Abstract
Owing to their high affinities and specificities, rabbit monoclonal antibodies (mAbs) have demonstrated value and potential primarily as basic research and diagnostic reagents, but in some cases also as therapeutics. To accelerate access to rabbit mAbs bypassing immunization, we generated a large naïve rabbit antibody repertoire represented by a phage display library encompassing >10 billion independent antibodies in chimeric rabbit/human Fab format and validated it by next-generation sequencing. Panels of rabbit mAbs selected from this library against two emerging cancer targets, ROR1 and ROR2, revealed high diversity, affinity, and specificity. Moreover, ROR1− and ROR2-targeting rabbit mAbs demonstrated therapeutic utility as components of chimeric antigen receptor-engineered T cells, further corroborating the value of the naïve rabbit antibody library as a rich and virtually unlimited source of rabbit mAbs.
Keywords: Antibody engineering, ROR1, ROR2, chimeric antigen receptors, cancer therapy
Graphical abstract
Introduction
The rabbit antibody repertoire, which in the form of polyclonal antibodies (pAbs) has been widely utilized for immunological techniques, such as immunohistochemistry (IHC), Western blotting, enzyme-linked immunosorbent assay (ELISA), and flow cytometry, represents an outstanding source for antibodies of high affinity and specificity.1 In contrast to humans and mice, which rely almost exclusively on somatic hypermutation to further diversify antibody genes following VH-D-JH (heavy chain) and VL-JL (light chain) gene rearrangements, V-(D)-J recombination in rabbits is followed by both somatic gene conversion and somatic hypermutation to expand primary and secondary antibody repertoires.2,3 In addition, rabbits (Oryctolagus cuniculus) belong to the taxonomic order Lagomorpha and are evolutionarily distant from mice, rats, and other rodents which belong to the taxonomic order of Rodentia. Consequently, epitopes conserved between mouse and human antigens that are invisible to mouse and human mAbs due to immune tolerance can often be recognized by rabbit pAbs. However, rabbit pAbs are undefined reagents with finite supply. The ability to generate rabbit mAbs by either phage display4–6 or hybridoma technology7 has overcome this limitation, providing access to defined reagents of infinite supply from the rabbit antibody repertoire.
Rabbit mAbs generated by phage display offer additional advantages due to the fact that phenotype and genotype are selected at the same time.8 Knowledge of the rabbit mAb sequence allows the ready generation of a variety of mAb formats, including scFv, Fab, scFv-Fc, and IgG, and, importantly, humanization and affinity maturation.9–11 Consequently, rabbit mAbs generated by phage display have become promising reagents for diagnostic and therapeutic applications in humans. Over the past decade, we have developed methods for the generation and selection of immune rabbit antibody libraries by phage display6,8,9 and generated a variety of rabbit mAbs with broad utility for basic research and for diagnosis and therapy of human diseases,9,10,12–19 including hematologic malignancies.20 In addition to their high affinity (typically in the 0.1–10 nM range for Fab) and specificity, many of these rabbit mAbs cross-reacted with mouse and human antigens.
Immune rabbit antibody libraries, however, rely on immunizing rabbits with antigens of interest and are biased toward immunodominant epitopes. To curtail these limitations, we here report the generation, validation, and selection of a first-in-its-kind highly complex naïve rabbit antibody library. Using reverse transcribed mRNA from bone marrow and spleen of genetically diverse naïve rabbits and an extensive set of oligonucleotides for the amplification of light and heavy chains by PCR, we generated a phage display library encompassing >10 billion rabbit mAbs as validated by next-generation sequencing (NGS). This vast and unique antigen binding repertoire, which we anticipate to have virtually no bias for or against the recognition of any human antigen and epitope, constitutes the platform of the current study.
To validate the naïve rabbit antibody library, we focused on selecting rabbit mAbs to human cell surface antigens of potential utility for cancer therapy, including receptor tyrosine kinases RET, ROR1, and ROR2, complement protein C3d, and T-cell marker CD3. The panels of rabbit mAbs to ROR1 and ROR2 were then extensively characterized.
Receptor tyrosine kinases ROR1 and ROR2 are type I single-pass transmembrane proteins that share 58% amino acid (aa) sequence identity and that are composed of a unique extracellular region with one immunoglobulin (Ig), one frizzled (Fz), and one kringle (Kr) domain and an intracellular region that harbors a pseudokinase domain (Suppl. Fig. 1A). ROR1 and ROR2 are widely expressed in embryonic tissues but reveal highly restricted expression in postpartum and adult tissues.21 Aberrant expression of ROR1 and ROR2 is associated with a number of hematologic and solid malignancies, making them promising targets for cancer therapy in general and for antibody-based therapeutics in particular.22–24 For example, corroborating previous reports of ROR1 and ROR2 expression in breast cancer (BC),23,25,26 we mined a BC gene expression database27 to confirm widespread ROR1 and ROR2 mRNA expression (Suppl. Fig. 1B). Interestingly, ROR1+ samples were predominantly ROR2− and vice versa. ROR1 and ERBB2 mRNA expression was highly complementary, collectively covering >50% of all samples. In addition, we noted a striking co-expression of ROR1 and EGFR mRNA in HER2 mRNA-negative samples which backs previous reports of a functional and physical interaction of ROR1 and EGFR in cancer cells.28 In addition, cell surface expression of ROR1 and ROR2 in BC cell lines was analyzed by flow cytometry. Shown in Suppl. Fig. 1C are the profiles of two representative HER2-negative BC cell lines, MDA-MB-231 (ROR1+ ROR2−) and T47D (ROR1− ROR2+), which were used in the current study. Taken together, ROR1 and ROR2 are emerging targets for antibody-based therapeutics specifically in BC and also cancer in general. In fact, the development of ROR1- and ROR2-targeting antibody-based therapeutics, including mAbs, antibody-drug conjugates, bispecific antibodies, and chimeric antigen receptor-engineered T cells (CAR-T), would be highly relevant for the majority of BC patients who do not overexpress HER2. To this end, we here report the utilization of the naïve rabbit antibody library for the generation of a panel of 13 ROR1- and 12 ROR2-targeting mAbs. By demonstrating their potential therapeutic utility as CAR-T, we validate the naïve rabbit antibody library as a rich source for antibody-based therapeutics.
Results
Generation and validation of a naïve rabbit antibody library
The library’s antibody repertoire was derived from bone marrow and spleen harvested from naïve rabbits (n=9, ages 3–4 months). Five of these rabbits were of the New Zealand White (NZW) strain, with three obtained from Pocono Rabbit Farm & Laboratory (Canadensis, PA) and two obtained from R & R Research (Stanwood, WA). Four rabbits were derived from a separate R & R Research colony that originated from a pedigreed colony developed and characterized at the National Institute of Allergy and Infectious Diseases (NIAID), NIH.29 Rabbits of this colony have the b9 κ-light chain allotype, which we previously showed to be superior for the phage display selection of rabbit mAbs in chimeric rabbit/human Fab format.6 Taken together, the antibody repertoire for the library was genetically diverse and derived from primary (bone marrow) and secondary (spleen) lymphoid organs. To represent this antibody repertoire with as little bias as possible, we designed a new set of 41 oligonucleotides for the amplification of rabbit light and heavy chain variable domains (rbVκ, rbVλ, and rbVH) based on IMGT databases (www.imgt.org) updated with genome sequencing and annotation data (OryCun2.0) from the Rabbit Genome Project30 (Suppl. Table S1). This new set by far exceeds our previous set of 12 oligonucleotides8 with respect to rbVκ (14 vs. 4 forward; 5 vs. 1 reverse primers), rbVλ (8 vs. 1 forward; 1 vs. 1 reverse primer), and rbVH (11 vs. 3 forward primers; 2 vs. 2 reverse primers) (Fig. 1), for a total of 100 vs. 11 primer combinations. Although VH-D-JH recombination in rabbits predominantly uses one VH gene,31 our new set of oligonucleotides was designed to also amplify rarely used VH genes. Rabbit Vκ-Jκ and Vλ-Jλ recombinations are less biased in terms of VL gene usage and our new set of oligonucleotides captures a larger portion of this diversity. In fact, a retrospective analysis of rbVκ, rbVλ, and rbVH sequences of 50 unique chimeric rabbit/human Fab clones following stringent phage display selection against 6 human cell surface antigens, including ROR1 and ROR2 reported here, revealed that 9/14 rbVκ forward, 4/5 rbVκ reverse, 4/8 rbVλ forward, 1/1 rbVλ reverse, 7/11 rbVH forward, and 2/2 rbVH reverse primers, i.e. 27/41 (66%) of all oligonucleotides, collectively contributed to the amplification.
Figure 1. Generation of the naïve rabbit antibody library.
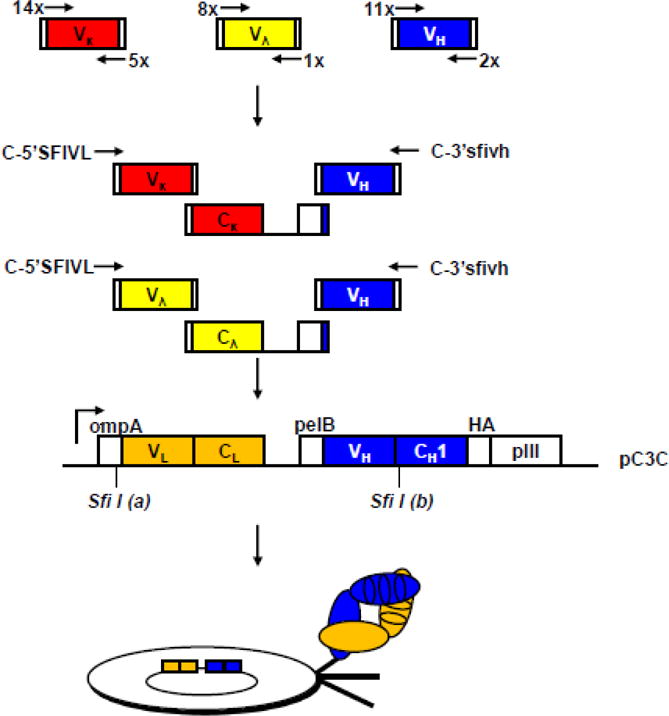
Chimeric rabbit/human Fab consist of rabbit variable domains (rbV) and human constant domains (huC). (Top) Amplification of rbV by PCR. The numbers of forward and reverse primers for amplification of rbVκ (red), rbVλ (yellow) and rbVH (blue) encoding sequences are shown. (Upper middle) Assembly of rbVκ/huCκ/rbVH and rbVλ/huCλ/rbVH expression cassettes by fusion PCR. (Lower middle) Asymmetric SfiI sites labeled as “(a)” and “(b)” facilitate the cloning of the rbVL/huCL/rbVH expression cassette into phage display vector pC3C. The design of pC3C17 is based on phagemids from the pComb3 series. A single lacZ promoter drives the synthesis of a dicistronic transcript. Two ribosome binding sites initiate the translation of two separate polypeptide chains, chimeric light chain rbVL-huCL (orange) and chimeric heavy chain fragment rbVH-huCH1 (blue) fused to a hemagglutinin (HA) decapeptide and the C-terminal pIII protein domain; pIII is the minor coat protein of filamentous phage and is displayed in low copy number at one end of the phage. Through leader peptides ompA and pelB both polypeptides are transported to the periplasm of E. coli, where they associate and form a natural disulfide bridge. Addition of helper phage leads to the incorporation of the fusion protein into phage particles (bottom) that display one chimeric rabbit/human Fab copy linked to the phage surface by the C-terminal pIII protein domain as their phenotype and, as their genotype, contain the corresponding single-stranded phagemid that encodes the Fab.
