Abstract
WRKY transcription factors (TFs) are involved in regulating a range of biological processes such as growth, development, and the responses to biotic and abiotic stresses. Genome-wide expression profiling of OsWRKY TF superfamily genes in rice after infection with Xanthomonas oryzae pv. oryzae (Xoo) was performed to elucidate the function of OsWRKY TFs in the interaction between rice and Xoo. Of the 111 OsWRKY TF genes tested, the transcription of 94 genes changed after Xoo infection. The OsWRKY TF genes were classified into eight types according to their expression profiles. Eighty-two genes in Groups I, II, III, IV, VII were up-regulated after exposure to a compatible or an incompatible race of Xoo. Examination of salicylic acid (SA)-deficient rice lines revealed that SA was involved in Xa1-mediated resistance to Xoo infection. OsWRKY TF genes involved in Xa1-mediated resistance were classified according to their SA-dependent or -independent expression. In SA-deficient rice, the expression of 12 of 57 OsWRKY TF genes involved in Xa1-mediated resistance was compromised. Of these six OsWRKY TF genes were induced by SA. OsWRKY88, an example of a gene possibly involved in SA-dependent Xa1-mediated resistance, activated defense related genes and increased resistance to Xoo. Thus, expression profiling of OsWRKY TF genes may help predict the functions of OsWRKY TF genes involved in Xa1-mediated resistance.
Keywords: OsWRKY transcription factors superfamily, expression profiling, Xanthomonas oryzae pv. oryzae
Introduction
WRKY transcription factors (TFs) are characterized by the presence of one or two 60-amino-acid WRKY motifs, a very highly conserved WRKYGQK sequence, and zinc finger-like motifs [Cys(2)-His(2) or Cys(2)-HisCys] that bind to the W-box [TTGAC(C/T)] cis-element in target gene promoters (Sun et al., 2003; Cai et al., 2008; Ciolkowski et al., 2008; Pandey and Somssich, 2009). WRKY TFs are classified into three groups (Groups I–III) according to the similarity of their WRKY motifs (Zhang and Wang, 2005). WRKY TF families in Arabidopsis and rice contain 72 and 125 members, respectively (Eulgem et al., 2000; Rice WRKY Working Group., 2012).
Numerous WRKY TFs are involved in responding to biotic stresses in rice and Arabidopsis (Dong et al., 2003; Ryu et al., 2006; Berri et al., 2009). Several studies described expression profiling of OsWRKY TF genes upon infection with pathogens such as Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae (Xoo) in rice (Ryu et al., 2006; Bagnaresi et al., 2012; Wei et al., 2013). The first such study examined the expression of 45 OsWRKY TF genes in response to M. oryzae challenge and found that 15 OsWRKYs were up-regulated upon infection (Ryu et al., 2006). Later research with a custom OsWRKYARRAY found that 18 of 104 OsWRKY TF genes were up-regulated upon M. oryzae infection (Berri et al., 2009). A recent transcriptome study in rice noted that OsWRKY TF genes were significantly enriched in the group of genes up-regulated during the early defense response to M. oryzae infection and that OsWRKY47 played a critical role in rice blast resistance (Wei et al., 2013).
To date, relatively few studies have examined OsWRKY TF superfamily gene expression upon infection with Xoo, but several studies have examined individual OsWRKY TF genes in Xoo-mediated resistance. One study reported that 9 of 45 OsWRKY genes, namely, OsWRKY7, 10, 11, 30, 32, 67, 70, 83 [renamed 94 by the Committee on Gene Symbolization, Nomenclature, and Linkage (CGSNL)], and 85 (renamed 96 by CGSNL) were up-regulated upon infection with Xoo (Ryu et al., 2006). The expression profiles of some OsWRKY genes were summarized by Jimmy and Babu (Jimmy and Babu, 2015). Other studies observed up-regulation of OsWRKY6, 12, 13, 30, 45-1, 71, and 76 in response to Xoo infection (Liu et al., 2005, 2007; Chujo et al., 2008; Qiu and Yu, 2009; Tao et al., 2009; Hwang et al., 2011; Lee et al., 2013; Choi et al., 2015). However, no comprehensive genome-wide expression data are available for all 125 known OsWRKY superfamily genes.
SA has been shown to play a key role in plant immunity (Delaney et al., 1994; Jimmy and Babu, 2015). Many WRKY genes from a number of plants have also been shown to be induced by SA (Dong et al., 2003; Ryu et al., 2006). Transgenic plants expressing the bacterial NahG gene encoding a salicylate hydroxylase that converts SA to catechol are often used to evaluate the role of SA in plant immunity (Delaney et al., 1994; Yang et al., 2004). Pathogen mediated WRKY gene expression has also been examined in NahG Arabidopsis plants (Dong et al., 2003). To date there is no report on SA effect using NahG transgenic rice plants in pathogen mediated OsWRKY gene expression yet.
Nine resistance (R) genes for Xoo, namely, Xa1, Xa3/26, xa5, Xa10, xa13, Xa21, Xa23, xa25, and Xa27, were cloned previously (Song et al., 1995; Yoshimura et al., 1998; Iyer and McCouch, 2004; Sun et al., 2004; Gu et al., 2005; Chu et al., 2006; Liu et al., 2011; Tian et al., 2014; Wang C. et al., 2015). Most proteins encoded by plant disease R genes share common structural features such as nucleotide-binding site leucine-rich repeat (NBS-LRR) regions. Of the nine Xoo R genes cloned to date, only Xa1 encodes a typical NBS-LRR-type protein. The mechanisms underpinning Xa1-mediated resistance remain largely unknown. In this study, genome-wide expression profiling of OsWRKY superfamily genes was used to investigate Xa1-mediated defense mechanisms and the function of OsWRKY88, an example of a gene possibly involved in SA-dependent Xa1-mediated resistance, was analyzed.
