Abstract
Exosomes are carriers of pro-tumorigenic factors that participate in glioblastoma (GBM) progression, and many fusion genes are strong driver mutations in neoplasia and are involved in tumorigenesis. However, the ability of fusion genes to be transduced by exosomes is unknown. We characterized exosomes from GBM cells harbouring and not harbouring PTPRZ1–MET fusion (ZM fusion). We also determined the effect of the exosomes from ZM fusion cells (ZM exosomes) on pro-oncogenic secretions and showed that ZM exosomes are internalized by the recipient cells. In addition, we studied the effect of ZM exosome-mediated intercellular communication in the GBM microenvironment. MET proto-oncogene expression was higher in ZM exosomes. Moreover, phosphorylated MET was detected only in ZM exosomes and not in exosomes released by non-ZM fusion GBM cells. ZM exosomes transferred to non-ZM fusion GBM cells and normal human astrocytes altered gene expression and induced epithelial–mesenchymal transition. The uptake of ZM exosomes also induced an exosome-dependent phenotype defined by GBM cell migration and invasion, neurosphere growth and angiogenesis. In addition, ZM exosomes conferred temozolomide resistance to the GBM cells, and exosome-derived ZM fusion network proteins targeted multiple pro-oncogenic effectors in recipient cells within the GBM microenvironment. Our findings show that exosomes mediate the aggressive character of GBM and demonstrate the role of ZM fusion in the exacerbation of this effect. These findings have possible implications for the foundation of gene fusion-based therapy for managing GBM.
Introduction
Glioblastoma (GBM) is characterized by highly infiltrative growth and invariably aggressive biological features.1, 2, 3 Despite treatment consisting of surgery combined with radiotherapy and chemotherapy, the prognosis of patients with GBM remains poor due to the malignant nature and poor response to therapy of this disease.2, 4, 5
Fusion genes combine parts of ⩾2 original genes and can be generated from chromosomal rearrangement or abnormal transcription, and these fusion genes have an important impact on the initial steps of tumorigenesis and cancer progression.6, 7, 8 Our RNA-sequencing study of 272 gliomas identified a novel, recurrent PTPRZ1–MET fusion (ZM fusion) transcript in secondary GBM. Specifically, ZM fusion was found in grade III astrocytomas (1/13; 7.7%) and secondary GBMs (3/20; 15.0%). We identified four ZM fusion transcripts involving four different breakpoints within the PTPRZ1 coding sequence, and the breakpoints in the MET gene were located at the same site.7 Furthermore, previous findings indicate that ZM fusions retain the fundamental properties of wild-type MET regarding processing and dimerization, and promote phosphorylation in a hepatocyte growth factor-dependent and -independent manner. ZM fusion can induce the development of glioma by increasing the expression and phosphorylation of the MET oncoprotein, whereas endogenously expressed MET is not phosphorylated in glioma cells.7, 9 Clinically, the survival of patients with GBM harbouring ZM fusion is poorer than that of patients harbouring disease without ZM fusion.7
The coexistence of complex cell types within the same tumour requires high-level coordination, which is managed by molecular mechanisms of intercellular communication.10, 11 The most intriguing of these mechanisms is cellular communication mediated by membrane-derived extracellular vesicles (EVs).12, 13, 14, 15, 16 Specifically, exosomes are 30–100 nm-wide EVs enclosed by a bilayer membrane that carry a unique cargo of proteins, lipids and RNAs.12, 13, 16, 17, 18 The release and uptake of exosomes containing cellular proteins and RNAs comprise a crucial form of cell–cell communication in tumours12, 17, 19, 20 because cells acquire a malignant phenotype by taking up exosomes that deliver tumour-derived oncogenic factors.21, 22, 23 Accordingly, a growing body of research also suggests an important role for EV communication in GBM.22, 24, 25 These studies reflect the need to evaluate the functional contribution of ZM fusion to the GBM phenotype and its role in exosome-associated cell interaction with the tumour microenvironment.
Results
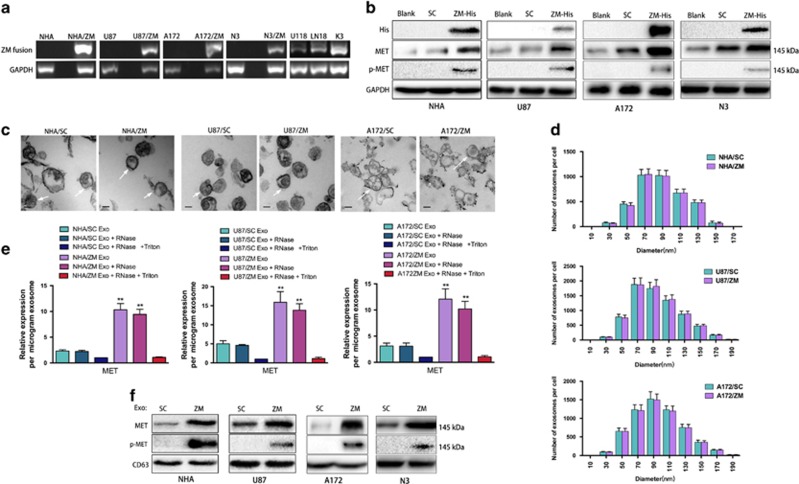
GBM cells harbouring ZM fusion secrete MET and phosphorylated MET via exosomes
The normal human astrocytes (NHAs) and six GBM cell lines were screened using fusion-specific PCR primers, and the ZM fusion sequence was detected in three cell lines (U118, LN18 and one primary GBM line (K3)) (Figure 1a). The ZM-harbouring GBM specimen CGGA_14757 harboured a ZM fusion that fused exon 2 of PTPRZ to exon 2 of MET. We cloned a His-tagged version of CGGA_1475 ZM fusion7 into an adenovirus vector and stably expressed the fusion gene in the NHA/ZM, U87/ZM, A172/ZM and N3/ZM cell lines (Figure 1a). Anti-His tag or anti-MET antibody probes against the protein also indicated stable expression (Figure 1b). The anticipated molecular weights of exons 1 and 2 of PTPRZ are 2.3 and 2.7 kDa, respectively. Exon 1 of MET encodes the 59 untranslated sequence (394 bp), and the molecular weight of MET is ~145 kDa. Therefore, the anticipated molecular weight of the ZM fusion gene in CGGA_1475, where exons 1 and 2 of PTPRZ are fused to exon 2 of MET, approximates that of native MET (~145 kDa). Thus, these two species cannot be discriminated based on SDS–polyacrylamide gel electrophoresis. Given this ambiguity, the strong 145 kDa MET band in Figure 1b cannot be conclusively identified as indicating MET or ZM fusion protein expression. Nevertheless, endogenously expressed MET in U87 cells is not phosphorylated at residue 1234/5,26, 27 and this phosphorylation event only occurs when MET is dimerized and activated.28 Exogenously expressed ZM fusion resulted in phosphorylated MET (p-MET) expression, indicating that ZM fusion activated MET7 (Figure 1b). Moreover, exosomes from cells harbouring and not harbouring ZM fusion were observed by electron microscopy and found to be of similar exosomal size and morphology (Figure 1c). The NanoSight results also showed similar exosome number and size distributions (10–210 nm wide) between the two groups (Figure 1d). Similar to their parental cells, ZM exosomes contained more MET than SC exosomes (Figure 1e). To ascertain whether this enriched MET was derived from intact ZM exosomes, samples were mixed with RNase A in the presence or absence of Triton X-100. RNase A+Triton X-100 substantially reduced the MET levels, indicating that the secreted MET was encapsulated within a membrane (Figure 1e). These results were consistent with previous reports describing the exosome-mediated secretion of MET by melanoma cells.29, 30 In addition, the protein expression of MET was significantly higher in U87/ZM exosomes than in U87/SC exosomes; we only detected p-MET in the U87/ZM exosome fractions and not in the U87/SC exosomes (Figure 1f), and these results were validated in NHA, A172 and N3 cells. We also analysed the levels of MET and p-MET in exosomes released from U118 and K3 GBM lines harbouring an endogenous ZM translocation. As expected, p-MET were detected in U118 and K3 exosomes (Supplementary Figures S1A and B). Moreover, the depletion of endogenous ZM fusion in U118 and K3 cells using short hairpin RNA that targets ZM fusion (shZM; Supplementary Figures S1A and B) significantly decreased the MET and p-MET protein levels in U118/shZM and K3/shZM exosomes compared with these levels in the shControl groups (Supplementary Figure S1C). Taken together, these data indicate that ZM exosomes contain p-MET and that ZM fusion in GBM cells upregulates MET in the corresponding exosomes.