Rabbit variable domain-encoding sequences rbVκ, rbVλ, and rbVH were amplified by PCR from reverse transcribed mRNA prepared from the 18 rabbit tissues (nine bone marrows and nine spleens) in approximately 2,000 independent reactions and then further assembled to chimeric rabbit/human Fab-encoding sequences in phage display vector pC3C as shown in Fig.1. Consistent with our previous immune rabbit antibody libraries,6,8,9,17,20 we chose a chimeric rabbit/human Fab format for the naïve rabbit antibody library. Chimeric rabbit/human Fab consist of rabbit variable domains fused to human constant domains. This is a preferred format for phage display for the following reasons: (i) Fab, unlike scFv, are less likely to multimerize and interfere with selection for high affinity; (ii) human but not rabbit constant domains are well expressed in E. coli; and (iii) our established phage display vector pC3C17 (Fig. 1) incorporates human constant domains. Using optimized ligation and electroporation protocols, we achieved a phage display library size of 1.2 × 1010 independent chimeric rabbit/human Fab clones. Thus, in terms of size, our naïve rabbit antibody library is comparable to large naïve and synthetic human antibody libraries previously generated and successfully mined in the industry.32–34
We next validated the naïve rabbit antibody library by NGS. For this we used an Ion Torrent PGM instrument for ~400-bp sequence reads35 starting from a reverse primer downstream of rbVH. The bioinformatic analysis of 2,756,201 rbVH sequences revealed 2,705,332 (98.2%) unique clones with ≥ 2 bp differences. As expected, the most frequent repeated clone had the rbVH sequence of parental pC3C,17 attesting a low percentage of incomplete SfiI digestion events. To further document the diversity of the naïve rabbit antibody library, we analyzed its most variable segments, i.e. the third complementarity determining region of the heavy chain (HCDR3). We found 1,112,793 unique HCDR3 sequences with a mean ± standard deviation (SD) length of 13.3 ± 3.3 aa and a mode length of 13 aa (Fig. 2A). This is in line with a previous study that found a mean ± SD length of 14.8 ± 3.6 aa and a mode length of 13 aa when analyzing (29,439) rbVH sequences from three naïve rabbits of the NZW strain by NGS.36 Thus, in terms of length, rabbit HCDR3s are closer to human HCDR3s (mean ± SD = 15.3 ± 4.0 aa; mode = 15 aa) than to mouse HCDR3s (mean ± SD = 11.1 ± 2.0 aa; mode = 10 aa),36 confirming our pre-NGS analysis of rabbit antibody repertoires.6 Computing the composition of all HCDR3s in the naïve rabbit antibody library, we found that glycine (G; 13.5%), alanine (A; 11.1%), arginine (R; 9.0%), tyrosine (Y; 8.9%), aspartic acid (D; 8.5%), leucine (L; 7.6%), and serine (S; 7.5%) were the most frequent aa, whereas glutamine (Q; 0.4%), lysine (K; 0.6%), cysteine (C; 1.2%), glutamic acid (E; 1.3%), histidine (H; 1.4%), and methionine (M; 1.7%) were the least frequent aa (Fig. 2B). Except for a higher frequency of R and L in the HCDR3s of our naïve rabbit antibody library, this composition is in line with what was found in the previous NGS analysis that was based on a smaller data set of only one rabbit strain.36 Notably, with this higher frequency of R and L, the composition of HCDR3s in our naïve rabbit antibody library is very similar to the composition of human HCDR3s.36
Figure 2. Validation of the naïve rabbit antibody library.
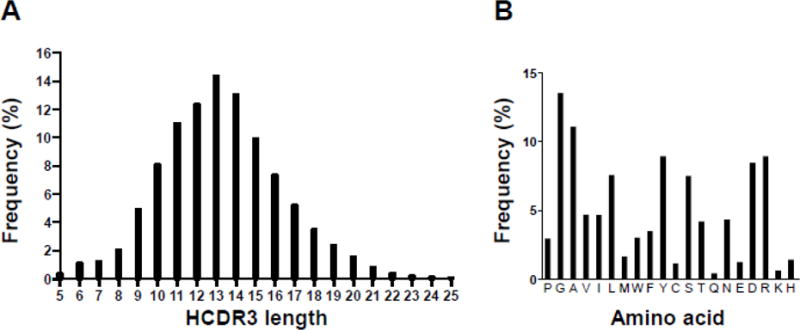
(A) HCDR3 lengths distribution and (B) HCDR3 amino acid composition as determined by NGS of 2,756,201 rbVH encoding sequences in the unselected chimeric rabbit/human Fab library. NGS data were analyzed with N2GSAb software.52
Selection of C3d-, RET-, and CD3-targeting mAbs
The naïve rabbit antibody library was selected by phage display using our established protocols for biotinylated antigen capture with streptavidin-coated magnetic beads.8 Enriched binders after four rounds of panning against human complement protein C3d and recombinant portions of the extracellular domains (ECDs) of human receptor tyrosine kinase RET and human T-cell marker CD3 were identified by ELISA. Three different C3d binders, three different RET binders, and one CD3 binder were expressed and purified as chimeric rabbit/human Fab (C3d) or IgG1 (RET and CD3). As shown by ELISA, the seven unique rabbit mAbs selectively bound to their respective antigens (Suppl. Fig. 2), providing initial proof-of-concept for the ability to select rabbit mAbs to human cell surface antigens from the naïve rabbit antibody library. To further demonstrate the therapeutic utility of rabbit mAbs derived from the naïve rabbit antibody library, we focused on selecting and extensively characterizing ROR1 and ROR2 binders.
Selection and characterization of ROR1-targeting mAbs
Using the same procedure, enriched binders after three and four rounds of panning against the ECD of human ROR1 were identified by ELISA screening and by NGS. We discovered 13 clones with highly diverse rbVH, rbVλ (11), and rbVκ (2) sequences (Suppl. Fig. S3A and S3B). HCDR3 and LCDR3 lengths ranged from 7 to 16 aa and from 9 to 12 aa, respectively (Table 1).
Table 1.
Properties of chimeric rabbit/human anti-human ROR1 Fabs.
| HCDR3 | Isotype | LCDR3 | Kd(nM)a | Epitopeb | mROR1c | Westernd | IHCe | |
|---|---|---|---|---|---|---|---|---|
| XBR1-402 | DHPTYGMDL | λ | QVWDSSAYV | 5.81 | Ig+Fz | − | ++ | + |
| ERR1-301 | DHPSYGMDL | λ | QLWDSSAVAYV | 118 | Ig+Fz | − | NA | NA |
| ERR1-306 | WVYGVDDYGDGNWLDL | λ | GTDYSGGYV | 9.66 | Ig+Fz | + | NA | ++ |
| ERR1-316 | DHPNYGMDL | λ | QLWDSSTGAYA | 121 | Ig+Fz | − | NA | NA |
| ERR1-324 | GTVSSDI | κ | LGGYVSQSYRAA | 108 | Ig+Fz | − | + | NA |
| ERR1-403 | GSDYFDL | λ | QLWDSSAGAYV | 41.6 | Ig+Fz | − | NA | NA |
| ERR1-409 | SYPGWTTEPYFDI | λ | LSSDSSAYV | 281 | Ig+Fz | − | NA | NA |
| ERR1-TOP4 | GWATDGIFDYDDTFNL | λ | QLWDGSAYV | 240 | Ig+Fz | − | NA | NA |
| ERR1-TOP15 | DGWGSDV | λ | QLWDSSAGAYV | 139 | Ig+Fz | − | NA | NA |
| ERR1-TOP22 | DLDYDMDL | λ | QLWDNNAAV | 110 | Ig+Fz | + | NA | NA |
| ERR1-TOP40 | LYVSDSTANL | κ | LGGYPNYFHRTA | 636 | Ig+Fz | − | NA | NA |
| ERR1-TOP43 | DVHSTATDL | λ | QLWDSSAGAYV | 1.11 | Ig+Fz | − | +++ | + |
| ERR1-TOP54 | DFGLSTGGL | λ | QLWDSSARAFV | 145 | Ig+Fz | − | NA | NA |
Determined by ELISA using hFc fusion proteins of hROR1 whole ECD (Ig+Fz+Kr), tandem domains (Ig+Fz and Fz+Kr), and single domains (Ig, Fz, and Kr).20
Cross-reactivity with mouse ROR1; Suppl. Fig. S4.
Determined with chimeric rabbit/human IgG1 using purified hROR1-HIS protein and hROR1-expressing K562 cells; purified hROR2-HIS protein and parental K562 cells were used as controls; NA, not analyzed.
All 13 Fabs were expressed and purified as soluble chimeric rabbit/human Fabs37 and revealed selective binding to the ECD of human ROR1 but not ROR2 as tested by ELISA (Suppl. Fig. S4). Two Fabs, ERR1-306 and ERR1-TOP22, cross-reacted with the ECD of mouse ROR1. As analyzed by flow cytometry and shown in Fig. 3A, all 13 Fabs recognized cell surface human ROR1, but not ROR2, in stably transfected mouse pre-B cell lines. We also confirmed their binding to human cancer cell lines expressing ROR1 endogenously, including MDA-MB-231 cells (data not shown). The kinetic and thermodynamic binding parameters of the interaction with human ROR1 were determined by surface plasmon resonance (SPR). The dissociation constants (Kd) of three Fabs, ERR1-TOP43 (1.1 nM), XBR1-402 (5.8 nM), and ERR1-306 (9.7 nM), were in the single digit nanomolar range, the Kd of the remaining ten Fabs ranged from 41.6 to 636 nM (Table 1 and Suppl. Fig. S5). As determined by NGS after three rounds of panning, the most frequent binder, ERR1-316 (44%; 121 nM), revealed only modest affinity. By contrast, the strongest binder, ERR1-TOP43 (0.3%; 1.1 nM), was much less enriched. Domain mapping by ELISA revealed that all 13 Fabs required the presence of Ig and Fz domain for binding (Table 1), similar to what we had observed for mAb R12, which had been selected by phage display from an immune rabbit antibody library.20 Further analysis by SPR showed that R12’s epitope indeed overlapped with the epitopes of the strongest binders, ERR1-TOP43, XBR1-402, and ERR1-306. Interestingly, ERR1-324 bound to a non-overlapping epitope (Suppl. Fig. S6). As expected, mAb R11, which binds to the Kr domain,20 was also not competed by any of the new Fabs.
Figure 3. Analysis of selected chimeric rabbit/human anti-human ROR1 and ROR2 Fabs by flow cytometry.
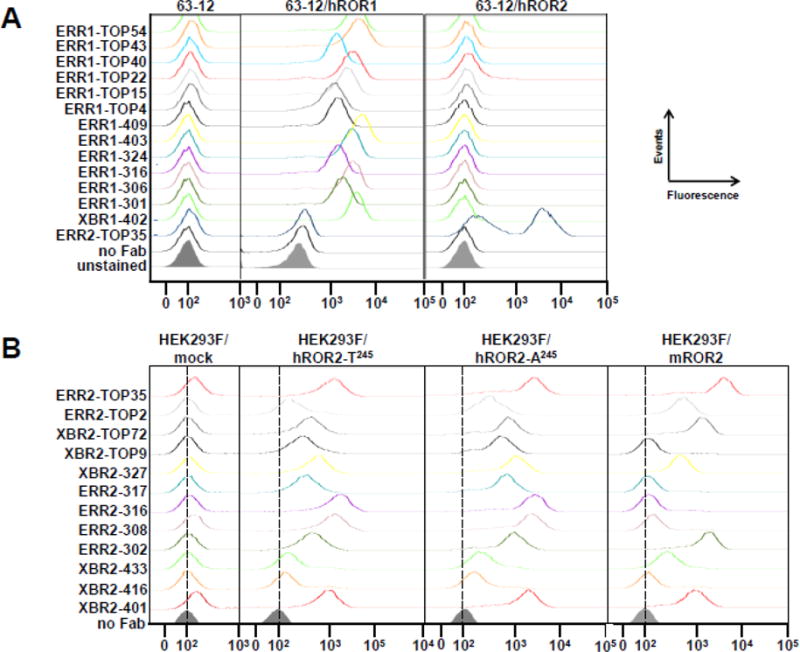
(A) The binding of 13 purified chimeric rabbit/human anti-human ROR1 Fabs (10 μg/mL) to ectopically and stably expressed human ROR1 or human ROR2 on mouse pre-B cell line 63-12 and to parental 63-12 cells was analyzed by flow cytometry. Chimeric rabbit/human anti-human ROR2 Fab ERR2-TOP35 was used as control. (B) The binding of 12 purified chimeric rabbit/human anti-human ROR2 Fabs (10 μg/mL) to ectopically and stably expressed human ROR2 allotypes T245 and A245 and mouse ROR2 on HEK293F cells was analyzed by flow cytometry. Mock transfected HEK293F cells were used as control.
Several chimeric rabbit/human Fabs were converted to the chimeric rabbit/human IgG1 format, expressed in mammalian cells, and purified by Protein A affinity chromatography. Conversion from monovalent Fab to bivalent IgG1 further increased the avidity as analyzed by ELISA, SPR, and flow cytometry (data not shown). The specificity of one rabbit mAb in chimeric rabbit/human IgG1 format, XBR1-402, was tested using the Retrogenix cell microarray platform. This cell microarray uses spotted HEK293 cells that were transiently transfected to ectopically express one of 4,336 human cell surface proteins and arrayed in duplicate. XBR1-402 was found to strongly and exclusively stain ROR1 (Suppl. Fig. S7).
