Abstract
Soluble epoxide hydrolase (sEH) is a potential pharmacological target for treating hypertension, vascular inflammation, cancer, pain and multiple cardiovascular related diseases. A variable domain of the heavy chain antibody (termed sdAb, nanobody or VHH) possesses advantages of small size, high stability, ease of genetic manipulation, and ability for continuous manufacture, making such nanobody a superior choice as an immunoreagent. In this work, we developed an ultrasensitive nanobody based immunoassay for human sEH detection using polymeric horseradish peroxidase (PolyHRP) for signal enhancement. Llama nanobodies against human sEH were used as the detection antibody in sandwich ELISAs with polyclonal anti-sEH as the capture antibody. A conventional sandwich ELISA using a HRP labeled anti-HA tag as the tracer showed a marginal sensitivity (0.0015 OD•mL/ng) and limit of detection (LOD) of 3.02 ng/mL. However, the introduction of the PolyHRP as the tracer demonstrated a 141-fold increase in the sensitivity (0.21 OD•mL/ng) and 57-fold decrease in LOD (0.05 ng/mL). Systematic comparison of three different tracers in four ELISA formats demonstrated the overwhelming advantage of PolyHRP as a label for nanobody based immunoassay. This enhanced sEH immunoassay was further evaluated in terms of selectivity against other epoxide hydrolases and detection of the target protein in human tissue homogenate samples. Comparison with an enzyme activity based assay and a Western-blot for sEH detection reveals good correlation with the immunoassay. This work demonstrates increased competiveness of nanobodies for practical sEH protein detection utilizing PolyHRP. It is worthwhile to rediscover the promising potential of PolyHRP in nanobody and other affinity based methods after its low-profile existence for decades.
Keywords: Nanobody, Soluble epoxide hydrolase, PolyHRP, Immunoassay, Signal amplification, VHH
Graphical Abstract
INTRODUCTION
Enzyme linked immunosorbent assays (ELISA) have the advantages of high sensitivity, speed, simplicity, general applicability, low cost, high throughput and safety. They have been widely used for the detection of numerous analytes. Two key components that relate directly to the sensitivity of the immunoassay are the antibody and the tracer used for signal transduction. The surprising discovery of functional heavy-chain-only antibodies in camelids and cartilaginous fish1–3, led to recombinant expression of these heavy chain variable domains and the emergence of single domain antibodies (sdAb, VHH or nanobody)4. Nanobody based assays are increasingly attractive due to their small size (~15 kDa), monoclonal nature and high specificity. The single domain nature of nanobodies devoid of light chains possess many advantages over other antibodies, including ease of genetic manipulation, high thermostability, resistance to proteolysis, excellent solubility, ease of expression in various expression systems and relatively low cost for both discovery and continuous manufacture4. Moreover, nanobodies can be promising solutions to the reproducibility crisis of antibodies that both the academic and industrial communities are suffering from5. There is pressure that “all antibody reagents should be sequenced and then produced from the genetic code obtained”6. This drives increasing interest in nanobodies since they are easily sequenced and resynthesized. Nanobodies hold great potential as reagents for immunoanalytical applications. Immunoassays based on nanobodies have been developed for both small molecules and large analytes. Also, the simplicity of genetic manipulation allows production of nanobodies biotinylated in vivo7 or fused to enzyme tracers with the nanobody-enzyme fusion protein expressed8,9, resulting in fewer steps in the immunoassay. For small molecule detection, competitive immunoassays based on nanobodies generally give comparable sensitivity and detectability to that using the corresponding conventional IgG antibodies, with the half-maximum signal inhibition concentration (IC50) at sub ng/mL levels and LOD even lower, e.g. 3-phenoxybenzoic acid4, aflatoxin10, ochratoxin8, and tetrabromobisphenol A9,11. However, for analysis of large analytes, a number of sandwich immunoassays using nanobodies showed moderate performance for procalcitonin12, epidermal growth factor receptor13, Bacillus thuringiensis Cry1Ac and Cry1Fa toxins14,15, influenza H3N2 and H5N116,17, and hepatitis B surface antigen18, with the LOD in the ng/mL range or even much higher. Thus, it is often important to increase the sensitivity of nanobody based immunoassays.
Two basic approaches are increasingly used to improve sensitivity of ELISAs through enhancing loading of enzyme labels. One is to employ the classical multi-cycle addition of signal molecules, e.g. enzyme-anti-enzyme/dye complex19, biotin-(strept)avidin complex20, and immuno-polymerase chain reaction21. This approach often requires multiple labels and multi-cycle operations for introducing signal molecules (i.e., addition, incubation and separation), thus becoming relatively complicated, labor-intensive and time-consuming. The other approach is to use nanomaterials as the carriers of signal molecules due to their advantage of high surface to volume ratio, e.g., gold nanoparticles22, magnetic beads23, carbon nanotubes24, silica beads25, quantum dots26, nanospherical brushes27. Despite the increased loading area of signal molecules or detection antibodies, the activity of biomolecules may be compromised due to the steric-hindrance effect and random orientation when complexed to nanomaterials. Moreover, compared to the classical enzyme labeled antibodies, practical issues of nanomaterial complexes, including their complicated bioconjugation and separation, relatively low yield, slow diffusion kinetics, poor re-dispersion, long-term stability or storage, nanomaterial cost and potential environmentally unfriendly impact, limit their commercialization in immunoassay kits.
