Abstract
Major progress has been made in understanding the epidemiology of bovine Cryptosporidium in China in the past 30 years. The overall infection rate in that period was 14.50% (5265/36316), with different prevalence being observed among dairy cattle, yaks, beef cattle, and buffalo. The infection rate declined as the animals’ ages increased and the lowest prevalence occurred in winter. Ten Cryptosporidium species and two genotypes have been found in cattle, with Cryptosporidium parvum, C. andersoni, C. bovis, and C. ryanae being the commonest species. Cryptosporidium bovis rather than C. parvum predominated in preweaned dairy cattle, and C. parvum IIdA15G1 and IIdA19G1 were the only subtypes detected in dairy cattle. Two subtype families, IIa and IId, were found in yaks. Population genetic analysis detected an epidemic population structure in C. andersoni, which suggested that the prevalence of C. andersoni in China is not attributable to the introduction of dairy cattle. Moreover, C. parvum IId subtypes probably dispersed from western Asia to other geographic regions based on population genetic analysis of isolates from China, Sweden, and Egypt. Therefore, we hypothesize that Cryptosporidium was introduced into China in the past, and different populations formed progressively in various hosts in response to diverse factors, including the transmission dynamics, geographic isolation, host specificity, and large-scale farming. More epidemiological studies are required to test this hypothesis and to clarify the prevalence and transmission of Cryptosporidium species in China.
Keywords: Cryptosporidium, cattle, genotype, subtype, population structure, China
Introduction
Cryptosporidium spp. are important zoonotic agents infecting a wide spectrum of vertebrate hosts (Xiao et al., 2004; Wang et al., 2011b). There is extensive genetic variation within the genus Cryptosporidium. To date, thirty-one recognized species and more than 60 Cryptosporidium genotypes have been discovered (Xiao et al., 2004; Wang et al., 2010; Ryan et al., 2014). Recent studies into the global causes of severe diarrhea in children suggested that Cryptosporidium is the second most important diarrheal pathogen after Rotavirus (Kotloff et al., 2013; Checkley et al., 2015; Vinayak et al., 2015).
Members of the genus Cryptosporidium complete all developmental stages in a single host. Sporulated oocysts, containing four sporozoites, are released from an infected host upon defecation (O’Hara and Chen, 2011). Following ingestion by a suitable host, the motile, infective sporozoites are released through a suture in the oocyst wall and parasitize epithelial cells of the gastrointestinal tract or other tissues (Reduker et al., 1985; O’Hara and Chen, 2011). In these cells, the parasites undergo asexual multiplication, which produces Type-I and Type-II merozoites (Current and Reese, 1986). Type-II merozoites ultimately produce either male or female equivalent sexual reproductive stages, microgametocytes and macrogametocytes, respectively (O’Hara and Chen, 2011). After the macrogamonts are fertilized by the microgametes, oocysts are formed and then sporulated in the infected host. Thick-walled and thin-walled representing two different types of oocysts are produced. The former is commonly excreted from the host, and the latter is primarily involved in autoinfection. Cryptosporidium can be transmitted by the fecal-oral route, via either direct contact or ingestion of contaminated food or water (Xiao et al., 2004; Wang et al., 2011c).
Cattle are the mammals in which Cryptosporidium infection is most commonly found, and preweaned calves are considered the most important reservoir for zoonotic infection (Wang et al., 2011b; Li F. et al., 2016). Cryptosporidium parvum, C. bovis, C. andersoni, and C. ryanae are predominantly responsible for bovine cryptosporidiosis, although several other Cryptosporidium species and genotypes are also discovered in cattle, including C. felis, C. hominis, C. suis, C. scrofarum, C. meleagridis, and C. suis-like genotype (Trout and Santín, 2008; Wang et al., 2011b; Zhang et al., 2013; Huang et al., 2014; Robertson et al., 2014). Studies conducted in numerous industrialized nations have suggested that C. parvum is the species most often found in preweaned calves and that it is a significant cause of diarrhea (Wang et al., 2011b). Cryptosporidium bovis and C. ryanae usually infect weaned calves and yearlings, although C. bovis is more commonly seen than C. ryanae, but neither are associated with the occurrence of diarrhea (Santín et al., 2008; Wang et al., 2011b). In contrast, C. andersoni is commonly observed in adult cattle and has been associated with gastritis, reduced milk yield, and poor weight gain (Esteban and Anderson, 1995; Wang et al., 2011b).