Materials and methods
Plant material and pathogen treatment
Rice seedlings (Oryza sativa L. Japonica cv. Ilmi) carrying the Xa1 R gene were grown in a greenhouse at 28°C for 3 weeks. Seedlings were inoculated with compatible (KACC10859) or incompatible (KXO42) Xoo strains grown on PSA medium (10 g/L peptone, 10 g/L sucrose, 1 g/L sodium glutamate, and 15 g/L agar) for 2 days until OD600 = 0.5 (Kim et al., 2013). Leaves were harvested at 0, 6, 12, and 24 hpi. Seedlings were treated with 1 mM SA, and leaves were harvested at 0, 12, 24 h after treatment (hat). All samples were frozen in liquid nitrogen immediately after harvesting and stored at −80°C until used.
Quantitative RT-PCR
The 125 OsWRKY TF superfamily genes were reported by CGSNL (Rice WRKY Working Group., 2012; Table S1). Among them we could not obtain the data for the 14 OsWRKY TFs in this study due to various reasons. Primers for qRT-PCR were designed using the rice genome annotation database (http://rice.plantbiology.msu.edu; Tables S1, S2). Nucleotide sequences for OsWRKY44 and OsWRKY59 were not found in the database. Some OsWRKY TFs are duplicated genes: OsWRKY46 (91), 61 (103), 4 (122). NCBI primer-blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to optimize the design of specific primers for each OsWRKY TF gene. For nine OsWRKY TFs (OsWRKY24, 33, 35, 57, 61, 92, 99, 116, 124) we could not optimize the condition to get PCR products. Specific qRT-PCR primers for OsWRKY TF genes are listed in Table S1.
Total RNA was extracted from harvested leaves using Trizol reagent (Invitrogen, USA). M-MLV reverse transcriptase (Promega, USA) was used to synthesize cDNA from total RNA (2 μg). qRT-PCR was conducted as described by Kim et al. (2013). OsActin was used as a calibration control to determine OsWRKY gene expression levels (Schmittgen and Livak, 2008). The 2−ΔΔct method was used for analysis of relative gene expression. Three biological replicates were performed for each qRT-PCR analysis. Above 2-fold difference at each time point in each expression profile was considered as either increase or decrease of OsWRKY expression.
Generation of transgenic rice plants
Gateway cloning using pB2GW7 (Karimi et al., 2002) was used to generate constructs 35S::NahG from Arabidopsis NahG plants and 35S::OsWRKY88. Agrobacterium tumefaciens LBA4404 was transformed with the 35S::NahG construct and the 35S::OsWRKY88 construct. A. tumefaciens carrying both constructs was used to transform rice embryogenic calli (Kim et al., 2009). Transgenic rice T0 plants were selectively screened using 0.1% Bastar™ spray. Integration of the construct was confirmed by PCR of genomic DNA with bar-specific primer sets (Table S2). Homozygous NahG transgenic rice plants were confirmed by 0.1% Bastar™ spray through several generations and used for disease assays and qRT-PCRs. In the case of OsWRKY88 ox lines we used T2 lines selected by 0.1% Bastar™ spray.
Plant disease assays
Xoo KXO42 and KACC10859 bacteria were grown overnight at 28°C in PSA medium and resuspended at 108 cells/mL in 10 mM MgCl2. The scissors-dip method (Kauffman et al., 1973) was used to challenge WT and ten SA-deficient rice plants with Xoo (KXO42) and WT and ten OsWRKY88 ox lines (T2) with Xoo (KACC10859). Disease was assessed as the average lesion length in leaves of individual plants 14 days post-inoculation (dpi). Bacterial growth was assessed at 0, 7, and 14 dpi (Shimono et al., 2007).
Promoter transient expression assays
A 2 kb region of the CHITINASE 2 (CHIT2; Os04g41620) promoter was amplified by PCR using promoter-specific primers (Table S2) and introduced into an entry vector, pENTR/d-TOPO (Invitrogen, Carlsbad, CA, USA). The pCHIT2::GFP-GUS construct was made an LR reaction between the entry clones containing the promoter and the destination vector pBGWFS7 (Karimi et al., 2002). Transient expression assays in Nicotiana benthamiana were performed using the protocol reported previously (Li, 2011). N. benthamiana was infiltrated with an Agrobacterium carrying pCHIT2::GFP-GUS and pPR10a::GFP-GUS (Hwang et al., 2008) constructs alone, or with a mixture of Agrobacterium carrying 35S::OsWRKY88 and either pCHIT2::GFP-GUS or pPR10a::GFP-GUS. Infiltrated leaves were collected 2 days post-infiltration, and promoter activities in each sample were visualized using β-glucuronidase (GUS) activity assay and staining.
