Abstract
Diverse leaf forms can be seen in nature. In Medicago truncatula, PALM1 encoding a Cys(2)His(2) transcription factor is a key regulator of compound leaf patterning. PALM1 negatively regulates expression of SGL1, a key regulator of lateral leaflet initiation. However, how PALM1 itself is regulated is not yet known. To answer this question, we used promoter sequence analysis, yeast one-hybrid tests, quantitative transcription activity assays, ChIP-PCR analysis, and phenotypic analyses of overexpression lines and mutant plants. The results show that M. truncatula AUXIN RESPONSE FACTOR3 (MtARF3) functions as a direct transcriptional repressor of PALM1. MtARF3 physically binds to the PALM1 promoter sequence in yeast cells. MtARF3 selectively interacts with specific auxin response elements (AuxREs) in the PALM1 promoter to repress reporter gene expression in tobacco leaves and binds to specific sequences in the PALM1 promoter in vivo. Upregulation of MtARF3 or removal of both PHANTASTICA (PHAN) and ARGONAUTE7 (AGO7) pathways resulted in compound leaves with five narrow leaflets arranged in a palmate-like configuration. These results support that MtARF3, in addition as an adaxial-abaxial polarity regulator, functions to restrict spatiotemporal expression of PALM1, linking auxin signaling to compound leaf patterning in the legume plant M. truncatula.
Keywords: auxin signaling, compound leaf development, adaxial-abaxial polarity regulation, MtARF3, Medicago truncatula
Introduction
Leaves are initiated from the flanks of the shoot apical meristem (SAM). The class I Knotted-like homeobox transcription factor genes (KNOXIs), which are required to promote and maintain the meristematic activity of SAM, are downregulated at the incipient sites of leaf primordia (Jackson et al., 1994; Lincoln et al., 1994; Long et al., 1996). In simple leaf species, KNOXI genes are permanently downregulated (Long et al., 1996). On the other hand, in most eudicot species with compound leaves, including tomato (Solanum lycopersicum) and Cardamine hirsuta, KNOXI genes are reactivated in developing leaf primordia, and this reactivation is required for compound leaf development (Hareven et al., 1996; Hay and Tsiantis, 2006; Barkoulas et al., 2008; Shani et al., 2009). In the subclade of legumes (Fabaceae), the inverted repeat lacking clade (IRLC), including pea (Pisum sativum) and Medicago (Wojciechowski et al., 2000), KNOXI genes are not reactivated during compound leaf development (Hofer et al., 2001; Champagne et al., 2007; Peng et al., 2011). Instead, UNIFOLIATA (UNI) and SINGLE LEAFLET1 (SGL1), orthologs of the Arabidopsis thaliana LEAFY (LFY) gene, are required for the initiation of lateral leaflet primordia (Hofer et al., 1997; Champagne et al., 2007; Wang et al., 2008).
It has been reported that PALMATE-LIKE PENTAFOLIATA1 (PALM1) acts as a key determinant of trifoliate leaves in Medicago truncatula. Loss of function mutations of PALM1 result in palmate-like pentafoliate leaves in contrast to trifoliate leaves in WT plants (Chen et al., 2010). PALM1 gene encodes a novel Cys(2)His(2) zinc finger transcription factor, which can binds to the SGL1 promoter sequence and negatively regulates SGL1 transcription (Chen et al., 2010; Ge et al., 2010).
Plant leaves usually exhibit a flat, bifacial structure with distinct adaxial and abaxial identities (Kidner and Timmermans, 2010; Yamaguchi et al., 2012; Nakata and Okada, 2013; Fukushima and Hasebe, 2014). The adaxial side of the leaf develops tightly packed layers of palisade mesophyll cells, while the abaxial side of the leaf develops loosely packed spongy mesophyll cells and a high density of stomata. The proper specification of the adaxial/abaxial domains in leaf primordia is critical for leaf blade expansion (Emery et al., 2003). PHANTASTICA (PHAN) was first identified as a key adaxial-abaxial polarity gene in Antirrhinum majus (Waites et al., 1998). In A. majus, phan mutant produces radialized leaves, in which the adaxial identity was abaxialized (Waites et al., 1998). Interestingly, loss of function mutations in the PHAN orthologs, ASYMMETRIC LEAVES1 (AS1) and ROUGH SHEATH2 (RS2) in A. thaliana and maize (Zea mays), respectively, only have mild effects on the adaxial-abaxial polarity of leaves (Schneeberger et al., 1998; Byrne et al., 2000). In M. truncatula, phan mutant displays minor leaf polarity defects especially at a late developmental stage (Ge and Chen, 2014). Independent studies have shown that AUXIN RESPONSE FACTOR3 (ARF3)/ARF4 play redundant roles in determining the leaf abaxial identity (Fahlgren et al., 2006; Garcia et al., 2006; Hunter et al., 2006). In Arabidopsis, single arf3 mutants display partial loss of the abaxial identity in petals, whereas arf3;arf4 double mutants display severe polarity defects, including narrow, upwardly curled leaves, and ectopic blade outgrowth on the abaxial side (Pekker et al., 2005). In Arabidopsis, both ARF3 and ARF4 RNAs are targeted for degradation by the trans-acting short-interfering RNA (ta-siRNA), tasiR-ARF. tasiR-ARF is derived from the non-coding TAS3 gene, whose RNA is initially targeted by miR390 for cleavage (Allen et al., 2005). Biogenesis and stabilization of tasiR-ARF requires TAS3, miR390, and AGONAUTE7 (AGO7), SUPPRESSOR OF GENE SILENCING3 (SGS3), RNA-DEPENDENT RNA POLYMERASE6 (RDR6), and DICER-LIKE4 (DCL4) proteins (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006). Loss of function mutations of ago7, sds3, or rdr6 result in reduction of tasiR-ARF and over accumulation of ARF3 and ARF4 transcripts. Mutant plants exhibit leaf polarity and heteroblastic defects. Genetic studies show that impairment of the tasiR-ARF pathway enhances the defects of the asymmetric leaves1 (as1) mutant in leaf adaxial-abaxial partitioning and margin development (Garcia et al., 2006; Xu et al., 2006). AS1 can bind to the ARF3 promoter and repress ARF3 transcription in Arabidopsis (Iwasaki et al., 2013). Thus, AS1 and the tasiR-ARF pathway negatively regulate ARF3 expression at the transcriptional and post-transcriptional level, respectively.
It is still not clear whether and how ARF3 regulates leaf patterning in M. truncatula that forms dissected leaves. In this study, we show that M. truncatula ARF3 acts as a negative transcriptional repressor of PALM1 gene expression. We show that ARF3 binds to putative auxin response elements (AuxREs) in the promoter region of PALM1 and represses expression of reporter gene activities in transcription activity assays in tobacco leaves.
Materials and Methods
Plant Materials
Medicago truncatula phan (mtphan), palm1 (palm1-5), mtago7 (mtago7-1), and sgl1 (sgl1-2) mutants were described previously (Wang et al., 2008; Chen et al., 2010; Zhou et al., 2013; Ge et al., 2014). Medicago plants were grown in a greenhouse under the following conditions: 24°C/20°C day/night temperature, 16 h/8 h photoperiod and 150 uE/m2/sec light intensity.
Scanning Electron Microscopy
Shoot apices of 2- to 4-week-old seedlings, or rachis and pulvini were subjected to vacuum infiltration in a fixative solution (3% glutaraldehyde in 25 mM phosphate buffer, pH 7.0) for 1 h and then incubated at 4°C overnight. Plant tissues were further fixed with 1.0% osmium tetroxide in the same phosphate buffer overnight and dehydrated in a graded ethanol series. Scanning electron microscopy (SEM) was performed as described previously (Wang et al., 2008).
Real-time RT-PCR
Total RNA was prepared using the RNeasy Plant Mini Kit (Qiagen). Residual genomic DNA was removed using a DNA-free Kit (Ambion). cDNA synthesis was performed using SuperScript III reverse transcriptase (Invitrogen), starting with 2 μg of total RNA in a 20-μL reaction mix with oligo(dT)15 primer (Promega). Real-time RT-PCR was performed using a 7900HT fast real-time PCR system (Applied Biosystems), as previously described (Peng et al., 2011). SDS2.2.1 software (Applied Biosystems) was used to analyze the melting curve to confirm single amplification. PCR efficiency was estimated by using the LinRegPCR program (Ramakers et al., 2003), and transcript levels were determined in reference to the expression of the M. truncatula ACTIN gene Medtr3g095530.
