Abstract
Nanomedicine provides a unique opportunity for promoting drug efficacy through enhanced delivery mechanisms. However, its translation into the clinics has been relatively slow compared with the large amount of research occurring in laboratory settings. Given the limitations of conventional cell culture models and preclinical animal models, we discuss the potential utility of recently developed cancer-on-a-chip platforms, which maximally replicate the pathophysiology of the human tumor microenvironments, as alternatives for effective evaluation of nanomedicine. We begin with a brief discussion of nanomedicine, then chart the history of organ-on-a-chip platform development and their recent evolution as tools for modeling different cancers for assessing nanomedicine efficacy, concluding with future perspectives for the field.
Keywords: organ-on-a-chip, cancer-on-a-chip, microfluidics, nanomedicine, nanoparticles
Introduction
The blooming field of nanomedicine has achieved significant advances in the use of nanocarrier formulations for delivering therapeutic drugs and diagnostic agents to tumor sites. Compared with the systemic administration of free molecules, the utilization of nanomedicine presents unique advantages in terms of improved protection of the biological activities of the agents in the serum-rich environment, prolonged circulation periods in the bloodstream, reduced adverse effects, enhanced permeability and retention effects, improved tumor-targeting efficiency, increased release profiles, and possible integration of stimuli responsiveness for on-demand therapeutics, among others [1–3]. Nanomaterials can be designed with favorable properties for facilitating their delivery in a variety of situations. For example, they can be fabricated from materials of different origins, including both inorganic (e.g., metals, silica, carbon, and their respective oxides) and organic (e.g., polymers and lipids) materials; their sizes can be manipulated to fall within a wide range, from a few nanometers to no more than 1 μm; their shapes can be tuned to be smooth or sharp; their plasticity can be altered to be stiff or deformable; and their surfaces can be functionalized with many different characteristics and moieties of interest (Figure 1) [1]. Compared with all these successes in the laboratory, the translation of nanomedicine into clinical practice for cancer theranostics has been limited, with only 175 nanomedicine products commercialized so far [4]. It was also recently reported that only 0.7% of injected nanoparticles (NPs) accumulate in the tumor regardless of how the physicochemical properties of the nanocarriers are changed [5]. More surprisingly, this delivery efficiency has not improved over past decade [5].
Figure 1.
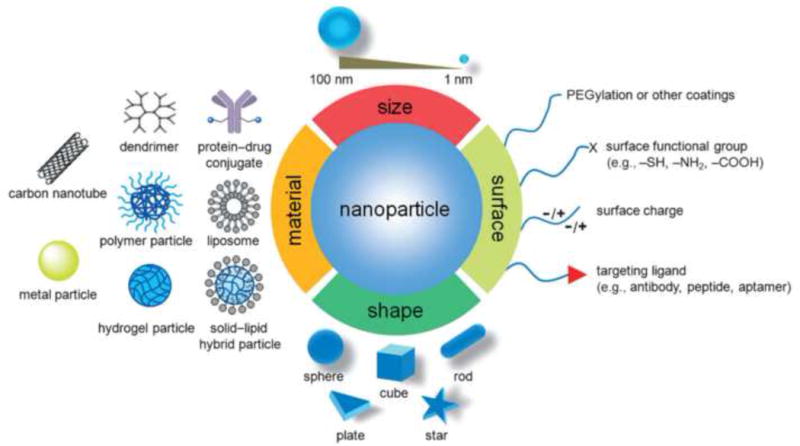
Illustration of nanomedicine with precisely engineered physicochemical properties for cancer therapy. Reprinted, with permission, from [1].
Preclinical vertebrate models, including rodents, large animals, and nonhuman primates, have served as gold standards for evaluating cancer nanomedicine. However, their typically high economic and ethical burdens, along with the fact that they often fail to predict human responses during clinical trials [6], have significantly impacted the efficacies of these animal models. In addition, direct translation of results obtained from cell culture studies in vitro into these preclinical models at earlier stages is difficult because of the inability of these oversimplified models to simulate the complex tissue architectures of their counterparts in vivo. Fortunately, advances in tissue-engineering strategies have assisted the development of functional human healthy or diseased organs, and knowledge gained in the field of stem cells allows for derivation of patient-specific cell populations to achieve personalized approaches; in combination with microfluidics technologies, physiological relevance can be further built into the systems to model the dynamic microenvironment and interorgan interactions [7–12]. These platforms, called ‘organ-on-a-chip’ systems, are anticipated to have important roles in bridging the gaps between conventional cell cultures, preclinical animal models, and clinical human trials not only in testing pharmaceutical compounds, biological species, and environmental toxins, but also with a potential to facilitate the evaluation and translation of nanomedicine.