Results
Expression profiles of OsWRKY TF superfamily genes during Xoo infection were analyzed
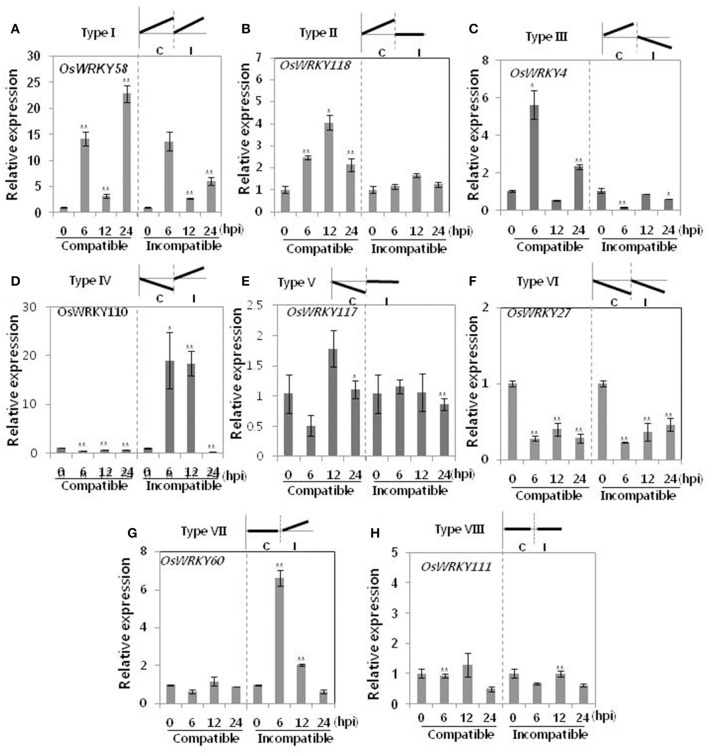
The roles of OsWRKY TF superfamily genes in the defense pathway mediated by Xa1, a bacterial blight resistance protein (Yoshimura et al., 1998), were investigated using expression profiling. Oryza sativa L spp. japonica cv. Ilmi, which carried the Xa1 gene, was profiled after challenge with an incompatible (KXO42) or a compatible (KACC10859) race of Xoo. OsWRKY TF superfamily genes used in this study were named according to recently re-defined nomenclature (Rice WRKY Working Group., 2012; Table S1). Expression profiles of OsWRKY TF superfamily genes were shown in eight different types and the results were summarized in Table 1 (Types I–VIII). The expression profile of a representative OsWRKY TF in each type was shown in Figure 1 and the rest of them were shown in Figure S1. Most of the OsWRKY TFs (57 genes) were up-regulated in response to both the compatible and incompatible Xoo races (Type I). Of these, OsWRKY58 transcription was particularly elevated upon challenge with Xoo (Figure 1A). OsWRKY TFs (OsWRKY107, 118) showed Type II pattern that were up-regulated only in response to the compatible Xoo race (Table 1). Of these, transcription of OsWRKY118 was noticeably elevated 6 hpi (Figure 1B). Fourteen OsWRKY TFs (OsWRKY4, 5, 14, 19, 28, 48, 78, 79, 82, 85, 86, 98, 101, 121) showed Type III pattern that up-regulated in response to the compatible Xoo race and down-regulated in response to the incompatible Xoo race. Of these transcription of OsWRKY4 was noticeably elevated 6 hpi (Figure 1C). Six OsWRKY TFs (OsWRKY74, 77, 110, 119, 120) showed Type IV pattern that were down-regulated in response to the compatible Xoo race and up-regulated in response to the incompatible Xoo race. Of these OsWRKY110 was decreased at 3 hpi upon the infection of the compatible Xoo race and increased at 12 hpi upon the infection of the incompatible Xoo race (Figure 1D). Two OsWRKY TFs showed Type V pattern that were down regulated only upon infection of the compatible Xoo race. Of these OsWRKY117 was decreased at 6 hpi (Figure 1E). Twenty two OsWRKY TFs showed Type VI pattern that was down regulated both upon infection of the compatible and incompatible Xoo race. Of these, OsWRKY27 was dramatically down-regulated 6 hpi with both the compatible and incompatible interactions (Figure 1F). Type VII contained three OsWRKY TFs (OsWRKY32, 46, 60) that were up-regulated only in response to only the incompatible Xoo race. Of these, the OsWRKY60 transcript reached maximum levels 6 h post-inoculation (hpi; Figure 1G). Finally, Type VIII contained five OsWRKY TFs (OsWRKY53, 72, 102, 111, 125) whose transcription profiles were unaffected by Xoo infection (Figure 1H). For the reasons described in the Materials and Methods section of qRT-PCR and Table 1, no expression profile data were obtained for 14 OsWRKY genes (OsWRKY24, 33, 35, 44, 57, 59, 61, 91, 92, 99, 103, 116, 122, 124).
Table 1.
Expression profiles of OsWRKY TF genes in response to Xoo infection were summarized.