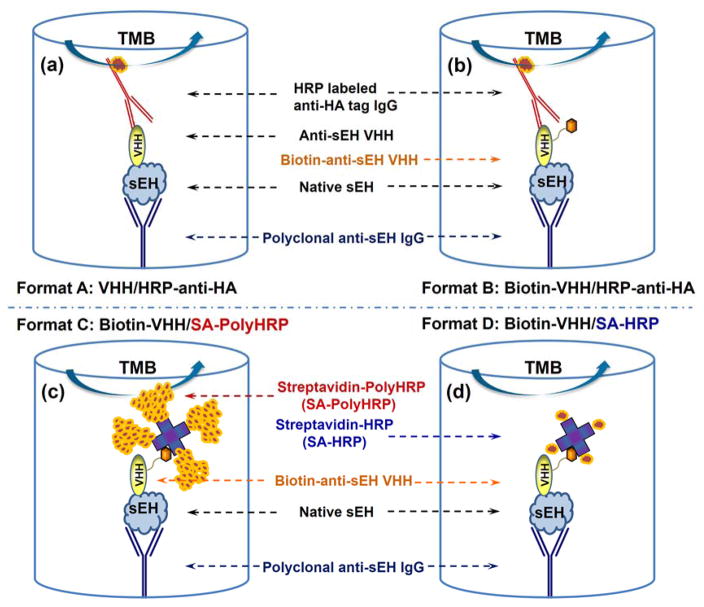
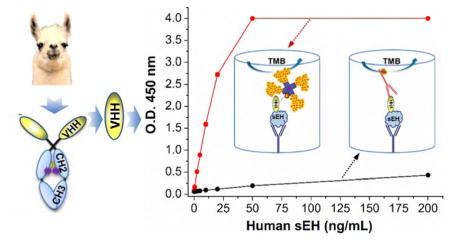
As a model to test ways of improving sensitivity for a protein target, we utilized a nanobody based sandwich immunoassay to detect the soluble epoxide hydrolase (sEH) protein. sEH protein in mammals is a 125 kDa dimer composed of two identical 62.5 kDa monomers. It is broadly distributed in multiple human tissues including brain, liver, kidney, heart and lungs. This enzyme is a potential pharmacological target for treating hypertension, vascular inflammation, cancer, pain and multiple cardiovascular related diseases28. Thus, the detection of sEH may represent an important diagnostic tool for human health. However, more information is needed on possible variations of sEH levels in different disease conditions and its diagnostic value. This work has been hampered by the lack of quantitative tests other than the semi-quantitative Western blots. Our group recently screened ten clones generating haemagglutinin (HA)-tagged nanobodies against human sEH from a llama VHH phage display library and developed a polyclonal/nanobody sandwich immunoassay for human sEH detection using a nanobody as the detection antibody29. Its format is shown in Figure 1a and this ELISA (format A) is termed conventional ELISA throughout the paper. Still, these nanobody based sandwich ELISAs showed moderate performance with LOD at ng/mL levels or higher. Our subsequent collaborative work developed a novel screening platform. Using the same VHH library as the previous work, new nanobodies were selected and paired for the sandwich detection of human sEH that resulted in a 100-fold increase in sensitivity30. Without rescreening the VHH library, and instead using the original ten nanobodies first acquired, in this work we developed an ultrasensitive nanobody based immunoassay for sEH using PolyHRP as the tracer with sensitivity increased over 100-fold. The assay format (termed PolyHRP ELISA) is illustrated in Figure 1c. PolyHRP is a supermolecular polymer of HRP containing up to 400 enzyme molecules at maximum that can be conjugated with various ligands and receptors (e.g. streptavidin) as shown in Figure S-1. This improved method is applicable to the nanobody based immunoassay with no need for further genetic manipulation. It can be helpful to all immunoassay practitioners not only with nanobodies but also other antibody fragment or full monoclonal or polyclonal antibodies. This paper investigates the factors that contributed to high sensitivity of the nanobody based PolyHRP ELISA through systematic comparison of four immunoassay formats, using native nanobody or biotinylated nanobody as the detection antibody and HRP labeled anti-HA tag, streptavidin-HRP conjugate or streptavidin-PolyHRP conjugate as the tracer (see Figure 1). Further evaluation of the nanobody based PolyHRP ELISA indicated significant advantages and promising potential for using PolyHRP in nanobody based immunoassays.
Figure 1.
Schematic comparison of four different sandwich ELISA formats for human sEH detection using the same rabbit polyclonal anti-sEH as capture antibody, (a) format A: native nanobody and HRP labeled anti-HA tag IgG (VHH/HRP-anti-HA), (b) format B: biotinylated nanobody and HRP labeled anti-HA tag IgG (Biotin-VHH/HRP-anti-HA), (c) format C: biotinylated nanobody and streptavidin-polyHRP conjugate (Biotin-VHH/SA-PolyHRP), (d) format D: biotinylated nanobody and streptavidin-HRP conjugate (Biotin-VHH/SA-HRP), as detection antibody and tracer, respectively.
EXPERIMENTAL SECTION
Safety
Personal protection equipment, especially safety glasses, should be worn when freezing aliquots of sEH stock solution with liquid nitrogen. The microtubes in liquid nitrogen can be cracked and the caps may fly with high velocity leading to injury.
Materials
Anti-human sEH VHHs and rabbit polyclonal antibody were prepared as described in our previous work29,31. Streptavidin-HRP conjugate (SA-HRP) was purchased from SouthernBiotech (Birmingham, Al). HRP labeled goat anti-HA tag polyclonal antibody conjugate (HRP-anti-HA) was purchased from Abcam (Cambridge, MA). Streptavidin PolyHRP40 conjugate (SA-PolyHRP) originally produced by Stereospecific Detection Technologies (SDT) GmbH (Baesweiler, Germany) was purchased from Fitzgerald Industries International (Concord, MA). Sources of other chemicals and biotinylation of anti-human sEH VHHs are detailed described in the Supporting Information.
Nanobody based sandwich ELISA for human sEH detection using PolyHRP as the tracer
The high-binding microplate was coated with anti-human sEH rabbit serum or purified rabbit polyclonal antibody in 0.05 M pH 9.6 carbonate-bicarbonate buffer (100 μL/well) overnight at 4 °C. After washing, the plate was blocked with 3% (w/v) skim milk (300 μL/well) in phosphate buffered saline (PBS) for 1 h and subsequently washed. Serial concentrations of human sEH standards in PBS containing 0.1 mg/mL bovine serum albumin (BSA) were then added to the wells (100 μL/well), followed immediately by biotinylated nanobodies (biotin-VHHs) in PBS (100 μL/well). The immunoreaction was allowed to proceed for 1 h. After washing, the microplate was incubated with SA-PolyHRP in PBS (100 μL/well) for 30 min. After the final washing, 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (100 μL/well) was added and the plate was incubated for 10 min. The optical density (O.D.) was recorded at 450 nm within 10 min after stopping the color development with 2 M sulfuric acid (100 μL/well). All incubations unless otherwise specified were conducted at room temperature (RT) with shaking (600 rpm) on the MTS 2/4 digital microtiter shaker of 4-plate capacity (IKA, Germany) and each washing step involved three washings with PBS containing 0.05% Tween-20 (PBST, 300 μL/well) using the plate washer.
