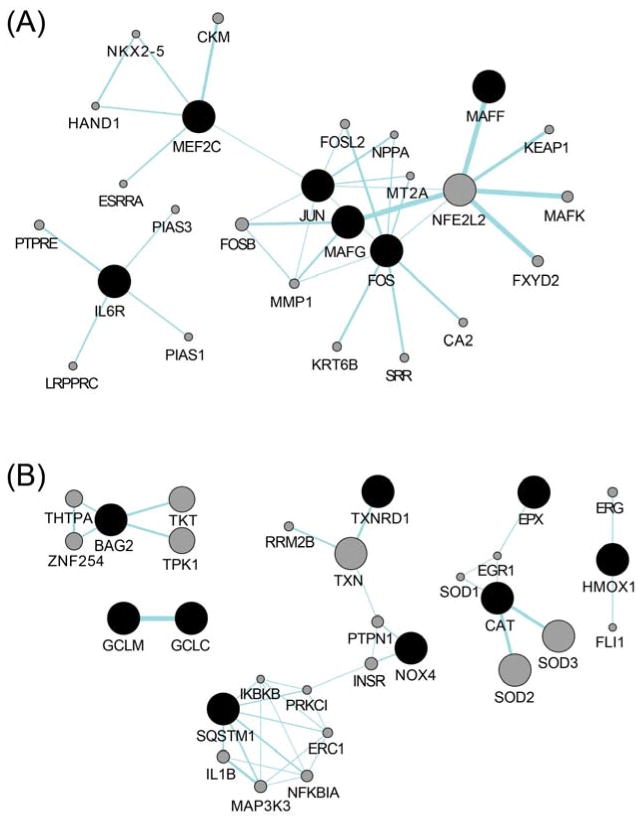
In this study, mRNA levels of 84 oxidative stress and 249 inflammation-associated genes were profiled to gain insight into potential mechanisms leading to adverse health effects induced by isoprene SOA. Expression of 29 genes was found to be significantly modified (p < 0.05), with 22 genes passing FDR<0.3 following exposure to isoprene SOA under conditions of minimal cytotoxicity (16/22 genes derived from the human oxidative stress RT2 Profiler, and 6/22 genes derived from the NanoString human inflammation platform, respectively). Notably, while only minimal inflammatory responses were observed under the given exposure condition, the differentially expressed genes identified through the NanoString platform, including FBJ murine osteosarcoma viral oncogene homolog (FOS), jun proto-oncogene (JUN), v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (MAFF), v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (MAFG), and v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (MAFK) closely mirror the oxidative stress response via Nrf2 pathway (Figure 4A).
Figure 4.
Networks of gene-gene functional interactions for differentially expressed genes (FDR<0.3) derived from (A) NanoString human inflammation platform, and (B) human oxidative stress plus RT2 Profiler. Black nodes represent the input genes which were significantly altered upon isoprene SOA exposure, and gray nodes represent predicted related genes. Links denote associated pathways. The interactive gene network construction was performed and visualized using the GeneMANIA Cytoscape app.
The identification of Nrf2 pathway in cells exposed to isoprene SOA is in accordance with our previous findings using the DTT assay,24 which measures the thiol reactivity of PM samples as a surrogate for their ROS generation potential. This is further confirmed with our measurement of Nrf2 activity through the reporter cell line exposed to isoprene SOA (Figure 5). The activity of Nrf2 is primarily regulated via its interaction with Keap1 (Kelch-like ECH-associated protein 1), where Keap1 binds to Nrf2 and tethers its nuclear translocation under basal conditions. The sulfhydryl groups within Keap1 act as sensors for oxidants and electrophiles. In the presence of ROS, critical cysteine residues (C151, C273 and C288) in Keap1 become oxidized and cause a conformational change of Keap1.54 As a consequence, Nrf2 is released from Keap1, and nuclear translocation of Nrf2 occurs. Thus, the results from the acellular DTT assay provide a plausible explanation to the cysteine-thiol modifications within cells that lead to activation of Nrf2-related gene expression. Significant upregulation of both glutamate-cysteine ligase catalytic subunit (GCLC) and glutamate-cysteine ligase modifier subunit (GCLM) genes identified via human oxidative stress RT2 Profiler (Figure 4B) also points to the activation of cysteine metabolism and glutathione biosynthesis.