Furthermore, conversion of chimeric/rabbit Fabs XBR1-402, ERR1-306, and ERR1-TOP43 to full rabbit IgG, expression in mammalian cells, and purification by Protein A affinity chromatography yielded reagents capable of detecting cell surface ROR1 on ectopically and endogenously expressing cells by IHC (Fig. 4).
Figure 4. Analysis of selected full rabbit anti-human ROR1 IgG1s by IHC.
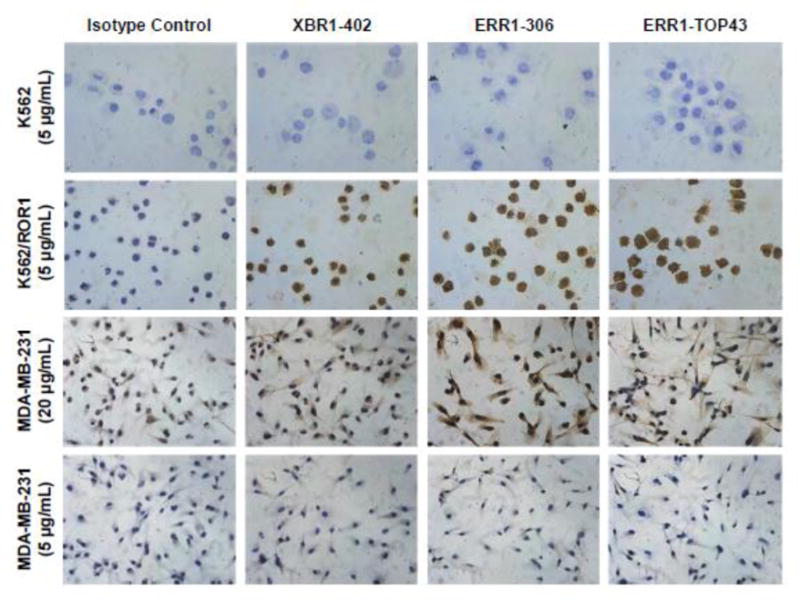
Ectopically expressed ROR1 (K562/ROR1 vs. K562 cells) and endogenously expressed ROR1 (MDA-MB-231 cells) was detected by IHC using full rabbit IgG that were generated from XBR1-402, ERR1-316, and ERR1-TOP43. Note that the IHC detection of endogenously expressed ROR1 on the cell surface of fixed human cells and tissues by mAbs to the ECD of ROR1 is known to be difficult.58 Fittingly, a high concentration of rabbit mAbs (20 μg/mL) was required for staining the MDA-MB-231 cells. All light microscopy images were taken at a magnification of 40× (total magnification of 400×).
Selection and characterization of ROR2-targeting mAbs
Next, we used the same selection and screening methods to mine the naïve rabbit antibody library for ROR2-targeting mAbs. We discovered 12 clones with highly diverse rbVH, rbVλ (4), and rbVκ (8) sequences (Suppl. Fig. S3C and S3D). HCDR3 and LCDR3 lengths ranged from 7 to 16 aa and from 9 to 13 aa, respectively (Table 2).
Table 2.
Properties of chimeric rabbit/human anti-human ROR2 Fabs.
| HCDR3 | Isotype | LCDR3 | Kd (nM)a | Epitopeb | hROR2 SNPc | mROR2d | cROR2e | Westernf | |
|---|---|---|---|---|---|---|---|---|---|
| XBR2-401 | DWTSLNI | κ | LGGYADASYRTA | 3.87 | Kr | + | + | + | +++ |
| XBR2-416 | GIGGAADL | κ | LGGYPNTSYRSA | 348 | Kr | + | − | NA | NA |
| XBR2-433 | DGYYSSGWGPYFNI | λ | ATSGGSGSNPQYV | 2280 | Fz | + | + | NA | NA |
| XBR2-327 | DYGNWAFDP | κ | LGGYASASYRTA | 57.2 | Kr | + | + | + | - |
| XBR2-TOP9 | GGYASGFNL | κ | LGWHSWSDDGWA | 142 | Kr | + | − | NA | NA |
| XBR2-TOP72 | AGTTYYTSFNL | λ | GADYSGGYV | 4150 | Fz+Kr | + | + | NA | NA |
| ERR2-302 | SPYGYVSAWGYHRLDL | λ | QLWDGSDVV | 52.5 | Ig+Fz+Kr | + | + | + | NA |
| ERR2-308 | GASNGCDL | κ | LGGYSNSDIV | 10.6 | Kr | + | − | + | NA |
| ERR2-316 | YFGWNTGFDI | κ | LGFSVVSNDGWA | 5.34 | Kr | + | − | + | NA |
| ERR2-317 | GSSWTGDI | κ | LGGYASASYQTA | 159 | Kr | + | _ | + | NA |
| ERR2-TOP2 | ELVAGGGSDL | κ | LGGYLSASYQTA | 81.1 | Fz+Kr | + | + | NA | NA |
| ERR2-TOP35 | DGYYSSGWGPYFSI | λ | ATSDGSGSRYYV | 43.3 | Fz+Kr | + | + | + | NA |
Determined by flow cytometry using cell surface hROR2 whole ECD (Ig+Fz+Kr), tandem domains (Ig+Fz and Fz+Kr), and single domains (Ig, Fz, and Kr) expressed on HEK293 cells.
Cross-reactivity with human ROR2 allotypes; Fig. 3B.
Cross-reactivity with mouse ROR2; Fig. 3B.
Cross-reactivity with cynomolgus ROR2; determined with chimeric rabbit/human IgG1; NA, not analyzed.
Determined with chimeric rabbit/human IgG1 using purified hROR2-HIS protein and hROR2-expressing 63-12 cells; purified hROR1-HIS protein, hROR1-expressing 63-12, and parental 63-12 cells were used as controls; NA, not analyzed.
As done for the ROR1-targeting mAbs, all 12 Fabs were expressed and purified as soluble chimeric rabbit/human anti-human ROR2 Fabs. Initial testing by ELISA (Suppl. Fig. S8A) and flow cytometry (Fig. 3B and Suppl. Fig. S8B) showed selective binding to human ROR2 but not to human ROR1. We also confirmed their binding to cancer cell lines expressing ROR2 endogenously, including T47D cells (data not shown). Flow cytometry was also used to show (i) that all 12 Fabs bound to both human ROR2 ECD allotypes, which arise from single nucleotide polymorphism (SNP) rs10820900 and are characterized by either having a tyrosine or an alanine residue at position 245 in the Fz domain38 and (ii) that half of the Fabs were cross-reactive with mouse ROR2 (Fig. 3B). All ROR2-targeting mAbs we tested were also cross-reactive with cynomolgus ROR2 (Table 2). SPR revealed that the Kd of three Fabs, XBR2-401 (3.9 nM), ERR2-316 (5.3 nM), and ERR2-308 (10.6 nM), were in or close to the single digit nanomolar range (Table 2 and Suppl. Fig. S9). Domain mapping by flow cytometry and epitope binning by SPR revealed that the majority of Fabs bound to non-overlapping epitopes in the Fz and Kr domains (Table 2 and Suppl. Fig. S10). Interestingly, ERR2-302 required the presence of the entire ECD (Ig, Fz, and Kr domain) for binding, suggesting that this mAb recognizes a unique conformational epitope shaped by all three domains.
Several chimeric rabbit/human anti-human ROR2 Fabs were also converted to the chimeric rabbit/human IgG1 format, revealing increased avidity as analyzed by ELISA, SPR, and flow cytometry (data not shown). The specificity of one rabbit mAb in chimeric rabbit/human IgG1 format, XBR2-401, was tested by using a Retrogenix cell microarray assay and revealed strong and exclusive staining of ROR2 among 4,336 human plasma membrane proteins (Suppl. Fig. S7).
Collectively, our large naïve rabbit antibody library yielded ROR1− and ROR2-targeting mAbs with nanomolar affinities and exceptional specificities.
Generation of ROR1- and ROR2-targeting CAR-T cells
To investigate the therapeutic utility of the ROR1− and ROR2-targeting mAbs, we engineered XBR1-402 and XBR2-401 CAR-T cells using methods previously described for ROR1-targeting mAbs R11 and R12.39 In brief, ex vivo expanded healthy donor CD8+ CD62L+ T cells were lentivirally transduced with an EF1α promoter-driven expression cassette containing either XBR1-402 or XBR2-401 in scFv format, followed by a short or long IgG4-derived spacer, the transmembrane domain of human CD28, the signaling domain of 4-1BB, the signaling domain of CD3ζ, and a T2A-separated transmembrane EGFR fragment with truncated ligand binding and tyrosine kinase domains.39,40 Magnetic isolation of EGFR+ transduced T cells revealed robust anti-ROR1 or anti-ROR2 recognition in >90% of CAR-T cells (data not shown).
We first tested the activity of the ROR1-targeting XBR1-402 CAR-T with a short spacer against BC cell lines MDA-MB-231 (ROR1+ ROR2−) and T47D (ROR1− ROR2+). In the presence of ROR1+ ROR2− but not ROR1− ROR2+ target cells, XBR1-402 CAR-T rapidly proliferated (Fig. 5A), secreted IFN-γ and IL-2 at high levels (Fig. 5B), and potently killed the target cells in vitro (Fig. 5C). Notably, in direct comparison, the XBR1-402 CAR-T was found to be equally or more potent than the already highly effective and clinically investigated R12 CART with the same short spacer and signaling domains.
Figure 5. Comparison of the activities of ROR1-targeting XBR1-402 CAR-T and R12 CAR-T.
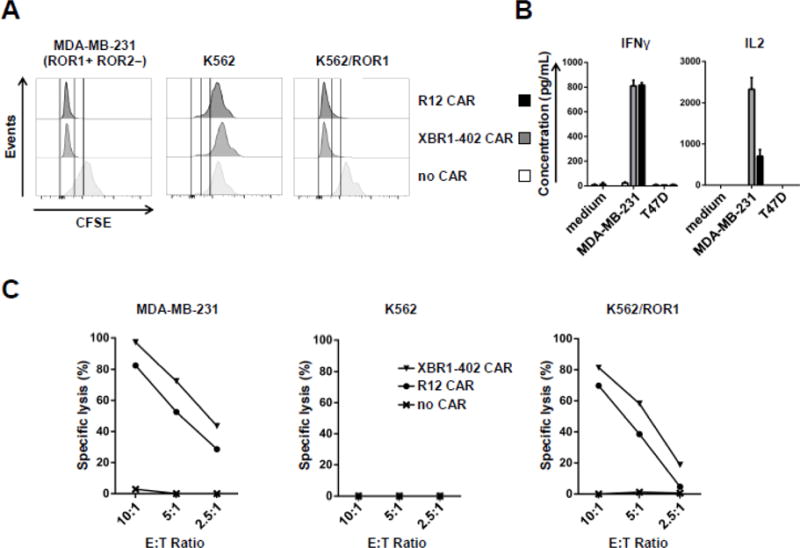
Ex vivo expanded primary human CD8+ CD62L+ T cells were lentivirally transduced with R12 or XBR1-402-derived CARs containing CD3ζ and 4-1BB signaling domains and a short IgG4 hinge-derived spacer. (A) CFSE-labeled CAR-Ts and untransduced T cells were incubated with the indicated target cell lines at an E:T ratio of 4:1 for 72 h and analyzed by flow cytometry. (B) CAR-Ts and untransduced T cells were incubated with the indicated target cell lines at an E:T ratio of 4:1 for 24 h. IFN-γ and IL-2 concentrations in the supernatant were measured by ELISA. (C) CAR-Ts and untransduced T cells were incubated with the indicated firefly luciferase-transduced target cell lines at the indicated E:T ratios for 7 h. Specific lysis was measured with a luciferase-based cytotoxicity assay.