Yeast One-Hybrid
The Matchmaker Gold System (Clontech) was used in yeast one-hybrid assays. The PALM1 promoter sequence was PCR-amplified, cloned into the pAbAi vector and integrated into the Y1H Gold genome to create the bait strain, according to the manufacturer’s instructions. Coding sequences of MtARF3 were PCR-amplified from cDNA templates, cloned into the pGADT7 vector and introduced into the bait strain Y1H Gold/PALM promoter. Positive clones grown on double selection media (SD-Ura-Leu) were tested on SD-Ura-Leu plates supplemented with Aureobasidin A (100 ng/ml). Primers are listed in Supplementary Table S1. Yeast transformation was performed as previously described (Peng et al., 2011).
Constructs and Plant Transformation
MtARF3 was generated by RT-PCR from total RNA extracted from vegetative shoot buds of M. truncatula. MtARF3m with mutations in tasiR-ARF recognition sites was created by overlapping PCR, cloned into the pENTR/D vector and then transferred into the pEARLEYGATE 202 vector using Gateway cloning technology. M. truncatula transformation was performed as previously described (Peng et al., 2011).
Transcription Activity Assays
Transcription activity assays were carried out essentially as described previously (Hellens et al., 2005), with the following modifications. E2, three times tandem repeats of the cis-element TGTCAA flanked by SpeI and NotI recognition sequences, was inserted into the 35S promoter at the -46 nucleotide position by overlapping PCR; the resulting 35S-E2 was cloned into pGREENII 0800-LUC vector between BamHI and NcoI restriction sites to create the 35S-E2::LUC construct. Three tandem repeats of other elements, E1, E3–E10 (Supplementary Figure S1), flanked by SpeI and NotI sequences, were synthesized, annealed, restriction-enzyme-digested and cloned into SpeI- and NotI-digested 35S-E2::LUC vector to generate the 35S-E::LUC reporter constructs. MtARF3m was cloned into the pMDC32 vector as the effector construct. Both the effector and reporter constructs were introduced into the Agrobacterium tumefaciens GV3101 strain. The reporter strains were infiltrated into Nicotiana benthamiana leaves, together with the control strain, which harbors GUS or the effector strain, which harbors MtARF3m. Firefly luciferase and Renillia luciferase were assayed using the dual luciferase assay reagents (Promega, Madison, WI, United States). Briefly, after inoculation and a transient incubation for 2 days, 1 cm leaf disks were harvested and ground in 100 μl of Passive Lysis Buffer. 5 μl of this crude extract was assayed in 100 μl of Luciferase Assay Buffer, and the first chemiluminescence was measured for firefly luciferase activities. 100 μl of Stop and GlowTM buffer was then added, and a second chemiluminescence measurement was made for Renillia luciferase activities. Absolute RLU was measured in a GloMax® 96 Microplate Luminometer with Dual Injectors, with 5-s delay and 15-s measurement. Data were collected as ratios of LUC/REN.
Phylogenetic Analysis
AUXIN RESPONSE FACTOR (ARF) protein sequences in A. thaliana from TAIR10 (The Arabidopsis Information Resource) were used to identify ARF protein sequences from M. truncatula genome project v4.01. Multiple sequences were aligned using ClustalX (Thompson et al., 1994). Phylogenetic trees were reconstructed using the maximum likelihood algorithm implemented in MEGA102, with 1,000 bootstrap replications (Tamura et al., 2007).
RNA In Situ Hybridization
RNA in situ hybridization was performed as described previously (Ferrandiz et al., 2000) with minor modifications. The MtAGO7 probes correspond to a 517-bp sequence of the MtAGO7 coding sequence. The MtARF3 probes correspond to a 511-bp sequence of the MtARF3 coding sequence. Eight-micrometer sections from shoot apices of 2- to 4-week-old seedlings were processed and hybridized with digoxigenin-labeled sense and antisense probes.
ChIP-PCR Analysis
ChIP was performed using FLAG-MtARF3m-overexpressing Medicago plants. ChIP-PCR analysis was done as previously described (Kaufmann et al., 2010; Zhu et al., 2012) with some modifications. Plant tissues were pulverized in liquid nitrogen. Powdered tissues were cross-linked in 10 mM Tris-HCl (pH 8.0) containing 1% formaldehyde, supplemented with 0.4 M sucrose, 5 mM β-mercaptoethanol (β-ME) and a complete proteinase inhibitor cocktail (Roche; PI tablets). Cross linking was done for 10 min at room temperature, and the reaction was quenched by the addition of glycine to a final concentration of 125 mM. The slurry was filtered through a mesh (55 μm) and precipitated by centrifugation at 4,000 g for 20 min; the pellet was then resuspended in extraction buffer 2 (0.25 M Sucrose, 10 mM Tris-Cl, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-ME and the PI cocktail). This was followed by centrifugation at 14,000 g for 20 min. The pellet was resuspended in extraction buffer 3 (1.7 M Sucrose, 10 mM Tris-Cl, pH 8, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-ME and the proteinase inhibitor cocktail) and centrifuged at 20,000 g for 40 min. The resulting pellet was resuspended in nuclei lysis buffer (10 mM Tris-HCl, pH 8.0, 20 mM EDTA, 400 mM NaCl, 1% Triton X-100 and the PI cocktail). Sonication, 6 X 15 pulses with a 1 min break between each 15 pulses, was used to shear the DNA. Cell debris was removed by centrifugation. ChIP was performed by incubation with rabbit anti-Flag polyclonal antibody (Abcam) and followed by incubation with Protein A/G magnetic beads (Pierce). The beads were washed twice with high-salt wash buffer (500 mM NaCl, 0.2% SDS, 0.5% Triton X-100, 20 mM Tris-HCl, pH 8.0, 2 mM EDTA and the PI tablet), and the chromatin was eluted in the elution buffer (1% SDS and 0.1 M NaHCO3) and de-cross-linked at 65°C for 16 h. Cellular RNA and protein were then removed by RNase (Qiagen) and proteinase-K (Applied Biosystems) treatments, respectively. DNA was purified through a purification column. Enrichment of DNA targets in each sample was determined by PCR using primers listed in Supplementary Table S1.
GenBank Accession Numbers
HM038482, PALM1; AY928184, SGL1; XM_003593616, MtARF3; DQ468322, MtPHAN; XM_003613868, MtAGO7; JF929904, MtNAM; XM_003602497, ACTIN.
Results
Medicago truncatula ARF3 Negatively Regulates PALM1 Expression
To identify potential regulators of PALM1 gene expression, we first analyzed whether any specific cis-elements are present within the promoter sequence of the PALM1 gene. This led to the discovery of 18 TGTCXX elements within the 2.5-kb promoter region of PALM (Supplementary Figure S1A and Figure 1A) (Li et al., 1994; Ulmasov et al., 1995, 1997a). These TGTCXX elements, representing 10 distinct groups (Supplementary Figure S1B), resemble the canonical auxin response element (AuxRE), TGTCTC, which interact with AUXIN RESPONSE FACTORs (ARFs) (Li et al., 1994; Ulmasov et al., 1995, 1997a). Next, we tested whether M. truncatula ARF3 (MtARF3) can interact with the PALM1 promoter sequence in the yeast one-hybrid system. The results show that MtARF3 interacted strongly with the PALM1 promoter sequence in yeast one-hybrid assays (Figure 1B).
FIGURE 1.
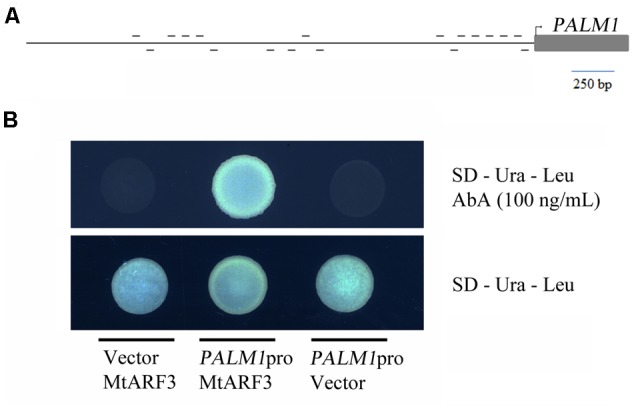
Yeast one-hybrid assays of interactions between MtARF3 and the PALM1 promoter sequence. (A) A schematic drawing of the promoter and coding regions of PALM1. Short horizontal lines represent putative auxin response elements (AuxREs; TGTCxx) on the plus (above) and minus (beneath) strands of the promoter sequence. (B) Yeast one-hybrid assays, showing interactions between MtARF3 and the PALM1 promoter sequence. SD, synthetic drop-out media; Ura, uridine; Leu, leucine; AbA, aureobasidin A.