| Types | Expression profile | OsWRKY TFs |
|---|---|---|
| I | 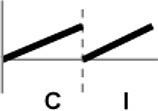 |
OsWRKY1, 2, 3, 6, 7, 8, 9, 10, 11, 12, 13, 16, 18, 22, 23, 25, 26, 29, 30, 36, 38, 40, 42, 43, 45, 47, 49, 50, 51, 52, 54, 55, 58, 63, 64, 65, 66, 69, 70, 71, 76, 80, 81, 84, 87, 88, 90, 94, 96, 105, 106, 108, 112, 113, 114, 115, 123 |
| II | 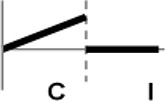 |
OsWRKY107, 118 |
| III | 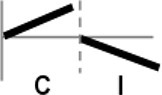 |
OsWRKY4, 5, 14, 19, 28, 48, 78, 79, 82, 85, 86, 98, 101, 121 |
| IV | 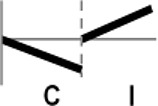 |
OsWRKY56, 74, 77, 110, 119, 120 |
| V | 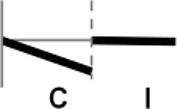 |
OsWRKY72, 117 |
| VI | 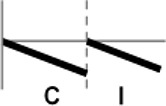 |
OsWRKY15, 17, 20, 21, 27, 31, 34, 37, 39, 41, 54, 56, 62, 67, 68, 73, 75, 83, 93, 95, 104, 109 |
| VII | 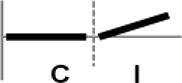 |
OsWRKY32, 46, 60 |
| VIII |  |
OsWRKY53, 72, 111, 102, 125 |
No data were generated for the following OsWRKY TF genes.
1. Nucleotide sequences for OsWRKY44 and OsWRKY59 were not found.
2. Duplicated genes: OsWRKY91 (46), 103 (61), 122 (4).
3. No PCR products were obtained: OsWRKY24, 33, 35, 57, 61, 92, 99, 116, 124.
Figure 1.
Expression profiling of OsWRKY TF genes upon infection with compatible and incompatible races of Xoo. (A) Type I: Genes up-regulated after challenge with both compatible and incompatible Xoo races. (B) Type II: Genes up-regulated only after challenge with a compatible Xoo race. (C) Type III: Genes up-regulated after challenge with a compatible Xoo race and down-regulated by infection of incompatible Xoo race. (D) Type IV: Genes down-regulated after challenge with a compatible Xoo race and up-regulated by infection of incompatible Xoo race. (E) Type V: Genes down-regulated only after challenge with an incompatible Xoo race. (F) Type VI: Genes up-regulated after challenge with both compatible and incompatible Xoo races. (G) Type VII: Genes up-regulated only after challenge with an incompatible Xoo race. (H) Type VIII: Genes exhibiting no expression response upon challenge with either a compatible or an incompatible Xoo race. Asterisks indicate significant differences (**P < 0.01; *P < 0.05).
Salicylic acid plays a role in Xa1-mediated resistance
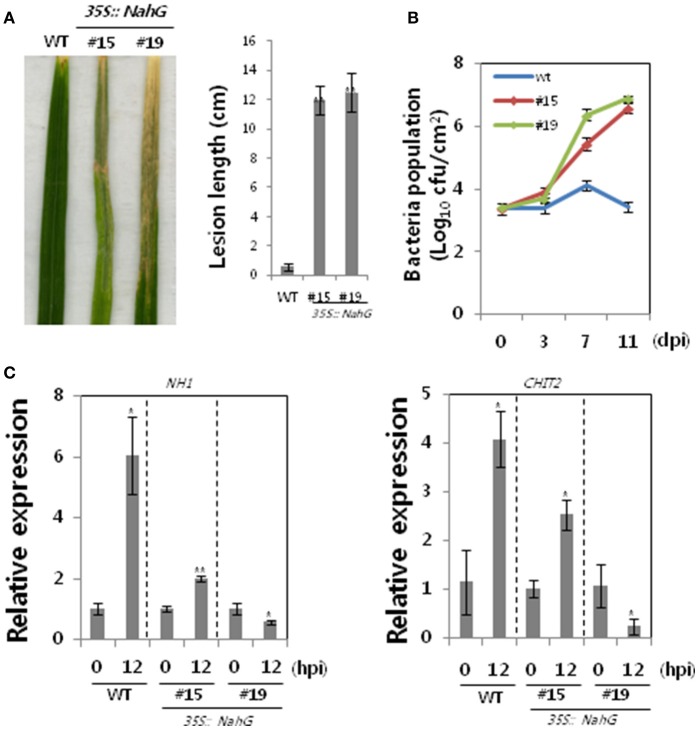
Transcription of 66 OsWRKY TF genes (Groups I, IV, and VII) was up-regulated during the incompatible interaction of rice cultivar Ilmi carrying Xa1 with Xoo, suggesting that many OsWRKY TFs could be involved in the Xa1-mediated defense pathway. Salicylic acid (SA) has a role in plant defense response, and SA-deficient transgenic rice plants were therefore used to investigate the involvement of SA in Xa1-mediated defense. Bacterial salicylate hydroxylase, which is encoded by the NahG gene, degrades SA to catechol. SA-deficient transgenic rice plants were generated by introduction of the NahG gene into rice via Agrobacterium-mediated transformation. Nineteen independent NahG-expressing transgenic lines were produced. Lines #15 and #19 were selected for further analysis based on their NahG transcript levels (data not shown). Transgenic lines challenged with incompatible Xoo race KXO42 exhibited longer lesions (~13 cm) than wild-type (WT) Ilmi plants (~0.2 cm; Figures 2A,B), indicating that Xa1-mediated resistance was compromised in the transgenic plants. Bacterial populations in the two SA-deficient lines were approximately higher than in the WT at 3, 7, and 11 dpi, respectively (Figure 2B). Activation of the Xa1-mediated defense pathway in the SA-deficient lines was examined using qRT-PCR transcriptional analysis of the genes encoding NH1/OsNPR1 and chitinase 2 (CHIT2). Compared with the WT, induction of both genes was partially compromised in line #15 and severely compromised in line #19 (Figure 2C). These observations correlated with lesion length and bacterial growth data. On the other hand, some studies claim that catechol results in susceptibility to Magnaporthe grisea but not to Striga hermonthica (Yang et al., 2004; Mutuku et al., 2015). We therefore examined the effect of catechol on Xoo infection (Figure S1), which revealed it had no effect on compatible or incompatible Xoo-rice interactions. Thus, impaired defense in SA-deficient lines is not due to catechol production. Overall, these results suggest that SA plays a role in Xa1-mediated resistance.