In our previous study,39 we observed that the R12 CAR-T was more active with a short rather than long spacer. By contrast, the R11 CAR-T was more active with the long rather than short spacer. We attributed these marked differences to the varying epitope locations in the ROR1 ECD. Whereas R12 binds to a membrane-distal epitope at the interface of Ig and Fz domains, R11 binds to a membrane-proximal epitope in the Kr domain. Based on this finding, we hypothesized that an optimal spacing between T cell and target cell membranes can be achieved by equipping CAR-T cells targeting membrane-distal epitopes with shorter spacers and vice versa. Based on this hypothesis, we predicted that XBR1-402 CAR-Ts, which have an overlapping epitope with R12, are more active when equipped with a short compared to a long spacer. Indeed, in the presence of ROR1+ ROR2− target cells, the XBR1-402 CAR-T with short spacer proliferated more rapidly than the XBR1-402 CAR-T with long spacer and also secreted significantly more IFN-γ and IL-2 (Fig. 6 and data not shown). In vitro cytotoxicity, however, was equivalent (Fig. 6).
Figure 6. Comparison of the activities of ROR1-targeting XBR1-402 CAR-Ts with short and long spacer.
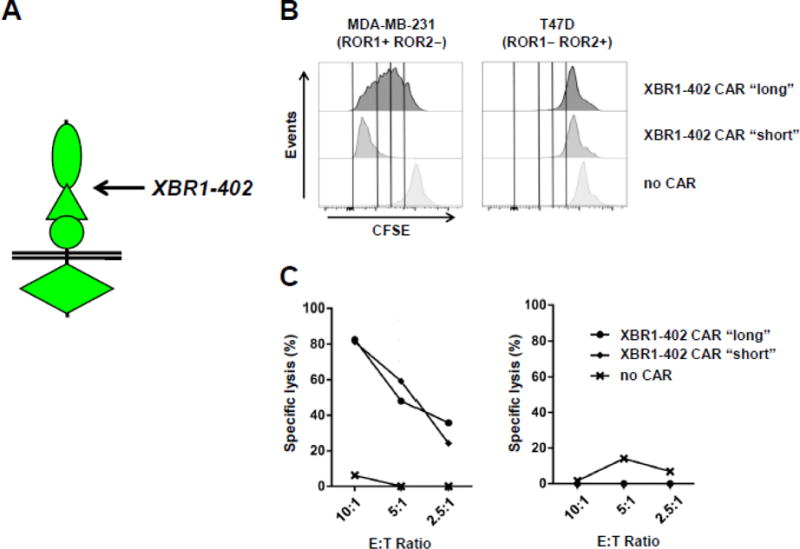
(A) Membrane distal epitope location of XBR1-402 on ROR1. Using XBR1-402-derived CARs containing CD3ζ and 4-1BB signaling domains and either a short (12 aa) or a long IgG4-derived spacer (229 aa), proliferation (B) and cytotoxicity studies (C) were carried out as described for Figure 5.
We next tested the activity of the ROR2-targeting XBR2-401 CAR-T with short and long spacer against BC cell lines MDA-MB-231 (ROR1+ ROR2−) and T47D (ROR1− ROR2+). Similar to anti-ROR1 mAb R11, XBR2-401 binds to a membrane-proximal epitope in the Kr domain of ROR2 (Table 2). Thus, we predicted that XBR2-401 CAR-Ts are more active with a long rather than a short spacer. This was confirmed with respect to proliferation, IFN-γ and IL-2 secretion (data not shown), and cytotoxicity in the presence of ROR1− ROR2+ target cells (Fig. 7 and data not shown).
Figure 7. Comparison of the activities of ROR2-targeting XBR2-401 CAR-T with short and long spacer.
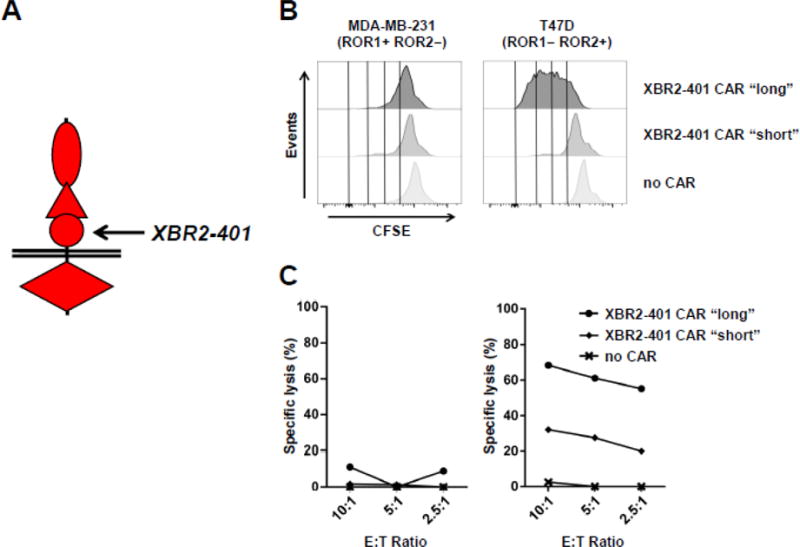
(A) Membrane proximal epitope location of XBR2-401 on ROR2. Using XBR2-401-derived CARs containing CD3ζ and 4-1BB signaling domains and either a short (12 aa) or a long IgG4-derived spacer (229 aa), proliferation (B) and cytotoxicity studies (C) were carried out as described for Figure 5.
Discussion
Recent discussions about the notorious unreliability of a substantial portion of mAbs and pAbs used in basic research have led to calls for a rapid switch to recombinant antibodies that are defined by their VL and VH amino acid sequences and extensively characterized in terms of affinity and specificity.41,42 Such standardization, which has been a sine qua non for therapeutic mAbs for decades, would improve the reproducibility of basic research that relies on antibody reagents. Here we provide a new tool that expands the access to recombinant antibodies to the naïve rabbit antibody repertoire.
The antibody repertoire in rabbits is different from that of mice and humans, reflecting the unique ontogeny of the rabbit B-cell repertoire.43 In addition to VH-D-JH and VL-JL recombination and somatic hypermutation, rabbits extensively use somatic gene conversion for the diversification of antibody sequences2. Importantly, both somatic gene conversion and somatic hypermutation evolve the neonatal B-cell repertoire to the primary B-cell repertoire during the first two months after birth in gut associated lymphoid tissue (GALT). Upon B-cell activation by immunogen binding, the primary B-cell repertoire then adapts to the secondary B-cell repertoire by using both somatic gene conversion and somatic hypermutation for affinity maturation. Considering the extensive antibody sequence diversification in the primary B-cell repertoire, we argued that naïve rabbits would provide an outstanding source for mAbs that bind human antigens with high affinity and specificity. We generated a naïve rabbit antibody library that was designed to further increase the complexity of antibody sequences by (i) harvesting spleen and bone marrow from nine rabbits belonging to two different strains and housed at two different facilities, (ii) RT-PCR amplifying VH, Vκ, and Vλ sequences with an expanded set of primers in approximately 2,000 individual reactions to overcome natural biases due to, e.g., dominant usage of the VH1 gene in VH-D-JH recombination,43 (iii) randomly combining VH/Vκ and VH/Vλ sequences to generate a total of >1010 independent antibodies. NGS analysis of the VH component of the naïve rabbit antibody library revealed a highly diverse antibody collection that retained key features of natural rabbit antibodies with respect to HCDR3 length distribution and amino acid sequence composition.
To test whether the naïve rabbit antibody library can be mined for binders of high affinity, specificity, and diversity to highly conserved human antigens, we chose the ECDs of human ROR1 and ROR2 as baits. The ECD of human ROR1 shares 100%, 97%, and 98% aa sequence identity with the ECDs of cynomolgus, mouse, and rabbit ROR1, respectively. The ECD of human ROR2 shares 99%, 93%, and 97% aa sequence identity with the ECDs of cynomolgus, mouse, and rabbit ROR2, respectively. These high homologies between the mammalian species have made it more difficult to raise a diverse panel of mAbs with high affinities by animal immunizations (data not shown). While the naïve rabbit antibody library is composed of in vivo vetted rabbit VH and VL sequences, we reasoned that their random combination bypasses immunological tolerance to epitopes conserved between human and rabbit antigens. Thus, we considered the ECDs of human ROR1 and ROR2, which share 63% aa sequence identity, ideal test antigens for probing the utility of the naïve rabbit antibody library. The panel of 13 rabbit anti-human ROR1 mAbs and 12 rabbit anti-human ROR2 mAbs we selected revealed a high degree of aa sequence, affinity, and epitope binding diversity. Five of these mAbs bound with single digit nanomolar affinity. Whereas none of the 25 mAbs were cross-reactive between human ROR1 and ROR2, 8 mAbs also bound to the respective mouse ortholog. This property makes these mAbs particularly attractive for preclinical investigations in mouse models of human cancer, where the presence or absence of on-target-off-tumor toxicities of candidate mAbs can provide important information prior to clinical translation.43 Collectively, our new panel of fully sequenced and extensively characterized 13 rabbit anti-human ROR1 mAbs and 12 rabbit anti-human ROR2 mAbs substantially expands the collection of antibody reagents available for studying and interrogating this family of receptor tyrosine kinases.
To further investigate the therapeutic utility of our new mAbs, we focused on two single-digit nanomolar binders, rabbit anti-human ROR1 mAb XBR1-402 and rabbit anti-human ROR2 mAb XBR2-401. Given the recent clinical translation of our rabbit anti-human ROR1 mAb R12 as CAR-T (ClinicalTrials.gov identifier NCT02706392), which was based on extensive preclinical studies,39,44 we wanted to compare our new mAbs in the CAR-T format. Thus, XBR1-402 and XBR2-401 were converted to CAR-Ts and their proliferation, cytokine secretion, and cytotoxicity in the presence of ROR1+ ROR2− and ROR1− ROR2+ BC cells were analyzed. In these in vitro experiments, the XBR1-402 CAR-T was found to be at least as potent and specific as the clinically investigated R12 CAR-T. In line with their overlapping epitopes, presumably at the interface of Ig and Fz domains, the XBR1-402 CAR-T also revealed the same preference for a short spacer as the R12 CAR-T.39 The XBR2-401 CAR-T, to our knowledge, represents the first published ROR2-targeting CAR-T. It potently and specifically killed ROR1− ROR2+ but not ROR1+ ROR2− BC cells. With its epitope located in the Kr domain of ROR2, we correctly predicted that the XBR2-401 CAR-T would be more potent with a long spacer. Notably, XBR2-401 is cross-reactive with mouse and cynomolgus orthologs of human ROR2 which will facilitate preclinical investigations toward its safety. While CAR-Ts are extraordinarily potent therapeutics, their safety relies in part on the specificity of the antibody component. Using a cell microarray assay, we screened both XBR1-402 and XBR2-401 against 4,336 human plasma proteins representing a large portion of the human surfectome. Strikingly, XBR1-402 only recognized ROR1 and, vice versa, XBR2-401 only recognized ROR2. Collectively, XBR1-402 and XBR2-401, whether as CAR-Ts or in other therapeutic formats, are highly attractive candidates for clinical translation. In breast cancer, they could provide treatment options for patients who do not benefit from HER2-targeting antibody therapeutics. Using established methods,43 we humanized both XBR1-402 and XBR2-401 to further facilitate clinical translation (data not shown).
Taken together, the selection of 13 new ROR1 mAbs and 12 new ROR2 mAbs, of which at least one mAb from each panel revealed therapeutic utility as CAR-T, suggests that naïve rabbit antibody repertoires in general and our naïve rabbit antibody library in particular provide an abundant source of recombinant antibodies of high quality for basic and applied research. In fact, beyond ROR1, ROR2, C3d, RET, and CD3, the naïve rabbit antibody library was used to successfully select panels of diverse mAbs against several additional human antigens (data not shown). Two of these mAbs are currently in the pipeline for clinical translation as CAR-T and antibody-drug conjugate for cancer therapy, respectively, and will be reported in due course.
Materials and Methods
Cell lines
BC cell lines MDA-MB-231 and T47D were purchased from ATCC and cultured in DMEM supplemented with 10% (v/v) heat inactivated FBS and 1 × penicillin-streptomycin (containing 100 U/mL penicillin and 100 mg/mL streptomycin; all from Thermo Fisher). Mouse RAG2−/− pre-B cell lines 63-1245 ectopically and stably expressing human ROR1 or human ROR2 were generated by transposition as described.46 Parental 63-12, 63-12/hROR1, and 63-12/hROR2 cells were cultured in IMDM supplemented with 10% (v/v) heat inactivated FBS, 0.1% (v/v) β-mercaptoethanol, and 1 × penicillin-streptomycin (all from Thermo Fisher). HEK293F cells (Thermo Fisher) were maintained in FreeStyle Medium supplemented with 1% (v/v) heat inactivated FBS to support adherent culture or without FBS for suspension culture, and 1 × penicillin-streptomycin (all from Thermo Fisher). Human chronic myelogenous leukemia cell line K562 was obtained from ATCC. A K562 cell line ectopically and stably expressing human ROR1 by retroviral transduction was kindly provided by Dr. Stanley R. Riddell (Fred Hutchinson Cancer Research Center).47 Both were cultured in RPMI-1640 supplemented with 10% (v/v) heat inactivated FBS and 1 × penicillin-streptomycin (all from Thermo Fisher).