Organ-on-a-chip platforms
Since the turn of this century, multiple research groups have focused on innovating organ-on-a-chip platforms that mimic both the biology and physiology of their counterparts in the human system. While full organ functions are still hard to model at this stage, most organ-on-a-chip platforms seek to emulate the important functions of tissues, or parts of organs, to satisfy the needs for particular applications through meticulous engineering of the hierarchical cell architectures, cell populations, and their dynamic microenvironments [13,14]. To date, most tissues and/or organ types have been successfully modeled to reproduce corresponding functional subunits (Figure 2), including for example, the brain [15], heart [16–18], lung [19–21], liver [22–25], intestine [26–29], vasculature [30–32], kidney [33,34], and musculoskeletal system [35,36]. These individual devices can be linked such that the integral platforms further mimic the physiology and compartmentalization of the human system, enabling investigations of pharmacokinetics and pharmacodynamics [8,10,11,37].
Figure 2.
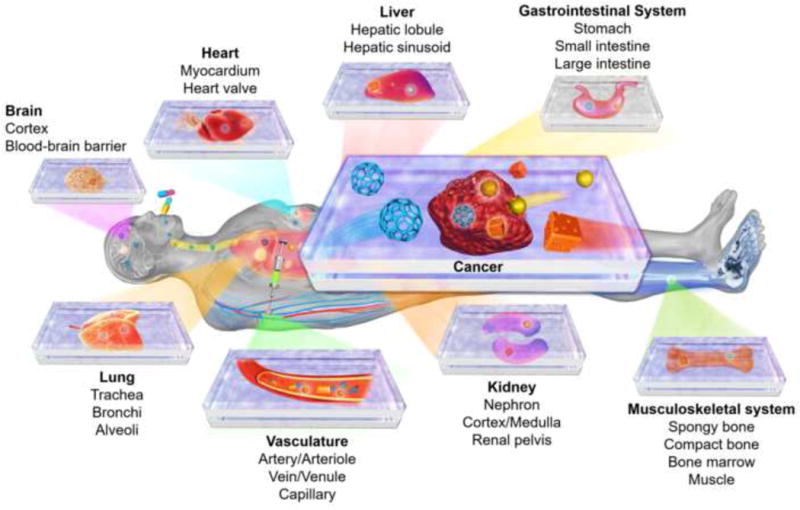
Organ-on-a-chip platforms for drug screening and their potential applications in evaluating nanomedicine. Major structures of the example organs are listed.
Modeling cancer biology and physiology in vitro
Tumorigenic microenvironments are complex, with hierarchically assembled multiple cell types and extracellular matrix (ECM) molecules, dynamic niches, and mass transport milieu, all potentially in a stage-dependent manner [38,39]. Such complexity can be partially reproduced using the concept of organ-on-a-chip platforms by replacing healthy cells and associated ECMs with those of cancer origins, so-called ‘cancer-on-a-chip’ systems [40–42]. Recent advances in cancer-on-a-chip platforms include high-throughput drug screening, therapy efficacy assessment, metastasis studies, and personalized medicine, among others [43,44]. For example, using a compartmentalized device, Huh and co-workers recently developed a ductal carcinoma microenvironment by first forming a confluent layer of mammary epithelial cells on a porous membrane immediately above another layer of fibroblast-containing hydrogel mimicking the surrounding matrix; then ductal carcinoma spheroids were inoculated on top of the epithelial cells to complete the model (Figure 3a,b) [45]. Using this model, the effects of anticancer drugs were evaluated: for example, paclitaxel induced negligible toxicity on the epithelial cells alone but showed pronounced toxicity towards the ductal carcinoma spheroids (Figure 3b). In addition, paclitaxel also effectively inhibited the progression of the ductal carcinoma spheroids, indicated by their maintained sizes compared with the significantly increased tumor volume without the drug (Figure 3d,e). In addition, a breast-cancer-microenvironment-on-a-chip model was developed by Imparato and colleagues to replicate the interactions of breast cancer cells with the stroma and ECM activation during tumor progression [46]. In another study, Varghese and co-workers encapsulated MCF7 breast cancer cell spheroids and human umbilical vein endothelial cells (HUVECs) within a gelatin methacryloyl (GelMA) hydrogel, and then drove HUVECs to migrate to the periphery of the GelMA to form endothelial barriers [47]. They authors then assessed the penetration of doxorubicin, a common anticancer drug, into the tumor spheroids and quantified its cytotoxic effect on the cells.
Figure 3.