Figure 2.
Disease assays of SA-deficient rice plants infected with an incompatible Xoo race. (A) WT (Ilmi cultivar carrying Xa1) and SA-deficient rice plants derived from the Ilmi cultivar were challenged with an incompatible race of Xoo. Lesion lengths were measured at 14 dpi. (B) Bacterial growth was assessed at 3 and 11 dpi. (C) qRT-PCR expression analysis of genes encoding NH1 and chitinase in WT and SA-deficient rice lines 12 h after challenge with an incompatible race of Xoo. Asterisks indicate significant differences (**P < 0.01; *P < 0.05).
SA-dependent transcriptional induction of OsWRKY TFs up-regulated in the Xa1-mediated defense pathway
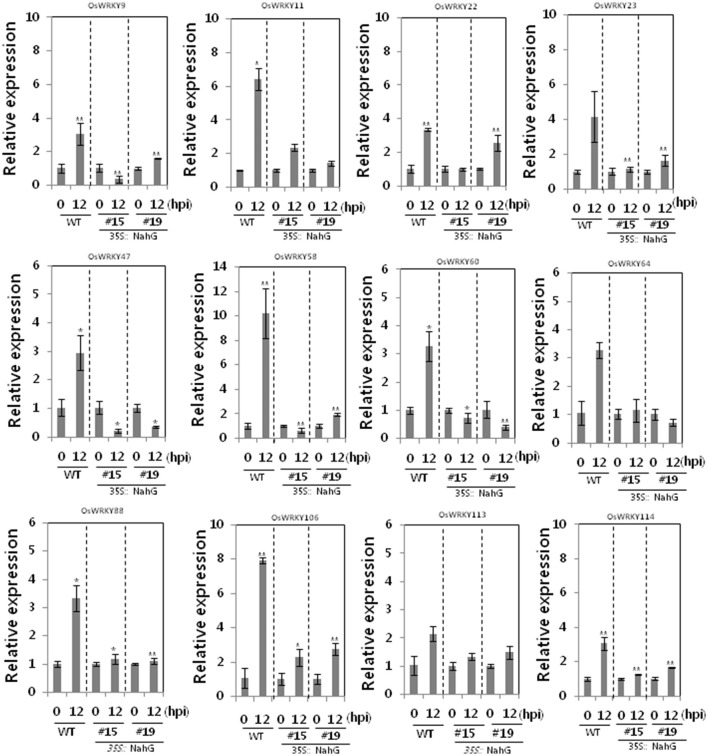
66 OsWRKY TF genes (Groups I, IV, and VII) were up-regulated after challenge with the incompatible Xoo race (Table 1). Type I expression pattern is further divided into Type Ia and Ib. OsWRKY TF genes in Type Ia exhibited more pronounced up-regulation (either a faster response or a more abundant response) with the incompatible interaction than with the compatible interaction, whereas OsWRKY TF genes in Type Ib exhibited a more pronounced effect with the compatible interaction than with the incompatible interaction. OsWRKY TF genes in Type Ia (Table 2) exhibited expression profiles typical of defense response genes, and this group was therefore used to further investigate SA-dependent Xa1-mediated resistance. Expression of OsWRKY TF genes in Type Ia was investigated in SA-deficient rice plants using qRT-PCR (Figure 3). Gene expression during the incompatible interaction was not compromised in the SA-deficient rice lines for the majority of Type Ia OsWRKY TF genes (34 genes). However, induction of OsWRKY9, 11, 22, 23, 47, 58, 60, 64, 88, 106, 113, and 114 expression during the incompatible interaction was reduced in the two SA-deficient rice lines. This suggests that OsWRKY9, 11, 22, 23, 47, 58, 60, 64, 88, 106, 113, and 114 were induced in a SA-dependent manner as part of the Xa1-mediated defense pathway but that other OsWRKY TF genes in Type 1a were induced as part of the Xa1 mediated response in a SA-independent manner.
Table 2.
Expression of OsWRKY TF genes in SA-deficient rice after challenge with an incompatible race of Xoo assessed by qRT-PCR.
| Type I | Expression pattern | Gene |
|---|---|---|
| OsWRKY2, 6, 7, 8, 9, 11, 12, 18, 22, 23,25 ,26, 29, 42, 45, 47, 50, 52, 55, 58, 60, 64, 65, 66,76, 80, 84, 88, 105, 106, 108, 112, 113, 114 | Compromised in SA-deficient rice compared with the WT | OsWRKY9, 11, 22, 23, 47, 58, 60, 64, 88, 106, 113, 114 |
| Unchanged in SA-deficient rice compared with the WT | OsWRKY2, 7, 12, 18, 25, 26, 42, 45, 50, 52, 55, 65, 66, 76, 80, 84, 105, 108, 112 |
Type I OsWRKY TF genes were further classified into Type Ia and Ib. Type Ia contained OsWRKY TF genes that exhibited more pronounced up-regulation with the incompatible interaction than with the compatible interaction. Type Ib contained OsWRKY TF genes that exhibited more pronounced up-regulation with the compatible interaction than with the incompatible interaction.