Comparison of four ELISA formats using different detection nanobodies and tracers
As shown in Figure 1, comparison of four sandwich ELISA formats using different tracers were performed. The ELISAs were run as described above for the PolyHRP based format with minor modifications. Anti-human sEH rabbit polyclonal serum (1:2,000 dilution) was coated as the capture antibody. Biotinylated nanobodies (biotin-VHH A1 or biotin-VHH B3, 1 μg/mL) were used as the detection antibody for format B (Figure 1b), C (Figure 1c) and D (Figure 1d), while unconjugated native nanobodies (VHH A1 or B3, 1 μg/mL) from the same batch of nanobodies used for biotinylation, was utilized for format A (Figure 1a). HRP-anti-HA (1:10,000 dilution, 100 ng/mL), SA-HRP (1:5,000 dilution, 80 ng/mL) and SA-PolyHRP (1:40,000 dilution, 25 ng/mL) were used as the tracer of the format A/B, D and C, respectively. To avoid plate to plate variability, the four formats were performed on the same plate. Each concentration had three replicates. Plate columns 1–3, 4–6, 7–9 and 10–12 were used for format D, C, B and A, respectively. The same reagents involved for similar steps of different formats were used from the same batch prepared. (i.e. buffers, capture serum, skim milk, TMB substrate, sulfuric acid for all formats). Capture serum, human sEH standards, native or biotinylated nanobody, and the tracers at working concentrations were freshly prepared from their stored aliquots within 30 min before addition. A 12-channel pipette and 12-channel pipette reservoir (Figure S-2) were used to ensure simultaneous addition of serial human sEH standards along the plate row direction, and various detection antibodies or tracers along the plate column direction. All same or similar steps of different formats were operated at the same time under the same conditions (e.g., coating, blocking, color development and stopping). A further evaluation using three plates simultaneously was done on another day with four ELISA formats performed as above on each plate. Addition of reagents from the same batch and incubation conditions for three plates were kept the same to minimize the plate-to-plate variation (e.g. simultaneous shaking on the same plate shaker).
Performance comparison using decreasing concentrations of antibodies
Comparisons of the four ELISA formats were done similarly to that described above, but with a serial 2-fold decrease in working concentrations of detection antibodies or tracers. Four plates were used. The human sEH standards were narrowed to concentrations of 0–100 ng/mL. Decreasing capture serum was coated on plate I (1:2,000–1:8,000 dilution) and plate II (1:16,000–1:64,000 dilution). Decreasing detection antibodies (biotinylated or native VHH A1) were used on plate III (1.0–1/4 μg/mL) and plate IV (1/8–1/32 μg/mL). The other unspecified conditions were the same as described above. Immunoassays were run simultaneously for four plates with incubation conditions kept the same.
Cross-reactivity
The cross-reactivity or selectivity of the nanobody based PolyHRP ELISA was determined by its selectivity coefficient (KA,I) with a group of epoxide hydrolases. The selectivity coefficient was calculated with the equation as follows: KA,I (%) =[k(interferent)/k(human sEH)]×100. It is a measure of the sensitivity of a method for an interferent relative to that for the analyte32. The working concentrations for capture serum, biotin-VHH A1 and SA-PolyHRP were 1:2,000 dilution, 1.0 μg/mL and 25 ng/mL, respectively.
Matrix Effects
The above nanobody based immunoassay using PolyHRP was applied to liver cytosol samples from sEH knockout (KO) mice. Evaluation of the matrix effect was performed following a simple dilution protocol. Briefly, the liver cytosol samples were diluted with PBS containing 0.1 mg/mL BSA to dilutions of 1:100, 1:500 and 1:1000. Then, a series of sEH solutions were spiked into each diluted samples. The resulting spiked samples were directly analyzed with the nanobody based PolyHRP ELISA.
Real Sample Analysis
Six tissue samples consisting of the S9 fractions of pooled (4–50 individuals) human tissues were taken to make serial dilutions using PBS containing 0.1 mg/mL BSA. Their sEH concentrations were determined using the nanobody based PolyHRP ELISA described above. Dilution conditions leading to the measured O.D. in the linear range of the generated calibration curve were used to calculate the human sEH content in the samples.
RESULTS AND DISCUSSION
Selection of SA-PolyHRP concentration
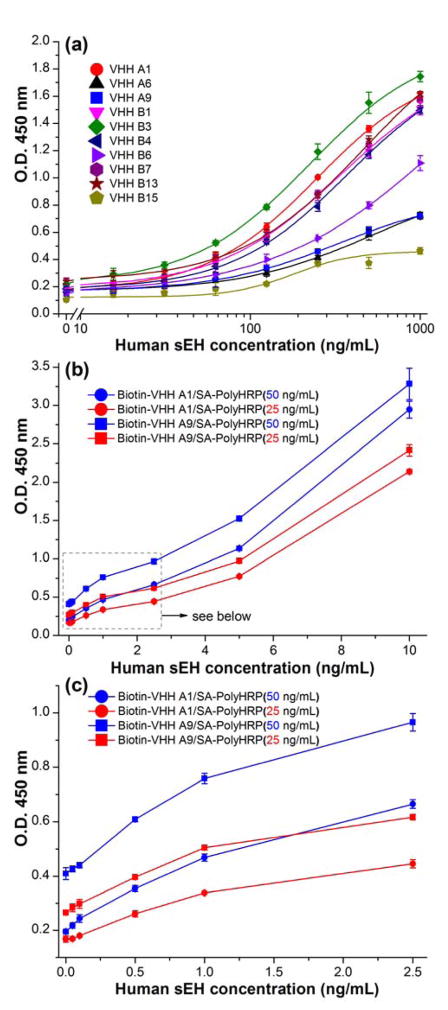
Figure 2a shows the signal response of our previously developed conventional ELISAs for human sEH using ten nanobodies (1.0 μg/mL) as the detection antibodies and HRP-anti-HA (200 ng/mL) as the tracer. It reveals values of O.D. signal below 2.0 at relatively high analyte concentration of 1,000 ng/mL for all the ten nanobodies based conventional ELISAs. However, the switch of tracer from the HRP-anti-HA (format A) to SA-PolyHRP (format C) results in a large improvement. As shown in Figure 2b, the nanobody (VHH A1 or A9, 1.0 μg/mL) based ELISAs plus SA-PolyHRP (25 or 50 ng/mL) gives much stronger signals, with O.D. values all over 2.0 at relative low human sEH concentration of 10 ng/mL. These signal responses are over 100-fold stronger than the ones in previous conventional ELISAs against the sEH of the same concentration. Virtually, similar signal enhancement was also observed for the other eight VHHs based immunoassays when PolyHRP was employed (Figure S-3). Moreover, in the lower human sEH range (0–2.5 ng/mL), Figure 2c shows appreciable signal response even below 0.1 ng/mL, indicating the high efficiency of the PolyHRP tracer. As expected, a higher concentration of tracer gave a higher O.D. response. The SA-PolyHRP 50 ng/mL (blue) generated signal higher than that of 25 ng/mL (red) but also rendered higher background. Considering the lower background and the adequate signal response, the 25 ng/mL was therefore adopted as the working concentration of SA-PolyHRP throughout. In addition, VHH A1 was selected as the detection nanobody for further comparison due to its high yield of expression, good affinity and relatively low background when used in the assay.