Cryptosporidium infections are frequently detected in humans and various domestic and wild animals in China (Wang et al., 2008, 2010, 2011c, 2014a; Feng et al., 2012). Among these, cattle are one of the major targets in which Cryptosporidium is studied. To date, 97 papers involving 24 provinces, autonomous districts, and municipalities of China have been published on Cryptosporidium infections in cattle since the first case was reported in Zhou et al. (1985). The present paper focuses on the advances in the molecular epidemiology of bovine Cryptosporidium that have occurred in China in the past 30 years.
Cryptosporidium Infection Rate
These data were calculated from 97 published papers reporting bovine Cryptosporidium infections in China. The overall infection rate was 14.50% (5265/36316), with a prevalence of 13.98% (4405/31504), 20.92% (667/3189), 10.47% (122/1165), and 15.50% (71/458) in dairy cattle, yaks, beef cattle, and buffalo, respectively (χ2 = 128.32; P < 0.01). The infection rate for Cryptosporidium species was 45.78% (141/308) in diarrheal calves (Zhou et al., 1985; Chen et al., 1992; Qin et al., 1994; Cui et al., 2014), which was significantly higher than the average prevalence in cattle (14.50%). A correlation between the prevalence and the age of the animals was observed. In general, the infection rate declined as the age of the animals increased (Guo et al., 1993; Chen et al., 2011; Wang et al., 2011a,b; Mi et al., 2013; Ma et al., 2014). In dairy cattle in Henan Province, the infection rates of Cryptosporidium species were 21.5% (172/801) in preweaned calves, 11.3% (86/758) in 3–11-month-old calves, 5.7% (5/262) in 12–24-month-old heifers, and 1.0% (3/295) in >24-month-old adult cattle (Wang et al., 2011a,b). In contrast, only two studies have determined the prevalence of Cryptosporidium across different seasons, and the prevalence was lowest in winter in both dairy cattle (Wang et al., 2011b) and yaks (Mi et al., 2013).
Cryptosporidium Species Distribution
A total of 1690 Cryptosporidium-positive isolates were genotyped and ten Cryptosporidium species and two genotypes were identified, including C. bovis, C. andersoni, C. ryanae, C. parvum, C. xiaoi, C. ubiquitum, C. meleagridis, C. hominis, C. tyzzeri, C. serpentis, C. suis-like genotype, and a new genotype (Table 1). Cryptosporidium bovis (129/364) rather than C. parvum (119/364) was the predominant Cryptosporidium species in preweaned dairy cattle (Wang et al., 2011b; Zhang et al., 2013; Cui et al., 2014; Huang et al., 2014; Qi et al., 2015). The earliest detection of C. bovis was in 1-week-old calves, indicating that the prepatent period was shorter than the previously recorded 10–12 days (Wang et al., 2011b). Cryptosporidium andersoni was most commonly found in heifers and adult cattle (Wang et al., 2011a). In contrast, another two common species, C. bovis and C. ryanae, occurred at different rates in different epidemiology studies. It is noteworthy that C. tyzzeri (formerly Cryptosporidium mouse genotype I) and C. serpentis probably arose from contamination (Chen and Huang, 2012), and the authors stated in the GenBank submissions that identical sequences (DQ855266 and DQ855267) were found in isolates from pigs with a reverse transcription–PCR analysis of the small subunit ribosomal RNA (Chen and Huang, 2007), and several SSU rRNA gene sequences of C. tyzzeri (EU369382, EF025503, EU369384, EU369381, and EU369383) previously isolated from bovine samples have also been deposited in GenBank. Neither C. tyzzeri nor C. serpentis is a known bovine parasite (Wang et al., 2011a,b).
Table 1.