Figure 3.
Expression of OsWRKY TF genes up-regulated by infection of an incompatible Xoo race in SA-deficient rice plants. qRT-PCR expression analysis of OsWRKY genes in WT and SA-deficient rice lines 12 h after challenge with an incompatible race of Xoo. Asterisks indicate significant differences (**P < 0.01; *P < 0.05).
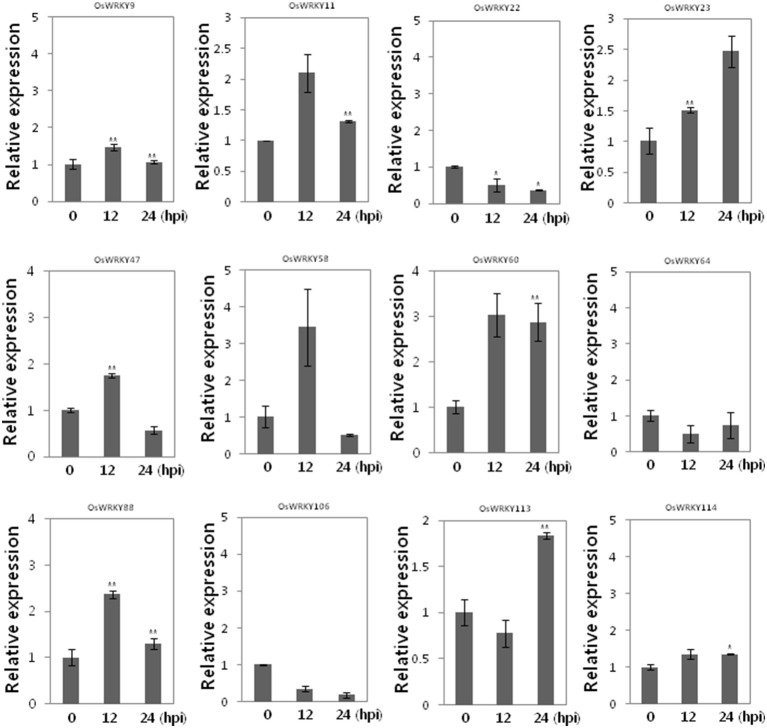
To investigate whether these OsWRKY TFs are induced by SA we performed qRT-PCR (Figure 4). Among 12 OsWRKY TFs OsWRKY11, 23, 47, 58, 60, 88 were induced by SA. However, OsWRKY9, 22, 64, 106, 113, 114 were not induced by SA. Taken together the six OsWRKY TFs appears to play roles in Xa1 mediated resistance.
Figure 4.
Expression of 12 OsWRKY TF genes in response to SA. Expression analysis of 12 OsWRKY genes in WT at 0, 12, 24 h after SA treatment. Asterisks indicate significant differences (**P < 0.01; *P < 0.05).
Ectopic expression of OsWRKY88, a type I OsWRKY TF that enhanced resistance to Xoo
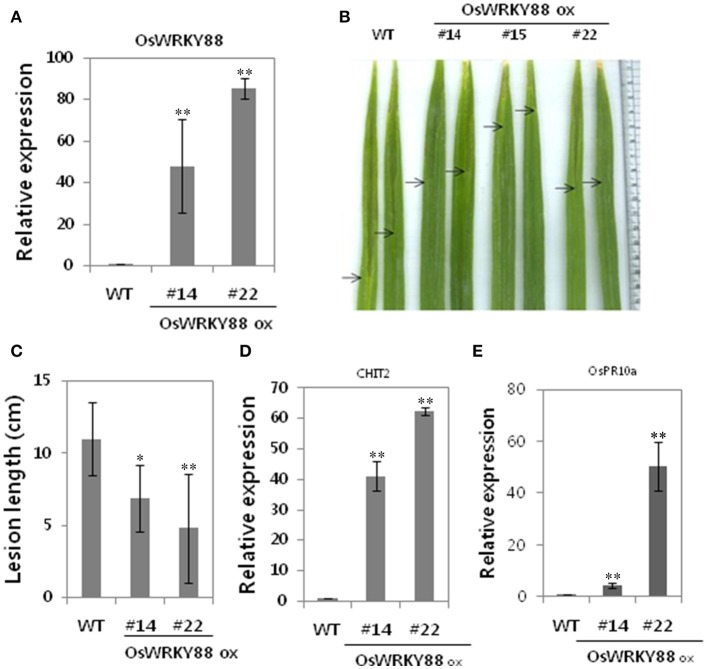
We showed that six OsWRKY TFs were induced as part of SA-dependent Xa1-mediated resistance. Among them the transgenic plants overexpressing OsWRKY88 were generated and analyzed. We found that OsWRKY88 transcript levels were higher in #22 lines than that in #14 lines (Figure 5A). OsWRKY88 ox lines were challenged with the compatible Xoo strain KACC10859 by the leaf clip method. After challenge, OsWRKY 88 ox lines #14 and #22 exhibited shorter lesion lengths (~4 cm) than those (~12c m) of WT plants (Figures 5B,C), suggesting that OsWRKY88 plays a role in Xoo resistance. We also performed qRT-PCR of defense related genes such as CHIT2 and OsPR10a. Unlike in WT plants, CHIT2 and OsPR10a were constitutively expressed in OsWRKY88 ox lines #14 and #22 (Figures 5D,E), indicating that OsWRKY88 positively regulates the defense response to Xoo.
Figure 5.