It is notable that the canonical AuxRE element (TGTCTC) is present at the -2129 position and a majority of TGTCXX elements are present in the proximity of the translation initiation codon (ATG) of PALM1 (Supplementary Figure S1). To test whether MtARF3 interacts with the TGTCXX elements in the PALM1 promoter in vivo, we first used quantitative luciferase (LUC) reporter activity assays in tobacco leaves (Hellens et al., 2005). For this, all 10 potential AuxRE elements in the PALM1 promoter were cloned as three tandem repeats, with the ‘ccttt’ spacer, which was used in the construction of the DR5 auxin response reporter (Ulmasov et al., 1997b), between repeats into the 35S promoter (designated as 35S-Es::LUC; Figures 2A,B). Because ARF3 is post-transcriptionally regulated by trans-acting short interfering RNA (tasiR-ARF) (Hunter et al., 2006; Montgomery et al., 2008), we generated a modified MtARF3 (MtARF3m) by mutating the tasiR-ARF targeting sites without changing the amino acid sequence (Figure 2C). To assess transcription repressor activities of the effector protein, we compared 35S-Es::LUC activities in the presence of the MtARF3m effector protein and the unrelated β-glucuronidase (GUS) protein (Figure 2D). The results show that (1) 35S::LUC activities were not significantly different; (2) several potential AuxRE elements, i.e., E3, E4, E9 and E10, exhibited moderate but not significant inhibitory effects on the luciferase activities; and (3) other AuxRE elements, namely E1, E2, E5, E7 and E8, significantly reduced the luciferase activities in the presence of the MtARF3m effector protein compared with that in the presence of the unrelated GUS protein control (Figure 2D). These results indicate that MtARF3 can recognize several putative AuxREs that are present in the PALM1 promoter and function as a transcription repressor in the transcription activity assay in tobacco.
FIGURE 2.
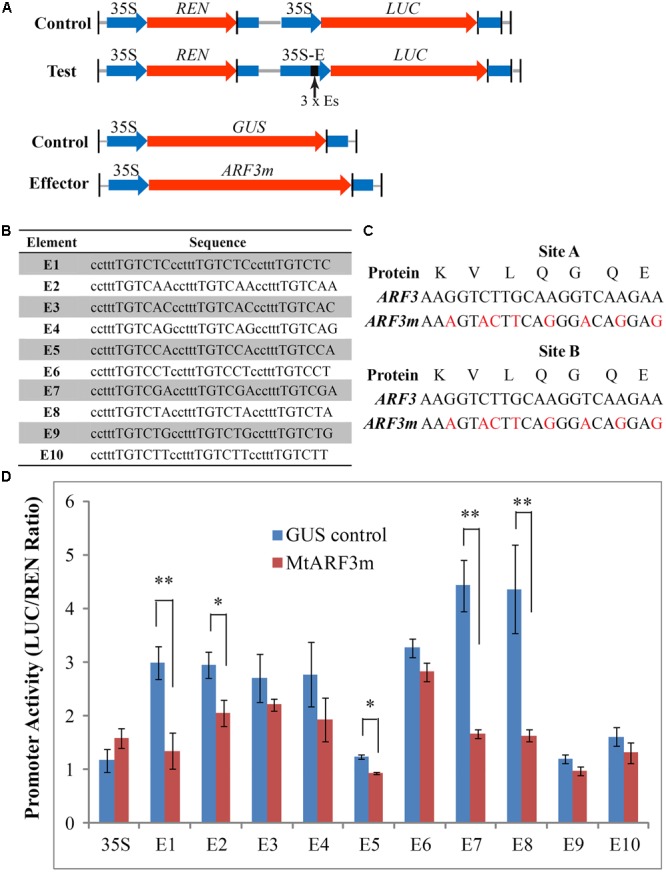
Transcription activity assays in tobacco leaves. (A) Schematic drawings of reporter and effector constructs used in quantitative luciferase activity assays in tobacco leaves. (B) Putative AuxRE sequences in the PALM1 promoter region were tested. Capital letters denote the cis-elements and lower case letters denote the spacer between cis-element repeats. (C) Construction of tasiR-ARF-resistant MtARF3m. Red letters denote synonymous mutations to disrupt tasiR-ARF recognition sites A and B. (D) Luciferase activity assays in tobacco leaves. Firefly luciferase (LUC) activities were normalized against Renillia luciferase (REN) activities. Shown are means ± SD; n > 6. Student’s t-test, ∗p < 0.05; Student’s t-test, ∗∗p < 0.01.
Next, we tested whether MtARF3 can bind to the PALM1 promoter in M. truncatula plants, using chromatin immunoprecipitation coupled with polymerase chain reaction (ChIP-PCR). For this, we first generated independent transgenic Medicago plants that overexpress MtARF3m-FLAG fusion proteins. ChIP-PCR analysis shows that P1, P2 and P4 regions, but not the P3 region of the PALM1 promoter were significantly enriched in MtARF3m-FLAG-bound chromatins (Figures 3A–C). Consistent with these results, P1, P2 and P4 regions, but not the P3 region, of the PALM1 promoter contain a number of putative AuxREs (Figure 3A and Supplementary Figure S1). Particularly, P1 and P4 contain several putative AuxREs, E1, E2, E5, E7, and E8 that exhibited significant inhibitory effects on luciferase activities in tobacco (Figures 2D, 3A). Although three different putative AuxRE elements (E4, E9, and E10) that are present in P2 exhibited moderate but not significant inhibitory effects on luciferase activities in the tobacco system (Figure 2D), the ChIP-PCR results clearly show that four copies of these elements in P2 are sufficient to be recognized by MtARF3 in vivo (Figures 3A–C). These results confirm that PALM1 is a direct target of ARF3 in M. truncatula plants
FIGURE 3.
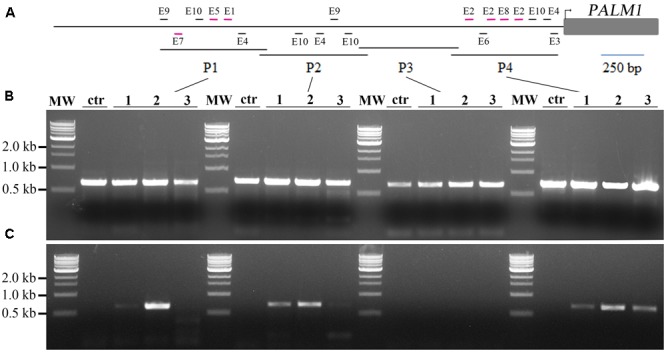
Chromatin immunoprecipitation (ChIP)-PCR assays of interactions between MtARF3 and the PALM1 promoter in vivo. (A) A schematic drawing of the PALM1 promoter, showing location of putative auxin response elements (AuxREs; short horizontal lines) and the promoter fragments, P1–P4. (B) PCR analysis shows equal inputs of the DNA samples. (C) ChIP-PCR analysis shows enrichment of the PALM1 promoter fragments, P1, P2 and P4, but not P3, in FLAG-MtARF3-bound chromatins. Lanes MW, 1 kb marker from NEB; ctr, negative control in which wild type plants were used; and 1–3, three independent 35S::FLAG-MtARF3m plants. P1, P2, and P4 each contain several putative AuxREs (short horizontal lines), whereas P3 does not contain any AuxREs. P1 and P4 contain several putative AuxREs (short horizontal lines in purple) that exhibited significant inhibitory effects on luciferase reporter gene expression in the presence of MtARF3m in transcription activity assays in tobacco. Although P2 contains four putative AuxREs that exhibited moderate but not significant inhibitory effects on reporter gene expression in the tobacco system, ChIP-PCR results showed clearly that they are sufficient to be recognized by ARF3 in Medicago truncatula plants.
Overexpression of MtARF3m Results in Palmate-Like Pentafoliate Leaves with Radialized Blades
ARF3 belongs to a large gene family in both A. thaliana and M. truncatula (Supplementary Figure S2). In M. truncatula, many of the ARF genes are duplicated, in contrast to the corresponding single genes in A. thaliana (Supplementary Figure S2), Because of gene redundancy, loss-of-function or reduced function mutants of single ARF genes may not exhibit visible phenotypes in M. truncatula. To test whether the interaction between ARF3 and the PALM1 promoter has physiological relevance, we analyzed transgenic Medicago plants, in which MtARF3m-FLAG is ectopically overexpressed by the 35S promoter. Out of four independent lines, three lines exhibited mild leaf phenotypes, including downward curling at the proximal leaf margin and deep serrations at the distal leaf margin, albeit to different degrees (Figures 4A,B). These phenotypes are consistent with the role of ARF3 in leaf adaxial-abaxial polarity regulation in Arabidopsis and resemble the phenotype of mtphan and mtago7 mutants (Zhou et al., 2013; Ge and Chen, 2014; Ge et al., 2014). Significantly, a strong MtARF3m overexpression line exhibited severe leaf phenotypes, including some compound leaves with five radialized leaflets arranged in a palmate-like configuration (Figures 4C,D). These results suggest that a high level of MtARF3 overexpression results in proliferation of compound leaves that resemble palmate-like pentafoliate leaves of the palm1 mutant (Chen et al., 2010), albeit with a severe adaxial-abaxial polarity defect.