Figure 1.
MET and p-MET proteins are secreted by ZM exosomes. (a) ZM fusion screening in NHA cells and six glioma cell lines. Three cell lines (U118, LN18 and K3) were positive for ZM fusion. A His-tagged version of the CGGA_1475 ZM fusion (ZM-His) transcript was cloned into an adenovirus vector and stably expressed in the four cell lines (NHA, U87, A172 and N3). (b) ZM fusion harbours p-MET; MET endogenously expressed by NHA cells and GBM cells was not phosphorylated. (c) Electron microscopy images of NHAs and GBM cell exosomes (SC vs ZM). Scale bars=50 nm. (d) Size distribution of NHAs and GBM cell exosomes (SC vs ZM) in culture supernatants obtained after 48 h. (e) TaqMan quantification of MET mRNA expression in NHA/SC, NAH/ZM, U87/SC, U87/ZM, A172/SC and A172/ZM exosomes (Exo) untreated or treated with RNase A and/or TritonX-100 (**P<0.01). (f) Exosomes (Exo) from NHA/ZM, U87/ZM, A172/ZM and N3/ZM harbour p-MET; NHA/SC, U87/SC, A172/SC and N3/SC exosomes lack p-MET.
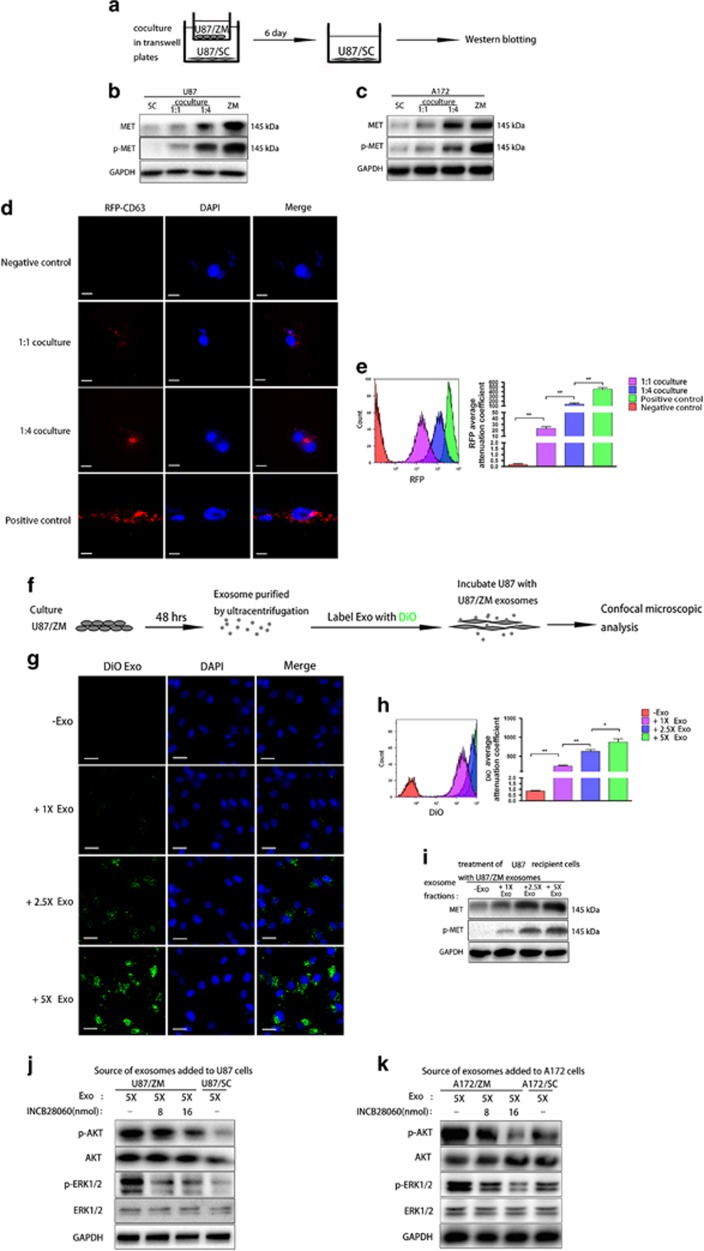
GBM cells not harbouring ZM fusion received MET and p-MET via exosomes
Exosomes have been reported to horizontally transfer MET to recipient cells in melanoma.29, 30 To test the ability of MET and p-MET to be transferred from U87/ZM to U87/SC, we co-cultured U87/ZM and U87/SC (Figure 2a). As expected, the U87/SC MET and p-MET protein levels were increased after 6 days of co-culture (Figure 2b). Western blotting also revealed increased MET and p-MET expression in A172/SC recipient cells (Figure 2c). To visualize the exosome transfer, we created U87/ZM cell lines expressing the red fluorescent protein (RFP)-labelled exosomal marker CD63 (U87/ZM/RFP-CD63) and used them as donor cells. Confocal microscopy revealed that U87/SC cells co-cultured at a 1:4 ratio (U87/SC:U87/ZM/RFP-CD63) harboured more RFP-labelled exosomes than cells co-cultured at a 1:1 ratio (Figure 2d). These results were confirmed by a flow cytometry analysis (Figure 2e). To test whether U87/ZM exosomes are required for MET and p-MET transfer, we blocked U87/ZM exosome secretion with dimethylamiloride (DMA), an exosome release inhibitor, or a short hairpin RNA targeting Rab27a/b (shRab27a/b), an exosome secretion mediator29 (Supplementary Figures S2A–D). Both strategies reduced U87/ZM-to-U87/SC MET and p-MET transfer (Supplementary Figures S2E and F). Using Vybrant cell-labelling solutions, we stained the U87/ZM exosomes with DiO-dye (Green) and visualized exosomes with confocal microscopy in the recipient cells after incubation with increased U87/ZM DiO-dyed exosomes (Figures 2f and g), as indicated by the elevated diffuse cytoplasmic green dots; flow cytometry confirmed these results (Figure 2h). Moreover, MET and p-MET were significantly increased in recipient cells after incubation with increased U87/ZM exosomes (Figure 2i). The transfer of exosome-mediated MET to recipient cells has been shown to have functional consequences in melanoma.29, 30 Thus, we then investigated the biological activity of exosome-transferred MET and p-MET. Specifically, the tyrosine phosphorylation of MET may contribute to the phosphorylation of downstream transducers such as ERK and AKT.31 The irreversible blockade of exosomal MET by pre-incubating this (cell-free) material with a highly selective MET inhibitor (INCB28060)32 markedly reduced its ability to trigger the phosphorylation of major downstream effectors of the MET pathway, including ERK1/2 and AKT31, 33 (Figures 2j and k). These results suggest that exosomal MET and p-MET transfer is required to trigger the activation of MET signalling in the recipient cell. To conclude, GBM cells not harbouring ZM fusion received biologically active MET and p-MET via exosomes.
Figure 2.
Functional MET and p-MET can be transferred via exosomes. (a) Schematic of U87/SC–U87/ZM co-culture. (b, c) MET and p-MET in U87/SC and A172/SC after 6 days of co-culture with GBM cells harbouring the ZM fusion gene in a 1:1 or 1:4 ratio. (d) Confocal microscopy visualization of exosomes taken up by U87/SC cells co-cultured with RFP-CD63-transfected U87/ZM cells at a 1:1 or 1:4 ratio over 24 h. Untreated U87/SC cells served as the negative control, whereas RFP-CD63-transfected U87/ZM cells served as the positive control. Scale bars=20 μm. (e) Quantification of RFP per cell in treatment groups. (f) Schematic of U87 cell incubation with DiO-dyed exosomes. (g) Confocal microscopy visualization of exosomes taken up by U87 cells treated with 1 ×, 2.5 × and 5 × DiO-dyed ZM exosomes over 24 h. Scale bars=50 μm. (h) Quantification of DiO-dyed exosomes per cell in treatment groups. (i) MET and p-MET in U87/SC in the presence of ZM exosomes. (j, k) Increase of AKT and ERK1/2 phosphorylation in U87 cells that had incorporated exosomes shed by U87/ZM cells. The pre-incubation of these exosomes for 2 h with an irreversible MET inhibitor (INCB28060) abrogated their ability to trigger AKT and ERK1/2 phosphorylation in a concentration-dependent manner. The same results were observed in A172 cells (*P<0.05 and **P<0.01).