Figure 5.
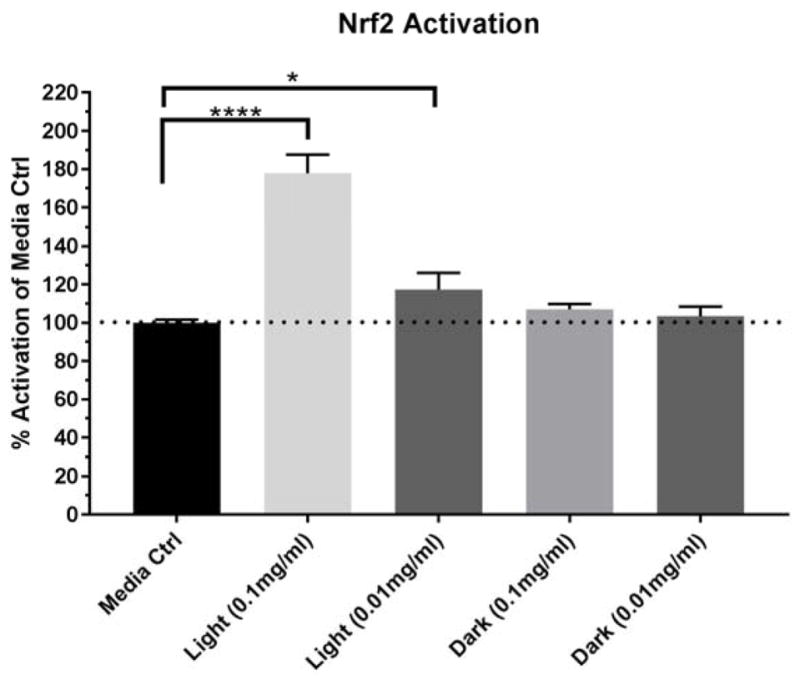
Isoprene SOA constituents-induced Nrf2 activity in the reporter cell line. Data is presented as mean ± SD (N=3). Statistical analyses were done using a one-way ANOVA and Dunnett’s multiple comparisons post-hoc test. Significance represented as *p<0.05 and ****p<0.0001.
Expression of the small musculoaponeurotic fibrosarcoma (sMAF) transcription factors has been linked to stress response and detoxification pathways. The sMAFs are bZIP proteins that can form heterodimeric complexes with the bZIP protein Nrf2 and bind at the antioxidant response element (ARE) to initiate transcription of target antioxidant genes.55, 56 Up-regulation of the three sMAF genes in this study agrees with our previous findings that isoprene SOA constituents induce oxidative stress responses in BEAS-2B cells,25 and further provides mechanistic insights into the cellular response to isoprene SOA exposure. Nrf2-sMAF complex has also been implicated in inflammasome activation.56 These findings are consistent with exposure to isoprene SOA leading to an increased burden of cellular oxidative stress which induces a variety of cellular responses, including activation of redox-sensitive transcription factors and up regulation of the expression of inflammatory response genes as observed here.
The induction of sulfiredoxin 1 (SRXN1) expression shown in this study is also in accord with the suggestion that the Nrf2 signaling is activated. SRXN1 is a target of Nrf2 regulation coding for a protein functioning in antioxidant defense,57 and regulated by the transcription factor activation protein-1 (AP-1).58 FOS and JUN are protooncogenes and members of the AP-1 family of bZIP transcription factors that function as heterodimers. They are transcribed in response to a wide variety of stimuli, including oxidative stress and inflammation,59 and up-regulation observed here is consistent with the generation of ROS by isoprene SOA. JUN is critical to maintenance of lung redox homeostasis, and deletion has been linked to progressive emphysema and consequent lung inflammation in mice exposed to cigarette smoke.60
Some caution is necessary in the extrapolation of our results to the in vivo situation. The lung is a complex organ. Lung lining fluid covering the respiratory epithelium contains proteins and antioxidants potentially mitigating the effects of inhaled chemicals. Such effects are not taken into consideration in this work. In addition, the BEAS-2B cell line is immortalized, and some biological functions may have been altered relative to primary cell lines. Although primary cells are more physiologically relevant, however, the impact of variability between donors could be significant and needs to be fully investigated. BEAS-2B is a stable proliferative cell line with significant antioxidative capacity.61 Reproducible results from the BEAS-2B cell line are critical to gaining initial insights into cellular effects of isoprene SOA exposure.