FIGURE 4.
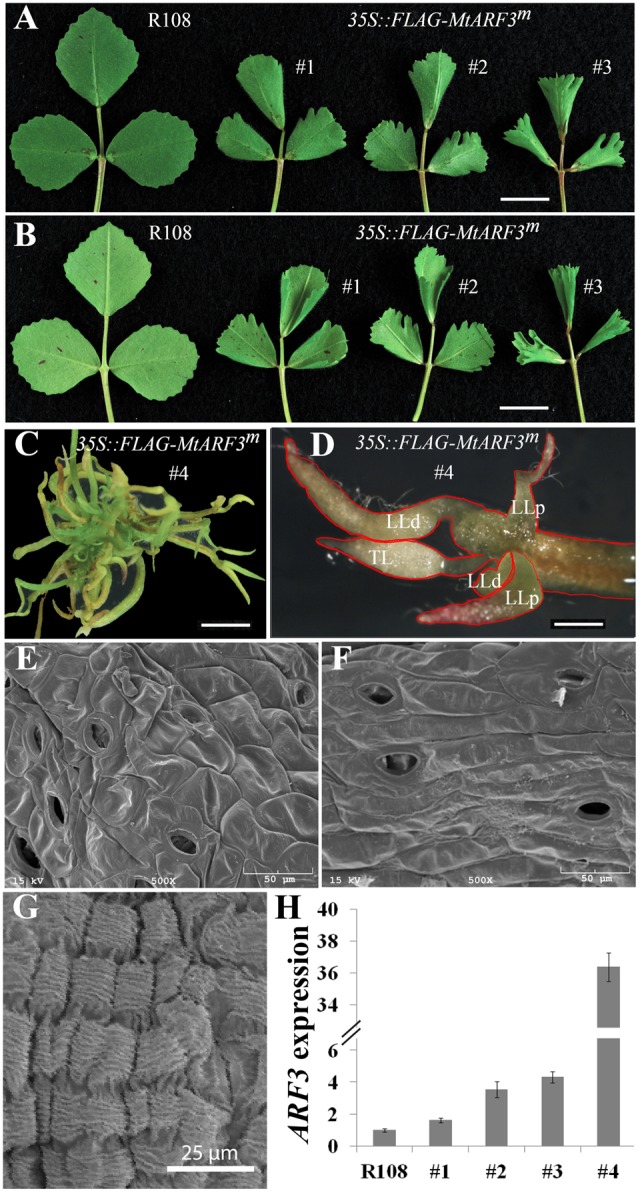
Overexpression of MtARF3m leads to altered compound leaf patterning in M. truncatula. Adaxial (A) and abaxial (B) views of compound leaves in wild type (R108) and three independent transgenic lines that overexpress MtARF3m (35S::FLAG-MtARF3m). Leaflets exhibited deep serration and curling at the distal and proximal margins, respectively, in transgenic plants compared with wild type. (C,D) Severe compound leaf phenotypes of a strong MtARF3m overexpressor. The transgenic plant exhibited pentafoliate leaves with needle-like blades (outlined in red). LLp, proximal lateral leaflets; LLd, distal lateral leaflets; TL, terminal leaflet. (E,F) SEM images of the adaxial (E) and abaxial (F) surface epidermal cells of TL (D). (G) A SEM image of epidermal cells of LLd (D). (H) Quantitative RT-PCR analysis of MtARF3 expression in young leaf tissues in MtARF3m overexpression lines. Shown are means ± SD; n = 3. Scale bars: 1 cm (A,B); 2 mm (C,D).
Scanning electron microscopy analysis showed that the needle-like structures in the strong MtARF3m overexpression line are leaflets, because (1) distinct epidermal cell shapes are present on the adaxial and abaxial surface of each needle-like structure (Figures 4E,F); (2) each needle-like structure has stomata and trichomes on its surface (Figures 4E,F); and (3) each needle-like structure is connected to a petiole or rachis through the motor organ, pulvinus (Chen et al., 2012) (Figure 4G).
Reverse transcription (RT)-PCR results show that MtARF3 transcript level was elevated in all overexpression lines compared with wild type plants (Figure 4H), and the severity of the leaf phenotype was correlated with the expression level of MtARF3 (Figure 4H). In the strong MtARF3m overexpression line, the MtARF3 transcript level was upregulated about 36 fold over the wild type level, supporting that a high level of MtARF3 expression is associated with the palmate-like pentafoliate leaf phenotype (Figure 4H).
mtphan;mtago7 Double Mutant Plants Exhibit Palmate-Like Pentafoliate Leaves
Since ARF3 transcript level is regulated both at the transcriptional level by the MYB domain protein, ASYMMETRIC LEAF1 (AS1) (Ikezaki et al., 2010; Kojima et al., 2011; Iwasaki et al., 2013) and at the post-transcriptional level by tasiR-ARF (Fahlgren et al., 2006; Montgomery et al., 2008) in Arabidopsis, we tested whether MtARF3 expression can be significantly upregulated in mtphan;mtago7 double mutants. To generate the mtphan;mtago7 double mutants, we used mtphan and mtago7 single mutants previously described (Zhou et al., 2013; Ge and Chen, 2014; Ge et al., 2014). Quantitative RT-PCR results show that MtARF3 transcripts accumulated four and two times higher in vegetative shoot buds in mtago7 and mtphan single mutants, respectively, than wild type (Figure 5K). And, the PALM1 transcript level was slightly but significantly reduced in both mtago7 and mtphan mutants compared with wild type (Figure 5L). Similar results on ARF3 gene expression were obtained in developing leaves in mtphan and mtago7 mutants (Figure 5K). Quantitative RT-PCR results further show that the MtARF3 transcript level was approximately eight times that of the wild type level in the mtago7;mtphan double mutant (Figure 5K). Most significantly, mtphan;mtago7 double mutants developed palmate-like pentafoliate leaves with narrow blades (Figure 5A). The palmate-like pentafoliate leaf phenotype of the double mutant resembles that of loss-of-function palm1 mutants, although the palm1 mutant did not exhibit adaxial-abaxial polarity defects (Figure 5A). Interestingly, in the R108 background, the petiolules of the ectopic lateral leaflets appear to diverge from secondary rachises in the mtphan;mtago7 double mutants and to a less degree also in the palm1 mutants (Figure 5A), whereas in the Jemalong A17 background, the petiolules of lateral leaflets appear to come from the same location on the petiole (Chen et al., 2010).
FIGURE 5.
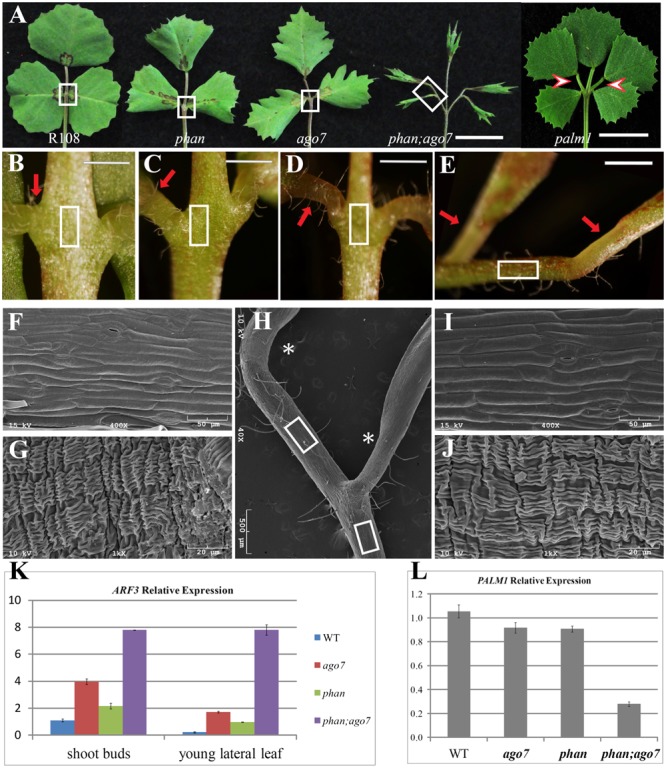
Altered compound leaf patterning in mtphan;mtago7 double mutant. (A) The adaxial view of mature leaves in wild type (R108), and mtphan, mtago7, mtphan;mtago7 and palm1 mutants (from left to right). The mtphan;mtago7 double-mutant leaves were palmate-like pentafoliate with narrow blades and elongated petiolules, in contrast to trifoliate leaves in wild type, and mtphan and mtago7 mutants. Scale bar, 1 cm. Red arrowheads indicate rachises in palm1 mutants. (B–E) Enlarged views of boxed regions in (A), showing the junction between lateral leaflets and petiole in R108 (B), mtphan (C), mtago7 (D) and mtphan;mtago7 (E). Scale bars: 2 mm. (F) A representative SEM image of adaxial epidermal cells of the petiole (boxed areas in B–D). (G) A representative SEM image of epidermal cells of pulvinus (red arrows in B–D). (H) SEM image of the boxed region from the mtphan;mtago7 mutant in (A). (I) SEM image of epidermal cells of petiolule (boxed regions in E,H) in the mtphan;mtago7 mutant. Note that epidermal cells of petiolule resemble epidermal cells of rachises in WT, and mtphan and mtago7 mutants. (J) SEM image of pulvinus (arrows in E; asterisks in H) in the mtphan;mtago7 mutant. (K,L) Quantitative RT-PCR analysis of expression of MtARF3 in vegetative shoot buds and young leaves (K) and PALM1 in vegetative shoot buds (L) in WT, mtago7, mtphan, and mtphan;mtago7 mutants. Note that the expression level of PALM1 is negatively correlated with that of MtARF3. Shown are means ± SD; n = 3.