Cryptosporidium species/genotypes identified in dairy cattle, yaks, beef cattle, and buffalo in China.
| Animal | Isolate no. | Cryptosporidium species/genotype | Reference |
|---|---|---|---|
| Dairy cattle | 1437 | C. andersoni (457), C. parvum (315), C. bovis (332), C. ryanae (86), C. tyzzeri (185), C. serpentis (4), C. hominis (24), C. meleagridis (5), C. bovis + C. ryanae (9), C. parvum + C. bovis (6), C. parvum + C. ryanae (4), C. parvum + C. andersoni (3) | Watanabe et al., 2005; Feng et al., 2007; Zhou et al., 2007; Liu et al., 2009; Su et al., 2011; Wang et al., 2011a,b; Chen and Huang, 2012; Zhang et al., 2013, 2015; Cui et al., 2014; Huang et al., 2014; Ma et al., 2015; Qi et al., 2015, 2016 |
| Yak | 337 | C. andersoni (75), C. parvum (28), C. bovis (143), C. ryanae (78), C. ubiquitum (2), C. xiaoi (1), C. suis-like genotype (2), C. parvum + C. bovis (2), C. bovis + C. ryanae (4), new genotype (2) | Mi et al., 2013; Ma et al., 2014; Qin et al., 2014; Qi et al., 2015; Li P. et al., 2016 |
| Beef cattle | 108 | C. andersoni (85), C. bovis (16), C. ryanae (6), C. bovis + C. ryanae (1) | Ma et al., 2015; Qi et al., 2016 |
| Buffalo | 40 | C. bovis (7), C. ryanae (33) | Ma et al., 2015 |
As in dairy cattle, C. parvum, C. andersoni, C. bovis, and C. ryanae were also the four Cryptosporidium species most commonly identified in yaks, although several other Cryptosporidium species and genotypes have been detected (Table 1). However, the distribution of different Cryptosporidium species according to yak age is still unclear. A study conducted in Qinghai suggested that C. bovis was the predominant species in yaks ≤ 2 years old, whereas C. parvum was more common in older yaks (Mi et al., 2013). In contrast, a recent study in Tibet showed that C. andersoni was predominant in 1–2-year-old yaks, whereas C. bovis was commonly found in older yaks (Li P. et al., 2016). Therefore, more studies are required to clarify the Cryptosporidium distributions according to age in different groups of animals. Two studies that genotyped Cryptosporidium-positive isolates from beef cattle and buffalo identified only C. andersoni, C. bovis, and C. ryanae (Ma et al., 2015; Qi et al., 2016).
Cryptosporidium Subtypes
Subtyping tools have been used extensively in studies of the transmission of C. hominis, C. parvum, and several other related Cryptosporidium species, including C. meleagridis and C. ubiquitum, in both humans and animals (Xiao, 2010; Li et al., 2014; Ryan et al., 2014). One of the most frequently used subtyping tools is a DNA sequence analysis of the 60-kDa glycoprotein (gp60, also known as gp40/15) gene (Ryan et al., 2014).
In dairy cattle, a total of 141 C. parvum isolates have been subtyped by sequencing of the gp60 gene (Table 2). Only the IId subtype family was identified, including IIdA15G1 in Gansu Province and the Ningxia Hui Autonomous Region (Cui et al., 2014; Huang et al., 2014; Zhang et al., 2015) and IIdA19G1 in Henan and Heilongjiang Provinces (Wang et al., 2011b; Zhang et al., 2013). Another study described a cryptosporidiosis outbreak caused by C. parvum subtype IIdA15G1 on a dairy farm in northwestern China (Cui et al., 2014). Three C. meleagridis isolates from preweaned calves were subtyped as IIIeA22G2R1, which was not identical to any known C. meleagridis subtype (Zhang et al., 2013). The subtypes of C. parvum in yaks appear to be more heterogeneous than those in dairy cattle. Thirteen Cryptosporidium-positive isolates from yaks in Qinghai Province were identified as family IIa and five subtypes were detected (Mi et al., 2013). In another study, three IId subtypes (IIdA15G1, IIdA18G1, and IIdA19G1) were detected in five yaks from Qinghai, Gansu, and Tibet, and one C. ubiquitum isolate belonged to zoonotic XIIa subtype 2 (Qi et al., 2015). To date, both C. parvum subtype IIdA19G1 and C. ubiquitum XIIa subtype 2 have also been found in humans in China (Wang et al., 2013; Li et al., 2014).
Table 2.