Taken together, this study characterizes differential gene expression in response to SOA derived from isoprene as a single source and to apply genomics assessment as a tool to gain insight into potential response mechanisms. The work is directly relevant to exposures in the southeastern US, where isoprene is a major source of SOA during the summer months.9, 10, 46, 47 Activation of Nrf2-associated genes has been identified with responses to oxidative stress and linked to traffic-related air pollution exposure in both toxicological and epidemiological studies,62, 63 where repeated low-dose exposures are proposed to lead to lung oxidative damage and systemic inflammatory reactions in vivo.64 Their implicit involvement in this study suggests biological significance with exposures to all sorts of PM types.43, 62, 65 Future studies are warranted to further understand its potential clinical significance and the link to health outcomes.
Supplementary Material
Acknowledgments
Research described in this article was conducted under contract to the Health Effects Institute (HEI), an organization jointly funded by the United States Environmental Protection Agency (EPA) (Assistance Award No. R-82811201) and certain motor vehicle and engine manufacturers. This research was supported in part by a grant from the National Institute of Environmental Health Sciences (T32ES007018). The contents of this article do not necessarily reflect the views of HEI, or its sponsors, nor do they necessarily reflect the views and policies of the EPA or motor vehicle and engine manufacturers.
Footnotes
The authors declare no conflict of interest.
Supporting Information Available. Gene symbols and full names of 84 oxidative stress-associated genes and housekeeping genes included in RT2 Profiler™ PCR Array Human Oxidative Stress Pathway Plus (PAHS-065Y) are provided in Table S1. Gene symbols and full names of 249 Inflammation-associated genes and housekeeping genes included in nCounter® GX Human Inflammation Kit are provided in Table S2. Pathways associated with isoprene SOA exposure are listed in Table S3. Venn diagram of two gene expression platforms profiled in this study is provided in Figure S1. Visualization of differentially expressed genes that pass various FDR thresholds is shown in Figure S2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hallquist M, Wenger JC, Baltensperger U, Rudich Y, Simpson D, Claeys M, Dommen J, Donahue NM, George C, Goldstein AH, et al. The formation, properties and impact of secondary organic aerosol: current and emerging issues. Atmos Chem Phys. 2009;9(14):5155–5236. [Google Scholar]
- 2.Guenther A, Karl T, Harley P, Wiedinmyer C, Palmer PI, Geron C. Estimates of global terrestrial isoprene emissions using MEGAN (Model of Emissions of Gases and Aerosols from Nature) Atmos Chem Phys. 2006;6(11):3181–3210. [Google Scholar]
- 3.Carlton AG, Wiedinmyer C, Kroll JH. A review of secondary organic aerosol (SOA) formation from isoprene. Atmos Chem Phys. 2009;9(14):4987–5005. [Google Scholar]
- 4.Pöschl U, Shiraiwa M. Multiphase chemistry at the atmosphere–biosphere interface influencing climate and public health in the anthropocene. Chem Rev. 2015;115(10):4440–4475. doi: 10.1021/cr500487s. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson R, Arey J. Atmospheric degradation of volatile organic compounds. Chem Rev. 2003;103(12):4605–4638. doi: 10.1021/cr0206420. [DOI] [PubMed] [Google Scholar]
- 6.Edney EO, Kleindienst TE, Jaoui M, Lewandowski M, Offenberg JH, Wang W, Claeys M. Formation of 2-methyl tetrols and 2-methylglyceric acid in secondary organic aerosol from laboratory irradiated isoprene/NOX/SO2/air mixtures and their detection in ambient PM2.5 samples collected in the eastern United States. Atmos Environ. 2005;39(29):5281–5289. [Google Scholar]