Scanning electron microscopy analysis shows that all five leaflets of pentafoliate leaves in the mtphan;mtago7 double mutant were subtended by the motor organ, pulvinus, similarly as in WT (R108), and mtphan and mtago7 mutants (Figures 5B–J). In addition, the two lateral leaflets had elongated petiolules, resembling the phenotype of the palm1 mutant, particularly in the R108 background (Figures 5A,E,H) (Chen et al., 2010). Consistent with the leaf patterning phenotype, PALM1 expression was significantly downregulated in the vegetative shoot buds in the mtphan;mtago7 double mutants (Figure 5L). These results suggest that transcriptional and post-transcriptional mechanisms act synergistically in restricting the expression of MtARF3, which, in turn; negatively regulate the expression of PALM1.
RNA in situ hybridization results show that in the vegetative shoot apex in wild type, MtARF3 transcripts were detected in the SAM and the abaxial domain of leaf primordia (Figures 6B,C). Its expression domain is opposite from MtAGO7 (Figure 6A) (Hunter et al., 2006; Iwasaki et al., 2013; Zhou et al., 2013; Ge et al., 2014). In mtphan and mtago7 mutants, MtARF3 transcripts were detected in the adaxial domain as well as the abaxial domain (Figures 6D–G), consistent with the notion that both AGO7 (tasiR-ARF pathway) and PHAN restrict the ARF3 expression in the abaxial domain. Similarly, in the mtphan;mtago7 double mutants, MtARF3 transcripts were detected in both adaxial and abaxial domains of developing leaf primordia; and MtARF3 transcripts appeared much higher in the adaxial domain than the abaxial domain and corresponding domains in wild type, and mtphan and mtago7 single mutants (Figure 6).
FIGURE 6.
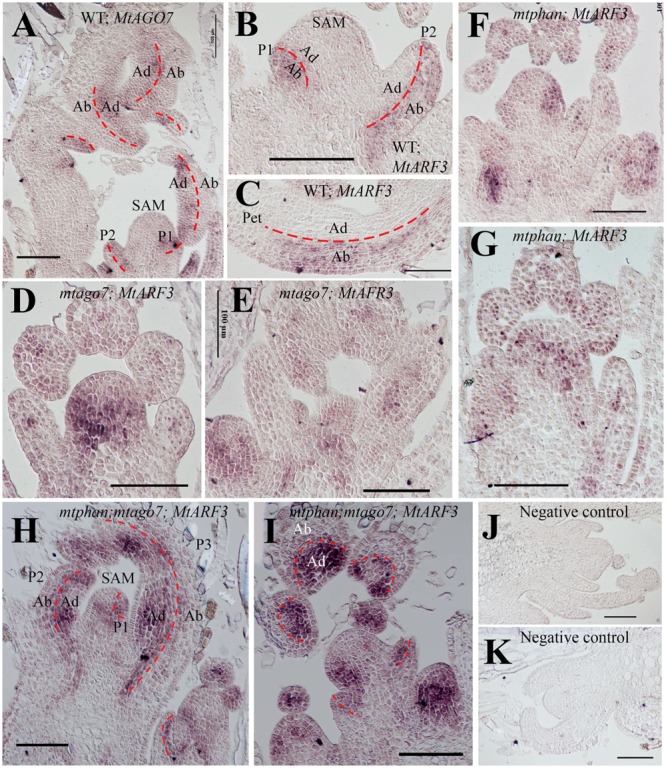
In situ hybridization analyses of MtARF3 and MtAGO7 gene expression in M. truncatula. (A) Localization of MtAGO7 transcripts in vegetative shoot buds in WT. (B,C) Localization of MtARF3 in vegetative shoot bud (B) and petiole of a developing leaf (C) in WT. Note that in wild type plants, MtAGO7 and MtARF3 expressed in opposite domains in leaf primordia, with MtAGO7 being expressed in the leaf adaxial domain and MtARF3 in the leaf abaxial domain. (D,E) Localization of MtARF3 gene expression in mtago7 mutant. MtARF3 gene expression was detected in both abaxial and adaxial domains of leaf primordia in the mtago7 mutant. (F,G) Localization of MtARF3 gene expression in mtphan mutant. MtARF3 gene expression was detected in both abaxial and adaxial domain of leaf primordia in the mtphan mutant. (H,I) Localization of MtARF3 gene expression in mtphan;mtago7 double mutant. MtARF3 gene expression was detected in both abaxial and adaxial domains of leaf primordia in the mtphan;mtago7 double mutant, with stronger signals in the adaxial domain. (J,K) Negative controls using sense probes in neighboring sections for detection of MtAGO7 (J) and MtARF3 (K). Scale bars: 100 μm. Dashed lines mark the boundary between the adaxial and abaxial domains. SAM, shoot apical meristem; P, plastochron; Ab, abaxial domain; Ab, abaxial domain.
Ectopic Lateral Leaflets Are Initiated at a Late Developmental Stage Similarly As in the palm1 Mutants
In the palm1 mutant, leaf developmental process is indistinguishable from wild type until the P3 stage, when an additional pair of lateral leaflet primordia (LLp) starts to initiate at the base of the lateral leaflet primordia (LLd) (Figures 7A–F) (Wang et al., 2008; Chen et al., 2010). We investigated the timing of initiation of the ectopic pair of lateral leaflets in the mtphan;mtago7 double mutant. SEM analysis shows that the common leaf primordia initiated normally in the mtphan;mtago7 double mutant as in wild type (Figure 7G) (Wang et al., 2008; Chen et al., 2010). However, subsequently, trichomes developed precociously before lateral leaflet and stipule primordia initiate. At the P2 stage, lateral leaflet primordia (LL) initiated at the marginal blastozone of the common leaf primordia (Figures 7G,H). And, lateral leaflets expanded and trichomes covered all of the terminal and lateral leaflet primordia (Figure 7H). Subsequently, at the P3 stage, an additional pair of lateral leaflet primordia (LLp) initiated at the base of the distal lateral leaflet primordia (LLd; Figure 7I). Although LLp initiated at similar position and time as in the palm1 mutant, they were narrower than those formed in the palm1 mutant starting from the P2 stage. In addition, stipule primordia were sometimes not initiated in the mtphan;mtago7 mutant. These results suggest that the initiation of ectopic lateral leaflets was similar between the mtphan;mtago7 double mutant and the palm1 mutant.
FIGURE 7.
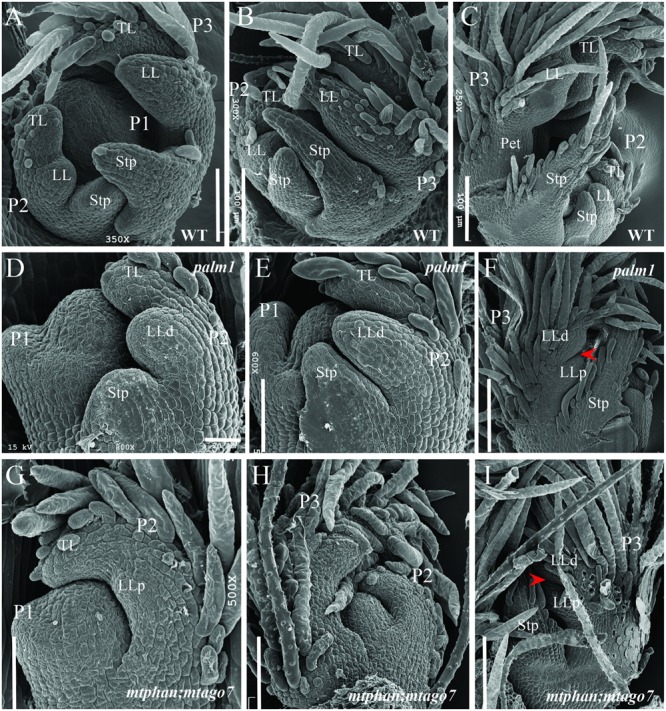
Scanning electron microscopic analyses of leaf primordia development in WT, and palm1 and mtphan;mtago7 mutants. SEM images of leaf development in WT (A–C), palm1 (D–F), and mtphan;mtago7 (G–I). Some trichomes were removed in order to view the boundary between LLd and LLp (I). Arrowheads (F,I) denote the boundary between LLd and LLp. P, plastochron; TL, terminal leaflet; LL, lateral leaflet; Stp, stipule; LLd, distal lateral leaflet; LLp, proximal lateral leaflet.