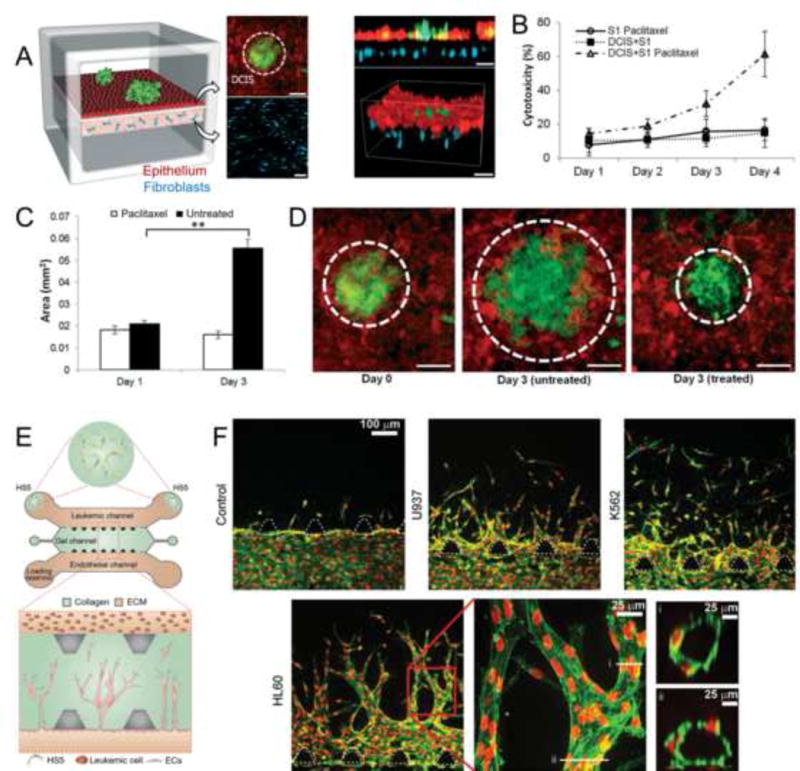
Cancer-on-a-chip platforms for modeling solid and liquid tumors. (a–c) A microengineered ductal carcinoma in situ (DCIS)-on-a-chip platform. (A) DCIS spheroids embedded in a layer of normal epithelium sitting on top of a stromal microenvironment containing mammary fibroblasts. (b) Cytoxocitiy of the anticancer drug paclitaxel on cells in the DCIS-on-a-chip platform. (c,d) Quantification and confocal images showing the effect of paclitaxel on the progress of the ductal carcinoma spheroids in the DCIS-on-a-chip platform. (e,f) Angiogenesis in liquid tumors. (e) Schemtaic showing biomimetic angiogenesis devices in response to leukemic cells. (f) Confocal images showing directional angiogenic sprouting towards a leukemic channel under different leukemic cell stimulations. Adapted, with permission, from [45] (c,d) and [54] (f).
Various other types of solid-tumor-on-a-chip platform have been developed. Gervais and colleagues cultured microdissected tissues from patients with oophoroma on microfluidic chips and obtained patient-specific carboplatin treatment-response data with high clinical relevance [48]. Guenat and co-workers developed two microfluidic systems for assessment of the chemosensitivity of non-small cell lung cancer (NSCLC) and malignant pleural mesothelioma [49,50]. They co-cultured primary lung cancer epithelial cell spheroids and lung pericytes, which were isolated from lung cancer specimens, on chips, and found a higher chemoresistance to cisplatin in co-culture conditions compared with monocultured spheroids. Alternatively, Wang and colleagues fabricated a 3D co-culture microfluidic device to monitor tumor cell invasion in a real-time manner, and proposed that cancer-associated fibroblasts could have a key role in the promotion of the invasive capacity of NSCLC cells by upregulating the expression of glucose-regulated protein 78 [51]. Akay and co-workers demonstrated the potential of a brain-cancer-on-a-chip device in the formation of spheroids from U87 glioblastoma cells, which was further used for high-throughput screening of simultaneously administrated drugs, pitavastatin and irinotecan [52]. Niu and colleagues established a microfluidic co-cultured platform to simulate the bladder cancer microenvironment [53]. They cultured stromal cells, fibroblasts, endothelial cells, and macrophages together with bladder cancer cells on this platform and tested neoadjuvant chemotherapy sensitivity using drugs including methotrexate, vincristine, doxorubicin, and cis-diammineplatinum(II) dichloride.
Not only have these platforms been used for reproducing the solid tumor microenvironment, but they can also be adapted to model liquid tumors. In a recent example, Fu and co-workers developed an in vitro assay to evaluate leukemic cell-induced bone marrow angiogenesis [54]. By modifying an established angiogenesis microchip device, leukemic cells were infused into one side of the chip, while endothelial cells were seeded on the other side to allow them to sprout into the central chamber filled with collagen; when necessary, HS5 human bone marrow stromal cells could also be inoculated at the two ends of the leukemic channel for realizing the co-culture (Figure 3e). The presence of leukemic cells induced directional sprouting of endothelial cells into the collagen matrix towards the leukemic channel, suggesting the angiogenic potential of these leukemic cell lines, whereas the control group without any leukemic cells showed minimal invasion of endothelial cells into the collagen matrix (Figure 3f). Different leukemic cells also indicated different degrees of angiogenic induction, potentially because of the different amounts of angiogenic factors produced, and the sprouted endothelial cells were observed to form lumen structures, both of which could be further enhanced by co-culturing the leukemic cells with the stromal cells. In another study, bone marrow plasma cells from patients with myeloma were cultured on a microfluidic platform; chemosensitivity and resistance assays were carried out to evaluate the ex vivo responses of these primary CD138+ multiple myeloma cells to bortezomib-containing therapies [55].