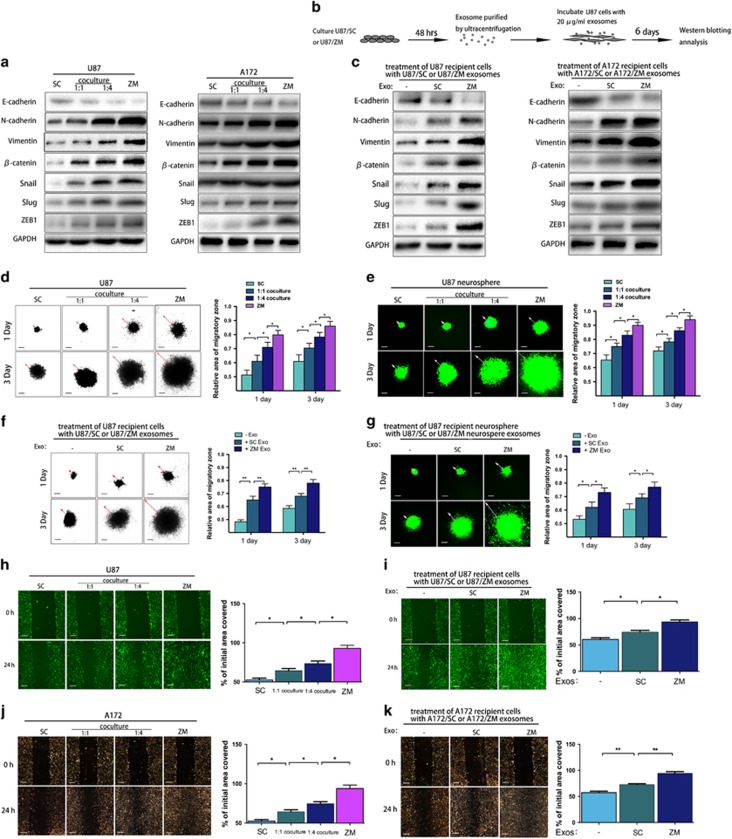
ZM exosomes induce epithelial–mesenchymal transition and promote cell migration and invasion
Many studies have shown that c-MET mediates epithelial–mesenchymal transition (EMT)-promoting signals in various cancers.34, 35 To test whether U87/ZM exosomes containing MET and p-MET induce EMT in U87/SC cells, we co-cultured U87/ZM and U87/SC cells. Specifically, the cells expressed more activated EMT-promoting signals at a co-culturing ratio of 1:4 than at a ratio of 1:1 (Figure 3a), as confirmed by immunofluorescence staining for EMT-associated proteins (E-cadherin, N-cadherin, vimentin, β-catenin, Snail, Slug and ZEB1; Supplementary Figure 3A). We also co-cultured U87/ZM or U87/SC with NHA at 1:1 or 1:4 ratios (NHA:U87/ZM or U87/SC) (Supplementary Figure 3B) to assess the ability of U87/ZM to induce EMT in NHA cells. As expected, changes in the expression of EMT-promoting signals were greater in the NHA–U87/ZM co-culture than in the NHA–U87/SC co-culture (Supplementary Figure 3C), as indicated by immunoblotting. These data were confirmed with immunofluorescence studies of the EMT-associated proteins (Supplementary Figure 3D). We next explored the EMT phenotype of GBM cells incubated with ZM exosomes (Figure 3b). As expected, ZM exosomes more significantly enhanced EMT in both U87 and A172 recipient cells (Figure 3c). NHA cells incubated with U87/ZM exosomes also exhibited more activated EMT-promoting signals than cells incubated with U87/SC exosomes (Supplementary Figure 3E), and adding more ZM exosomes to U87 and NHA cells enhanced these changes in EMT (1 ×, 2.5 × or 5 × ; Supplementary Figures 3F, G and I). Moreover, dose–response experiments using SC exosomes convincingly demonstrated an increase in EMT markers (Supplementary Figure 3H). These data indicate that ZM fusion can promote EMT in glioma via exosomes.
Figure 3.
ZM fusion and ZM exosomes induce EMT and promote GBM cell migration and invasion. (a) Staining the EMT-associated proteins in U87/SC and A172/SC cells after 6 days of co-culture. (b) Schematic of U87 cells incubated with U87/SC or U87/ZM exosomes. (c) EMT-associated protein staining in U87 cells after 6 days of incubation with U87/SC or U87/ZM exosomes. The same staining was also carried out in A172 cells. (d) Spheroid dispersal assay measurement of U87 cell migration. Representative images of the spheroid migration of U87/ZM(ZM), untreated U87/SC(SC) and U87/SC cells co-cultured with U87/ZM at a 1:1 or 1:4 ratio are shown. Scale bars=250 μm. (e) Spheroid dispersal assay of U87 stem-like neurosphere migration. Scale bars=250 μm. (f) Spheroid dispersal assay of U87 cell migration in the presence of exosomes. Scale bars=250 μm. (g) Spheroid dispersal assay of U87 neurosphere migration in the presence of exosomes. Scale bars=250 μm. (h, j) Wound-healing assay of U87 cell migration. Scale bars=250 μm. (i, k), Wound-healing assay to assess U87 cell migration in the presence of exosomes. These findings were also validated in A172 cells. Scale bars=250 μm (*P<0.05 and **P<0.01).
We next explored the effect of ZM fusion on phenotypic changes in GBM migration. Specifically, U87/ZM showed increasing infiltration in a three-dimensional (3D) collagen matrix: cells migrated out of the cell mass, exhibiting a more invasive morphology (Figure 3d), and this behaviour was echoed by green fluorescent protein (GFP)-expressing U87/ZM neurospheres (U87/ZM/GFP) seeded in a 3D collagen matrix (Figure 3e). U87 recipient cells displayed much ability of GBM cell migration at a co-culturing ratio of 1:4 than at a ratio of 1:1(Figures 3d and e). Cells incubated with U87/ZM exosomes also exhibited a more invasive morphology than cells incubated with U87/SC exosomes (Figure 3f), and these results were validated by U87/GFP-derived neurospheres seeded in a 3D collagen matrix incubated with U87/SC or U87/ZM exosomes (Figure 3g). The impact of ZM fusion on migration was also assessed with a wound-healing assay (Figure 3h), and U87/ZM exosomes more significantly promoted migration (Figure 3i); these results were validated in A172 cells (Figures 3j and k). Furthermore, U87 cells (Supplementary Figure 3J) and U87/GFP-derived neurospheres (Supplementary Figure 3K) exhibited higher migration capacity in a 3D collagen matrix upon incubation with more U87/ZM exosomes, and these results were validated the wound-healing assay using U87 cells grown in serum culture (Supplementary Figure 3L). In addition, we observed persistent promotion of migration and invasion in U118/shControl exosomes but not exosomes released from U118/shZM cells (Supplementary Figures 3M–O). Thus, we concluded that MET and p-MET in ZM exosomes are important for promoting cancer invasion and migration, and ZM exosomes promote GBM cell migration and invasion.