Leaves of the mtphan;mtago7 Double Mutant Were Abaxialized
Both PHAN and AGO7 regulate leaf adaxial-abaxial polarity (Xu et al., 2006; Iwasaki et al., 2013). In A. majus, loss of function phan mutant develops abaxialized needle-like leaves (Waites and Hudson, 1995). In crispa, a phan mutant in pea (Pisum sativum), compound leaves remain pinnate, with individual leaflets abaxialized (Tattersall et al., 2005). It has been shown that the mtphan mutant displays a mild polarity defect at a late developmental stage (Ge et al., 2014). SEM observation shows that the epidermal cells of the mtphan mutant is similar as wild type, with abaxial epidermal cells exhibiting the jigsaw puzzle-shaped border and adaxial epidermal cells exhibiting smoother border (Supplementary Figures S3A–C,F,G) (Ge et al., 2014). In the mtago7 mutant, both the adaxial and abaxial epidermal cells are slightly elongated compared with wild type, suggesting that the adaxial domain is abaxialized in the mtago7 mutant (Supplementary Figures S3A,B,D,F,H). In the mtphan;mtago7 double mutant, epidermal cells on the adaxial surface were slightly elongated, similar as those of the mtago7 mutant (Supplementary Figures S3A,B,D,E); epidermal cells in the proximal region on the abaxial surface were elongated with smooth borders, which are reminiscent of epidermal cells of midveins (Supplementary Figures S3I,J). Epidermal cells at the distal margin are radialized with elongated and smooth cells (Supplementary Figures S3K–M). These results are consistent with a role of PHAN and tasiR-ARF pathways in leaf adaxial-abaixal polarity regulation in plants.
SGL1 and NAM Are Required for Lateral Leaflet Development in mtphan;mtago7 Double Mutant
The Medicago LFY/UNI ortholog SGL1 is required for lateral leaflet initiation in M. truncatula (Wang et al., 2008; Chen et al., 2010; Peng et al., 2011). To test genetically the requirement of SGL1 in the development of pentafoliate leaves in the mtphan;mtago7 double mutant, we generated sgl1;mtphan;mtago7 triple mutant. Phenotypic analysis shows that sgl1;mtphan;mtago7 triple mutant plants developed simple leaves with narrow lamina (Figures 8A–D). Thus, the genetic interaction results confirm that the pentafoliate leaf phenotype of the mtphan;mtago7 double mutant is dependent on SGL1. Thus, the epistatic interaction between sgl1 and mtphan;mtago7 is similar to that between sgl1 and palm1 (Chen et al., 2010).
FIGURE 8.
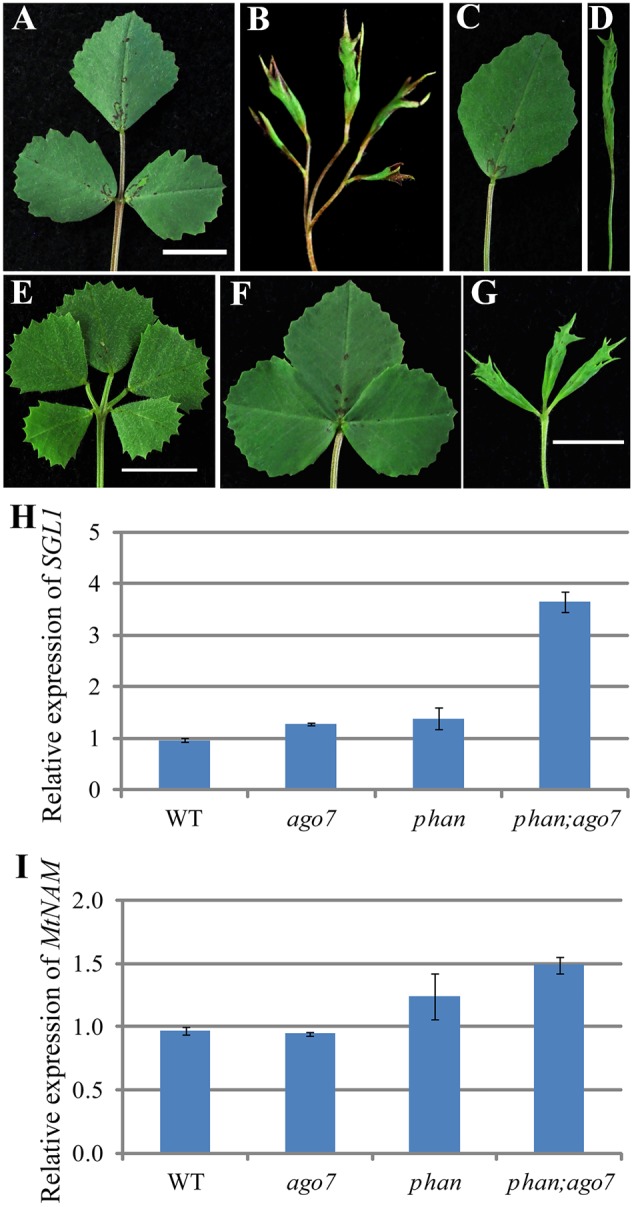
SGL1 and NAM are required for the development of ectopic lateral leaflets in the mtphan;mtago7 double mutant. Images of mature leaves in WT (A), mtphan;mtago7 (B), sgl1 (C), mtphan;mtago7;sgl1 (D), palm1 (E), mtnam (F), and mtphan;mtago7;mtnam (G). (E) Was the same as in Figure 5A. The mtphan;mtago7;sgl1 triple mutant develops simple leaves similarly as sgl1, and the mtphan;mtago7;mtnam triple mutant develops trifoliate leaves with greatly reduced rachis, resembling leaf phenotype of the mtnam mutant. Quantitative RT-PCR analyses of transcript levels of SGL1 (H) and MtNAM (I) in WT, and mtago7, mtphan, and mtphan;mtago7 mutants. Shown are means ± SD; n = 3.
In M. truncatula, NO APICAL MERISTEM (NAM)/CUP-SHAPED COTYLEDON2 (CUC2) has been shown to regulate organ boundary establishment (Cheng et al., 2012). Our phenotypic analysis shows that mtnam;palm1 double mutant exhibited primarily the phenotype of the mtnam/cuc2 mutant, indicating that nam/cuc2 is epistatic to palm1 (Supplementary Figure S4) To further test genetic interactions, we also generated mtnam;mtphan;mtago7 triple mutant. Phenotypic analysis shows that the mtnam;mtphan;mtago7 triple mutant developed trifoliate leaves with greatly reduced rachis, resembling the mtnam/cuc2 mutant phenotype (Figures 8E–G). The epistatic interaction shows that MtNAM/CUC2 is required for the elaboration of palmate-like pentafoliate leaves in the mtphan;mtago7 double mutant, similar to that in the palm1 mutant (Figures 8E–G and Supplementary Figure S4). Consistent with the phenotypic analyses, the transcript level of SGL1 is slightly increased in mtago7 and mtphan single mutants and greatly increased in the mtphan;mtago7 double mutant (Figure 8H), and the transcript level of MtNAM/CUC2 was significantly increased in the mtphan mutant and even more so in the mtphan;mtago7 double mutant (Figure 8I).
Discussion
In both simple and compound leaf species, initiation of leaf primordia requires downregulation of the class I knotted-like homeodomain proteins (KNOXI) in incipient leaf primordia. In compound leaf species, including tomato and C. hirsuta, the KNOXI genes are reactivated in developing leaf primordia and this reactivation is required for compound leaf development in these species. In some legume species including pea and M. truncatula, the KNOXI genes are not reactivated in developing leaf primordia and thus not likely involved in the development of compound leaves. Instead, UNI in pea and SGL1 in M. truncatula act in place of KNOXI in mediating the initiation of leaflet primordia in these legume species. In both simple and compound leaf species, it has also been shown that local gradients of auxin (auxin maxima) mediated by the auxin transport PIN-FORMED (PIN) proteins are required for the initiation and positioning of leaf primordia along the periphery of the SAM (Koenig et al., 2009; Peng and Chen, 2011; Vernoux et al., 2011). In compound leaf species, auxin maxima are also required for development of leaf margin serrations and initiation of leaflets (Peng and Chen, 2011; Ben-Gera et al., 2012). However, the underlying molecular mechanisms remain largely elusive.