Cancer-on-a-chip platforms for evaluating nanomedicine
Given their close mimicry of in vivo biology and physiology, biomimetic cancer-on-a-chip platforms are anticipated to surpass the accuracy of conventional planar, static cell culture models for evaluating pharmaceutical agents. The ability to couple these systems with cells of human origin, with the potential for personalization, further renders them more attractive than animal models in many cases. However, the use of these platforms for assessing nanomedicine (Figure 2), although not new, has not been intensively implemented.
An early example dates back to 2005, when Langer and co-workers adopted a simple microfluidic device, the bottom of which was coated with a monolayer of cancer cells, to study the dynamic interactions of perfused nano-and microparticles with the cancer cells (Figure 4a) [56]. The two prostate cancer cell lines, PC3 and LNCaP, cultured in this microfluidic platform, showed differential uptake behaviors towards poly(lactic acid) (PLA) particles in a size- and flow-dependent manner, for particles functionalized with and without aptamers recognizing prostate-specific membrane antigen (PSMA) (Figure 4b,c).
Figure 4.
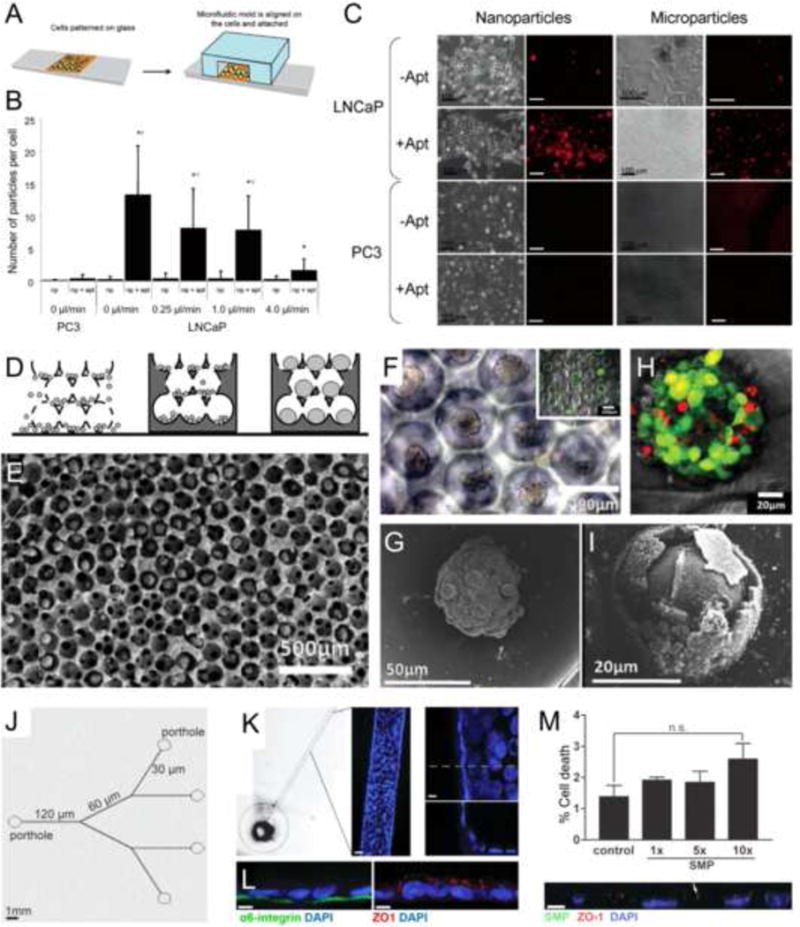
Cancer-on-a-chip platforms for evaluating nanomedicine. (a–c) A microfluidic planar culture model for evaluating nanoparticle (NP) interactions with cancer cells. (a) Schematic of the device fabrication. (b,c) Quantification and images showing the uptake of prostate-specific membrane antigen (PSMA)-functionalized poly(lactic acid) (PLA) nano- and microparticles by PC3 and LNCaP prostate cancer cells cultured in the microfluidic device. (d–i) An inverted opal 3D cancer model for evaluating NP interactions with cancer cells. (d) Schematic of the model fabrication. (E–G) SEM and optical microscopy images showing the liver cancer spheroids formed in the inverse opal scaffolds. Inset in (f) shows the viability of the spheroids. (h,i) Viability analysis and SEM image showing the liver cancer spheroids after treatment with CdTe semiconductor NPs. (j–l) A biomimetic mammary duct-on-a-chip platform. (j) Schematic of the branching microchannel system built in a polydimethylsiloxane (PDMS) chip. (k) Formation of mammary epithelium surrounding the periphery of the microchannel. (l) Confocal micrographs showing staining for both basal (α6 integrin) and apical (ZO-1) polarity markers. (m) Dose-dependent cell death caused by injected submicron magnetic particles (SMPs); the bottom panel shows a confocal image of the co-registration of the SMPs with the epithelium. Adapted, with permission, from [56] (b,c), [57] (h,i),and [58] (m).