Cryptosporidium subtypes identified in dairy cattle and yaks in China.
| Animal | Cryptosporidium species | Subtypes | Reference |
|---|---|---|---|
| Dairy cattle | C. parvum | IIdA15G1 (86), IIdA19G1 (55) | Wang et al., 2011b; Zhang et al., 2013, 2015; Cui et al., 2014; Huang et al., 2014 |
| C. meleagridis | IIIeA22G2R1 (3) | Zhang et al., 2013 | |
| Yak | C. parvum | IIdA15G1 (3), IIdA18G1 (1), IIdA19G1 (1), IIaA15G2R1 (8), IIaA16G2R1 (2), IIaA14G1R1 (1), IIaA14G2R1 (1), IIaA16G3R1 (1) | Mi et al., 2013; Qi et al., 2015 |
| C. ubiquitum | XIIa subtype 2 (1) | Qi et al., 2015 | |
Population Genetics
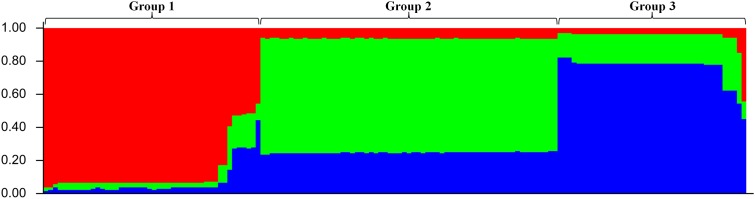
With the development of Cryptosporidium subtyping tools, it has become possible to assess the genetic and population structures of Cryptosporidium species (Xiao and Ryan, 2008). A total of 149 C. andersoni isolates from beef cattle (n = 38) and dairy cattle (n = 111) were subtyped with a multilocus sequence typing (MLST) tool based on the MS1, MS2, MS3, and MS16 loci. Fourteen MLST subtypes were identified and A4,A4,A4,A1 was the predominant subtype (Wang et al., 2012; Zhao et al., 2013; Qi et al., 2016). To test the possibility that this linkage disequilibrium (LD) was attributable to the clonal expansion of one or more subtype, masking the underlying equilibrium, an LD analysis was conducted using only the MLST subtypes (by considering each group of isolates with the same MLST subtype as one individual (Wang et al., 2012), 14 MLST subtypes were used in this analysis), with LIAN version 3.7. This analysis suggested that the C. andersoni population in cattle in China had an epidemic population structure (ISA = 0.0010, VD < L). An analysis using STRUCTURE 2.3.4 with K-means partitional clustering and the admixture model revealed three ancient lineages among the 149 C. andersoni specimens (Figure 1).
FIGURE 1.
Population structure inferred by Bayesian clustering using multilocus subtype informations of C. andersoni. K-means partitional clustering and the admixture mode were used in STRUCTURE 2.3.4 and the most appropriate number of K was calculated using an ad hoc statistic-based approach implemented in Structure Harvester v0.6.94 (http://taylor0.biology.ucla.edu/struct_harvest/).
Cryptosporidium parvum is another species that has been targeted for the population genetic analysis in cattle. Cryptosporidium parvum IId isolates (n = 111) from several species of animals in China, Sweden, and Egypt (Wang et al., 2014b) were subtyped with an MLST tool based on 12 microsatellite, minisatellite, and single-nucleotide polymorphism loci (Wang et al., 2014b). Host adaptation and significant geographic segregation were both observed in the MLST subtypes. A clonal population structure was detected in the C. parvum IId isolates from China and Sweden. Three ancestral lineages and the same RPGR (retinitis pigmentosa GTPase regulator) sequence were shared by the isolates examined (Wang et al., 2014b). The authors concluded that the C. parvum IId subtypes probably dispersed from western Asia to other geographic regions (Wang et al., 2014b; Zahedi et al., 2016).
Conclusion and Perspectives
Epidemiological data suggest that Cryptosporidium infections are commonly found in cattle in China. The infection rate declines as the age of the animals increases, and the lowest infection rate occurs in winter. Similar to the species distributions reported in other countries and areas of the world, C. parvum, C. bovis, C. andersoni and C. ryanae are the four commonest Cryptosporidium species in cattle. Cryptosporidium bovis rather than C. parvum was the dominant Cryptosporidium species in preweaned dairy cattle, which differs from the dominant species in this age group in other countries and areas of the world. Uniquely, the C. parvum subtypes identified in dairy cattle were all zoonotic IIdA15G1 or IIdA19G1 (Wang et al., 2014b).