Auxin signaling is dependent on AUXIN RESPONSE FACTORs (ARFs), which activate or repress downstream gene expression, and Aux/IAA transcription repressors that interact with ARF proteins. At a high auxin level, Aux/IAA proteins are ubiquitinated and degraded by the SCFTIR1/AFBs 26S proteasomes, and thus releasing their inhibitory effects on ARF’s transcriptional activities (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). In tomato that forms dissected leaves, the IAA9 protein, ENTIRE (E) accumulates between initiating leaflet primordia and is required for leaflet separation, since the loss-of-function e mutant exhibits simplified leaves compared with dissected leaves in wild type plants (Ben-Gera et al., 2012). In M. truncatula that forms trifoliate leaves, a loss-of-function mutation of PIN10, encoding an auxin efflux protein, results in altered positioning and fusion of common leaf primordia at the periphery of SAM and smooth leaflet margins (Peng and Chen, 2011; Zhou et al., 2011).
It has been shown that loss-of-function sgl1 or uni mutants develop simplified leaves (Hofer et al., 1997; Champagne et al., 2007; Wang et al., 2008). On the other hand, loss-of-function mutations in PALM1 result in dissected leaves with five leaflets in contrast to dissected leaves with three leaflets in wild type plants (Chen et al., 2010; Ge et al., 2010). PALM1 has been shown to directly interact with the SGL1 promoter sequence and negatively regulate transcription of SGL1, thus serving as a negative regulator of dissected leaf development in M. truncatula (Chen et al., 2010; Ge et al., 2010).
Here, we show that PALM1 gene expression is negatively regulated by AUXIN RESPONSE FACTOR3 (ARF3) in M. truncatula. Our bioinformatics analysis shows that there are 18 TGTCXX cis-elements, resembling the canonical auxin response element (AuxRE; TGTCTC) in the 2.5 kb promoter region of the PALM1 gene. We show that in the presence of three tandem repeats of several TGTCXX elements, ARF3 significantly repressed reporter gene expression in the transient transcription activity assay in tobacco. We noticed that there are different background activities when different elements were assayed in tobacco. We speculated that this may be due to the activity of endogenous ARF3 proteins in tobacco cells. By comparing the reporter gene expression in the presence of ARF3 and the unrelated β-glucuronidase (GUS) protein, we reasoned that any influence of background activity on the assay should be minimized. In M. truncatula, chromatin immunoprecipitation (ChIP) assay shows that P1, P2 and P4 regions, which harbor the TGTCXX elements, but not P3 region, which does not contain any TGTCXX elements, of the PALM1 promoter sequence were enriched in ARF3-bound chromatin. The P2 region of the PALM1 promoter sequence contain four TGTCXX elements. However, in the transcription activity assay in tobacco, three tandem repeats of individual elements did not confer ARF3 inhibitory effects on reporter gene expression. There may be several possibilities to account for the discrepancy between the two assay results: (1) three tandem repeats of the same element may not be optimal for the binding of ARF3 in tobacco cells; (2) the spacing between tandem repeats may not be optimal for ARF3 binding in tobacco; and (3) the heterologous system (tobacco cells) may not be optimal for ARF3 binding of the TGTCXX elements. Nevertheless, the combined results strongly support that M. truncatula ARF3 interacts with the TGTCXX cis-elements in the PALM1 promoter sequence in vivo.
Consistent with previous reports, the MYB domain protein, PHANTASTICA (PHAN), negatively regulates the expression of AUXIN RESPONSE FACTOR3 (ARF3) in Medicago plants and the trans-acting short interfering RNA, tasiR-ARF pathway negatively regulates the transcript level of ARF3 (Figure 9) (Iwasaki et al., 2013). Both PHAN and tasiR-ARF pathways mediate adaxial-abaxial polarity in diverse species. When both pathways are compromised as in the mtphan;mtago7 double mutant, the dissected leaf patterning changes from trifoliate to palmate-like pentafoliate leaves (Figure 5), in addition to narrow blades. The change in the dissected leaf patterning is associated with a drastically increased level of ARF3, decreased level of PALM1 (Figure 5) and a greatly increased level of SGL1 (Figures 8H, 9). Independent evidence that supports this conclusion includes (1) when the tasiR-ARF resistant ARF3m is highly overexpressed, dissected leaf patterning changes from trifoliate to palmate-like pentafoliate leaves, albeit with radialized blades (Figure 4); (2) sgl1;mtphan;mtago7 triple mutant exhibits simple leaves with narrow blades, similar to sgl1;palm1 double mutant (Figure 8) (Chen et al., 2010); and (3) nam;mtphan;mtago7 triple mutant exhibits trifoliate leaves with greatly reduced rachis, indicating that NAM is required for the development of ectopic lateral leaflets in the mtphan;mtago7 double mutant as in the nam;palm1 mutant (Figure 8 and Supplementary Figure S4).
FIGURE 9.
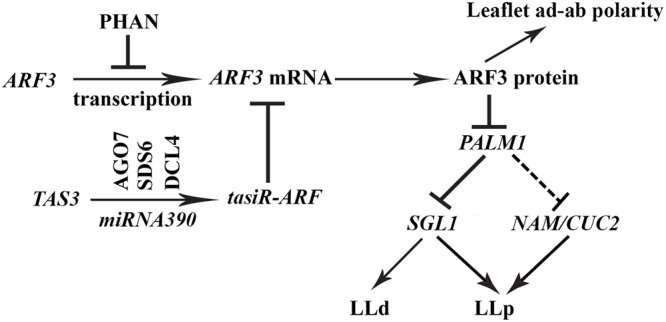
A genetic model illustrating the role of the ARF3-PALM1 module in dissected leaf development in M. truncatula. The MYB domain protein PHAN negatively regulates transcription of ARF3 and the trans-acting short interfering RNA, tasiR-ARF, whose biogenesis is dependent on the AGO7-SDS6-DCL4-miRNA390 pathway, negatively regulates the transcript level of ARF3. ARF3 in turn negatively regulate the expression of PALM1, a negative regulator of the Medicago LEAFY ortholog, SGL1, which is required for the initiation of lateral leaflets in M. truncatula. A fine-tuned expression of PALM1 and SGL1 is crucial for the development of trifoliate leaves in M. truncatula. NAM/CUC2, whose expression is repressed directly or indirectly by PALM1, is required for the initiation of proximal lateral leaflets (LLp) in the palm1 mutant. Solid and dashed lines denote confirmed and unconfirmed molecular interactions, respectively. LLd, distal lateral leaflets.
The PALM1-SGL1 module is important for the development of trifoliate leaves in M. truncatula. Our results that the PHAN and tasi-ARF pathways, which are known adaxial-abaxial regulators, are recruited to fine tune the expression level and domain of PALM1 and therefore regulate the compound leaf patterning in M. truncatula (Figure 9) are consistent with the observation that auxin and AS1 converge at the incipient leaf primordia to control leaf development in A. thaliana that has simple leaves (Hay et al., 2006). In conclusion, the multilayered regulation of gene expression suggests that a properly balanced expression of key regulators including PALM1, SGL1 and likely NAM/CUC2 is critical for the development of trifoliate leaves in the legume plant, and this process requires auxin signaling (Figure 9).
Author Contributions
JP and RC: designed experiments; JP and AB: performed the experiments; JP, FM, and RC: analyzed data; JP and RC: wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Preston Larson for assistance in SEM, Yewei Wang, Chunxiang Fu, and Qingzhen Jiang for assistance in plant transformation, and Guangming Li and Colleen Elles for assistance in genetic crossing and plant care.