Interactions between NPs and cancerous cells were further developed as 3D configurations. In a study conducted by Kotov and colleagues in 2009, inverse opal-structured hydrogel scaffolds were used to form spheroids of HepG2 hepatocellular carcinoma cells (Figure 4d,e) [57]. The pristine spheroids formed retained high viability as well as expressing intact cell junctions (Figure 4f,g). It was shown that, when these spheroid-based liver cancer organoids were subjected to treatment with semiconductor CdTe NPs, they showed reduced viability (Figure 4h) and impaired functions of cells close to the surface (Figure 4i). Nevertheless, the 3D form of the organoids could buffer the toxicity of the NPs, limiting it to the peripheries, unlike the significant toxicity of these same NPs towards these cells even at lower doses.
More recently, with microfluidic on-chip tumor models, the advantages shown in the two previous examples could be further integrated: not only can the mass transport of NPs with varying parameters, such as size, shape, and surface characterizations, be emulated, but the geometry of the relevant architectures can also be reproduced. In an interesting study by Lelièvre and co-workers in 2011 using human mammary epithelial cells (non-neoplastic), a microfluidic channel containing hierarchical branches with gradually tapering sizes from 120 to 30 Mm was coated with these cells (Figure 4j) [58]. Importantly, the epithelial cells formed a confluent layer surrounding the entire circumference of the microchannels by expressing both basal and apical biomarkers to emulate the structure of mammary ducts (Figure 4k,l). The platform was subsequently adopted to assess the toxicity of superparamagnetic submicron particles (SMPs), which are clinically used as magnetic resonance imaging contrast agents. Simulating nipple delivery, the SMPs were injected from the inlet porthole connected to the 120-Mm microchannel and allowed to move through the branches under magnetic guidance. Dose-dependent cytotoxicity of these NPs on the mammary ducts was observed with their co-localization and potential update by the epithelial cells (Figure 4m).
Various other prototype cancer-on-a-chip platforms have been subsequently developed for NP testing. Wang and co-workers customized a microfluidic-based 3D breast cancer tissue model with MCF7 cells and primary adipose-derived stromal cells [59]. The effective evaluation of photodynamic therapy (PDT) agents, (i.e., photosensitizer and gold NPs) was achieved using this model. The results indicated that MCF7 spheroids exhibited more resistance to PDT compared with those in monolayer cultures. Pant and colleagues modeled the 3D cervical cancer microenvironment with a physiologically and morphologically relevant microvasculature [60]. Based on this model, two nanosized polymeric vehicles were characterized to predict in vivo drug delivery efficiencies. Chan and colleagues reported a tumor-on-a-chip system for the study of NP accumulation, as well as their transport kinetics and mechanisms, in a microfluidic model containing melanoma spheroids [61]. The authors demonstrated that not only could the interstitial flow rate affect the accumulation of PEGylated gold NPs at the spheroid/fluid interface, but the physicochemical properties (i.e., size and surface chemistries) of these NPs could also influence the interaction of the NPs with tumor spheroids, results that were further validated in vivo in a mouse model. Arvanitis and colleagues presented a novel acoustofluidic 3D tumor platform to investigate the localized release of temperature-sensitive liposomal doxorubicin [62]. They locally activated the focused ultrasound-triggered doxorubicin-encapsulating liposomes and studied their release profile and chemotherapeutic efficacy on a glioblastoma model.
Conclusions and perspectives
As a result of technological advancements in microfluidics and tissue engineering, biomimetic human cancer models with increasingly improved architectural and functional similarity to their in vivo counterparts have been designed and fabricated. Although they show huge promise in bridging the gaps between conventional planar, static cell cultures, preclinical animal models, and the human body for drug screening, their effective utilization in assessing nanomedicine has been limited. The clinical translation of such devices is still premature, although multiple agencies, including the US Food and Drug Administration (FDA), are making significant efforts to explore how these platforms can lead to improved testing for toxicity and to potentially reduce the need for animal trials [63].
However, we envisage the fast progression of this field in the near future to address several major challenges associated with the cancer-on-a-chip systems. To unveil the potential adverse effects of a nanomedicine while assessing its therapeutic effect on the tumorous tissue, it is necessary to further connect the cancerous organoids with healthy ones that are interconnected by the flow of body fluids, to realize the ultimate goal of building a living ‘cancer-patient-on-a-chip’ system (Figure 2) [64]. Integrating functional vascular networks into these systems is also crucial to achieve the accurate assessment of the pharmacokinetics and pharmacodynamics of nanomedicine [65,66]. In addition, designs that include immunocompetent microenvironments are of particular interest because of the significant interactions of NPs with the immune system, such as phagocytes in the circulation [67] and immune cells residing in the target tissues [68]. Other factors to consider include the common media that support the viability and functions of all integrated organoids [9,69,70], as well as the proper scaling of the different organoids to ensure a human effect [71].