- 7.Surratt JD, Murphy SM, Kroll JH, Ng NL, Hildebrandt L, Sorooshian A, Szmigielski R, Vermeylen R, Maenhaut W, Claeys M, et al. Chemical composition of secondary organic aerosol formed from the photooxidation of isoprene. J Phys Chem A. 2006;110(31):9665–9690. doi: 10.1021/jp061734m. [DOI] [PubMed] [Google Scholar]
- 8.Hu WW, Campuzano-Jost P, Palm BB, Day DA, Ortega AM, Hayes PL, Krechmer JE, Chen Q, Kuwata M, Liu YJ, et al. Characterization of a real-time tracer for isoprene epoxydiols-derived secondary organic aerosol (IEPOX-SOA) from aerosol mass spectrometer measurements. Atmos Chem Phys. 2015;15(20):11807–11833. [Google Scholar]
- 9.Budisulistiorini SH, Baumann K, Edgerton ES, Bairai ST, Mueller S, Shaw SL, Knipping EM, Gold A, Surratt JD. Seasonal characterization of submicron aerosol chemical composition and organic aerosol sources in the southeastern United States: Atlanta, Georgia, and Look Rock, Tennessee. Atmos Chem Phys. 2016;16(8):5171–5189. [Google Scholar]
- 10.Xu L, Guo H, Boyd CM, Klein M, Bougiatioti A, Cerully KM, Hite JR, Isaacman-VanWertz G, Kreisberg NM, Knote C, et al. Effects of anthropogenic emissions on aerosol formation from isoprene and monoterpenes in the southeastern United States. Proc Natl Acad Sci USA. 2015;112(1):37–42. doi: 10.1073/pnas.1417609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkins CK, Clausen PA, Wolkoff P, Larsen ST, Hammer M, Larsen K, Hansen V, Nielsen GD. Formation of strong airway irritants in mixtures of isoprene/ozone and isoprene/ozone/nitrogen dioxide. Environ Health Perspect. 2001;109(9):937–941. doi: 10.1289/ehp.01109937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle M, Sexton KG, Jeffries H, Bridge K, Jaspers I. Effects of 1,3-Butadiene, Isoprene, and Their Photochemical Degradation Products on Human Lung Cells. Environ Health Perspect. 2004;112(15):1488–95. doi: 10.1289/ehp.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins CK, Wolkoff P, Clausen PA, Hammer M, Nielsen GD. Upper airway irritation of terpene/ozone oxidation products (TOPS). Dependence on reaction time, relative humidity and initial ozone concentration. Toxicol Lett. 2003;143(2):109–114. doi: 10.1016/s0378-4274(03)00115-2. [DOI] [PubMed] [Google Scholar]
- 14.Paulot F, Crounse JD, Kjaergaard HG, Kürten A, StClair JM, Seinfeld JH, Wennberg PO. Unexpected epoxide formation in the gas-phase photooxidation of isoprene. Science. 2009;325(5941):730. doi: 10.1126/science.1172910. [DOI] [PubMed] [Google Scholar]
- 15.Krechmer JE, Coggon MM, Massoli P, Nguyen TB, Crounse JD, Hu W, Day DA, Tyndall GS, Henze DK, Rivera-Rios JC, et al. Formation of low volatility organic compounds and secondary organic aerosol from isoprene hydroxyhydroperoxide low-no oxidation. Environ Sci Technol. 2015;49(17):10330–10339. doi: 10.1021/acs.est.5b02031. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, D’Ambro EL, Lee BH, Lopez-Hilfiker FD, Zaveri RA, Rivera-Rios JC, Keutsch FN, Iyer S, Kurten T, Zhang Z, et al. Efficient isoprene secondary organic aerosol formation from a non-IEPOX pathway. Environ Sci Technol. 2016;50(18):9872–9880. doi: 10.1021/acs.est.6b01872. [DOI] [PubMed] [Google Scholar]
- 17.Riva M, Budisulistiorini SH, Chen Y, Zhang Z, D’Ambro EL, Zhang X, Gold A, Turpin BJ, Thornton JA, Canagaratna MR, et al. Chemical characterization of secondary organic aerosol from oxidation of isoprene hydroxyhydroperoxides. Environ Sci Technol. 2016;50(18):9889–9899. doi: 10.1021/acs.est.6b02511. [DOI] [PubMed] [Google Scholar]