Figure 2.
Signal response of nanobody based ELISA formats toward human sEH using rabbit anti-sEH polyclonal antibody (4.5 μg/mL) as capture antibody: (a) format A: conventional ELISA format using ten native VHH clones (1 μg/mL) as detection antibody and HRP labeled anti-HA tag IgG (200 ng/mL) as the tracer. This graph is plotted based on the data from work of ref 29; (b) format C: PolyHRP based format using biotinylated nanobody (A1 or A9, 1 μg/mL) as the detection antibody and SA-PolyHRP (25 or 50 ng/mL) as the tracer; (c) expanded low concentration range of Figure 2b. Error bars indicate standard deviations (n = 3).
Experimental comparison of the performance of four ELISA formats
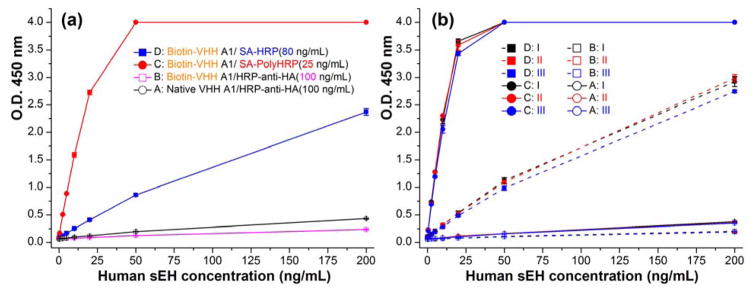
“A major problem that has plagued the literature concerning immunoassays is a lack of standardized terminology and practices when comparing the performance characteristics of various assay designs, and in particular estimates of their sensitivity”33. Loose use of terms like sensitivity often leaded to confusion34. In immunoassay, sensitivity may be described in a variety of ways35. Classically, for a dose–response curve, sensitivity is the change in response (dR) per unit of dose (dC) and equals dR/dC (not necessarily constant)34. This definition is used throughout this paper and should not be confused with the limit of detection (LOD). The LOD is the smallest amount of analyte that can be determined with confidence and therefore is a statistical parameter32. The LOD = 3SB/k refers to the computed analyte concentration corresponding to signal response of the blank plus three times of its standard deviation and is used throughout this work, where k is the slope or sensitivity and i is the intercept and SB is the standard deviation of the blank, for a linear regression analysis expressed as y = kx + i. This should be distinguished from the LOD estimated based on a specified signal-to-noise ratio36. Moreover, comparison between assays based on literature reports can be difficult since the sensitivity is influenced by assay parameters such as volume, incubation time, numbers of replicates or integration of signal detection33. Thus, assay conditions in this work were rigorously controlled so that meaningful comparison of the four tested ELISA formats could be made. Due to the limited supply of the purified polyclonal antibody, serum was thereafter used as the capture antibody considering the adequate signal and lower background generated. Figure 3a shows the calibration curves of the four formats. The PolyHRP based format C (Biotin-VHH/SA-PolyHRP) demonstrated the strongest signal response, with the O.D. saturated at 50 ng/mL of analyte (Note: The SpectraMax microplate reader is capable of reporting a maximum O.D. of 4.0). The conventional ELISA (format A) showed an O.D. around 0.4 at 200 ng/mL of human sEH, higher than format B (Biotin-VHH/HRP-anti-HA) with O.D. around 0.2 at the same target concentration. Format D (Biotin-VHH/SA-HRP) gave a signal response between the conventional ELISA and the PolyHRP based assay, generating an O.D. close to 2.4 at 200 ng/mL. The sensitivity trend for the four formats is C>D>A>B. To systematically evaluate variation within and between plates, the repetition of the ELISAs comparison on three plates further demonstrated the trend. As shown in Figure 3b, similar responses to those described above were obtained for the four formats in all the plates, indicating good reproducibility of the comparison results. For quantitative comparison of the performances between the assays in terms of sensitivity and LOD, data processing and analysis including curve-fitting is of great importance. As the simplest regression model, the linear model can be influenced by many factors, e.g. concentration range covered, number and distribution of standards, replication and preparation of standards, calibration model with non-zero or zero intercept, and weighting procedures. However, this aspect is rarely given in detail for many research papers. In this paper, an unweighted linear model was used for fitting the data. Figure S-5 shows the resulting linear-fitting curves with related parameters generated for the four ELISA formats on three plates. As summarized in Table S-2 and Figure S-6, the average sensitivity (coefficient of variation, CV) for format A, B, C, and D is 0.0015 (5%), 0.0007 (7%), 0.21 (6%), and 0.0196 (7%) OD•mL/ng, respectively. The corresponding average LOD (CV) for format A, B, C, and D is 3.02 (30%), 13.4 (88%), 0.05 (1%), and 0.56 (60%) ng/mL, respectively. The format B showed a 0.45 fold sensitivity and 0.22 fold lower LOD compared to the conventional ELISA (format A), suggesting partial inactivation of the detection nanobody or partial loss of epitopes for HRP-anti-HA after the biotinylation. Still, the introduction of SA-HRP (format D) demonstrated a 13 fold higher sensitivity and 5.4 fold lower LOD than the conventional ELISA. The extraordinarily high affinity (1015 M−1) between the streptavidin-biotin complexes likely accounts for the improved assay performance, favoring the efficient binding of HRP conjugated to the streptavidin to the biotinylated detection nanobody. Moreover, the sensitivity difference between formats A, B and D also demonstrated the relative inefficiency of the HRP-anti-HA as the tracer for