As well as designing factors of the cancer-on-a-chip platforms themselves, characterization of these cancer organoids posts additional challenges because of the inability of current imaging modalities to obtain volumetric information of the 3D tissues. To address this issue, Chan and colleagues recently developed an imaging-on-a-chip system to achieve optical clearing and high-resolution imaging of optically transparent structures of intact tumor spheroids [72]. With the built-in fluid exchange capability of this microfluidic chip, multiple tumor spheroids could be cleared within 1 day by removing the scattering lipid molecules post crosslinking, allowing for high-throughput tissue imaging of intact organoids, analyses of tumor biomarkers and microstructures, and, potentially, investigations of nanomedicine-cancer organoid interactions. Alternatively, sensing units can be built into the organoids for conformal and real-time behavioral monitoring [73].
Overall, cancer-on-a-chip platforms are sophisticated microsystems that simulate their human counterparts. However, they are also simple enough to decompose the complex in vivo biological environments, offering a unique opportunity to improve our fundamental understanding of nanomedicine. We believe that the continued development of these platforms, along with advances in cancer biology and bioanalysis, will likely boost the translation of nanomedicine in the foreseeable future [4].
Highlights.
Nanomedicine promotes drug efficacy through enhanced delivery mechanisms.
The translation of nanomedicine is slow with current cell culture and preclinical models.
Organ-on-a-chip platforms better replicate the biology and physiology of the human system.
Cancer-on-a-chip platforms have potentials to use as alternatives for effective evaluation of nanomedicine.
Teaser.
We discuss the potential utility of the emerging cancer-on-a-chip platforms that better replicate the pathophysiology of the human cancer microenvironments, as alternatives or supplements to conventional planar cell culture and animal models for effective evaluation of nanomedicine.
Acknowledgments
Y.S.Z. acknowledges supports from the National Cancer Institute Pathway to Independence Award (K99CA201603) and the LUSH Award. Y.N.Z acknowledges supports from the Ontario Graduate Scholarship, the Wildcat Foundation graduate scholarship, the Paul and Sally Wang distinguished graduate scholarship from University of Toronto, and an Alexander Graham Bell Canada graduate scholarship from the Natural Sciences and Engineering Research Council of Canada. W.Z. acknowledges supports from NSFC (31501555), STCSM (16391903900), the 1000 Young Talents Program of China, and a Young Eastern Professorship Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sun T, et al. Engineered nanoparticles for drug delivery in cancer therapy. Angew Chem Int Ed. 2014;53:12320–12364. doi: 10.1002/anie.201403036. [DOI] [PubMed] [Google Scholar]
- 2.Xia Y. Nanomaterials at work in biomedical research. Nat Mater. 2008;7:758–760. doi: 10.1038/nmat2277. [DOI] [PubMed] [Google Scholar]
- 3.Farokhzad OC, Langer R. Impact of nanotechnology on drug delivery. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 4.Jang HL, et al. Boosting clinical translation of nanomedicine. Nanomedicine. 2016;11:1495–1497. doi: 10.2217/nnm-2016-0133. [DOI] [PubMed] [Google Scholar]
- 5.Wilhelm S, et al. Analysis of nanoparticle delivery to tumours. Nat Rev Mater. 2016;1:16014. [Google Scholar]
- 6.McGonigle P, Ruggeri B. Animal models of human disease: challenges in enabling translation. Biochem Pharmacol. 2014;87:162–171. doi: 10.1016/j.bcp.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Bhise NS, et al. Organ-on-a-chip platforms for studying drug delivery systems. J Control Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YS, Khademhosseini A. Seeking the right context for evaluating nanomedicine: from tissue models in petri dishes to microfluidic organs-on-a-chip. Nanomedicine. 2015;10:685–688. doi: 10.2217/nnm.15.18. [DOI] [PubMed] [Google Scholar]
- 9.Maschmeyer I, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab on a Chip. 2015;15:2688–2699. doi: 10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 10.Esch M, et al. The role of body-on-a-chip devices in drug and toxicity studies. Ann Rev Biomed Eng. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 11.Sung JH, et al. Using physiologically-based pharmacokinetic-guided body-on-a-chip’ systems to predict mammalian response to drug and chemical exposure. Exp Biol Med. 2014;239:1225–1239. doi: 10.1177/1535370214529397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esch EW, et al. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov. 2015;14:248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotech. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 14.Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell. 2016;164:1105–1109. doi: 10.1016/j.cell.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 15.Park J, et al. Microfluidic compartmentalized co-culture platform for CNS axon myelination research. Biomed Microdevices. 2009;11:1145–1153. doi: 10.1007/s10544-009-9331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosberg A, et al. Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab on a Chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal A, et al. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab on a Chip. 2013;13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YS, et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016;110:45–59. doi: 10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huh D, et al. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc Natl Acad Sci. 2007;104:18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douville NJ, et al. Combination of fluid and solid mechanical stresses contribute to cell death and detachment in a microfluidic alveolar model. Lab on a chip. 2011;11:609–619. doi: 10.1039/c0lc00251h. [DOI] [PubMed] [Google Scholar]
- 22.Kane BJ, et al. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem. 2006;78:4291–4298. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- 23.Carraro A, et al. In vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomed Microdevices. 2008;10:795–805. doi: 10.1007/s10544-008-9194-3. [DOI] [PubMed] [Google Scholar]
- 24.Bhise NS, et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8:014101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 25.Lee PJ, et al. An artificial liver sinusoid with a microfluidic endothelial- like barrier for primary hepatocyte culture. Biotechnol Bioeng. 2007;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 26.Imura Y, et al. A microfluidic system to evaluate intestinal absorption. Analyt Sci. 2009;25:1403–1407. doi: 10.2116/analsci.25.1403. [DOI] [PubMed] [Google Scholar]
- 27.Kimura H, et al. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip. 2008;8:741–746. doi: 10.1039/b717091b. [DOI] [PubMed] [Google Scholar]
- 28.Kim HJ, et al. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab on a Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W, et al. Elastomeric free-form blood vessels for interconnecting organs on chip systems. Lab on a Chip. 2016;16:1579–1586. doi: 10.1039/c6lc00001k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas J, et al. Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl in vitro Toxicol. 2016;2:82–96. doi: 10.1089/aivt.2016.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasotharan S, et al. Artery-on-a-chip platform for automated, multimodal assessment of cerebral blood vessel structure and function. Lab on a Chip. 2015;15:2660–2669. doi: 10.1039/c5lc00021a. [DOI] [PubMed] [Google Scholar]
- 33.Jang K-J, et al. Fluid-shear-stress-induced translocation of aquaporin-2 and reorganization of actin cytoskeleton in renal tubular epithelial cells. Integr Biol. 2011;3:134–141. doi: 10.1039/c0ib00018c. [DOI] [PubMed] [Google Scholar]
- 34.Jang K-J, Suh K-Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab on a Chip. 2010;10:36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 35.Nesmith AP, et al. Human airway musculature on a chip: an in vitro model of allergic asthmatic bronchoconstriction and bronchodilation. Lab on a chip. 2014;14:3925–3936. doi: 10.1039/c4lc00688g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J-H, et al. Microfluidic 3D bone tissue model for high-throughput evaluation of wound-healing and infection-preventing biomaterials. Biomaterials. 2012;33:999–1006. doi: 10.1016/j.biomaterials.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 37.Moraes C, et al. Organs-on-a-chip: a focus on compartmentalized microdevices. Ann Biomed Eng. 2012;40:1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 38.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turley SJ, et al. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 40.Wlodkowic D, Cooper JM. Tumors on chips: oncology meets microfluidics. Curr Opin Chem Biol. 2010;14:556–567. doi: 10.1016/j.cbpa.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Young EWK. Cells, tissues, and organs on chips: challenges and opportunities for the cancer tumor microenvironment. Integr Biol. 2013;5:1096–1109. doi: 10.1039/c3ib40076j. [DOI] [PubMed] [Google Scholar]
- 42.Vidi P-A, et al. Disease-on-a-chip: mimicry of tumor growth in mammary ducts. Lab on a Chip. 2014;14:172–177. doi: 10.1039/c3lc50819f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner K, Gottesman MM. Beyond 3D culture models of cancer. Sci Transl Med. 2015;7:283ps9. doi: 10.1126/scitranslmed.3009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvalho MR, et al. Evaluating biomaterial- and microfluidic-based 3D tumor models. Trends Biotechnol. 2015;33:667–678. doi: 10.1016/j.tibtech.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Choi Y, et al. A microengineered pathophysiological model of early-stage breast cancer. Lab on a Chip. 2015;15:3350–3357. doi: 10.1039/c5lc00514k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gioiella F, et al. An engineered breast cancer model on a chip to replicate ECM- activation in vitro during tumor progression. Adv Healthcare Mater. 2016;5:3074–3084. doi: 10.1002/adhm.201600772. [DOI] [PubMed] [Google Scholar]