- 18.Surratt JD, Chan AWH, Eddingsaas NC, Chan M, Loza CL, Kwan AJ, Hersey SP, Flagan RC, Wennberg PO, Seinfeld JH. Reactive intermediates revealed in secondary organic aerosol formation from isoprene. Proc Natl Acad Sci USA. 2010;107(15):6640–6645. doi: 10.1073/pnas.0911114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y-H, Zhang Z, Docherty KS, Zhang H, Budisulistiorini SH, Rubitschun CL, Shaw SL, Knipping EM, Edgerton ES, Kleindienst TE, et al. Isoprene epoxydiols as precursors to secondary organic aerosol formation: Acid-catalyzed reactive uptake studies with authentic compounds. Environ Sci Technol. 2012;46(1):250–258. doi: 10.1021/es202554c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y-H, Zhang H, Pye HOT, Zhang Z, Marth WJ, Park S, Arashiro M, Cui T, Budisulistiorini SH, Sexton KG, et al. Epoxide as a precursor to secondary organic aerosol formation from isoprene photooxidation in the presence of nitrogen oxides. Proc Natl Acad Sci USA. 2013;110(17):6718–6723. doi: 10.1073/pnas.1221150110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjaergaard HG, Knap HC, Ørnsø KB, Jørgensen S, Crounse JD, Paulot F, Wennberg PO. Atmospheric Fate of Methacrolein. 2. Formation of Lactone and Implications for Organic Aerosol Production. J Phys Chem A. 2012;116(24):5763–5768. doi: 10.1021/jp210853h. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen TB, Bates KH, Crounse JD, Schwantes RH, Zhang X, Kjaergaard HG, Surratt JD, Lin P, Laskin A, Seinfeld JH, et al. Mechanism of the hydroxyl radical oxidation of methacryloyl peroxynitrate (MPAN) and its pathway toward secondary organic aerosol formation in the atmosphere. Phys Chem Chem Phys. 2015;17(27):17914–17926. doi: 10.1039/c5cp02001h. [DOI] [PubMed] [Google Scholar]
- 23.Riva M, Budisulistiorini SH, Zhang Z, Gold A, Thornton JA, Turpin BJ, Surratt JD. Multiphase reactivity of gaseous hydroperoxide oligomers produced from isoprene ozonolysis in the presence of acidified aerosols. Atmos Environ. 2017;152:314–322. [Google Scholar]
- 24.Kramer AJ, Rattanavaraha W, Zhang Z, Gold A, Surratt JD, Lin YH. Assessing the oxidative potential of isoprene-derived epoxides and secondary organic aerosol. Atmos Environ. 2016;130:211–218. [Google Scholar]
- 25.Lin Y-H, Arashiro M, Martin E, Chen Y, Zhang Z, Sexton KG, Gold A, Jaspers I, Fry RC, Surratt JD. Isoprene-derived secondary organic aerosol induces the expression of oxidative stress response genes in human lung cells. Environ Sci Technol Lett. 2016;3(6):250–254. [Google Scholar]
- 26.de Bruijne K, Ebersviller S, Sexton KG, Lake S, Leith D, Goodman R, Jetters J, Walters GW, Doyle-Eisele M, Woodside R, et al. Design and testing of electrostatic aerosol in vitro exposure system (EAVES): An alternative exposure system for particles. Inhal Toxicol. 2009;21(2):91–101. doi: 10.1080/08958370802166035. [DOI] [PubMed] [Google Scholar]
- 27.Lichtveld KM, Ebersviller SM, Sexton KG, Vizuete W, Jaspers I, Jeffries HE. In vitro exposures in diesel exhaust atmospheres: Resuspension of PM from filters versus direct deposition of PM from air. Environ Sci Technol. 2012;46(16):9062–9070. doi: 10.1021/es301431s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebersviller S, Lichtveld K, Sexton KG, Zavala J, Lin Y-H, Jaspers I, Jeffries HE. Gaseous VOCs rapidly modify particulate matter and its biological effects – Part 1: Simple VOCs and model PM. Atmos Chem Phys. 2012;12(24):12277–12292. doi: 10.5194/acpd-12-5065-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arashiro M, Lin Y-H, Sexton KG, Zhang Z, Jaspers I, Fry RC, Vizuete WG, Gold A, Surratt JD. In vitro exposure to isoprene-derived secondary organic aerosol by direct deposition and its effects on COX-2 and IL-8 gene expression. Atmos Chem Phys. 2016;16(22):14079–14090. [Google Scholar]
- 30.Peng H, Chen P, Cai Y, Chen Y, Wu Q-h, Li Y, Zhou R, Fang X. Endothelin-1 increases expression of cyclooxygenase-2 and production of interlukin-8 in human pulmonary epithelial cells. Peptides. 2008;29(3):419–424. doi: 10.1016/j.peptides.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Lin YH, Zhang H, Surratt JD, Ball LM, Gold A. Technical Note: Synthesis of isoprene atmospheric oxidation products: isomeric epoxydiols and the rearrangement products cis- and trans-3-methyl-3,4-dihydroxytetrahydrofuran. Atmos Chem Phys. 2012;12(18):8529–8535. [Google Scholar]