Population genetic analyses of 149 C. andersoni isolates in three published studies confirmed an epidemic population structure, and as proposed in a previous study, these data suggest that the prevalence of C. andersoni in China is not attributable to the introduction of dairy cattle (Wang et al., 2012). According to a population genetic analysis, the C. parvum IId subtypes probably dispersed from western Asia to other geographic regions (Wang et al., 2014b). Therefore, we hypothesize that Cryptosporidium was introduced into China at some time in the past, and then different Cryptosporidium populations developed progressively in various hosts in response to diverse factors, including the transmission dynamics, geographic isolation, host specificity, and large-scale farming. More epidemiological studies are required to confirm this hypothesis and to clarify the transmission and public-health impact of Cryptosporidium species in China.
Author Contributions
LZ had the ideal for the review and revised the manuscript. RW wrote the paper. RW, GZ, and YG reviewed and abstracted data from each selected article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was supported in part by the Key Program of the National Natural Science Foundation of China (31330079), the National Natural Science Foundation of China (31672548, 31302079, and 31572509), the Program for Science and Technology Innovation Talents in Universities of Henan Province (16HASTIT018), and the Natural Science Foundation of Henan Province (162300410129).
References
- Checkley W., White A. C., Jr., Jaganath D., Arrowood M. J., Chalmers R. M., Chen X. M., et al. (2015). A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 15 85–94. 10.1016/S1473-3099(14)70772-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Huang K. (2007). Prevalence and phylogenetic analysis of Cryptosporidium in pigs in eastern China. Zoonoses Public Health 54 393–400. 10.1111/j.1863-2378.2007.01078.x [DOI] [PubMed] [Google Scholar]
- Chen F., Huang K. (2012). Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle from farms in China. J. Vet. Sci. 13 15–22. 10.4142/jvs.2012.13.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xu S. Z., Shan H., Wang H. (2011). Investigation on cow cryptosporidiosis in some regions of Yantai. Progress Vet. Med. 32 114–117. [Google Scholar]
- Chen Y. G., Li H. S., Dai M. X., Zhang H. R. (1992). Investigation on Cryptosporidium spp. from animals in Xuzhou area. Chin. J. Zoonoses 8 39–40. [Google Scholar]
- Cui Z., Wang R., Huang J., Wang H., Zhao J., Luo N., et al. (2014). Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasit. Vectors 7:529 10.1186/s13071-014-0529-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Current W. L., Reese N. C. (1986). A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J. Protozool. 1986 98–108. 10.1111/j.1550-7408.1986.tb05567.x [DOI] [PubMed] [Google Scholar]
- Esteban E., Anderson B. C. (1995). Cryptosporidium muris: prevalence, persistency, and detrimental effect on milk production in a drylot dairy. J. Dairy Sci. 7 1068–1072. 10.3168/jds.S0022-0302(95)76723-6 [DOI] [PubMed] [Google Scholar]
- Feng Y., Ortega Y., He G., Das P., Xu M., Zhang X., et al. (2007). Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 144 1–9. 10.1016/j.vetpar.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Feng Y., Wang L., Duan L., Gomez-Puerta L. A., Zhang L., Zhao X., et al. (2012). Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg. Infect. Dis. 18 312–314. 10.3201/eid1802.110666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S. L., Yuan F. S., Yu H., Yu A. K., Han Y. M., Wang C. L., et al. (1993). Investigation on cryptosporidiosis in cattle in Jinan area. Chin. J. Infect. Dis. Livest. poult. 1 34–35. [Google Scholar]
- Huang J., Yue D., Qi M., Wang R., Zhao J., Li J., et al. (2014). Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet. Res. 10:292 10.1186/s12917-014-0292-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff K. L., Nataro J. P., Blackwelder W. C., Nasrin D., Farag T. H., Panchalingam S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382 209–222. 10.1016/S0140-6736(13)60844-2 [DOI] [PubMed] [Google Scholar]