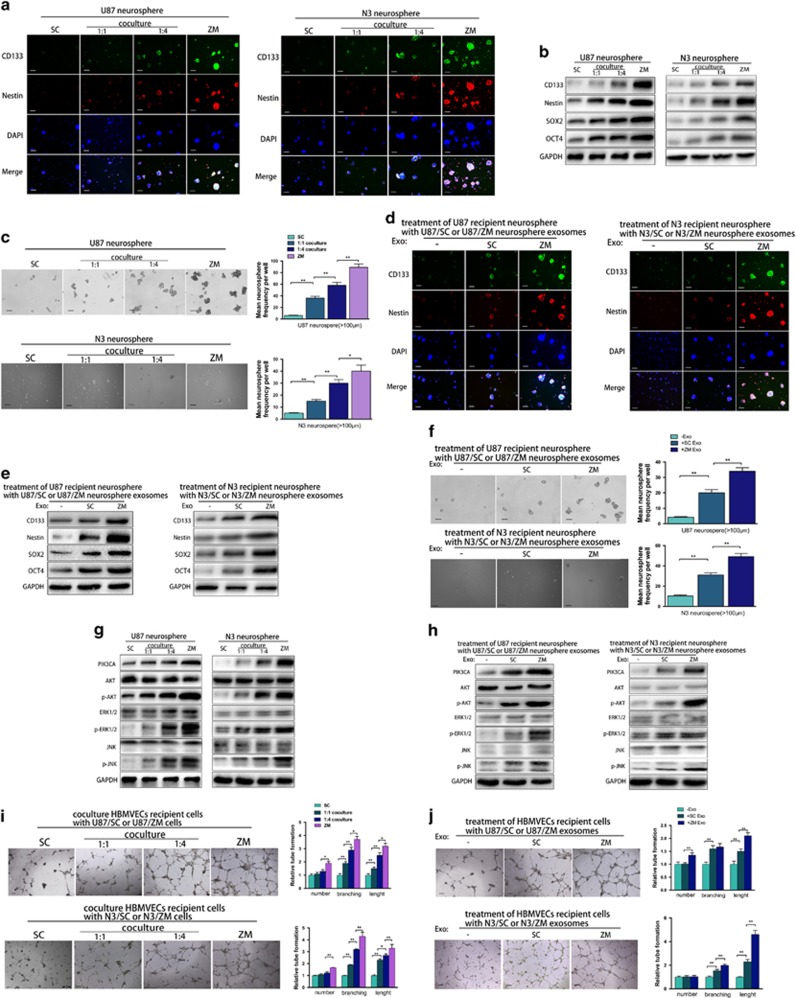
ZM exosomes promote neurosphere formation and angiogenesis
We enriched CD133-positive neurospheres from the U87 and N3 primary GBM cell line by growing them in stem cell-like conditions (Figure 4a). Immunofluorescence showed that ZM fusion significantly increased the protein levels of the putative GBM stem cell marker CD133 and nestin (Figure 4a). In the 1:4 co-culture, the cells expressed more CD133 and nestin than in the 1:1 co-culture (Figure 4a). Moreover, ZM fusion in stem-like culture conditions not only significantly increased the CD133 and nestin levels but also increased the expression of the stem cell self-renewal factors SOX2 and OCT436 (Figure 4b). The neurosphere formation assay was then used to assess the impact of ZM fusion on the self-renewal ability of GBM cells. ZM fusion promoted U87 and N3 neurosphere formation in stem-like conditions (Figure 4c). As expected, the cells in the 1:4 co-culture exhibited more significant self-renewal than cells in the 1:1 co-culture (Figure 4c). To compare their effects, U87/SC and U87/ZM exosomes were added separately to the stem-like U87 neurosphere culture conditions. The levels of CD133, nestin, SOX2 and OCT4 were significantly increased in U87 neurospheres incubated with U87/ZM exosomes compared with the expression in those incubated with U87/SC exosomes, and these results were validated in the N3-derived neurospheres (Figures 4d and e). ZM exosomes also more significantly promoted neurosphere formation in stem-like conditions (Figure 4f). We speculated that ZM fusion upregulates multiple signalling pathways involved in GBM progression and consequently investigated oncogenic signalling in GBM cells with or without ZM fusion cultured under stem-like neurosphere conditions. Compared to the levels in U87/SC cells, the levels of cellular p-AKT, p-ERK1/2 and p-JNK were significantly increased in U87/ZM cells (Figure 4g). ZM exosomes also produced greater increases in p-AKT, p-ERK1/2 and p-JNK expression in neurospheres than SC exosomes (Figure 4h). Furthermore, adding more U87/ZM exosomes to the U87 neurosphere culture increased GBM stem cell marker expression (Supplementary Figures 4A and B), which consequently increased neurosphere formation (Supplementary Figure 4C). The changes in GBM cell oncogenic signalling were consistent with the above results (Supplementary Figure 4D). Moreover, a tube formation assay showed that ZM fusion-transfected human brain microvessel endothelial cells (HBMVECs) produced longer tubes, indicating that angiogenesis was inactivated (Figure 4i). During tumour progression, microparticles can stimulate vascular formation and normally quiescent vasculature to sprout new vessels that help sustain expanding neoplastic growths.25, 37 As shown in Figure 4j, ZM exosomes promoted HBMVEC angiogenesis more significantly than SC exosomes, and internalizing more U87/ZM exosomes also increased HBMVEC angiogenesis (Supplementary Figure 4E), as expected. In addition, we observed a significant decrease in the expression of stem cell markers in U118 neurospheres incubated with U118/shZM exosomes compared with the expression in those incubated with U118/shControl exosomes (Supplementary Figures 4F and G), and these findings were echoed by the neurosphere formation of U118 (Supplementary Figures 4H and I). Silencing ZM in U118 cells also inhibited the ability of their exosomes to promote angiogenesis (Supplementary Figure 4J). These results indicate that ZM exosomes facilitate GBM neurosphere formation and angiogenesis in vitro.
Figure 4.
ZM fusion promotes GBM cell neurosphere formation and angiogenesis via exosomes. (a) CD133, nestin and 4′,6-diamidino-2-phenylindole (DAPI) immunostaining in U87/ZM neurospheres (ZM), untreated U87/SC neurospheres (SC) and U87/SC neurospheres after 6 days of co-culture with U87/ZM neurospheres at a 1:1 or 1:4 ratio. These findings were also validated in N3 cells. Scale bars=100 μm. (b) Western blotting of stemness factors in U87 and N3 cells cultured as stem cell-like neurospheres. (c) Self-renewal assay of U87/ZM neurospheres (ZM), untreated U87/SC neurospheres (SC) and U87/SC neurospheres after 6 days of co-culture with U87/ZM neurospheres at a 1:1 or 1:4 ratio. These findings were also validated in N3 cells. Scale bar=100 μm. (d) CD133, nestin and DAPI immunostaining in U87 and N3 neurospheres. Representative images of untreated (−) neurospheres or neurospheres cultured in the presence of ZM exosomes (ZM) or SC exosomes (SC) over 6 days are shown. Scale bars=100 μm. (e) ZM exosomes promoted glioma cell stemness more significantly than U87/SC or N3/SC exosomes. (f) Self-renewal assay to measure neurosphere formation capacity. Scale bars=100 μm. (g, h) Western blotting of cellular signalling mediators in U87 or N3 cells cultured as stem cell-like neurospheres. (i) Tube formation by HBMVEC transfected with ZM fusion (ZM) or scramble control (SC) construct and after 6 days of co-culture with U87/ZM cells at a 1:1 or 1:4 ratio. Scale bars=250 μm. (j) Tube formation by HBMVEC in the presence of U87/ZM exosomes (ZM) or U87/SC exosomes (SC). These findings were also validated in N3 cells. Scale bars=250 μm (*P<0.05 and **P<0.01).