- 32.Budisulistiorini SH, Li X, Bairai ST, Renfro J, Liu Y, Liu YJ, McKinney KA, Martin ST, McNeill VF, Pye HOT, et al. Examining the effects of anthropogenic emissions on isoprene-derived secondary organic aerosol formation during the 2013 Southern Oxidant and Aerosol Study (SOAS) at the Look Rock, Tennessee ground site. Atmos Chem Phys. 2015;15(15):8871–8888. [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, Bhattacharya S, Kandjanapa K, Soontararuks S, Nookabkaew S, et al. Activation of Inflammation/NF-κB Signaling in Infants Born to Arsenic-Exposed Mothers. PLOS Genet. 2007;3(11):e207. doi: 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 36.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37(suppl 1):D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39(suppl 1):D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41(D1):D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhavoronkov A, Buzdin A, Garazha A, Borissoff N, Moskalev A. Signaling pathway cloud regulation for in silico screening and ranking of the potential geroprotective drugs. Front Genet. 2014;5(49) doi: 10.3389/fgene.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(suppl 2):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen T, Yang CS, Pickett CB. The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radic Biol Med. 2004;37(4):433–441. doi: 10.1016/j.freeradbiomed.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 43.Li N, Alam J, Venkatesan MI, Eiguren-Fernandez A, Schmitz D, Di Stefano E, Slaughter N, Killeen E, Wang X, Huang A, et al. Nrf2 is a key transcription factor that regulates antioxidant defense in macrophages and epithelial cells: Protecting against the proinflammatory and oxidizing effects of diesel exhaust chemicals. J Immunol. 2004;173(5):3467–3481. doi: 10.4049/jimmunol.173.5.3467. [DOI] [PubMed] [Google Scholar]
- 44.Kroll JH, Ng NL, Murphy SM, Flagan RC, Seinfeld JH. Secondary organic aerosol formation from isoprene photooxidation. Environ Sci Technol. 2006;40(6):1869–1877. doi: 10.1021/es0524301. [DOI] [PubMed] [Google Scholar]
- 45.Riedel TP, Lin Y-H, Budisulistiorini SH, Gaston CJ, Thornton JA, Zhang Z, Vizuete W, Gold A, Surratt JD. Heterogeneous reactions of isoprene-derived epoxides: Reaction probabilities and molar secondary organic aerosol yield estimates. Environ Sci Technol Lett. 2015;2(2):38–42. [Google Scholar]
- 46.Lin Y-H, Knipping EM, Edgerton ES, Shaw SL, Surratt JD. Investigating the influences of SO2 and NH3 levels on isoprene-derived secondary organic aerosol formation using conditional sampling approaches. Atmos Chem Phys. 2013;13(16):8457–8470. [Google Scholar]
- 47.Rattanavaraha W, Chu K, Budisulistiorini SH, Riva M, Lin Y-H, Edgerton ES, Baumann K, Shaw SL, Guo H, King L, et al. Assessing the impact of anthropogenic pollution on isoprene-derived secondary organic aerosol formation in PM2.5 collected from the Birmingham, Alabama, ground site during the 2013 Southern Oxidant and Aerosol Study. Atmos Chem Phys. 2016;16(8):4897–4914. doi: 10.5194/acp-16-4897-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiarelli N, Carini G, Zoppi N, Dordoni C, Ritelli M, Venturini M, Castori M, Colombi M. Transcriptome-wide expression profiling in skin fibroblasts of patients with joint hypermobility syndrome/ehlers-danlos syndrome hypermobility type. PLOS ONE. 2016;11(8):e0161347. doi: 10.1371/journal.pone.0161347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kutmon M, Riutta A, Nunes N, Hanspers K, Willighagen Egon L, Bohler A, Mélius J, Waagmeester A, Sinha Sravanthi R, Miller R, et al. WikiPathways: Capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44(D1):D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishimura D. BioCarta. Biotech Software & Internet Report. 2001;2(3):117–120. [Google Scholar]