ROR1 antigens
Fc-hROR1 and other Fc fusion proteins
The cloning, expression, purification, and biotinylation of human Fc fusion proteins containing different domains of human ROR1 or mouse ROR1 were described.20 hROR1-HIS protein. To equip the ECD of human ROR1 (hROR1) with a His6 tag for purification, its encoding cDNA (aa 24-403) was PCR-amplified with primers SP-hROR1-F and hROR1-HIS-R using plasmid pCEP4-hFc-hROR120 as template, and cloned into mammalian cell expression vector pCEP4 (Thermo Fisher) via KpnI/XhoI. The resulting plasmid was confirmed by DNA sequencing and transiently transfected into HEK293F cells (Thermo Fisher) using 293fectin (Thermo Fisher). Transfected cells were cultured in FreeStyle protein-free medium (Thermo Fisher) and the hROR1-HIS protein was purified from supernatants by Immobilized Metal Ion Affinity Chromatography (IMAC) using a 1-mL HisTrap column (GE Healthcare) as described.37 The quality and quantity of purified hROR1-HIS was analyzed by SDS-PAGE and A280 absorbance, respectively. hROR1-AVI-HIS protein. To equip the ECD of hROR1 with an Avi tag (GLNDIFEAQKIEWHE) for enzymatic biotinylation by BirA and a His6 tag for purification, its encoding cDNA (24-403) was PCR-amplified with primers pCEP4-hROR1-F and pCEP4-hROR1-AVI-R again using plasmid pCEP4-hFc-hROR120 as template, PCR extended with primers pCEP4-SP-F-KpnI and pCEP4-HIS-R-XhoI, and cloned into pCEP4 via KpnI/XhoI. The resulting plasmid was confirmed by DNA sequencing and transiently transfected into HEK293F cells as described above. The hROR1-AVI-HIS protein was then purified by IMAC as described above. The quality and quantity of purified hROR1-AVI-HIS was analyzed by SDS-PAGE and A280 absorbance, respectively. Subsequently, 2 mg hROR1-AVI-HIS was biotinylated using the BirA Biotin-Protein Ligase kit (Avidity) followed by IMAC purification as described above. Oligonucleotides: SP-hROR1-F: 5′-gctgggtaccggcgcgccaccatggactggacttggagaatcctgtttctcgtagctgctgcaactggagcacactccgcccggggcgccgccgcccag-3′; hROR1-HIS-R: 5′-cggcctcgagtcagtgatggtgatggtggtgctccatcttgttcttctcctt-3′; pCEP4-hROR1-F: 5′-atcctgtttctcgtagctgctgcaactggagcacactccgcccggggcgccgccgcccag-3′; pCEP4-hROR1-AVI-R: 5′-ccactcgatcttctgggcctcgaagatgtcgttcaggccctccatcttgttcttctcctt-3′; pCEP4-SP-F-KpnI: 5′-gctgggtaccggcgcgccaccatggactggacttggagaatcctgtttctcgtagctgct-3′; pCEP4-HIS-R-XhoI: 5′-gccggcctcgagtcagtgatggtgatggtggtgctcgtgccactcgatcttctgggcctc-3′.
ROR2 antigens
Fc-hROR2 protein
Full-length human ROR2 cDNA (clone ID 40146553; GE Healthcare Dharmacon) was used as template for PCR amplifications. ROR2 has an SNP at amino acid position 245. As part of the cloning, we mutated the less common alanine at position 245 of clone 40146553 to the more frequent threonine. Two cDNA fragments encoding N-terminal and C-terminal portions of ECD of human ROR2 (hROR2) were PCR-amplified with (i) primers SP-hROR2-F and hROR2-A245T-R and (ii) primers hROR2-A245T-F and hROR2-R. Subsequently, the whole hROR2 ECD (aa 34-394) encoding cDNA was assembled by overlap extension PCR using the flanking primers SP-hROR2-F and hROR2-R and cloned into pCEP4-hFc48 downstream of human Fc via HindIII/XhoI. The resulting pCEP4-hFc-hROR2 plasmid was confirmed by DNA sequencing and transiently transfected into HEK293F cells using 293fectin. Transfected cells were cultured in FreeStyle protein-free medium and the Fc-hROR2 protein was purified from supernatants by Protein A affinity chromatography as described.20 The quality and quantity of purified Fc-hROR2 was analyzed by SDS-PAGE and A280 absorbance, respectively. Subsequently, 300 μg Fc-hROR2 was biotinylated using the Biotin-Tag Micro Biotinylation kit (Sigma-Aldrich). hROR2-HIS protein. To equip the ECD of hROR2 with a His6 tag for purification, the ECD of hROR2 (aa 34-394) was PCR-amplified with primers SP-hROR2-F and hROR2-HIS-R using plasmid pCEP4-hFc-hROR2 as template and cloned into pCEP4 via KpnI/XhoI. The resulting plasmid was confirmed by DNA sequencing and used to express hROR2-HIS protein as described above for hROR1-HIS protein. Oligonucleotides: SP-hROR2-F: 5′-gcctaagcttgtctccgggtgccgaagtggaggttctggatccgaacg-3′; hROR2-A245T-R: 5′-gctcacgcggcttgggtgtccgggagcgcgcgtcgc-3′; hROR2-A245T-F: 5′-gcgacgcgcgctcccggacacccaagccgcgtgagc-3′; hROR2-R: 5′-agctctcgagtcaccccatcttgctgctgtctcggggactacacgagg-3′; SP-hROR2-F: gctgggtaccggcgcgccaccatggactggacttggagaatcctgtttctcgtagctgctgcaactggagcacactccgaagtggaggttctggatccg-3′; hROR2-HIS-R: 5′-cggcctcgagtcagtgatggtgatggtggtgccccatcttgctgctgtctcg-3′. Cell surface ROR2. Extracellular domains of human ROR2 with Thr at position 245 (hROR2-T245) (aa 55-394) and human ROR2 with the less frequent SNP hROR2-A245 (aa 55-394), as well as mouse ROR2 (aa 34-403), were separately fused to a (G4S)3 linker followed by an HA tag (YPYDVPDYAS) and a PDGFRB segment (aa 513-561) that included the transmembrane domain (aa 533-553), and were stably displayed on HEK293F cells. Using the same (G4S)3-HA-PDGFRB anchor, different compositions of the three extracellular domains of hROR2-T245 − Ig (aa 55-145), Fz (aa 169-303), Kr (aa 316-394), Ig+Fz (aa 55-303), and Fz+Kr (aa 169-394) − were also stably displayed on HEK293F cells. The cell surface densities of these ROR2 variants were compared by flow cytometry (see below) using a 1:500 dilution of the biotinylated rat anti-HA mAb 3F10 (Roche) in conjunction with 2 μg/mL PE-conjugated streptavidin (BD Biosciences). cROR2. The ECD encoding sequence of cynomolgus ROR2 (XP_005582291.1) was fused to an N-terminal signal sequence (MNFGLRLIFLVLTLKGVQC) encoding sequence and a C-terminal sequence encoding a twin strep tag (SAWSHPQFEKGGGSGGGSGGSAWSHPQFEKG), custom synthesized (GenScript) with flanking NotI/HindIII sites, and assembled in mammalian cell expression vector pCB14. This vector, a derivative of pCEP4, carries the EBV replication origin, encodes the EBV nuclear antigen (EBNA-1) to permit extrachromosomal replication, and contains a puromycin selection marker in place of the original hygromycin B resistance gene. The resulting pCB14-cROR2 plasmid was transfected into HEK293T cells using Lipofectamine LTX with PLUS Reagent (Thermo Fisher Scientific). After 24 h culture in DMEM supplemented with high glucose (4.5 g/L), L-glutamine, 10% FCS, and antibiotics (all from BioConcept), the transfected cells were expanded under selection conditions in 2 μg/mL puromycin (Sigma-Aldrich). Cells were further expanded in 14-cm cell culture dishes coated with poly-L-lysine and maintained in DMEM/F12 serum-free medium (Thermo Fisher Scientific) containing 161 μg/mL N-acetyl-L-cysteine, 10 mg/mL L-glutathione, and 1 μg/mL puromycin (all from Sigma-Aldrich). Supernatants were harvested twice a week, sterile-filtered, and stored at 4°C until purification. Twin strep tagged cynomolgus ROR2 ECD was purified from supernatants using a Strep-Tactin Superflow High Capacity cartridge column (IBA) according to the manufacturer’s directions, and its quality and quantity was analyzed by SDS-PAGE and A280 absorbance, respectively.
RET, CD3, and C3d antigens
RET
The sequence encoding the cysteine-rich domain (CRD; aa 514-645) of human RET isoform 4 (NCBI Reference Sequence NM_020630.4) fused downstream of a human IgG1 Fc encoding sequence was custom synthesized as gBlock (Integrated DNA Technologies). Oncogene mutation C634R in the RET CRD was introduced by overlap extension PCR. Both wild-type and mutated expression cassettes were AscI/XhoI-cloned into mammalian expression vector pCEP4 and expressed and purified as described for the Fc-hROR1 and Fc-hROR2 fusion proteins. CD3. The sequences encoding the ECDs of human CD3δ (aa 22-105; NM_000732.4) and CD3ε (aa 23-126; NM_000733.3) were custom synthesized as gBlocks. To obtain heterodimerized CD3 δ/ε, the “knobs-into-holes” technology was applied.49 Two human IgG1 Fc encoding sequences with an N297A mutation and with either knob mutations S354C and T366W or with hole mutations Y349C, T366S, L368A, and Y407V were synthesized as gBlocks, assembled into CD3δ-Fc knob and CD3ε-Fc hole expression cassettes, AscI/XhoI-cloned into mammalian expression vector pCEP4 and expressed, purified, and biotinylated as described for the Fc-hROR1 and Fc-hROR2 fusion proteins. C3d. Human complement protein C3d derived from human complement protein C3 after a series of proteolytic cleavages was purchased from Complement Technology and biotinylated using the Biotin-Tag Micro Biotinylation kit.
Library generation
All rabbit handling was carried out by veterinary personnel at Pocono Rabbit Farm & Laboratory (Canadensis, PA) or R & R Research (Stanwood, WA). Spleen and bone marrow from nine rabbits (five NZW and four b9 κ-light chain allotype rabbits; ages 3–4 months) were collected and separately processed for total RNA preparation and RT-PCR amplification of rbVκ, rbVλ, and rbVH encoding cDNA using established protocols.8 NZW rbVκ, rbVλ, and rbVH cDNAs were kept in separate pools from b9 rbVκ, rbVλ, and rbVH cDNAs. Using three-fragment overlap extension PCR, rbVκ-huCκ-rbVH (library κ) and rbVλ-huCλ-rbVH (library λ) segments were assembled as previously described.8,50 To generate high combinatorial diversity in this step, the NZW rbVH cDNA pool was separately combined with the NZW rbVκ, the NZW rbVλ, the b9 rbVκ, and the b9 rbVλ cDNA pools, and the b9 rbVH cDNA pool was separately combined with the b9 rbVκ and the b9 rbVλ cDNA pools. SfiI-cut segments were ligated into SfiI-cut phage display vector pC3C17 at 16°C for 24 h. Subsequently, 15 μg purified pC3C-rbVκ/hCκ/rbVH ligated products were transformed into E. coli strain SR320 (a lytic phage resistant variant of SS32051 kindly provided by Dr. Sachdev S. Sidhu, University of Toronto) by 30 separate electroporations (each using 0.5 μg DNA in 50 μL electrocompetent cells) and yielded 7.5 × 109 independent transformants for library κ. For library λ, 4.8 × 109 independent transformants were obtained using the same procedure. Using VCSM13 helper phage (Stratagene), the phagemid libraries were converted to phage libraries and stored at −80°C as described.8 For selection by phage display, phage libraries κ and λ were separately re-amplified using E. coli strains XL1-Blue (Agilent) or ER2738 (Lucigen)8 and mixed equally.