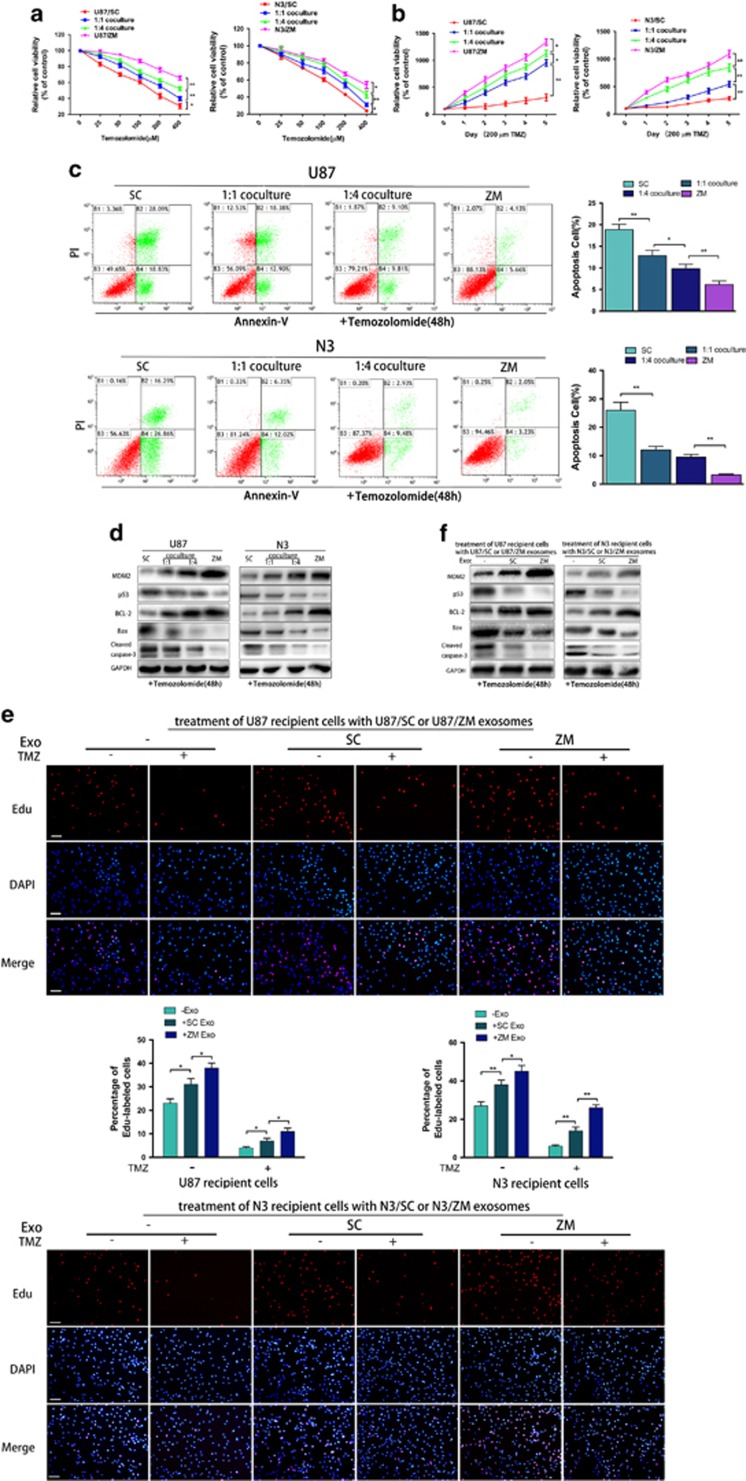
ZM exosomes confer temozolomide resistance
ZM fusion significantly decreased U87 and N3 cell temozolomide chemosensitivity (Figures 5a and b), as confirmed by the flow cytometry (Figure 5c). An increasing number of studies have indicated that EVs are involved in drug resistance in various cancers.13 To test the ability of exosomes to transfer drug resistance from U87/ZM to U87/SC cells, we co-cultured U87/ZM and U87/SC cells at 1:1 or 1:4 ratios, and U87/SC and N3/SC were more temozolomide resistant at the 1:4 ratio than at the 1:1 ratio (Figures 5a and b). Our previous report indicated that TP53 is an activated canonical GBM signalling pathway in sGBM harbouring ZM fusion.7 Here we found that temozolomide-treated U87/ZM and N3/ZM expressed higher levels of MDM2, a key protein that mediates P53 function,38 and the apoptosis inhibitor BCL-2 but lower levels of P53 and the pro-apoptosis factors Bax and cleaved caspase-3 than temozolomide-treated U87/SC and N3/SC cells (Figure 5d). At the 1:4 ratio, temozolomide-treated U87/SC expressed more BCL-2 and less Bax and cleaved caspase-3 than U87/SC cells co-cultured at the 1:1 ratio (Figure 5d). We next investigated the modulatory role of ZM exosomes in DNA synthesis with and without temozolomide. ZM exosomes promoted GBM cell resistance more significantly than SC exosomes (Figure 5e), as confirmed by immunoblotting (Figure 5f), and these results were also validated in N3 primary GBM cells. Furthermore, adding more U87/ZM exosomes to U87 conditions enhanced the temozolomide resistance of GBM cells (Supplementary Figures 5A and B). To identify the mechanisms that mediate temozolomide resistance in cells treated with ZM exosomes, O-6-methylguanine-DNA methyltransferase gene methylation and protein expression were assessed in untreated and treated cells, but these markers did not significantly differ between these groups (data not shown). Thus, ZM-inducing TMZ resistance was independent of O-6-methylguanine-DNA methyltransferase expression. In addition, ZM silencing abrogated the ability of exosomes released from U118 to promote temozolomide resistance (Supplementary Figures 5C–E). These data demonstrate that ZM exosomes confer temozolomide resistance to GBM cells.
Figure 5.
ZM fusion and ZM exosomes confer temozolomide resistance to GBM cells. (a, b) Cell proliferation of temozolomide-treated U87 cells was examined every 24 h. Temozolomide resistance in U87/ZM, untreated U87/SC and U87/SC co-cultured with U87/ZM at a 1:1 or 1:4 ratio is shown. These findings were also validated in N3 cells. (c) Fluorescence-activated cell sorting of temozolomide-treated U87 and N3 cells. (d) Western blotting of U87/ZM(ZM), untreated U87/SC(SC) and U87/SC co-cultured with U87/ZM at a 1:1 or 1:4 ratio. These results were also validated in N3 cells. (e) Images and quantification of ethynyl deoxyuridine-labelled untreated (−) U87 cells or U87 cells cultured in the presence of U87/SC exosomes (SC) or U87/ZM exosomes (ZM) with dimethyl sulphoxide (DMSO, −) or temozolomide (+, 200 μM). These findings were also validated in N3 cells. Scale bars=100 μm. (f) Western blotting to assess temozolomide resistance in untreated (−) U87 cells or U87 cells cultured in the presence of ZM exosomes (ZM) or SC exosomes (SC) is shown. These results were also validated in N3 cells (*P<0.05 and **P<0.01).
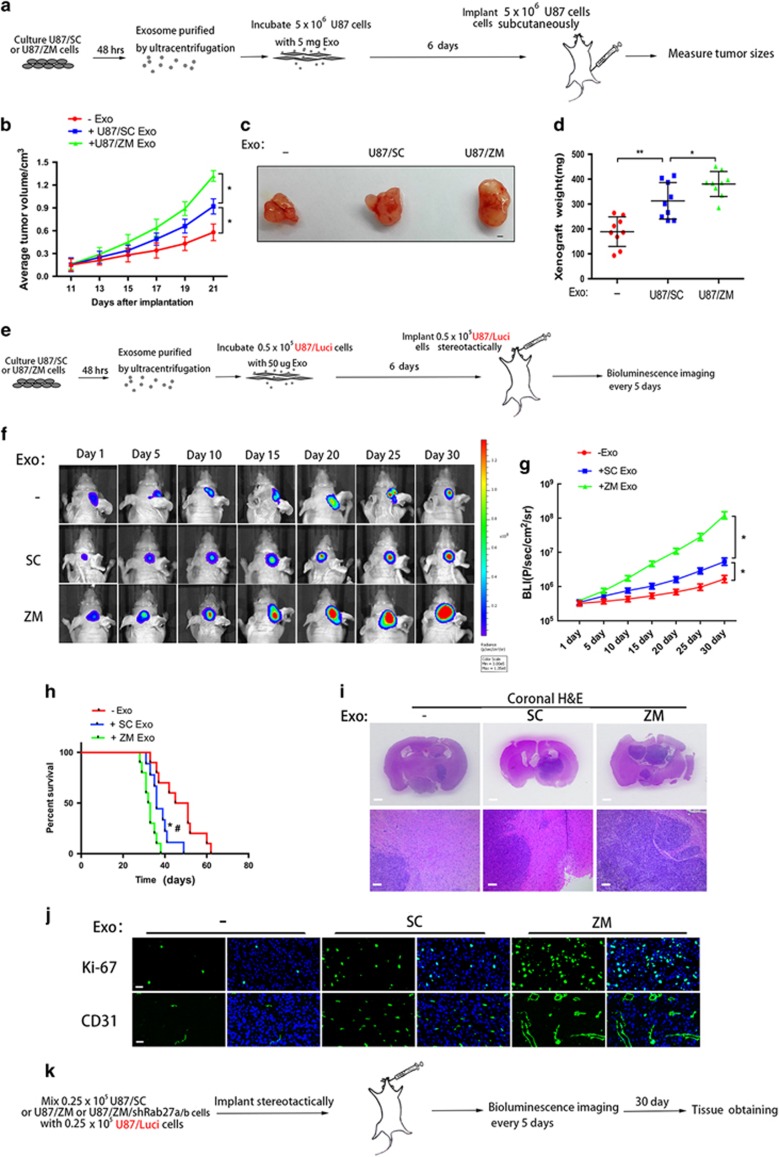
ZM exosomes enhance tumorigenicity, invasion and angiogenesis in vivo
We next subcutaneously injected mice with U87 cells that had been incubated with U87/SC or U87/ZM exosomes for 6 days (Figure 6a). Xenografts of U87 cells incubated with ZM exosomes (U87+ZM Exo) were markedly larger than those from cells incubated with SC exosomes (U87+SC Exo) (Figure 6b). On day 21, the flanks of mice harbouring U87+ZM Exo and U87+SC Exo xenografts were significantly larger than those of mice harbouring tumours consisting of non-incubated U87 control cells (Figure 6c). Moreover, the final tumour weight of the U87+ZM Exo group was significantly higher than that of the U87+SC Exo group and approximately twofold higher than that of the control group (Figure 6d). Furthermore, bioluminescence imaging of the U87 GBM intracranial model (Figure 6e) revealed faster tumour growth in the U87+ZM Exo group than in the U87+SC Exo group (Figures 6f and g); survival also significantly differed between the two groups (Figure 6h). Whole-brain sections revealed that the U87+ZM Exo xenograft tumours were considerably larger than the U87+SC Exo tumours (Figure 6i), and the magnified patterns of the intracranial xenografts showed that more U87 cells migrated out of the tumour core in the U87+ZM Exo group than in the U87+SC Exo group (Figure 6i). Furthermore, immunofluorescence revealed that tumour proliferation and neovascularization at 6 weeks after tumour implantation were higher in the U87+ZM Exo group (Figure 6j), and the expression levels of EMT-promoting proteins were simultaneously increased in the U87+ZM Exo group relative to the U87+SC Exo group (Supplementary Figure 6A). In addition, pre-incubating U87/ZM exosomes with INCB28060 significantly decreased ZM exosome-promoted tumorigenicity in vivo (Supplementary Figures 6B–D).