- 51.Pope CA. Epidemiology of fine particulate air pollution and human health: biologic mechanisms and who’s at risk? Environ Health Perspect. 2000;108(Suppl 4):713–723. doi: 10.1289/ehp.108-1637679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang H, Jang M, Yu Z. Dithiothreitol activity by particulate oxidizers of SOA produced from photooxidation of hydrocarbons under varied NOx levels. Atmos Chem Phys Discuss. 2017;2017:1–24. [Google Scholar]
- 54.Bryan HK, Olayanju A, Goldring CE, Park BK. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol. 2013;85(6):705–717. doi: 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Kannan MB, Solovieva V, Blank V. The small MAF transcription factors MAFF, MAFG and MAFK: Current knowledge and perspectives. Biochim Biophys Acta - Molecular Cell Research. 2012;1823(10):1841–1846. doi: 10.1016/j.bbamcr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 56.Ma Q. Role of Nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor RC, Acquaah-Mensah G, Singhal M, Malhotra D, Biswal S. Network inference algorithms elucidate Nrf2 regulation of mouse lung oxidative stress. PLOS Comput Biol. 2008;4(8):e1000166. doi: 10.1371/journal.pcbi.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soriano FX, Baxter P, Murray LM, Sporn MB, Gillingwater TH, Hardingham GE. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol Cells. 2009;27(3):279–282. doi: 10.1007/s10059-009-0050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park HS, Kim SR, Lee YC. Impact of oxidative stress on lung diseases. Respirology. 2009;14(1):27–38. doi: 10.1111/j.1440-1843.2008.01447.x. [DOI] [PubMed] [Google Scholar]
- 60.Reddy NM, Vegiraju S, Irving A, Paun BC, Luzina IG, Atamas SP, Biswal S, Ana N-A, Mitzner W, Reddy SP. Targeted deletion of Jun/AP-1 in alveolar epithelial cells causes progressive emphysema and worsens cigarette smoke–induced lung inflammation. Am J Pathol. 2012;180(2):562–574. doi: 10.1016/j.ajpath.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devlin RB, McKinnon KP, Noah T, Becker S, Koren HS. Ozone-induced release of cytokines and fibronectin by alveolar macrophages and airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 1994;266(6):L612–L619. doi: 10.1152/ajplung.1994.266.6.L612. [DOI] [PubMed] [Google Scholar]
- 62.Pardo M, Shafer MM, Rudich A, Schauer JJ, Rudich Y. Single exposure to near roadway particulate matter leads to confined inflammatory and defense responses: Possible role of metals. Environ Sci Technol. 2015;49(14):8777–8785. doi: 10.1021/acs.est.5b01449. [DOI] [PubMed] [Google Scholar]
- 63.Wittkopp S, Staimer N, Tjoa T, Stinchcombe T, Daher N, Schauer JJ, Shafer MM, Sioutas C, Gillen DL, Delfino RJ. Nrf2-related gene expression and exposure to traffic-related air pollution in elderly subjects with cardiovascular disease: An exploratory panel study. J Expos Sci Environ Epidemiol. 2016;26(2):141–149. doi: 10.1038/jes.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pardo M, Porat Z, Rudich A, Schauer JJ, Rudich Y. Repeated exposures to roadside particulate matter extracts suppresses pulmonary defense mechanisms, resulting in lipid and protein oxidative damage. Environ Pollut. 2016;210:227–237. doi: 10.1016/j.envpol.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 65.Pardo M, Katra I, Schaeur JJ, Rudich Y. Mitochondria-mediated oxidative stress induced by desert dust in rat alveolar macrophages. GeoHealth. 2017 doi: 10.1002/2016GH000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.