Funding. Funding of this work was provided in part by The Samuel Roberts Noble Foundation and by grants from the Oklahoma Center for Advancement of Science and Technology (OCAST; PS12-036 and PS16-034) and the National Science Foundation (IOS-1127155). The laboratory of FM was funded by the Spanish Ministerio de Economía y Competitividad and FEDER (BIO2015-64307-R) and the Generalitat Valenciana (ACOMP2012-099).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01630/full#supplementary-material
References
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasciolli V., et al. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. 10.1016/j.cub.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A. M., Carrington J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40 1136–1141. 10.1038/ng.189 [DOI] [PubMed] [Google Scholar]
- Ben-Gera H., Shwartz I., Shao M. R., Shani E., Estelle M., Ori N. (2012). ENTIRE and GOBLET promote leaflet development in tomato by modulating auxin response. Plant J. 70 903–915. 10.1111/j.1365-313X.2012.04939.x [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Barley R., Curtis M., Arroyo J. M., Dunham M., Hudson A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. 10.1038/35050091 [DOI] [PubMed] [Google Scholar]
- Champagne C. E., Goliber T. E., Wojciechowski M. F., Mei R. W., Townsley B. T., Wang K., et al. (2007). Compound leaf development and evolution in the legumes. Plant Cell 19 3369–3378. 10.1105/tpc.107.052886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Moreau C., Liu Y., Kawaguchi M., Hofer J., Ellis N., et al. (2012). Conserved genetic determinant of motor organ identity in Medicago truncatula and related legumes. Proc. Natl. Acad. Sci. U.S.A. 109 11723–11728. 10.1073/pnas.1204566109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yu J., Ge L., Wang H., Berbel A., Liu Y., et al. (2010). Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 107 10754–10759. 10.1073/pnas.1003954107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Peng J., Ma J., Tang Y., Chen R., Mysore K. S., et al. (2012). NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytol. 195 71–84. 10.1111/j.1469-8137.2012.04147.x [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Emery J. F., Floyd S. K., Alvarez J., Eshed Y., Hawker N. P., Izhaki A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. 10.1016/j.cub.2003.09.035 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T. A., Howell M. D., Allen E., Dvorak S. K., Alexander A. L., et al. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. 10.1016/j.cub.2006.03.065 [DOI] [PubMed] [Google Scholar]
- Ferrandiz C., Gu Q., Martienssen R., Yanofsky M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 725–734. [DOI] [PubMed] [Google Scholar]
- Fukushima K., Hasebe M. (2014). Adaxial-abaxial polarity: the developmental basis of leaf shape diversity. Genesis 52 1–18. 10.1002/dvg.22728 [DOI] [PubMed] [Google Scholar]
- Garcia D., Collier S. A., Byrne M. E., Martienssen R. A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. 10.1016/j.cub.2006.03.064 [DOI] [PubMed] [Google Scholar]
- Ge L., Chen J., Chen R. (2010). Palmate-like pentafoliata1 encodes a novel Cys(2)His(2) zinc finger transcription factor essential for compound leaf morphogenesis in Medicago truncatula. Plant Signal. Behav. 5 1134–1137. 10.4161/psb.5.9.12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Chen R. (2014). PHANTASTICA regulates leaf polarity and petiole identity in Medicago truncatula. Plant Signal. Behav. 9:e28121 10.4161/psb.28121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L., Peng J., Berbel A., Madueno F., Chen R. (2014). Regulation of compound leaf development by PHANTASTICA in Medicago truncatula. Plant Physiol. 164 216–228. 10.1104/pp.113.229914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hareven D., Gutfinger T., Parnis A., Eshed Y., Lifschitz E. (1996). The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84 735–744. 10.1016/S0092-8674(00)81051-X [DOI] [PubMed] [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133 3955–3961. 10.1242/dev.02545 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38 942–947. 10.1038/ng1835 [DOI] [PubMed] [Google Scholar]
- Hellens R. P., Allan A. C., Friel E. N., Bolitho K., Grafton K., Templeton M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1 13 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J., Gourlay C., Michael A., Ellis T. H. (2001). Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Mol. Biol. 45 387–398. 10.1023/A:1010739812836 [DOI] [PubMed] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., et al. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7 581–587. 10.1016/S0960-9822(06)00257-0 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M. R., Wu G., Yoshikawa M., de la Luz Gutierrez-Nava M., Poethig S. R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981. 10.1242/dev.02491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezaki M., Kojima M., Sakakibara H., Kojima S., Ueno Y., Machida C., et al. (2010). Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 61 70–82. 10.1111/j.1365-313X.2009.04033.x [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Takahashi H., Iwakawa H., Nakagawa A., Ishikawa T., Tanaka H., et al. (2013). Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 140 1958–1969. 10.1242/dev.085365 [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize KNOTTED 1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Kaufmann K., Muino J. M., Osteras M., Farinelli L., Krajewski P., Angenent G. C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5 457–472. 10.1038/nprot.2009.244 [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Kidner C. A., Timmermans M. C. (2010). Signaling sides adaxial-abaxial patterning in leaves. Curr. Top. Dev. Biol. 91 141–168. 10.1016/S0070-2153(10)91005-3 [DOI] [PubMed] [Google Scholar]
- Koenig D., Bayer E., Kang J., Kuhlemeier C., Sinha N. (2009). Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136 2997–3006. 10.1242/dev.033811 [DOI] [PubMed] [Google Scholar]
- Kojima S., Iwasaki M., Takahashi H., Imai T., Matsumura Y., Fleury D., et al. (2011). Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 52 1259–1273. 10.1093/pcp/pcr083 [DOI] [PubMed] [Google Scholar]
- Li Y., Liu Z. B., Shi X., Hagen G., Guilfoyle T. J. (1994). An auxin-inducible element in soybean SAUR promoters. Plant Physiol. 106 37–43. 10.1104/pp.106.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Long J., Yamaguchi J., Serikawa K., Hake S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. 10.1105/tpc.6.12.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. A., Moan E. I., Medford J. I., Barton M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. 10.1038/379066a0 [DOI] [PubMed] [Google Scholar]
- Montgomery T. A., Howell M. D., Cuperus J. T., Li D., Hansen J. E., Alexander A. L., et al. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133 128–141. 10.1016/j.cell.2008.02.033 [DOI] [PubMed] [Google Scholar]
- Nakata M., Okada K. (2013). The leaf adaxial-abaxial boundary and lamina growth. Plants (Basel) 2 174–202. 10.3390/plants2020174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I., Alvarez J. P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17 2899–2910. 10.1105/tpc.105.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Chen R. (2011). Auxin efflux transporter MtPIN10 regulates compound leaf and flower development in Medicago truncatula. Plant Signal. Behav. 6 1537–1544. 10.4161/psb.6.10.17326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Yu J., Wang H., Guo Y., Li G., Bai G., et al. (2011). Regulation of compound leaf development in Medicago truncatula by fused compound leaf1, a class M KNOX gene. Plant Cell 23 3929–3943. 10.1105/tpc.111.089128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339 62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Schneeberger R., Tsiantis M., Freeling M., Langdale J. A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125 2857–2865. [DOI] [PubMed] [Google Scholar]
- Shani E., Burko Y., Ben-Yaakov L., Berger Y., Amsellem Z., Goldshmidt A., et al. (2009). Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21 3078–3092. 10.1105/tpc.109.068148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tattersall A. D., Turner L., Knox M. R., Ambrose M. J., Ellis T. H., Hofer J. M. (2005). The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. Plant Cell 17 1046–1060. 10.1105/tpc.104.029447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T. J. (1997a). ARF1, a transcription factor that binds to auxin response elements. Science 276 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T. J. (1997b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z. B., Hagen G., Guilfoyle T. J. (1995). Composite structure of auxin response elements. Plant Cell 7 1611–1623. 10.1105/tpc.7.10.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Brunoud G., Farcot E., Morin V., Van den Daele H., Legrand J., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7 508 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Waites R., Selvadurai H. R., Oliver I. R., Hudson A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 779–789. 10.1016/S0092-8674(00)81439-7 [DOI] [PubMed] [Google Scholar]
- Wang H., Chen J., Wen J., Tadege M., Li G., Liu Y., et al. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146 1759–1772. 10.1104/pp.108.117044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski M. F., Sanderson M. J., Steele K. P., Liston A. (2000). Molecular phylogeny of the “temperate herbaceous tribes” of papilionoid legumes: a supertree approach. Adv. Legume Syst. 9 277–298. [Google Scholar]
- Xu L., Yang L., Pi L., Liu Q., Ling Q., Wang H., et al. (2006). Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol. 47 853–863. 10.1093/pcp/pcj057 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Nukazuka A., Tsukaya H. (2012). Leaf adaxial-abaxial polarity specification and lamina outgrowth: evolution and development. Plant Cell Physiol. 53 1180–1194. 10.1093/pcp/pcs074 [DOI] [PubMed] [Google Scholar]
- Zhou C., Han L., Fu C., Wen J., Cheng X., Nakashima J., et al. (2013). The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell 25 4845–4862. 10.1105/tpc.113.117788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Han L., Hou C., Metelli A., Qi L., Tadege M., et al. (2011). Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant Cell 23 2106–2124. 10.1105/tpc.111.085464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Y., Sun Y., Wang Z. Y. (2012). Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). Methods Mol. Biol. 876 173–188. 10.1007/978-1-61779-809-2_14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