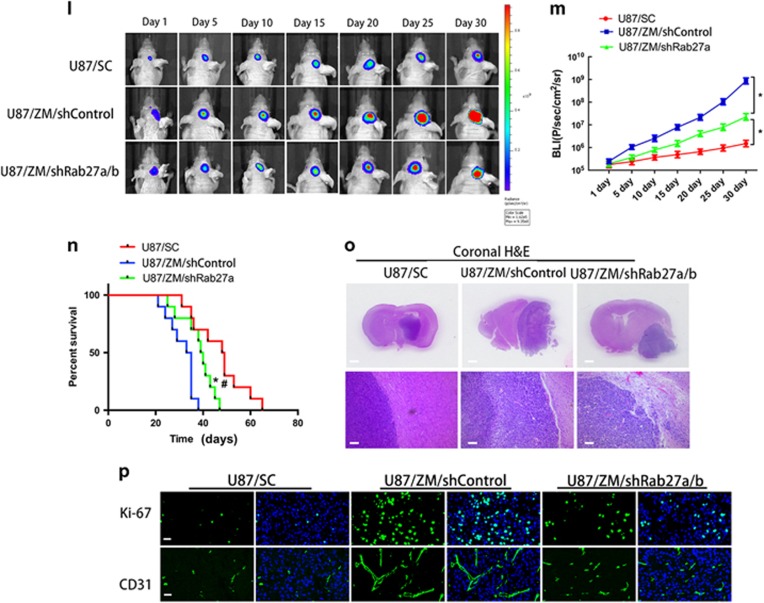
Figure 6.
ZM exosomes enhance GBM cell tumorigenicity, invasion and angiogenesis in vivo. (a) Schematic of in vivo tumorigenicity assay. (b–d) Effect of exosomes on the growth of U87 cells inoculated into BALB/c mice. Representative images of untreated (−) U87 cells or U87 cells cultured in the presence of ZM exosomes (ZM) or SC exosomes (SC) are shown. Scale bars=1 mm. (e) Schematic of in vivo tumorigenicity assay. (f) Representative images of mice harbouring intracranial U87 tumours on days 1, 5, 10, 15, 20, 25 and 30. The intracranially implanted U87 cells were untreated (−) or cultured in the presence of ZM exosomes (ZM) or SC exosomes (SC). (g) Activity plot obtained by bioluminescence imaging for intracranial tumours. (h) Kaplan–Meier survival curve of animals injected with untreated (−) U87 cells or U87 cells cultured in the presence of ZM exosomes (ZM) or SC exosomes (SC). *Significant difference compared to untreated cells, #significant difference compared to SC exosomes at P<0.05. (i) Representative haematoxylin–eosin (H&E) staining of tumour cytostructure and cytomorphology analysis in tumours. Scale bars=1 mm (top panels) and 100 mm (bottom panels). (j) Cell proliferation and angiogenesis in vivo were assessed by Ki-67 and CD31 immunostaining, respectively. Scale bars=25 μm. (k) Schematic showing the imaging of the in vivo tumorigenicity assay. Luminescence was measured to assess the tumorigenicity of the U87/Luci cells. (l) Representative luminescent images of mice after intracranial injection. (m) Activity plot generated by bioluminescence imaging of intracranial tumours. (n) Kaplan–Meier survival curve of animals injected with a mixture of U87/Luci cells and U87/SC, U87/ZM/shControl or U87/ZM/shRab27a/b cells. *Significant difference compared to U87/SC, #significant difference compared to U87/ZM/shRab27a/b cells at P<0.05. (o) Representative H&E staining of tumour cytostructure and cytomorphology analysis in tumours. Scale bars=1 mm (top panels) and 100 μm (bottom panels). (p) Cell proliferation and angiogenesis in vivo were assessed by Ki-67 and CD31 immunostaining, respectively. Scale bars=25 μm. Data are presented as the mean±s.d. from three independent experiments.
To test whether U87/ZM exosomes are required for enhancing GBM cell tumorigenicity in vivo, a short hairpin RNA targeting Rab27a/b, an exosome secretion mediator (shRAb27a/b), was transfected into U87/ZM cells (U87/ZM/shRab27a/b), and U87 cells stably expressing the luciferase reporter (U87/Luci) were generated. We then intracranially injected a mixture of 0.25 × 105 U87/Luci cells and 0.25 × 105 U87/SC, U87/ZM/shControl or U87/ZM/shRab27a/b cells into nude mice and measured the luminescence to assess the tumorigenicity of the U87/Luci cells (Figure 6k). Bioluminescence intensity was lowest in the U87/SC tumour-bearing mice (Figures 6l and m). As expected, the U87/Luci cells in the U87/ZM/shRab27a/b tumour-bearing mice exhibited lower-intensity bioluminescence than cells in the U87/ZM/shControl tumour-bearing mice (Figure 6m). Overall, the U87/ZM/shRab27a/b tumour-bearing mice had a better prognosis than the U87/ZM/shControl tumour-bearing mice (Figure 6n). Whole-brain sections from U87/ZM/shRab27a/b tumour-bearing mice contained considerably smaller xenograft tumours than sections from U87/ZM/shControl tumour-bearing mice (Figure 6o), and immunofluorescence revealed decreased Ki-67 and CD-31 staining (Figure 6p) and EMT-promoting signals in the U87/ZM/shRab27a/b tumour-bearing mice (Supplementary Figure 6E) compared with U87/ZM/shControl tumour-bearing mice. These results indicate that U87/ZM exosomes within a primary tumour can enhance the spontaneous tumorigenicity, invasion and angiogenesis of aggressive GBM. Taken together, our data show that U87/ZM exosomes enhance GBM cell tumorigenicity, invasion and angiogenesis in vivo.
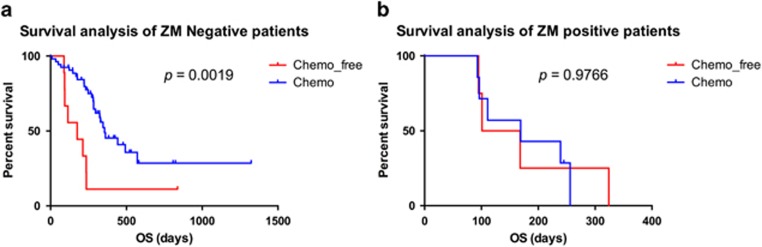
Tumours from patients harbouring ZM-fused GBM are chemoresistant to temozolomide therapy
A cohort of 73 patients with GBM was investigated to assess the association between ZM fusion and TMZ resistance. As expected, a Kaplan–Meier survival analysis showed that patients harbouring non-ZM fusion tumours receiving temozolomide therapy (n=53) had prolonged overall survival (OS) compared with patients who did not receive temozolomide treatment (n=9; Figure 7a; OS, days 281 vs 175 days, P=0.0019). However, the prognosis of patients harbouring ZM-fused GBM who received temozolomide therapy (n=7) did not significantly differ from that of patients not treated with temozolomide (n=4; Figure 7b; OS, days 169 vs 135 days, P=0.9766). These findings shed light on the clinical correlation between ZM fusion and the acquisition of temozolomide resistance.