Library validation
To analyze the diversity of the chimeric rabbit/human Fab library, rbVH encoding sequences were PCR-amplified with primers VHF-trP and VHR-A and sequentially purified first with the Montage Gel Extraction Kit (EMD Millipore) and then with the Agencourt AMPure XP kit (Beckman Coulter) before NGS using an Ion PGM instrument (Thermo Fisher) and an Ion 318 Chip Kit v2 (Thermo Fisher). NGS data were analyzed with N2GSAb software kindly provided by Novimmune.52 Oligonucleotides: VHF-trP: 5′-CCTCTCTATGGGCAGTCGGTGATCTGCCCAACCAGCCATGGCC-3′; VHR-A: 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGGATGGGCCCTTGGTGGAGGC-3′. In the diversity analysis, different clones were defined by having ≥ 2 bp differences.
Library selection
The re-amplified phage display library was selected by four rounds of panning against biotinylated Fc-hROR1, hROR1-AVI-HIS, or Fc-hROR2 in solution through capture with streptavidin-coated magnetic beads as described.8 Briefly, 5 μg/mL biotinylated antigen was pre-incubated with Dynabeads MyOne Streptavidin C1 magnetic beads (Thermo Fisher) at 37°C for 30 min and then binders from the phage library were captured in the presence of 1 mg/mL unspecific polyclonal human IgG (Thermo Fisher). Starting from the third round of panning, the input phage was background depleted by incubation with empty beads before selection against protein-loaded beads. Following selection, supernatants of isopropyl β-D-1-thiogalactopyranoside (IPTG)-induced bacterial colonies were analyzed by ELISA and by flow cytometry for ROR1 and ROR2 binding. Repeated clones were identified by DNA fingerprinting with AluI, and the rbVκ, rbVλ, and rbVH sequences of unique clones were determined by DNA sequencing. To find less enriched binders after three rounds of panning, rbVH encoding sequences with ≥ 2 bp differences were compiled by NGS as described above. The most abundant clones ranked in the top 100 that were different from clones identified by screening bacterial colonies were retrieved by two-fragment overlap PCR with complementary primers (HCDR3-F and HCDR3-R) corresponding to the unique sequences of the HCDR3 segment of rbVH as well as primers flanking the chimeric rabbit/human Fab encoding cassette region. Subsequently, the reassembled cassette was SfiI-cloned into a compatible bacterial expression vector pET11a variant18 and confirmed by DNA sequencing. Using this strategy, the overall success rate for retrieving clones with rbVH encoding sequences identified by NGS was >90%. Supernatants of pET11a-transformed and IPTG-induced bacterial colonies were analyzed by ELISA and flow cytometry for ROR1 and ROR2 binding. The nomenclature of selected binders included the name of the E. coli strain used for re-amplification (XB for XL1-Blue and ER for ER2738), followed by the name of the antigen (R1 for ROR1 and R2 for ROR2), followed by the clone ID which, for binders identified by screening bacterial colonies, started with either 3 or 4 if identified after three or four rounds of panning, respectively. For example, XBR1-402 was found after four rounds of XL1-Blue-based panning against ROR1. For binders identified by NGS, the clone ID indicated their ranking in the top 100. For example, ERR2-TOP35 was the 35th most frequent clone identified by NGS after three rounds of ER2738-based panning against ROR2. Interestingly, as noted previously18 and regardless of whether the panning was based on XL1-Blue or ER2738, a number of clones (7/13 of ROR1 binders and 4/12 ROR2 binders) contained a TAG codon (amber stop codon) in place of the first native aa codon of rbVH. The amber stop codon is suppressed by genotype supE44 enabling glutamine incorporation to compete with termination of translation. We hypothesize that the lower expression levels of chimeric rabbit/human Fab with this unintended mutation afforded a competitive advantage in their selection. The TAG codon was replaced with the canonical glutamine codon CAG for Fab and IgG1 expression. In addition to biotinylated Fc-hROR1, hROR1-AVI-HIS, and Fc-hROR2, the re-amplified phage display library was also selected by four rounds of panning against biotinylated human C3d, Fc-hRET (C634R), and Fc-hCD3δε using similar procedures. As described in detail for the ROR1 and ROR2 binders, three different C3d, three different RET, and one CD3 binder were identified by DNA fingerprinting and DNA sequencing, expressed and purified as chimeric rabbit/human Fab (C3d) and IgG1 (RET and CD3), and analyzed by ELISA using biotinylated human antigens captured by coated streptavidin and Peroxidase-AffiniPure F(ab′)2 Fragment Goat Anti-Human IgG, F(ab′)2 Fragment Specific pAbs (Jackson ImmunoResearch Laboratories) for detection.
Chimeric rabbit/human Fab cloning, expression, and purification
Chimeric rabbit/human Fab XBR1-402, XBR2-401, XBR2-416, and XBR2-433 were cloned into bacterial expression vector pC3C-His,53 transformed into E. coli strain XL1-Blue, and IMAC-purified from supernatants as described.37 All other chimeric rabbit/human Fab were cloned into bacterial expression vector pET11a,18 transformed into E. coli strain Rosetta(DE3) (EMD Millipore), and IMAC-purified from supernatants as described.37 The quality and quantity of purified Fab was analyzed by SDS-PAGE and A280 absorbance, respectively. Yields varied widely depending on the clone and ranged from 0.1 to 5 mg per liter culture.
Chimeric rabbit/human IgG1 cloning, expression, and purification
For conversion of chimeric rabbit/human Fab to chimeric rabbit/human IgG1, XBR1-402, XBR2-401, XBR2-416, and XBR2-433 were cloned into mammalian cell expression vector PIGG54 by SacI/ApaI-replacing the rbVH-encoding sequence and HindIII/XbaI-replacing the light chain (rbVκ-huCκ)-encoding sequence of PIGG-R11.20 The resulting plasmids were transiently transfected into HEK293F cells using 293fectin and purified with a 1-mL recombinant Protein A HiTrap column as described.55 For conversion of all other chimeric rabbit/human Fab to chimeric rabbit/human IgG1 (ERR1-324, ERR1-TOP43, ERR1-TOP54, XBR2-327, ERR2-308, ERR2-317, XBR2-TOP72, and ERR2-302), their light and heavy chains were cloned separately into pCEP4. For the light chains, the signal peptide-rbVκ-huCκ-encoding segments or signal peptide-rbVλ-huCλ-encoding segments were PCR-amplified using the phagemids as templates and cloned via AscI/XhoI into pCEP4. For the heavy chain of ERR2-302, a signal peptide-rbVH- huCH1(aa 1-49)-encoding segment was custom synthesized as gBlock and fused by overlap extension PCR to a huCH1(aa 50-88)-hinge-huCH2-huCH3-encoding genomic segment with three introns that had been PCR-amplified from PIGG. The resulting heavy chain-encoding segment was cloned via AscI/XhoI into pCEP4. In this first heavy chain construct, we engineered a silent EheI restriction site in the 5′ region of the huCH1-encoding cDNA, allowing AscI/EheI-replacement of the signal peptide-rbVH-encoding segment for all subsequent heavy chains generated by PCR amplification using the phagemids as templates. The same human heavy chain signal peptide (MDWTWRILFLVAAATGAHS) was used for all constructs. The resulting plasmids were transiently co-transfected (1:1 molar ratio) into HEK293F cells using 293fectin and purified with a 1-mL recombinant Protein A HiTrap column as described.55 The quality and quantity of purified IgG1 was analyzed by SDS-PAGE and A280 absorbance, respectively. Yields for pCEP4-based expression ranged from 8 to 15 mg per liter culture.
Rabbit IgG cloning, expression, and purification
For conversion of chimeric rabbit/human Fabs (XBR1-402, ERR1-306, and ERR1-TOP43) to full rabbit IgG, light and heavy chains were cloned separately into pCEP4. For the light chains, the signal peptide-rbVλ-encoding segments were PCR-amplified using the phagemids as templates, fused to the constant domain of the rabbit λ light chain (rbCλ, custom synthesized as gBlock) by overlap extension PCR, and cloned into pCEP4 via AscI/XhoI. For the heavy chain of XBR1-402, the signal peptide-rbVH-encoding segment was PCR-amplified from the phagemid and fused to the constant domains (CH1-hinge-CH2-CH3) of a rabbit heavy chain encoding genomic segment with three introns that was amplified from rabbit DNA. Note that rabbits only have one IgG isotype. The entire rabbit heavy chain-encoding segment was then cloned via AscI/XhoI into pCEP4. In this XBR1-402 heavy chain construct, we engineered a silent EheI restriction site in the 5′ region of the rbCH1-encoding cDNA, allowing AscI/EheI-replacement of the signal peptide-rbVH-encoding segment for the ERR1-306 and ERR1-TOP43 heavy chains generated by PCR amplification using the phagemids as templates. The same human heavy chain signal peptide (MDWTWRILFLVAAATGAHS) was used for all constructs. Expression, purification, and analysis of rabbit IgG was done as described for chimeric rabbit/human IgG1.
ELISA
Fab binding assay to captured Fc fusion proteins
Each well of a 96-well Costar 3690 plate (Corning) was coated with 100 ng goat anti-human IgG Fcγ pAbs (Jackson ImmunoResearch) in 25 μL coating buffer (0.1 M Na2CO3, 0.1 M NaHCO3, pH 9.6) for 1 h at 37°C. After blocking with 150 μL 3% (w/v) BSA/TBS for 1 h at 37°C, 100 ng Fc fusion proteins of ROR1 and ROR2 in 50 μL 1% (w/v) BSA/TBS were captured by incubation for 1 h at 37°C. The wells were washed three times with 150 μL TBS. Next, 100 ng Fab in 50 μL 1% (w/v) BSA/TBS was applied to each well. Following incubation for 2 h at 37°C and washing as before, 50 μL of a 1:1,000 dilution of a mouse anti-His tag mAb conjugated to horse radish peroxidase (HRP) (R&D Systems) in 1% (w/v) BSA/TBS was added and incubated for 1 h at 37°C. The wells were washed four times and colorimetric detection was performed using 2,2′-azino-bis(3-ethylbenzothiazoline)-6-sulfonic acid (Roche) as substrate according to the manufacturer’s directions. The absorbance was measured at 405 nm using a SpectraMax M5 microplate reader (Molecular Devices) and SoftMax Pro software (Molecular Devices). Fab binding assay to directly coated ROR1 and ROR2 antigens. Each well of a 96-well Costar 3690 plate was coated with 100 ng ROR1 or ROR2 antigen in 25 μL coating buffer (0.1 M Na2CO3, 0.1 M NaHCO3, pH 9.6) for 1 h at 37°C before proceeding with blocking, primary antibody, washing, secondary antibody, washing, and colorimetric detection as described above.
Western blotting
Cells or proteins were lysed by 1× NuPAGE LDS sample buffer (Thermo Fisher) containing 1% β-mercaptoethanol and boiled for 10 min before running on NuPAGE Novex 4-12% Bis-Tris gels (Thermo Fisher). After transfer to a PVDF membrane (Millipore) and blocking by 5% (w/v) dry milk in PBS, the membrane was incubated with 2 μg/mL chimeric rabbit/human IgG1 in 1% (w/v) milk in PBS at 4°C overnight, followed by incubation with a 1:1,000 dilution of HRP-conjugated goat anti-human IgG Fcγ pAbs (Jackson ImmunoResearch) before developing using ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Immunohistochemistry
K562 and K562/ROR1 cells grown in suspension (3–5 × 106 cells/mL) were washed with PBS, adhered to slides by Cytospin centrifugation (Shandon Cytospin 4) for 5 min at 1,000 rpm, and then air-dried for 15 min at room temperature. Adherent MDA-MB-231 cells were grown on pre-coated coverslips in a 24-well tissue culture plate and rinsed twice with PBS before staining. All cells were fixed with 2% (v/v) formaldehyde, permeabilized with 0.3% (v/v) Tween-20 in PBS on ice, and further treated according to the Ready-to-Use IHC/ICC Kit (BioVision). Briefly, cells were blocked with Peroxidase Block (H2O2), followed by Protein Blocking Solution, then incubated for 30 min with isotype control or primary rabbit IgG in Primary Antibody Dilution Buffer, followed by 30-min incubation with HRP-Anti-Rabbit Polymer, and finally stained with a few drops (~30 μL) of freshly prepared DAB Reagent for 5 min. The slides were then counterstained with hematoxylin (ScyTek Laboratories), dehydrated by passing through a graded alcohol solution, cleared, and mounted with DPX Mountant (Sigma-Aldrich). Slides were analyzed and imaged with a Leica DM5000 light microscope.