immunoassays using a nanobody as the detection antibody. With respect to format C, this PolyHRP ELISA displayed the best performance. It gave a 141 fold higher sensitivity and 57 fold lower LOD than the conventional ELISA. This superior performance was likely due to both the high affinity of the streptavidin-biotin complex and the high HRP loading density of PolyHRP. Considering the relatively low protein working concentration (25 ng/mL) used, SA-PolyHRP appears to be a superior tracer compared to the others tested here. In addition, the assay precision was evaluated for well-to-well and plate-to-plate variation as shown in Table S-2. The well-to-well precision was 4.2%, 3.5%, 1.5%, and 3.7% for format A, B, C, and D, respectively. The plate-to-plate precision was 23.4%, 29.1%, 5.5%, and 25.5% for format A, B, C, and D, respectively. Interestingly, the PolyHRP ELISA displayed the highest reproducibility in terms of both well-to-well (1.5%) and plate-to-plate (5.5%) precision. It also showed smallest CV in LOD (1%) and acceptable CV in sensitivity (6%) on the three plates. The high precision of the PolyHRP ELISA may be attributed to the high signal-to-noise ratio because the high HRP loading gave a large signal and the relatively low protein working concentrations used resulted in a low background. Also, the performance comparison of different assay formats under the exact the same conditions can be meaningful, because small difference can be seen even in the same formats between different days (Figure 3a,b). In addition, as summarized in Table S-2 and Figure S-6, the CV of sensitivity among the three plates for format A, B, C and D is 5%, 7%, 6% and 7%, respectively, while the CV of LOD among the three plates for the format A, B, C and D is 30%, 88%, 1% and 60%, respectively. This suggests selection of sensitivity as the comparison index between different assays can be more robust and meaningful since it shows the small CV range (5–7%) compared to that (1–88%) of LOD. LOD is not only related to the assay sensitivity but also to the variation in blank background readings, which may vary largely between different plates. Thus, the performance comparison between different assays based on the ratio of LOD may result in relatively high uncertainty.
Figure 3.
(a) Calibration curves of four ELISA formats on the same plate using anti-sEH polyclonal serum (1:2,000 dilution) as the capture antibody, native or biotinylated VHH A1 (1 μg/mL) as the detection antibody, and HRP-anti-HA (100 ng/mL), SA-HRP (80 ng/mL) and SA-PolyHRP (25 ng/mL) as the tracer of the format A/B, D and C, respectively; (b) Repeat of the comparison of the four ELISA formats on three plates (plate I, II, III). The same reagents involved for similar steps of different formats are used from the same batch prepared. All same or similar steps of different formats were performed at the same time under the same conditions. Error bars indicate standard deviations (n = 3).
ELISAs comparison in decreasing working concentrations of antibodies
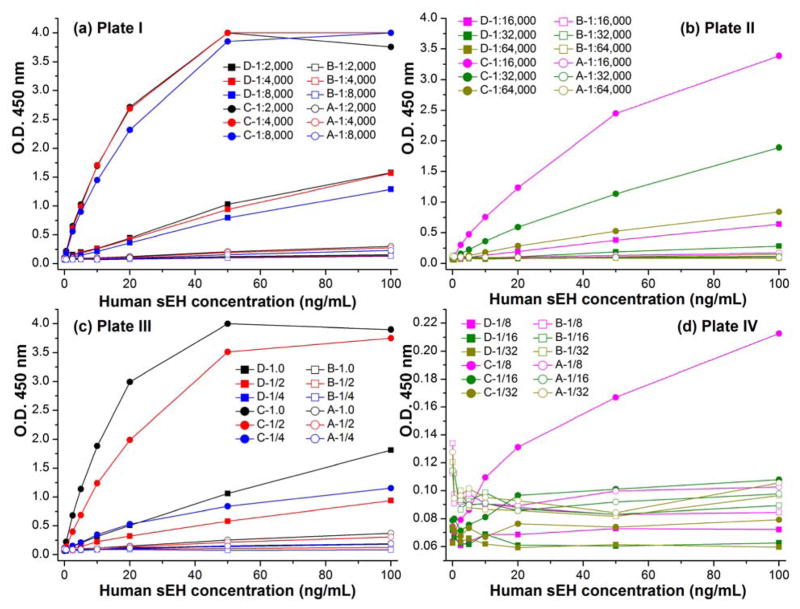
The nanobody based PolyHRP ELISA format revealed advantages in sensitivity, LOD and robustness. This can be vital in removing matrix effects for real sample detection since it allows relatively high dilution of samples due to its high sensitivity. Also, it conserves reagents. Figure 4a,b and Figure S-7 (plate I-II) showed the performance of the four ELISA formats with decreasing concentrations of the capture serum. As expected, the signal responses decreased with the decreasing capture serum in the serial calibrators range. The ELISA format A and B completely lost their signal response even with high calibrator concentration when the capture serum was diluted more than 1:8,000. Still, the PolyHRP based format C showed adequate and clearly increasing O.D. response with increasing sEH concentrations even in a dilution of 1:64,000 for capture serum. Its performance is also superior to that of the format D with the serum dilution of 1:16,000. Further dilution of capture serum for format D did not give an appreciable O.D. response trend with increasing sEH concentrations. On the other hand, as shown in Figure 4c,d and Figure S-7 (plate III–IV), four ELISA formats displayed decreasing O.D. response with decreasing working concentrations of the detection nanobody. Similarly, the O.D. response was lost for format A and B at high calibrator concentrations when the detection nanobody was diluted lower than 1/4 μg/mL. Format D demonstrated better performance. A clear and adequate signal response trend was seen when the working concentration of biotinylated VHH A1 was down to 1/2 μg/mL. However, no appreciable signal response trend was observed when the detection nanobody was further diluted lower than 1/4 μg/mL, while the PolyHRP based format C still showed quite a clear signal response trend at the relatively low nanobody concentration of 1/8 μg/mL. An appreciable signal response trend was also observed for format C when the detection nanobody further went down to 1/16 μg/mL. Thus, among the four ELISA formats the nanobody based PolyHRP ELISA demonstrated the best performance, which allowed the use of much less of both capture and detection antibodies. The corresponding potential for cost decrease can be very practical to the assay application.