Figure 7.
Clinical outcomes of ZM-negative and ZM-positive subgroups. (a) Kaplan–Meier survival plots for ZM-negative samples treated or not treated with TMZ. (b) Kaplan–Meier survival plots for ZM-positive samples treated or not treated with TMZ.
Discussion
The signal regulation and multiple interactions between parenchymal and tumour cells in the GBM microenvironment are not well understood.39 Exosomes function in intercellular communication by enabling tumour cells to interact with their surroundings to contribute to a malignant phenotype, and fusion genes act as drivers of malignant transformation and progression in many human cancers.8 Previously, we identified a novel, recurrent ZM fusion transcript using RNA sequencing of 272 gliomas.7 Because exosomes transport nucleic acids and proteins, the use of ZM fusion in donor cells may be applied by exosomes in the GBM microenvironment. Our observations demonstrate the biological activity of ZM fusion after exosomal delivery to recipient cells: ZM exosomes contain p-MET, and ZM fusion in GBM cells upregulates MET in the corresponding exosomes. The MET oncogene can activate multiple cellular signalling pathways in various cancers, including GBM.34, 40 Consistent with previous reports describing that the horizontal transfer of functional MET via exosomes induces a more malignant phenotype in recipient cells,29, 30 we find that increasing ZM exosomes resulted in the uptake of MET and p-MET from GBM cells harbouring the ZM fusion gene by the recipient cells, which significantly altered the behaviour of these cells, including the induction of EMT. The exosome-mediated phenotype observed in the recipient cells suggests an active role for the molecular cargo of the exosome. In the brain tumour microenvironment, reactive astrocytes are frequently associated with glioma cells, and we confirmed that ZM exosomes promote EMT in NHA. Our study is the first to analyse and reveal that ZM fusion and ZM exosomes in GBM induce EMT and promotes cell migration and invasion in vitro and in vivo. GBM exhibits a high level of intratumoural cellular heterogeneity and contains tumorigenic cell subpopulations with stem cell-like properties.41 The present study indicates that ZM fusion significantly increases stemness in neurospheres under stem-like culture conditions and inactivates HBMVEC angiogenesis in the ZM fusion-transfected group. In addition, ZM exosomes promoted GBM cell neurosphere formation and promoted angiogenesis. Temozolomide is the most potent chemotherapy agent for treating GBM. However, its efficacy in treating primary and recurrent GBM remains unsatisfactory because of inherent or acquired resistance in GBM cells.42, 43, 44 Here ZM fusion conferred temozolomide resistance via exosomes. Our research also indicates that ZM fusion promotes GBM tumorigenicity in vitro and in vivo. Incubation with ZM exosomes enhanced GBM cell tumorigenicity. In summary, all ZM exosome-mediated functional properties contribute to a more malignant phenotype and foster multiple hallmark functions of cancers to promote the development and progressive growth of GBMs at multiple levels.45, 46
However, our unresolved problem is whether exosomal MET and p-MET secretion is random, reflecting cellular expression patterns, or selective, reflecting an active sorting of specific mRNAs and proteins. In our research, the distribution of exosomal MET and p-MET paralleled their abundance in the cell of origin, suggesting the absence of a selective mechanism.
The major implication of the results of this study is that ZM fusion in GBM cells promotes tumour cell proliferation, EMT, migration, invasion, neurosphere formation and angiogenesis in vitro and in vivo, and confers temozolomide resistance. ZM exosomes caused the expected changes to the GBM cell phenotype in the same sets of recipient cells. Hence, ZM exosomes containing MET and p-MET may not only be a novel approach to biomarker detection but may also provide therapeutic intervention targets in aggressive GBM.
Materials and methods
Patients and samples
This study was approved by the institutional review boards of all hospitals involved in the study, and written informed consent was obtained from all patients. Patient sample information is detailed in the online Supplementary Material.
Cell culture
The human GBM cell lines U87, A172, U118 and LN18 (American Type Culture Collection, Manassas, VA, USA) were authenticated by American Type Culture Collection using the short tandem repeat genotyping method. Primary human N3 GBM cells were obtained from the Beijing Neurosurgical Institute, Capital Medical University. Primary human K3 GBM cells were obtained as reported.22 HBMVECs (ScienCell, San Diego, CA, USA) were grown in endothelial cell basal medium supplemented with 5% fetal bovine serum and 1% endothelial cell growth supplement. NHAs (ScienCell) were grown in astrocyte basal medium supplemented with 2% fetal bovine serum and 1% astrocyte growth supplement (ScienCell). For the neurosphere culture, U87 was cultured in stem cell medium consisting of Dulbecco’s modified Eagle’s medium /F12 (Gibco, Rockford, IL, USA) supplemented with 1% N2, 2% B27 (Invitrogen, Carlsbad, CA, USA), 20 ng ml−1 epidermal growth factor and fibroblast growth factor-2 (Invitrogen). K3, NHA and HBMVEC cells were authenticated by Genechem (Shanghai, China) using the short tandem repeat genotyping method. To maintain the authenticity of the cell lines, we prepared frozen stocks from initial stocks, and a new frozen stock was used for the experiments every 3 months.
Exosome size analysis and purification
Exosomes were isolated by the differential centrifugation of conditioned medium collected from U87 and A172 cells grown to 70–80% confluence. The medium was replaced with medium containing exosome-depleted fetal bovine serum media supplement (System Biosciences, Mountain View, CA, USA). After 48 h of incubation, the conditioned medium was cleared of cellular debris/dead cells at 3000 g for 30 min at 4 °C. The resulting supernatants were spun at 100 000 g for 70 min at 4 °C. The exosome pellet was washed with 1 × phosphate-buffered saline solution and centrifuged at 100 000 g for 70 min.47 The final pellet containing exosomes was re-suspended in phosphate-buffered saline and used for follow-up experiments (Supplementary Methods).
Cell co-culture
Cells with ZM fusion (U87/ZM, A172/ZM and N3/ZM) were plated in 0.4 μm porous Transwell inserts (Corning Inc., Corning, NY, USA) suspended over U87/SC, A172/SC, N3/SC and NHA cells plated at a 1:1 or 1:4 ratio and co-cultured for 6 days. In control wells containing only U87/ZM cells, no cells were detected at the bottom of the wells after 6 days, confirming that U87/ZM, A172/ZM and N3/ZM cells could not cross the Transwell membrane.
Western blotting
Protein extraction and western blotting were performed as described previously.7, 48 Representative images from two or three independent experiments are shown.
RNA extraction, reverse transcription–PCR and quantitative reverse transcription–PCR
RNA was isolated with TRIzol according to the manufacturer’s instructions (Invitrogen). SYBR-based quantitative reverse transcription–PCR was performed using pre-designed primers (Life Technologies, Carlsbad, CA, USA). The quantitative reverse transcription–PCR was performed using SYBR Premix Dimer Eraser (Takara, Dalian, China) on a 7900HT system (Applied Biosystems, Foster City, CA, USA). ZM mRNA amplification using fusion-specific PCR primers was performed as described previously.7
Lentiviral and adenovirus packaging and stable cell line establishment
The lentiviral packaging kit was obtained from Open Biosystems (Huntsville, AL, USA). Lentivirus was packaged in human embryonic kidney 293 T cells and collected from the supernatant according to the manufacturer’s instructions. Stable cell lines were established by infecting cells with lentivirus followed by puromycin selection. The lentiviral-based packaging vectors pCDH-RFP-CD63 and pTRIPZ-Rab27a/b short hairpin RNAs were procured from Genechem (Shanghai, China). The ZM fusion gene was packaged in an adenovirus as described previously.7
Exosome internalization and confocal analysis
Exosomes were suspended in phosphate-buffered saline and treated with DiO dye (Vybrant Multi-Color Cell Labeling Kit; Life Technologies) for 20 min at 37 °C. The DiO-dyed exosomes were incubated with recipient cells in a 35 mm culture plate (culture conditions as described above). Confocal microscopy images were obtained after using a Zeiss LSM 510 confocal microscope.
In vitro two-dimensional and 3D assays
We performed and quantified 3D spheroid migration assays in a collagen matrix.22 The vessel-forming ability of HBMVEC cells was characterized in vitro using a Matrigel assay,22 and a cell invasion assay was performed as described previously.5 The wound-healing assay and in vitro chemosensitivity assay were performed as previously described.5 The flow cytometry analysis of apoptosis and exosome interaction was carried out using the Becton Dickinson FACSCalibur system (San Jose, CA, USA). Immunostaining and the ethynyl deoxyuridine assay were visualized with a Leica DMI3000B microscope (Buffalo Grove, IL, USA; Supplementary Methods).