- Li F., Wang H., Zhang Z., Li J., Wang C., Zhao J., et al. (2016). Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet. Parasitol. 219 61–65. 10.1016/j.vetpar.2016.01.023 [DOI] [PubMed] [Google Scholar]
- Li N., Xiao L., Alderisio K., Elwin K., Cebelinski E., Chalmers R., et al. (2014). Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 20 217–224. 10.3201/eid2002.121797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Cai J., Cai M., Wu W., Li C., Lei M., et al. (2016). Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet. Parasitol. 215 58–62. 10.1016/j.vetpar.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Liu A., Wang R., Li Y., Zhang L., Shu J., Zhang W., et al. (2009). Prevalence and distribution of Cryptosporidium spp. in dairy cattle in Heilongjiang Province, China. Parasitol. Res. 105 797–802. 10.1007/s00436-009-1457-2 [DOI] [PubMed] [Google Scholar]
- Ma J., Cai J., Ma J., Feng Y., Xiao L. (2014). Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet. Parasitol. 202 113–118. 10.1016/j.vetpar.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Ma J., Li P., Zhao X., Xu H., Wu W., Wang Y., et al. (2015). Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 207 220–227. 10.1016/j.vetpar.2014.10.011 [DOI] [PubMed] [Google Scholar]
- Mi R., Wang X., Li C., Huang Y., Zhou P., Li Z., et al. (2013). Prevalence and genetic characterization of Cryptosporidium in yaks in Qinghai Province of China. PLOS ONE 8:e74985 10.1371/journal.pone.0074985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara S. P., Chen X. M. (2011). The cell biology of Cryptosporidium infection. Microbes Infect. 13 721–730. 10.1016/j.micinf.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Wang R., Jing B., Jian F., Ning C., Zhang L. (2016). Prevalence and multilocus genotyping of Cryptosporidium andersoni in dairy cattle and He cattle in Xinjiang, China. Infect. Genet. Evol. 44 313–317. 10.1016/j.meegid.2016.07.022 [DOI] [PubMed] [Google Scholar]
- Qi M. Z., Fang Y. Q., Wang X. T., Zhang L. X., Wang R. J., Du S. Z., et al. (2015). Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, north-western China. J. Med. Microbiol. 64 111–116. 10.1099/jmm.0.079327-0 [DOI] [PubMed] [Google Scholar]
- Qin J. H., Zhao Y. L., Hao C. W., Li C. X., Li D., Chen S. H., et al. (1994). Cryptosporidium infections in diarrheal calves in Zhangjiakou city. Chin. J. Vet. Sci. 2 150. [Google Scholar]
- Qin S. Y., Zhang X. X., Zhao G. H., Zhou D. H., Yin M. Y., Zhao Q., et al. (2014). First report of Cryptosporidium spp. in white yaks in China. Parasit. Vectors 7:230 10.1186/1756-3305-7-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reduker D. W., Speer C. A., Blixt J. A. (1985). Ultrastructure of Cryptosporidium parvum oocysts and excysting sporozoites as revealed by high resolution scanning electron microscopy. J. Protozool. 32 708–711. 10.1111/j.1550-7408.1985.tb03106.x [DOI] [PubMed] [Google Scholar]
- Robertson L. J., Björkman C., Axén C., Fayer R. (2014). “Cryptosporidiosis in farmed animals,” in Cryptosporidium: Parasite and Disease, eds Cacciò S. M., Widmer G. (Berlin: Springer; ), 149–236. [Google Scholar]
- Ryan U., Fayer R., Xiao L. (2014). Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141 1667–1685. 10.1017/S0031182014001085 [DOI] [PubMed] [Google Scholar]
- Santín M., Trout J. M., Fayer R. (2008). A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet. Parasitol. 155 15–23. 10.1016/j.vetpar.2008.04.018 [DOI] [PubMed] [Google Scholar]
- Su Y., Bai G. Y., Sun X. D., Liu Y., Wang C. R., Zhang J., et al. (2011). Characterization of Cryptosporidium spp. from preweaned calves of Jilin and Daqing area by 18S rRNA gene nested PCR-RFLP. Chin. J. Vet. Sci. 31 347–351. [Google Scholar]
- Trout J. M., Santín M. (2008). “Livestock,” in Cryptosporidium and Cryptosporidiosis, eds Fayer R., Xiao L. (Boca Raton, FL: CRC Press; ), 451–483. [Google Scholar]