Figure 4.
Comparison of the four ELISA formats with the decreasing working concentrations of (Figure 4a,b) capture serum (1:2,000–1:64,000 dilution) or (Figure 4c,d) detection nanobody (1.0–1/32 μg/mL). The O.D. decreased slightly below 4.0 in higher concentrations than the sEH saturated concentration due to the partial degradation of the over-saturated product of color development (see Figure 4a,c).
Cross-reactivity
The cross-reactivity was evaluated by comparing the selectivity coefficient value of human sEH with that of a group of epoxide hydrolases using PolyHRP ELISA with VHH A1 as the detection antibody and rabbit anti-human sEH polyclonal serum as the capture antibody (see Table 1). The assay showed slight cross-reactivity of 1.0%, 2.4% and 0.8% against affinity purified recombinant mouse sEH, rat sEH and denatured human sEH, respectively. Negligible cross-reactivity (<0.1%) was observed for horse sEH hepatic S9, dog sEH hepatic S9, liver cytosol from sEH KO mice, purified recombinant human mEH, and recombinant human EH3. Overall, the nanobody based PolyHRP ELISA demonstrated excellent selectivity for the target. In particular, the high specificity of the nanobodies used against undenatured human sEH allows detection of only the bioactive form with negligible interference from potentially co-occurring human mEH and human EH3.
Table 1.
Cross-reactivity of nanobody based PolyHRP ELISA for human sEH with sEH of other species and other human epoxide hydrolases.
| Epoxide hydrolases (puritya) | Cross-reactivity (%) | Epoxide hydrolases (purity) | Cross-reactivity (%) |
|---|---|---|---|
| Human sEH (>95%) | 100 | Dog sEH hepatic S9 (~20%) | <0.1 |
| Mouse sEH (>95%) | 1.0 | Liver cytosol from sEH KO mice | <0.1 |
| Rat sEH (>95%) | 2.4 | Human mEH (~80%) | <0.1 |
| Denatured human sEH (>95%) | 0.8 | Human EH3 (~4%) | <0.1 |
| Horse sEH hepatic S9 (~20%) | <0.1 | -- | -- |
purity was determined by densitometry on SDS-PAGE using recombinant affinity purified enzyme except for equine and canine enzyme partially purified by ion exchange chromatography from liver homogenates.
Matrix effects
Spike-and-recovery tests are a crucial method for validation and evaluation of the accuracy of ELISA for certain sample types. It can identify whether the analyte analysis is influenced by a difference between the diluent for calibration curve preparation and the biological sample matrix. One of the most common ways to minimize matrix effect is to dilute the sample with the assay buffer. The higher dilution of samples requires higher assay sensitivity since the target analytes in practical samples may be accordingly diluted. Varying dilutions of the samples may lead to different extents of removal of matrix effects. As shown in Table 2 and Figure S-8, the background decreased and the net signal increased with increasing dilutions of the sample matrix of sEH free liver cytosol. The recovery was 74–91%, 83–96% and 91–142% for the spiked samples prepared at 1:100, 1:500 and 1:1000 dilution of the sample matrix, respectively. The corresponding overall recovery (namely the slope of the linearly-fitted equation between the sEH spiked and that detected) was 83%, 85% and 91%, respectively. These data support the acceptance of nanobody based PolyHRP ELISA for sEH detection in biological samples.
Table 2.
Recovery of pure human sEH spiked into the hepatic cytosol samples from sEH knockout mice (n = 3)a.
| spiked (ng/mL) | 1:100 dilution | 1:500 dilution | 1:1000 dilution | |||
|---|---|---|---|---|---|---|
| detected (ng/mL) | recovery | detected (ng/mL) | recovery | detected (ng/mL) | recovery | |
| 0.00 | 0.78 ± 0.09 | -- | 0.16 ± 0.00 | -- | 0.06 ± 0.00 | -- |
| 0.25 | 0.96 ± 0.08 | 74% | 0.36 ± 0.01 | 83% | 0.41 ± 0.06 | 142% |
| 0.50 | 1.19 ± 0.02 | 84% | 0.64 ± 0.11 | 96% | 0.61 ± 0.06 | 111% |
| 1.00 | 1.59 ± 0.06 | 81% | 0.96 ± 0.01 | 81% | 1.26 ± 0.03 | 120% |
| 2.00 | 2.46 ± 0.16 | 84% | 2.03 ± 0.07 | 94% | 1.99 ± 0.16 | 97% |
| 4.00 | 4.43 ± 0.36 | 91% | 3.72 ± 0.11 | 89% | 4.08 ± 0.17 | 101% |
| 6.00 | 5.82 ± 0.41 | 84% | 5.30 ± 0.31 | 86% | 5.50 ± 0.26 | 91% |
| 8.00 | 7.33 ± 0.82 | 82% | 6.96 ± 0.56 | 85% | 7.48 ± 0.45 | 93% |
sEH activity was not detected in unspiked liver cytosol samples from sEH KO mice as described in ref 39.