In vivo studies
Male BALB/c nude mice (6 weeks old) were purchased from the Shanghai Laboratory Animal Center (Chinese Academy of Sciences) and maintained in specific pathogen-free conditions for 1 week. The animal handling and experimental procedures were in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the Nanjing Medical University Animal Experimental Ethics Committee. The mice were randomly divided into 10 mice per group, and the in vivo studies were performed as previously described.5, 49 U87 cells incubated with 5 mg SC Exos or ZM Exos were injected subcutaneously into both flanks of nude mice (5 × 106 cells in 100 μl), and tumour sizes were measured using a Vernier calliper every 2 days when the tumours were readily visualized. The tumour volume was calculated according to the following formula: volume=0.5 × length × width2. To establish intracranial gliomas, 0.5 × 105 U87 cells stably expressing the luciferase reporter were stereotactically implanted. Before the tumour cell implantation, U87 cells were treated with 50 μg Exos purified from the culture supernatants of U87/SC or U87/ZM cells and cultured for 6 days in Exo-free medium. The mice were imaged for Fluc activity using bioluminescence imaging. Before imaging, each mouse received an intraperitoneal injection of D-luciferin (10 μl g−1). Tumours from mouse flanks and brains were fixed in 4% paraformaldehyde for 24 h, followed by incubation in 30% sucrose for 48 h. The sections (20 μm) were evaluated for Ki-67 and CD31 fluorescence immunostaining, and paraffin-embedded tissue sections were stained with haematoxylin–eosin (H&E). Three sections per tumour were analysed to quantify staining.
Statistical analysis
All microscopy-based assays were edited/analysed using ImageJ (Bethesda, MD, USA). All experiments were performed thrice, and data were analysed with GraphPad Prism 6 (San Diego, CA, USA). The significance of differences was assessed using the two-tailed Student’s t-test, and the survival analysis was carried out using log-rank tests. The variance was similar between compared groups. P<0.05 was considered to indicate a significant difference (single asterisks or pound signs in the Figures), and P<0.01 was strongly significant (double asterisks).
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (No. 81402056 and 81472362), National High Technology Research, Development Program of China (863) (No.2012AA02A508) and the National Key Research and Development Plan (No. 2016YFC0902500).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A, Hu Q, Liu Y, Wang Z, Cui X, Li R et al. IDH1/2 mutation status combined with Ki-67 labeling index defines distinct prognostic groups in glioma. Oncotarget 2015; 6: 30232–30238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008; 359: 492–507. [DOI] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352: 987–996. [DOI] [PubMed] [Google Scholar]
- Wang L, Shi ZM, Jiang CF, Liu X, Chen QD, Qian X et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget 2014; 5: 5416–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett 2013; 340: 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res 2014; 24: 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer 2015; 15: 371–381. [DOI] [PubMed] [Google Scholar]
- Chen HM, Yu K, Tang XY, Bao ZS, Jiang T, Fan XL et al. Enhanced expression and phosphorylation of the MET oncoprotein by glioma-specific PTPRZ1-MET fusions. FEBS Lett 2015; 589: 1437–1443. [DOI] [PubMed] [Google Scholar]
- Ricklefs F, Mineo M, Rooj AK, Nakano I, Charest A, Weissleder R et al. Extracellular vesicles from high-grade glioma exchange diverse pro-oncogenic signals that maintain intratumoral heterogeneity. Cancer Res 2016; 76: 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, van Rheenen J. Implications of extracellular vesicle transfer on cellular heterogeneity in cancer: what are the potential clinical ramifications? Cancer Res 2016; 76: 2071–2075. [DOI] [PubMed] [Google Scholar]
- Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in tumour microenvironment: overview of the crosstalk between normal and cancer cells. Biomed Res Int 2014; 2014: 179486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol 2015; 40: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Asti E, Garnier D, Lee TH, Montermini L, Meehan B, Rak J. Oncogenic extracellular vesicles in brain tumor progression. Front Physiol 2012; 3: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol 2015; 40: 72–81. [DOI] [PubMed] [Google Scholar]
- Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol 2015; 77: 13–27. [DOI] [PubMed] [Google Scholar]
- Belting M, Wittrup A. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. J Cell Biol 2008; 183: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadou E, Slack FJ. Cancer. malicious exosomes. Science 2014; 346: 1459–1460. [DOI] [PubMed] [Google Scholar]
- Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N et al. Exosomal HIF1alpha supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 2014; 33: 4613–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josson S, Gururajan M, Sung SY, Hu P, Shao C, Zhau HE et al. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015; 34: 2690–2699. [DOI] [PubMed] [Google Scholar]
- Bronisz A, Wang Y, Nowicki MO, Peruzzi P, Ansari KI, Ogawa D et al. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res 2014; 74: 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Hamar P, Guo C, Basar E, Perdigao-Henriques R, Balaj L et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest 2014; 124: 5109–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Eaton SA, Young PE, Lee M, Shuttleworth R, Humphreys DT et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol 2013; 10: 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10: 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res 2009; 15: 2207–2214. [DOI] [PubMed] [Google Scholar]
- Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984; 311: 29–33. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe D, Kong-Beltran M. Met activation and receptor dimerization in cancer: a role for the Sema domain. Cell Cycle 2005; 4: 683–685. [DOI] [PubMed] [Google Scholar]
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012; 18: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi E, Sakai K, Nishiuchi T, Imamura R, Sato H, Matsumoto K. Different growth and metastatic phenotypes associated with a cell-intrinsic change of Met in metastatic melanoma. Oncotarget 2016; 7: 70779–70793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Du Z, Zhang M. Biomarker development in MET-targeted therapy. Oncotarget 2016; 7: 37370–37389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Q, Yang G, Marando C, Koblish HK, Hall LM et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin Cancer Res 2011; 17: 7127–7138. [DOI] [PubMed] [Google Scholar]
- Graziani A, Gramaglia D, Cantley LC, Comoglio PM. The tyrosine-phosphorylated hepatocyte growth factor/scatter factor receptor associates with phosphatidylinositol 3-kinase. J Biol Chem 1991; 266: 22087–22090. [PubMed] [Google Scholar]
- Lee JK, Joo KM, Lee J, Yoon Y, Nam DH. Targeting the epithelial to mesenchymal transition in glioblastoma: the emerging role of MET signaling. Onco Targets Ther 2014; 7: 1933–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol 2010; 11: 834–848. [DOI] [PubMed] [Google Scholar]
- Lopez-Bertoni H, Lal B, Li A, Caplan M, Guerrero-Cazares H, Eberhart CG et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 2015; 34: 3994–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MC, Andriantsitohaina R. Microparticles in angiogenesis: therapeutic potential. Circ Res 2011; 109: 110–119. [DOI] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia 2012; 60: 502–514. [DOI] [PubMed] [Google Scholar]
- Boccaccio C, Comoglio PM. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Res 2013; 73: 3193–3199. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Heddleston JM, Venere M, Rich JN. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell 2011; 8: 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Chen Z, Wang S, Zhang R, Qiu W, Zhao L et al. c-Myc-miR-29c-REV3L signalling pathway drives the acquisition of temozolomide resistance in glioblastoma. Brain 2015; 138: 3654–3672. [DOI] [PubMed] [Google Scholar]
- Messaoudi K, Clavreul A, Lagarce F. Toward an effective strategy in glioblastoma treatment. Part I: resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov Today 2015; 20: 899–905. [DOI] [PubMed] [Google Scholar]
- Messaoudi K, Clavreul A, Lagarce F. Toward an effective strategy in glioblastoma treatment. Part II: RNA interference as a promising way to sensitize glioblastomas to temozolomide. Drug Discov Today 2015; 20: 772–779. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev 2015; 29: 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Javeed N, Sagar G, Dutta SK, Smyrk TC, Lau JS, Bhattacharya S et al. Pancreatic cancer-derived exosomes cause paraneoplastic beta-cell dysfunction. Clin Cancer Res 2015; 21: 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang C et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol 2014; 16: 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Wei W, Chen X, Zhang Y, Wang Y, Zhang J et al. NF-kappaB/RelA-PKM2 mediates inhibition of glycolysis by fenofibrate in glioblastoma cells. Oncotarget 2015; 6: 26119–26128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.