- 47.Aung A, et al. Chemotaxis-driven assembly of endothelial barrier in a tumor-on-a-chip platform. Lab on a Chip. 2016;16:1886–1898. doi: 10.1039/c6lc00184j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Astolfi M, et al. Micro-dissected tumor tissues on chip: an ex vivo method for drug testing and personalized therapy. Lab on a Chip. 2016;16:312–325. doi: 10.1039/c5lc01108f. [DOI] [PubMed] [Google Scholar]
- 49.Ruppen J, et al. A microfluidic platform for chemoresistive testing of multicellular pleural cancer spheroids. Lab on a Chip. 2014;14:1198–1205. doi: 10.1039/c3lc51093j. [DOI] [PubMed] [Google Scholar]
- 50.Ruppen J, et al. Towards personalized medicine: chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab on a Chip. 2015;15:3076–3085. doi: 10.1039/c5lc00454c. [DOI] [PubMed] [Google Scholar]
- 51.Yu T, et al. Cancer-associated fibroblasts promote non-small cell lung cancer cell invasion by upregulation of glucose-regulated protein 78 (GRP78) expression in an integrated bionic microfluidic device. Oncotarget. 2016;7:25593–25603. doi: 10.18632/oncotarget.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Y, et al. Engineering a brain cancer chip for high-throughput drug screening. Sci Rep. 2016;6:25062. doi: 10.1038/srep25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu P-F, et al. A bladder cancer microenvironment simulation system based on a microfluidic co-culture model. Oncotarget. 2015;6:37695–37705. doi: 10.18632/oncotarget.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng Y, et al. Angiogenesis in liquid tumors: an in vitro assay for leukemic cell induced bone marrow angiogenesis. Adv Healthcare Mater. 2016;5:1014–1024. doi: 10.1002/adhm.201501007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pak C, et al. MicroC3: an ex vivo microfluidic cis-coculture assay to test chemosensitivity and resistance of patient multiple myeloma cells. Integr Biol. 2015;7:643–654. doi: 10.1039/c5ib00071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farokhzad OC, et al. Microfluidic system for studying the interaction of nanoparticles and microparticles with cells. Analyt Chem. 2005;77:5453–5459. doi: 10.1021/ac050312q. [DOI] [PubMed] [Google Scholar]
- 57.Lee J, et al. In vitro toxicity testing of nanoparticles in 3D cell culture. Small. 2009;5:1213–1221. doi: 10.1002/smll.200801788. [DOI] [PubMed] [Google Scholar]
- 58.Grafton MMG, et al. Breast on-a-chip: mimicry of the channeling system of the breast for development of theranostics. Integr Biol. 2011;3:451–459. doi: 10.1039/c0ib00132e. [DOI] [PubMed] [Google Scholar]
- 59.Yang Y, et al. Evaluation of photodynamic therapy efficiency using an in vitro three-dimensional microfluidic breast cancer tissue model. Lab on a Chip. 2015;15:735–744. doi: 10.1039/c4lc01065e. [DOI] [PubMed] [Google Scholar]
- 60.Prabhakarpandian B, et al. Synthetic tumor networks for screening drug delivery systems. J Control Release. 2015;201:49–55. doi: 10.1016/j.jconrel.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albanese A, et al. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat Commun. 2013;4:2718. doi: 10.1038/ncomms3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zervantonakis IK, Arvanitis CD. Controlled drug release and chemotherapy response in a novel acoustofluidic 3D tumor platform. Small. 2016;12:2616–2626. doi: 10.1002/smll.201503342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.FDA_Voice. FDA Voice Interviews Jesse Goodman, MD, MPH, on the DARPA and NIH Project Collaboration: Human on a Chip. FDA; 2012. [Google Scholar]
- 64.Skardal A, et al. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today. 2016;21:1399–1411. doi: 10.1016/j.drudis.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danquah MK, et al. Extravasation of polymeric nanomedicines across tumor vasculature. Adv Drug Deliv Rev. 2011;63:623–639. doi: 10.1016/j.addr.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 66.Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat Rev Clin Oncol. 2010;7:653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y-N, et al. Nanoparticle–liver interactions: cellular uptake and hepatobiliary elimination. J Control Release. 2016;240:332–348. doi: 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 68.Hirai T, et al. Metal nanoparticles in the presence of lipopolysaccharides trigger the onset of metal allergy in mice. Nat Nanotechnol. 2016;11:808–816. doi: 10.1038/nnano.2016.88. [DOI] [PubMed] [Google Scholar]
- 69.Huh D, et al. Microengineered physiological biomimicry: organs-on-chips. Lab on a Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 70.Zhang C, et al. Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments. Lab on a Chip. 2009;9:3185–3192. doi: 10.1039/b915147h. [DOI] [PubMed] [Google Scholar]
- 71.Wikswo JP, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab on a Chip. 2013;13:3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen YY, et al. Clarifying intact 3D tissues on a microfluidic chip for high-throughput structural analysis. Proc Natl Acad Sci U S A. 2016;113:14915–14920. doi: 10.1073/pnas.1609569114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang YS, Yu C. Towards engineering integrated cardiac organoids: beating recorded. J Thoracic Dis. 2016;8:E1683–E1687. doi: 10.21037/jtd.2016.12.37. [DOI] [PMC free article] [PubMed] [Google Scholar]