Analysis of OsWRKY88 ox plants. (A) Expression analysis of OsWRKY88 in OsWRKY88 ox rice lines and the WT (Ilmi cultivar carrying Xa1). (B,C) OsWRKY88 ox rice plants derived from the Ilmi cultivar were challenged with a compatible strain of Xoo. Photos were taken and lesion lengths were measured at 14 dpi. (D,E) Expression analysis of defense related genes (OsPR10a, CHIT2) in OsWRKY88 ox lines. Asterisks indicate significant differences (**P < 0.01; *P < 0.05).
OsWRKY88 directly regulates the OsPR10a promoter but not the CHIT2 promoter
Defense related genes OsPR10a and CHIT2 were expressed at a high level in OsWRKY88 ox lines. To examine whether OsWRKY88 directly regulates OsPR10a and CHIT2, we examined its effect on transcription from the OsPR10a and CHIT2 gene promoters using the promoter transient assay (Figure 6). Agrobacterium-mediated transient assays for pOsPR10a::GFP-GUS (Hwang et al., 2008) were performed in N. benthamiana leaves (Figures 6A,C). The trans-activation activity of OsWRKY88 at the OsPR10a promoter was assessed using GUS staining and GUS enzyme activity. GUS activity was stronger in leaves co-infiltrated with pOsPR10a::GFP-GUS and 35S::OsWRKY88 than in leaves infiltrated with either pOsPR10a::GFP-GUS or 35S::OsWRKY88. We also performed a CHIT2 promoter transient assay (Figures 6B,D), which revealed no significant difference in GUS staining and GUS enzyme activity between the sample co-infiltrated with a mixture of pCHIT2::GFP-GUS and 35S::OsWRKY88 and infiltrated with either pCHIT2::GFP-GUS or 35S::OsWRKY88. These results suggest that OsWRKY88 trans-activates the OsPR10a promoter but not the CHIT2 promoter.
Figure 6.
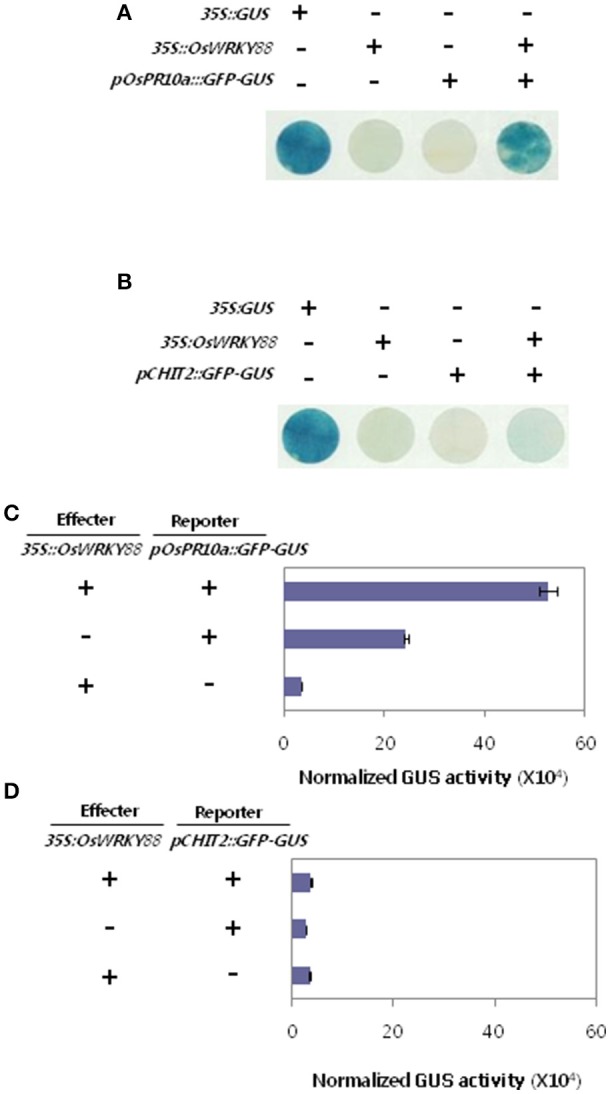
Promoter transient assays in N. benthamiana. (A,B) Agrobacterium carrying promoter-reporter constructs pOsPR10a::GFP-GUS or pCHIT2::GFP-GUS were co-infiltrated into leaves along with Agrobacterium carrying an effector construct (35S::OsWRKY88). Infiltrated leaves were removed at 24 hpi and visualized by GUS staining. (C,D) Infiltrated leaves used to measure GUS enzyme activity.
Discussion
Plants have developed unique immune systems for defense against a range of stresses. WRKY TFs have roles in biological processes such as growth, development, and responses to abiotic stress, but are also involved in regulating plant immune responses (Jimmy and Babu, 2015). In this study, expression of OsWRKY TF genes was profiled during pathogenic challenge with Xoo to elucidate the roles of OsWRKY TFs in the defense response.