Analysis of human tissue samples
A successful analytical method must meet the requirement of detecting real samples with high accuracy19. The nanobody based PolyHRP ELISA was employed to analyze the sEH in the S9 fractions of pooled (4–50 persons) human tissue samples. The proper dilution factor used in the assay for tissues of liver, kidney, lung (nonsmoker), lung (smoker), intestine and renal was 5000, 2000, 50, 50, 2000 and 1000 fold, respectively. The resultant data were compared with that obtained through the enzyme activity based radiometric assay, Western-blot, conventional ELISA and ELISA NbS7/NbS43 (Table 3). The PolyHRP ELISA, ELISA NbS7/NbS43, conventional ELISA and the Western-blot all showed good correlation to the classical enzyme activity based method in these tissue samples (Figure S-9) with correlation coefficient R2 > 0.93. Thus, the quantitation of the sEH protein content through these antibody based methods are good reflection of its enzyme activity. The assays in this study were optimized using affinity purified human recombinant enzyme. The sequence of this enzyme represents the most common form in the human population, however there are a number of other single nucleotide polymorphisms (SNPs) in the human population. All of these SNPs are relatively rare and the changes in catalytic rate with the sEH are small when screened with several substrates37,38. However, the nanobodies reported here have not yet been screened against human sEH enzymes with these SNPs, and this could result in a lack of correlation between the protein level determined by ELISA and the catalytic activity.
Table 3.
Comparison of PolyHRP ELISA and four other methods for the analysis of human sEH in the S9 fraction of pooled (4–50 persons) human tissues samples.
| Human tissues | sEH concentration (nM) estimateda | ||||
|---|---|---|---|---|---|
| Enzyme activityb | Western blotb | Conventional ELISAc | ELISA NbS7 /NbS43d | PolyHRP ELISA | |
| Liver | 420 ± 89 (21%) | 500 ± 150 (30%) | 386 ± 48 (12%) | 468 ± 22 (5%) | 691 ± 18 (3%) |
| Kidney | 44 ± 3 (7%) | 80 ± 15 (19%) | 44.4 ± 1.6 (4%) | 37 ± 3 (8%) | 76.3 ± 0.7 (1%) |
| Lung (nonsmoker) | 3.0 ± 0.3 (10%) | 22 ± 6 (27%) | 6.1 ± 0.3 (5%) | 2.9 ± 0.1 (3%) | 3.60 ± 0.12 (3%) |
| Lung (smoker) | 2.8 ± 0.3 (11%) | 21 ± 4 (19%) | 5.9 ± 0.1 (2%) | 2.1 ± 0.2 (10%) | 4.07 ± 0.03 (1%) |
| Intestine | 80 ± 9 (11%) | 110 ± 10 (9%) | -- | 121 ± 6 (5%) | 131 ± 2.5 (2%) |
| Renal | -- | -- | -- | -- | 44.2 ± 0.4 (1%) |
In addition, the PolyHRP ELISA revealed better precision (CV, 1–3%) than the other assays (enzyme activity, 7–21%; Western-blot, 9–30%; conventional ELISA, 2–12%; ELISA NbS7 /NbS43, 3–10%). The precision variation may be due to both the intrinsic limitations of the assay types and the difference in experience of the assay operators. The small CV of the PolyHRP ELISA demonstrated here was consistent with its performance in the aforementioned comparison of four ELISA formats, favoring its promise for practical detection with high accuracy. Also, the underlying value of PolyHRP as tracer was demonstrated. Despite being available for many years, PolyHRP has been vastly under-used an enzyme label in ligand binding assays. A summary of PolyHRP development was done in the Supporting Information.
CONCLUSION
In summary, we developed an ultrasensitive nanobody based immunoassay for human soluble epoxide hydrolase using PolyHRP for signal enhancement. Compared with the conventional nanobody based immunoassay employing the HRP labeled anti-HA tag as tracer, the PolyHRP based immunoassay showed a 141-fold higher sensitivity and 57-fold lower LOD of the conventional ELISA, with the sensitivity improved from 0.0015 OD•mL/ng to 0.21 OD•mL/ng and LOD from 3.02 ng/mL to 0.05 ng/mL for human sEH detection. Four ELISA formats using native or biotinylated nanobody as detection antibody and HRP-anti-HA tag, streptavidin-HRP or streptavidin-PolyHRP as tracer were systematically compared in terms of sensitivity, LOD and decreasing working concentrations of antibodies. The results revealed the difference of various tracers in capacity of signal production, with PolyHRP displaying the best performance. This further indicated the advantages of PolyHRP in immunoassays in terms of high-efficiency of signal enhancement, simplicity, robustness and availability. Also, the simple introduction of PolyHRP provided excellent competitiveness in both sensitivity and detectability for nanobody based immunoassays compared the classical IgG based format while maintaining benefits of the promising nanobody in gene engineering and thermo-chemical stability. Although only human sEH was detected in this work, the PolyHRP based immunoassays are readily extended to other nanobody based ELISA systems or other affinity based systems.
Supplementary Material
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (81402722) and National Institute of Health (Superfund P42ES04699, R01 MH106781, and R01 ES002710). The support from China Postdoctoral Science Foundation (2013T60601, 2012M521182) is also acknowledged. D.L. is an awardee of The International Postdoctoral Exchange Fellowship (06/2014–06/2015) by The Office of China Postdoctoral Council. C.S.B. received support from the UC Davis Environmental Health Sciences Core Center, P30 ES02351. We thank Mr. Sean D. Kodani for preparing the KO mouse sample and Dr. Todd Harris, Dr. Kin Sing Stephen Lee for helpful discussion through this work.
Footnotes
Notes: The authors declare no competing financial interest.
Supporting Information Available
Additional information as noted in text. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hammers C, Songa EB, Bendahman N, Hammers R. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 3.Steeland S, Vandenbroucke RE, Libert C. Drug Discovery Today. 2016;21:1076–1113. doi: 10.1016/j.drudis.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Kim H-J, McCoy MR, Majkova Z, Dechant JE, Gee SJ, Tabares-da Rosa S, González-Sapienza GG, Hammock BD. Anal Chem. 2012;84:1165–1171. doi: 10.1021/ac2030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker M. [accessed Feb 22, 2017]; http://www.nature.com/news/reproducibility-crisis-blame-it-on-the-antibodies-1.17586.
- 6.Arnaud CH. [accessed Feb 22, 2017]; http://cen.acs.org/articles/93/web/2015/02/Call-Antibody-Standardization.html.