Flow cytometry
Cells were stained using standard flow cytometry methodology. Briefly, for purified anti-ROR1 and anti-ROR2 Fabs, 0.1–1 × 106 cells were stained with 1 μg Fab in 100 μL ice-cold flow cytometry buffer (PBS containing 1% (w/v) BSA, 0.1% (w/v) sodium azide, and 1 mM EDTA) in a V-shaped 96-well plate (BrandTech) for 1 h on ice. After washing twice with 300 μL ice-cold flow cytometry buffer, the cells were incubated with a 1:1,000 dilution of an Alexa Fluor 488-conjugated mouse anti-His tag mAb (Qiagen) in 100 μL flow cytometry buffer for 30 min on ice. For purified anti-ROR1 and anti-ROR2 IgG1, 0.1–1 × 106 cells were first blocked with 4% (v/v) normal goat serum (Jackson ImmunoResearch) in 100 μL flow cytometry buffer for 15 min at room temperature and then incubated with 100 ng IgG1 in 100 μL flow cytometry buffer on ice for 1 h. After washing twice with 300 μL ice-cold flow cytometry buffer, cells were stained with a 1:500 dilution of APC-conjugated goat anti-human IgG (Fcγ) pAbs (Jackson ImmunoResearch) in 100 μL flow cytometry buffer for 30 min on ice. For commercial goat anti-human ROR1 and ROR2 pAbs (R&D Systems), 0.1–1 × 106 cells were stained with 200 ng pAbs in 100 μL flow cytometry buffer for 1 h on ice. After washing twice with ice-cold flow cytometry buffer, the cells were incubated with a 1:1,000 dilution of an Alexa Fluor 647-conjugated donkey anti-goat IgG (H+L) pAbs in F(ab′)2 format (Jackson ImmunoResearch) in 100 μL flow cytometry buffer on ice for 30 min. Finally, 4′,6-diamidino-2-phenylindole (DAPI; Cell Signaling) was added to a final concentration of 100 ng/mL to exclude dead cells. Cells were analyzed using a FACSCalibur instrument (BD Biosciences) and FlowJo analytical software (Tree Star).
Surface plasmon resonance
SPR for the measurement of kinetic and thermodynamic parameters of the binding of purified anti-ROR1 and anti-ROR2 Fabs to ROR1 and ROR2 antigens and for epitope binning studies were performed on a Biacore X100 instrument using Biacore reagents and software (GE Healthcare). A mouse anti-human IgG CH2 mAb was immobilized on a CM5 sensor chip using reagents and instructions supplied with the Human Antibody Capture Kit (GE Healthcare). hFc-hROR1 and hFc-hROR2 fusion proteins were captured at a density not exceeding 1,000 RU (Suppl. Figs. S3 and S7). Each sensor chip included an empty flow cell for instantaneous background depletion. All binding assays used 1× HBS-EP+ running buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA (pH 7.4), and 0.05% (v/v) Surfactant P20) and a flow rate of 30 μL/min. For affinity measurements, all Fabs were injected at five different concentrations, one of which was tested in duplicate. The sensor chips were regenerated with 3 M MgCl2 from the Human Antibody Capture Kit without any loss of binding capacity. Calculation of association (kon) and dissociation (koff) rate constants was based on a 1:1 Langmuir binding model. The equilibrium dissociation constant (Kd) was calculated from koff/kon. For epitope binning studies, each Fab was prepared at 500 nM alone or in a mixture in 1× HBS-EP+ running buffer.
Retrogenix cell microarray
Primary screen
XBR1-402 and XBR2-401 in chimeric rabbit/human IgG1 format were pooled to a concentration of 2 μg/mL each. The pool was screened for binding against a fixed HEK293 cell microarray as described.56 Briefly, 4,336 mammalian cell expression vectors, each encoding a full-length human plasma membrane protein including ROR1 and ROR2, were arrayed in duplicates across 13 slide sets using duplicate slides per set. HEK293 cells were grown over the vector microarray, leading to reverse transfection at each location. ZsGreen1 encoded on the vector was used to monitor the transfection efficiency and define the microarray position. All transfection efficiencies exceeded the minimum threshold. After fixing the cells, the interaction between the pooled mAbs and the cells presenting the human plasma membrane protein microarray was detected using AlexaFluor647-conjugated goat anti-human IgG Fcγ pAbs (Jackson ImmunoResearch) and analyzed using ImageQuant software (GE Healthcare) to identify primary hits (duplicate spots). Vectors encoding primary screen hits were sequenced to confirm their correct identities. Confirmation screen. Vectors encoding primary screen hits, plus control vectors, were spotted in duplicate on new slides, and used to reverse transfect HEK293 cells as before. All transfection efficiencies exceeded the minimum threshold. Duplicate fixed slides were treated with each of the two mAbs individually in addition to positive (rituximab biosimilar) and negative (secondary antibody alone) controls. The slides were analyzed as before.
CAR-T cells
Lentiviral vectors
ROR1− and ROR2-targeting CARs were constructed using methods previously described for rabbit anti-human ROR1 mAbs R11 and R12.39 Specifically, XBR1-402 and XBR2-401 in scFv format (VH-(G4S)3-VL) were fused to a long spacer comprising the human IgG4 hinge-CH2-CH3 segment (229 aa) or to a short spacer comprising the human IgG4 hinge segment (12 aa) alone. Both spacers contained an S108P substitution in the hinge segment and were fused to the 27-aa transmembrane segment of human CD28 and to a signaling module comprising the 42-aa cytoplasmic domain of human 4-1BB followed by the 112-aa cytoplasmic domain of isoform 3 of human CD3ζ. Downstream of the CAR constructs, the expression cassettes further encoded a T2A ribosomal skip element and a truncated EGF receptor (EGFRt) sequence.40 Codon-optimized DNA sequences encoding the expression cassettes were synthesized (GeneArt, Thermo Fisher) and cloned into the epHIV7 lentiviral vector under control of a human elongating factor 1-α (EF-1α) promoter.40 XBR1-402 CAR, XBR2-401 CAR, each with long and short spacer, and R12 CAR with short spacer39 were produced in HEK293T cells using the packaging vectors pCHGP-2, pCMV-Rev2, and pCMV-G. Generation of CAR-T cell lines. CD8+ CD62L+ naïve/central memory T cells were sorted from PBMC of healthy donors and then activated with anti-CD3/CD28 beads (Dynabeads Human T activator CD3/CD28, Thermo Fisher), transduced with lentiviral supernatant, expanded, enriched for EGFRt expression by magnetic selection, further expanded using a rapid expansion protocol, and cultured as described in detail previously.39 Subsequent flow cytometry analysis revealed that CD8+ and EGFRt+ purity was >95%. Proliferation. T cells (5 × 104/well) labeled with 0.2 μM carboxyfluorescein succinimidyl ester (CFSE; Thermo Fisher) were plated in triplicate with target cells at an effector:target (E:T) ratio of 4:1 in medium without exogenous cytokines. After 72-h incubation, cells were labeled with anti-CD8 mAb (BD Biosciences), and analyzed by flow cytometry to assess cell division of T cells. Cytokine secretion. T cells (5 × 104/well) were plated in triplicate with target cells at an E:T ratio of 4:1. IFN-γ and IL-2 concentrations were measured by ELISA (BioLegend) in supernatant removed after 24-h incubation. Cytotoxicity. T cells (5/2.5/1.25 × 104/well) were plated in triplicate with target cells transduced with firefly luciferase at E:T ratios of 10:1, 5:1, and 2.5:1. Specific lysis was measured with a luciferase-based cytotoxicity assay following 7–11 h of co-culture as described.57
Supplementary Material
Research Highlights.
Generation and validation of a highly diverse naïve rabbit mAb library
>10 billion chimeric rabbit/human Fab clones minable by phage display
Selection of ROR1 and ROR2 mAbs of high diversity, affinity, and specificity
Conversion of ROR1 and ROR2 mAbs to highly potent CAR-Ts
Acknowledgments
We thank Marco Stringhini and Theresa Pesch for their help with generating recombinant human ROR1 and ROR2, respectively, in the Rader laboratory, Benaja Stolz (NBE-Therapeutics) for generating cynomolgus ROR2, Fabian I. Wolter (NBE-Therapeutics) for generating the 63-12/hROR1 and 63-12/hROR2 cell lines, Dr. Sachdev S. Sidhu for providing E. coli strain SR320, Ulla Ravn (Novimmune) for providing the N2GSAb software, and Rader laboratory members Henry D. Wilson and Rebecca S. Goydel for reading and editing the manuscript. C.R. acknowledges support by NIH grants U01 CA174844 and R01 CA181258, the Lymphoma Research Foundation, the Klorfine Foundation, the Holm Charitable Trust, and NBE-Therapeutics. C.R., H.P., R.R.B., and U.G. are named inventors on a patent application claiming the ROR1 mAbs reported here. C.R., H.P., and X.L. are named inventors on a patent application claiming the ROR2 mAbs reported here. R.R.B. and U.G. are employees and C.R. is a member of the scientific advisory board of NBE-Therapeutics. J.S. and J.F. are employees of Retrogenix. This is manuscript # 29479 from The Scripps Research Institute.