- Vinayak S., Pawlowic M. C., Sateriale A., Brooks C. F., Studstill C. J., Bar-Peled Y., et al. (2015). Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523 477–480. 10.1038/nature14651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhang H., Zhao X., Zhang L., Zhang G., Guo M., et al. (2013). Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 51 557–563. 10.1128/JCM.02758-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Jian F., Zhang L., Ning C., Liu A., Zhao J., et al. (2012). Multilocus sequence subtyping and genetic structure of Cryptosporidium muris and Cryptosporidium andersoni. PLOS ONE 7:e43782 10.1371/journal.pone.0043782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Li G., Cui B., Huang J., Cui Z., Zhang S., et al. (2014a). Prevalence, molecular characterization and zoonotic potential of Cryptosporidium spp. in goats in Henan and Chongqing, China. Exp. Parasitol. 142 11–16. 10.1016/j.exppara.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Wang R., Ma G., Zhao J., Lu Q., Wang H., Zhang L., et al. (2011a). Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitol. Int. 60 1–4. 10.1016/j.parint.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Wang R., Qiu S., Jian F., Zhang S., Shen Y., Zhang L., et al. (2010). Prevalence and molecular identification of Cryptosporidium spp. in pigs in Henan, China. Parasitol. Res. 107 1489–1494. 10.1007/s00436-010-2024-6 [DOI] [PubMed] [Google Scholar]
- Wang R., Wang H., Sun Y., Zhang L., Jian F., Qi M., et al. (2011b). Characteristics of Cryptosporidium transmission in preweaned dairy cattle in henan, China. J. Clin. Microbiol. 49 1077–1082. 10.1128/JCM.02194-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang L., Axén C., Bjorkman C., Jian F., Amer S., et al. (2014b). Cryptosporidium parvum IId family: clonal population and dispersal from Western Asia to other geographical regions. Sci. Rep. 4:4208 10.1038/srep04208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang L., Feng Y., Ning C., Jian F., Xiao L., et al. (2008). Molecular characterization of a new genotype of Cryptosporidium from American minks (Mustela vison) in China. Vet. Parasitol. 154 162–166. 10.1016/j.vetpar.2007.12.038 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang X., Zhu H., Zhang L., Feng Y., Jian F., et al. (2011c). Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp. Parasitol. 127 42–45. 10.1016/j.exppara.2010.06.034 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Yang C. H., Ooi H. K. (2005). Cryptosporidium infection in livestock and first identification of Cryptosporidium parvum genotype in cattle feces in Taiwan. Parasitol. Res. 97 238–241. 10.1007/s00436-005-1428-1 [DOI] [PubMed] [Google Scholar]
- Xiao L. (2010). Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124 80–89. 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]
- Xiao L. H., Fayer R., Ryan U., Upton S. J. (2004). Cryptosporidium Taxonomy: recent advances and implications for public health. Clin. Microbiol. 17 72–97. 10.1128/CMR.17.1.72-97.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L. H., Ryan U. (2008). “Molecular epidemiology,” in Cryptosporidium and Cryptosporidiosis, eds Fayer R., Xiao L. (Boca Raton, FL: CRC Press; ), 119–172. [Google Scholar]
- Zahedi A., Phasey J., Boland T., Ryan U. (2016). First report of Cryptosporidium species in farmed and wild buffalo from the Northern Territory, Australia. Parasitol. Res. 115 1349–1353. 10.1007/s00436-016-4901-0 [DOI] [PubMed] [Google Scholar]
- Zhang W., Wang R., Yang F., Zhang L., Cao J., Zhang X., et al. (2013). Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China’s Heilongjiang Province. PLOS ONE 8:e54857 10.1371/journal.pone.0054857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. X., Tan Q. D., Zhou D. H., Ni X. T., Liu G. X., Yang Y. C., et al. (2015). Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol. Res. 114 2781–2787. 10.1007/s00436-015-4537-5 [DOI] [PubMed] [Google Scholar]
- Zhao G. H., Ren W. X., Gao M., Bian Q. Q., Hu B., Cong M. M., et al. (2013). Genotyping Cryptosporidium andersoni in cattle in Shaanxi province, northwestern China. PLOS ONE 8:e60112 10.1371/journal.pone.0060112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Li G., Xiao S., Xia Y., Guo Y. (2007). PCR amplification and sequence analyses of ITS-1 rDNA from Cryptosporidium andersoni in dairy cattle. Parasitol. Res. 100 1135–1138. 10.1007/s00436-006-0358-x [DOI] [PubMed] [Google Scholar]
- Zhou S. W., Zhang Y., Ji D. C., Qi T. Y. (1985). Investigation on cryptosporidiosis in calves and cross-transmission study. Chin. J. Prev. Vet. Med. 3 21–22. [Google Scholar]