- 7.Saerens D, Frederix F, Reekmans G, Conrath K, Jans K, Brys L, Huang L, Bosmans E, Maes G, Borghs G, Muyldermans S. Anal Chem. 2005;77:7547–7555. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Xu Y, Wan DB, Xiong YH, He ZY, Wang XX, Gee SJ, Ryu D, Hammock BD. Anal Chem. 2015;87:1387–1394. doi: 10.1021/ac504305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Majkova Z, Bever CRS, Yang J, Gee SJ, Li J, Xu T, Hammock BD. Anal Chem. 2015;87:4741–4748. doi: 10.1021/ac504735p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Li P, Majkova Z, Bever CRS, Kim HJ, Zhang Q, Dechant JE, Gee SJ, Hammock BD. Anal Chem. 2013;85:8298–8303. doi: 10.1021/ac4015885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Bever CR, Majkova Z, Dechant JE, Yang J, Gee SJ, Xu T, Hammock BD. Anal Chem. 2014;86:8296–8302. doi: 10.1021/ac5017437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan J, Wang P, Zhu M, Li G, Romão E, Xiong S, Wan Y. J Nanobiotechnol. 2015;13:33–43. doi: 10.1186/s12951-015-0091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegner KD, Lindén S, Jin Z, Jennings TL, Khoulati Re, van Bergen en Henegouwen PMP, Hildebrandt N. Small. 2014;10:734–740. doi: 10.1002/smll.201302383. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Zhu M, Zhang C, Liu X, Wan Y. Toxins. 2014;6:3208–3222. doi: 10.3390/toxins6123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P, Li G, Yan J, Hu Y, Zhang C, Liu X, Wan Y. Toxicon. 2014;92:186–192. doi: 10.1016/j.toxicon.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Zhu M, Gong X, Hu Y, Ou W, Wan Y. J Transl Med. 2014;12:352–361. doi: 10.1186/s12967-014-0352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu M, Hu Y, Li G, Ou W, Mao P, Xin S, Wan Y. Nanoscale Res Lett. 2014;9:528–537. doi: 10.1186/1556-276X-9-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serruys B, Van Houtte F, Verbrugghe P, Leroux-Roels G, Vanlandschoot P. Hepatology. 2009;49:39–49. doi: 10.1002/hep.22609. [DOI] [PubMed] [Google Scholar]
- 19.Hou L, Tang Y, Xu M, Gao Z, Tang D. Anal Chem. 2014;86:8352–8358. doi: 10.1021/ac501898t. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Shi Y, Lan F, Pan Y, Lin Y, Jingzhang L, Zhu Z, Jiang Q, Changqing Y. Chem Commun. 2014;50:1848–1850. doi: 10.1039/c3cc48417c. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Xu Y, Xiong YH, Tu Z, Li YP, He ZY, Qiu YL, Fu JH, Gee SJ, Hammock BD. Anal Chem. 2014;86:7471–7477. doi: 10.1021/ac501202d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambrosi A, Airo F, Merkoci A. Anal Chem. 2010;82:1151–1156. doi: 10.1021/ac902492c. [DOI] [PubMed] [Google Scholar]
- 23.Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai G, Yan F, Ju H. Anal Chem. 2009;81:9730–9736. doi: 10.1021/ac901996a. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y, Zhou Q, Lin Y, Tang D, Niessner R, Knopp D. Anal Chem. 2015;87:8531–8540. doi: 10.1021/acs.analchem.5b02253. [DOI] [PubMed] [Google Scholar]
- 26.Qian J, Zhang C, Cao X, Liu S. Anal Chem. 2010;82:6422–6429. doi: 10.1021/ac100558t. [DOI] [PubMed] [Google Scholar]
- 27.Qu Z, Xu H, Xu P, Chen K, Mu R, Fu J, Gu H. Anal Chem. 2014;86:9367–9371. doi: 10.1021/ac502522b. [DOI] [PubMed] [Google Scholar]
- 28.Imig JD, Hammock BD. Nat Rev Drug Discovery. 2009;8:794–805. doi: 10.1038/nrd2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Y, Li D, Morisseau C, Dong JX, Yang J, Wan D, Rossotti M, Gee S, González-Sapienza G, Hammock B. Anal Bioanal Chem. 2015;407:1–9. doi: 10.1007/s00216-015-8889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossotti MA, Pirez M, Gonzalez-Techera A, Cui Y, Bever CS, Lee KSS, Morisseau C, Leizagoyen C, Gee S, Hammock BD, González-Sapienza G. Anal Chem. 2015;87:11907–11914. doi: 10.1021/acs.analchem.5b03561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Z, Davis BB, Morisseau C, Hammock BD, Olson JL, Kroetz DL, Weiss RH. Am J Physiol Renal Physiol. 2004;286:F720–F726. doi: 10.1152/ajprenal.00165.2003. [DOI] [PubMed] [Google Scholar]
- 32.Harvey D. Modern analytical chemistry. 1. McGraw-Hill; New York: 2000. [Google Scholar]
- 33.Davies C. In: The Immunoassay Handbook. 4. Wild D, editor. Elsevier; Oxford: 2013. pp. 29–59. [Google Scholar]
- 34.Tijssen P. Practice and theory of enzyme immunoassays. Elsevier; Amsterdam: 1985. [Google Scholar]
- 35.Pardue HL. Clin Chem. 1997;43:1831–1837. [PubMed] [Google Scholar]
- 36.Gao Z, Hou L, Xu M, Tang D. Sci Rep. 2014;4:3966. doi: 10.1038/srep03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CR, North KE, Bray MS, Fornage M, Seubert JM, Newman JW, Hammock BD, Couper DJ, Heiss G, Zeldin DC. Hum Mol Genet. 2006;15:1640–1649. doi: 10.1093/hmg/ddl085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Przybyla-Zawislak BD, Srivastava PK, Vázquez-Matías J, Mohrenweiser HW, Maxwell JE, Hammock BD, Bradbury JA, Enayetallah AE, Zeldin DC, Grant DF. Mol Pharmacol. 2003;64:482–490. doi: 10.1124/mol.64.2.482. [DOI] [PubMed] [Google Scholar]
- 39.Luria A, Weldon SM, Kabcenell AK, Ingraham RH, Matera D, Jiang H, Gill R, Morisseau C, Newman JW, Hammock BD. J Biol Chem. 2007;282:2891–2898. doi: 10.1074/jbc.M608057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morisseau C, Wecksler AT, Deng C, Dong H, Yang J, Lee KSS, Kodani SD, Hammock BD. J Lipid Res. 2014;55:1131–1138. doi: 10.1194/jlr.M049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.