Abbreviations
- mAb
monoclonal antibody
- pAbs
polyclonal antibodies
- IHC
immunohistochemistry
- ELISA
enzyme-linked immunosorbent assay
- NGS
next-generation sequencing
- aa
amino acid
- Ig
immunoglobulin
- Fz
frizzled
- Kr
kringle
- BC
breast cancer
- CAR-T
chimeric antigen receptor-engineered T cells
- NZW
New Zealand White
- rb
rabbit
- hu or h
human
- CDR
complementary-determining region
- ECD
extracellular domain
- SPR
surface plasmon resonance
- IMAC
immobilized metal ion affinity chromatography
- SNP
single nucleotide polymorphism
- CRD
cysteine-rich domain
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- HRP
horse radish peroxidase
- E:T ratio
effector:target ratio
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mage RG, Lanning D, Knight KL. B cell and antibody repertoire development in rabbits: the requirement of gut-associated lymphoid tissues. Dev Comp Immunol. 2006;30:137–153. doi: 10.1016/j.dci.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Knight KL, Winstead CR. Generation of antibody diversity in rabbits. Curr Opin Immunol. 1997;9:228–232. doi: 10.1016/s0952-7915(97)80140-9. [DOI] [PubMed] [Google Scholar]
- 3.Winstead CR, Zhai SK, Sethupathi P, Knight KL. Antigen-induced somatic diversification of rabbit IgH genes: gene conversion and point mutation. J Immunol. 1999;162:6602–6612. [PubMed] [Google Scholar]
- 4.Ridder R, Schmitz R, Legay F, Gram H. Generation of rabbit monoclonal antibody fragments from a combinatorial phage display library and their production in the yeast Pichia pastoris. Biotechnology. 1995;13:255–260. doi: 10.1038/nbt0395-255. [DOI] [PubMed] [Google Scholar]
- 5.Lang IM, Barbas CF, 3rd, Schleef RR. Recombinant rabbit Fab with binding activity to type-1 plasminogen activator inhibitor derived from a phage-display library against human alpha-granules. Gene. 1996;172:295–298. doi: 10.1016/0378-1119(96)00021-2. [DOI] [PubMed] [Google Scholar]
- 6.Popkov M, Mage RG, Alexander CB, Thundivalappil S, Barbas CF, 3rd, Rader C. Rabbit immune repertoires as sources for therapeutic monoclonal antibodies: the impact of kappa allotype-correlated variation in cysteine content on antibody libraries selected by phage display. J Mol Biol. 2003;325:325–335. doi: 10.1016/s0022-2836(02)01232-9. [DOI] [PubMed] [Google Scholar]
- 7.Spieker-Polet H, Sethupathi P, Yam PC, Knight KL. Rabbit monoclonal antibodies: generating a fusion partner to produce rabbit-rabbit hybridomas. Proc Natl Acad Sci USA. 1995;92:9348–9352. doi: 10.1073/pnas.92.20.9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rader C. Generation and selection of rabbit antibody libraries by phage display. Methods Mol Biol. 2009;525:101–128. doi: 10.1007/978-1-59745-554-1_5. [DOI] [PubMed] [Google Scholar]
- 9.Rader C, Ritter G, Nathan S, Elia M, Gout I, Jungbluth AA, Cohen LS, Welt S, Old LJ, Barbas CF., 3rd The rabbit antibody repertoire as a novel source for the generation of therapeutic human antibodies. J Biol Chem. 2000;275:13668–13676. doi: 10.1074/jbc.275.18.13668. [DOI] [PubMed] [Google Scholar]
- 10.Steinberger P, Sutton JK, Rader C, Elia M, Barbas CF., 3rd Generation and characterization of a recombinant human CCR5-specific antibody. A phage display approach for rabbit antibody humanization. J Biol Chem. 2000;275:36073–36078. doi: 10.1074/jbc.M002765200. [DOI] [PubMed] [Google Scholar]
- 11.Borras L, Gunde T, Tietz J, Bauer U, Hulmann-Cottier V, Grimshaw JP, Urech DM. Generic approach for the generation of stable humanized single-chain Fv fragments from rabbit monoclonal antibodies. J Biol Chem. 2010;285:9054–9066. doi: 10.1074/jbc.M109.072876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jendreyko N, Popkov M, Beerli RR, Chung J, McGavern DB, Rader C, Barbas CF., 3rd Intradiabodies, bispecific, tetravalent antibodies for the simultaneous functional knockout of two cell surface receptors. J Biol Chem. 2003;278:47812–47819. doi: 10.1074/jbc.M307002200. [DOI] [PubMed] [Google Scholar]
- 13.Popkov M, Jendreyko N, Gonzalez-Sapienza G, Mage RG, Rader C, Barbas CF., 3rd Human/mouse cross-reactive anti-VEGF receptor 2 recombinant antibodies selected from an immune b9 allotype rabbit antibody library. J Immunol Methods. 2004;288:149–164. doi: 10.1016/j.jim.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Popkov M, Jendreyko N, McGavern DB, Rader C, Barbas CF., 3rd Targeting tumor angiogenesis with adenovirus-delivered anti-Tie-2 intrabody. Cancer Res. 2005;65:972–981. [PubMed] [Google Scholar]
- 15.Popkov M, Rader C, Barbas CF., 3rd Isolation of human prostate cancer cell reactive antibodies using phage display technology. J Immunol Methods. 2004;291:137–151. doi: 10.1016/j.jim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Jendreyko N, Popkov M, Rader C, Barbas CF., 3rd Phenotypic knockout of VEGF-R2 and Tie-2 with an intradiabody reduces tumor growth and angiogenesis in vivo. Proc Natl Acad Sci USA. 2005;102:8293–8298. doi: 10.1073/pnas.0503168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofer T, Tangkeangsirisin W, Kennedy MG, Mage RG, Raiker SJ, Venkatesh K, Lee H, Giger RJ, Rader C. Chimeric rabbit/human Fab and IgG specific for members of the Nogo-66 receptor family selected for species cross-reactivity with an improved phage display vector. J Immunol Methods. 2007;318:75–87. doi: 10.1016/j.jim.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stahl SJ, Watts NR, Rader C, DiMattia MA, Mage RG, Palmer I, Kaufman JD, Grimes JM, Stuart DI, Steven AC, Wingfield PT. Generation and characterization of a chimeric rabbit/human Fab for co-crystallization of HIV-1 Rev. J Mol Biol. 2010;397:697–708. doi: 10.1016/j.jmb.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci USA. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Baskar S, Kwong KY, Kennedy MG, Wiestner A, Rader C. Therapeutic potential and challenges of targeting receptor tyrosine kinase ROR1 with monoclonal antibodies in B-cell malignancies. PloS One. 2011;6:e21018. doi: 10.1371/journal.pone.0021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrester WC. The Ror receptor tyrosine kinase family. Cell Mol Life Sci. 2002;59:83–96. doi: 10.1007/s00018-002-8407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebagay G, Yan S, Liu C, Cheung NK. ROR1 and ROR2 in human malignancies: potentials for targeted therapy. Front Oncol. 2012;2:34. doi: 10.3389/fonc.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debebe Z, Rathmell WK. Ror2 as a therapeutic target in cancer. Pharmacol Ther. 2015;150:143–148. doi: 10.1016/j.pharmthera.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borcherding N, Kusner D, Liu GH, Zhang W. ROR1, an embryonic protein with an emerging role in cancer biology. Protein Cell. 2014;5:496–502. doi: 10.1007/s13238-014-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J, Tang L, Chen G, Basak GW, Kipps TJ. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PloS One. 2012;7:e31127. doi: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui B, Zhang S, Chen L, Yu J, Widhopf GF, 2nd, Fecteau JF, Rassenti LZ, Kipps TJ. Targeting ROR1 inhibits epithelial-mesenchymal transition and metastasis. Cancer Res. 2013;73:3649–3660. doi: 10.1158/0008-5472.CAN-12-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massagué J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi T, Yanagisawa K, Sugiyama R, Hosono Y, Shimada Y, Arima C, Kato S, Tomida S, Suzuki M, Osada H, Takahashi T. NKX2-1/TITF1/TTF- 1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell. 2012;21:348–361. doi: 10.1016/j.ccr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 29.McCartney-Francis N, Skurla RM, Jr, Mage RG, Bernstein KE. Kappa-chain allotypes and isotypes in the rabbit: cDNA sequences of clones encoding b9 suggest an evolutionary pathway and possible role of the interdomain disulfide bond in quantitative allotype expression. Proc Natl Acad Sci USA. 1984;81:1794–1798. doi: 10.1073/pnas.81.6.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carneiro M, Rubin CJ, Di Palma F, Albert FW, Alföldi J, Barrio AM, Pielberg G, Rafati N, Sayyab S, Turner-Maier J, Younis S, Afonso S, Aken B, Alves JM, Barrell D, Bolet G, Boucher S, Burbano HA, Campos R, Chang JL, Duranthon V, Fontanesi L, Garreau H, Heiman D, Johnson J, Mage RG, Peng Z, Queney G, Rogel-Gaillard C, Ruffier M, Searle S, Villafuertem R, Xiong A, Young S, Forsberg-Nilsson K, Good JM, Lander ES, Ferrand N, Lindblad-Toh K, Andersson L. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science. 2014;345:1074–1079. doi: 10.1126/science.1253714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinheiro A, Neves F, Lemos de Matos A, Abrantes J, van der Loo W, Mage R, Esteves PJ. An overview of the lagomorph immune system and its genetic diversity. Immunogenetics. 2016;68:83–107. doi: 10.1007/s00251-015-0868-8. [DOI] [PubMed] [Google Scholar]
- 32.de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruïne AP, Arends JW, Hoogenboom HR. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 33.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothe C, Urlinger S, Löhning C, Prassler J, Stark Y, Jäger U, Hubner B, Bardroff M, Pradel I, Boss M, Bittlingmaier R, Bataa T, Frisch C, Brocks B, Honegger A, Urban M. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J Mol Biol. 2008;376:1182–1200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 35.Glanville J, D’Angelo S, Khan TA, Reddy ST, Naranjo L, Ferrara F, Bradbury AR. Deep sequencing in library selection projects: what insight does it bring? Curr Opin Struct Biol. 2015;33:146–160. doi: 10.1016/j.sbi.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PloS One. 2014;9:e101322. doi: 10.1371/journal.pone.0101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwong KY, Rader C. E. coli expression and purification of Fab antibody fragments. Curr Protoc Protein Sci. 2009 doi: 10.1002/0471140864.ps0610s55. Chapter 6, Unit 6.10. [DOI] [PubMed] [Google Scholar]
- 38.Schwabe GC, Tinschert S, Buschow C, Meinecke P, Wolff G, Gillessen-Kaesbach G, Oldridge M, Wilkie AO, Kömec R, Mundlos S. Distinct mutations in the receptor tyrosine kinase gene ROR2 cause brachydactyly type B. Am J Hum Genet. 2000;67:822–831. doi: 10.1086/303084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudecek M, Lupo-Stanghellini MT, Kosasih PL, Sommermeyer D, Jensen MC, Rader C, Riddell SR. Receptor affinity and extracellular domain modifications affect tumor recognition by ROR1-specific chimeric antigen receptor T cells. Clin Cancer Res. 2013;19:3153–3164. doi: 10.1158/1078-0432.CCR-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Chang WC, Wong CW, Colcher D, Sherman M, Ostberg JR, Forman SJ, Riddell SR, Jensen MC. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood. 2011;118:1255–1263. doi: 10.1182/blood-2011-02-337360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 42.Bradbury AR, Plückthun A. Getting to reproducible antibodies: the rationale for sequenced recombinant characterized reagents. Protein Eng Des Sel. 2015;28:303–305. doi: 10.1093/protein/gzv051. [DOI] [PubMed] [Google Scholar]
- 43.Weber J, Peng H, Rader C. From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp Mol Med. 2017;49:e305. doi: 10.1038/emm.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berger C, Sommermeyer D, Hudecek M, Berger M, Balakrishnan A, Paszkiewicz PJ, Kosasih PL, Rader C, Riddell SR. Safety of targeting ROR1 in primates with chimeric antigen receptor-modified T cells. Cancer Immunol Res. 2015;3:206–216. doi: 10.1158/2326-6066.CIR-14-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 46.Waldmeier L, Hellmann I, Gutknecht CK, Wolter FI, Cook SC, Reddy ST, Grawunder U, Beerli RR. Transpo-mAb display: Transposition-mediated B cell display and functional screening of full-length IgG antibody libraries. mAbs. 2016;8:726–740. doi: 10.1080/19420862.2016.1160990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudecek M, Schmitt TM, Baskar S, Lupo-Stanghellini MT, Nishida T, Yamamoto TN, Bleakley M, Turtle CJ, Chang WC, Greisman HA, Wood B, Maloney DG, Jensen MC, Rader C, Riddell SR. The B-cell tumor-associated antigen ROR1 can be targeted with T cells modified to express a ROR1-specific chimeric antigen receptor. Blood. 2010;116:4532–4541. doi: 10.1182/blood-2010-05-283309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hofer T, Thomas JD, Burke TR, Jr, Rader C. An engineered selenocysteine defines a unique class of antibody derivatives. Proc Natl Acad Sci USA. 2008;105:12451–12456. doi: 10.1073/pnas.0800800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merchant AM, Zhu Z, Yuan JQ, Goddard A, Adams CW, Presta LG, Carter P. An efficient route to human bispecific IgG. Nat Biotechnol. 1998;16:677–681. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 50.Rader C. Generation of human Fab libraries for phage display. Methods Mol Biol. 2012;901:53–79. doi: 10.1007/978-1-61779-931-0_4. [DOI] [PubMed] [Google Scholar]
- 51.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Phage display for selection of novel binding peptides. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- 52.Ravn U, Didelot G, Venet S, Ng KT, Gueneau F, Rousseau F, Calloud S, Kosco-Vilbois M, Fischer N. Deep sequencing of phage display libraries to support antibody discovery. Methods. 2013;60:99–110. doi: 10.1016/j.ymeth.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Kwong KY, Baskar S, Zhang H, Mackall CL, Rader C. Generation, affinity maturation, and characterization of a human anti-human NKG2D monoclonal antibody with dual antagonistic and agonistic activity. J Mol Biol. 2008;384:1143–1156. doi: 10.1016/j.jmb.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rader C, Popkov M, Neves JA, Barbas CF., 3rd Integrin alpha(v)beta3 targeted therapy for Kaposi’s sarcoma with an in vitro evolved antibody. FASEB J. 2002;16:2000–2002. doi: 10.1096/fj.02-0281fje. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Rader C. Cloning, expression, and purification of monoclonal antibodies in scFv-Fc format. Methods Mol Biol. 2012;901:209–232. doi: 10.1007/978-1-61779-931-0_14. [DOI] [PubMed] [Google Scholar]
- 56.Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown CE, Wright CL, Naranjo A, Vishwanath RP, Chang WC, Olivares S, Wagner JR, Bruins L, Raubitschek A, Cooper LJ, Jensen MC. Biophotonic cytotoxicity assay for high-throughput screening of cytolytic killing. J Immunol Methods. 2005;297:39–52. doi: 10.1016/j.jim.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 58.Balakrishnan A, Goodpaster T, Randolph-Habecker J, Hoffstrom BG, Jalikis FG, Koch LK, Berger C, Kosasih PL, Rajan A, Sommermeyer D, Porter PL, Riddell SR. Analysis of ROR1 protein expression in human cancer and normal tissues. Clin Cancer Res. 2017;23:3061–3071. doi: 10.1158/1078-0432.CCR-16-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