Genes encoding WRKY superfamily TFs in Arabidopsis and rice were previously expression profiled in response to biotic stresses (Dong et al., 2003; Ryu et al., 2006). In this study, gene expression was modulated in 106 of the 111 OsWRKY TF genes examined in response to Xoo infection (Table 1). Approximately 96% of OsWRKY TFs were up- or down-regulated upon Xoo infection. The OsWRKY TF genes showed 8 different types of expression profiles (Table 1). Genes in type I, II, III, IV, and V (81 OsWRKY TF genes in total; ~73%of the 111 OsWRKY TF genes) were up-regulated after challenge with a compatible or an incompatible race of Xoo. In Arabidopsis leaves, gene expression was modulated in 49 of 53 AtWRKY TF genes (~90%) as assessed by northern blotting (Dong et al., 2003). Of these, 43 AtWRKY TF genes (~81%) were up-regulated after challenge with an incompatible pathogen. Research in rice reported that 9 of 15 (60%) OsWRKY TF genes tested were induced by an incompatible race of Xoo (Ryu et al., 2006). However, the study examined only 15 of the 125 known OsWRKY TF genes. In this study, 66 of 111 OsWRKY TF genes (type I, IV, and VII; 60%) were up-regulated by an incompatible race of Xoo. This discrepancy may be due to the sensitivity of methods such as RT-PCR and qRT-PCR when used for expression analysis. The previous study noted that OsWRKY7, 10, 11, 30, 32, 67, 70, 83 (renamed 94 by CGSNL), and 85 (renamed 96 by CGSNL) were induced by an incompatible race of Xoo (Ryu et al., 2006). Of these, OsWRKY7, 10, 11, 30, 32, 70, 94, and 96 were also induced by an incompatible race of Xoo in our results. However, in contrast with the previous study (Ryu et al., 2006), our results showed clear down-regulation of OsWRKY67 after exposure to incompatible and compatible Xoo races. This discrepancy is also predicted to be caused by the different methods used for expression analysis of OsWRKY TF genes in the two studies. Consistent with our data, previous studies showed that OsWRKY03 (renamed 12 by CGSNL), 6, 13, 30, 45, and 76 were induced by Xoo and were involved in regulation of Xoo-mediated resistance (Liu et al., 2005, 2007; Chujo et al., 2008; Qiu and Yu, 2009; Tao et al., 2009; Hwang et al., 2011; Han et al., 2013; Lee et al., 2013; Choi et al., 2015).
Studies exploring OsWRKY TF gene expression after infection with a compatible race of Xoo are scarce. Expression profiles of defense-related genes induced by compatible and incompatible pathogens have been identified (Jimmy and Babu, 2015). However, to our knowledge, the expression profiles of genes induced only by compatible pathogens have not been published. In this study, OsWRKY TF gene expression profiles were generated in response to challenge with a compatible race of Xoo. Sixteen OsWRKY TF genes were induced by compatible infection. Of these, 14 OsWRKY TFs (OsWRKY4, 5, 14, 19, 28, 48, 78, 79, 82, 85, 86, 98, 101 and 121) were down-regulated by incompatible infection, suggesting that some OsWRKY TFs might function as negative regulators in the defense response, resulting in increased disease susceptibility to Xoo. Previous research indicated that OsWRKY4 enhanced resistance to a necrotrophic pathogen, Rhizotonia solani, and was involved in the jasmonic acid (JA)-mediated pathway (Wang H. et al., 2015). This suggests that OsWRKY4 has a negative role in responding to challenge with biotrophic pathogens such as Xoo. Another TF, OsWRKY28, was found to act as a negative regulator in innate immunity, consistent with the gene expression profile seen in this study (Qiu et al., 2007). Xoo produces transcription activator-like (TAL) effectors that activate transcription of susceptibility (S) genes such as OsSWEET14 and OsTFX1 (Huang et al., 2016). Type II and III OsWRKY TFs might therefore be targets of TAL effectors.
Previous studies showed that SA was involved in Xoo-mediated resistance (Chujo et al., 2013; Choi et al., 2015). However, the incompatible interactions of Xoo with Mudanjiang 8 and Minghui 63 rice varieties were due to the Xa3/26 resistance factor, which is a typical pattern recognition receptor, rather than a NBS-LRR-type resistance factor. This resistance mechanism resembled pattern-triggered immunity, and it therefore remained unclear whether SA was involved in canonical R-gene-mediated resistance against Xoo in rice. In this study, we demonstrated that SA-deficient rice exhibited disease susceptibility when exposed to an incompatible race of Xoo, while the WT plant (Ilmi, carrying Xa1) exhibited hypersensitive-response-type resistance. This result suggested that SA was involved in Xa1-mediated resistance. OsWRKY TF genes induced during the Xa1-mediated resistance response were classified based on their expression profiles in response to SA. Of the 34 OsWRKY TF genes in Types I, III, and V, induction of 12 OsWRKY TF genes (OsWRKY9, 11, 22, 23, 47, 58, 60, 64, 88, 106, 113, and 114) by Xoo was compromised in SA-deficient rice lines generated by over-expression of NahG. Among them 6 OsWRKY TF genes (OsWRKY11, 23, 47, 58, 60, 88) were induced by SA. We therefore propose that Xa1-mediated resistance involves both SA-dependent and SA-independent pathways. Induction of OsWRKY TF genes such as OsWRKY10, 30, 45, 62, 82 (renamed 89 by CGSNL), and 83 (renamed 94 by CGSNL) by SA was reported previously (Ryu et al., 2006). An additional study reported that OsWRKY19, 45, 62, and 76 were induced by the SA analog BTH (Shimono et al., 2007). Studies of individual OsWRKY TF genes indicated that OsWRKY03 (renamed 12 by CGSNL), OsWRKY6, OsWRKY33 (renamed 81 by CGSNL), OsWRKY51, and OsWRKY77 were induced by SA (Liu et al., 2005; Koo et al., 2009; Hwang et al., 2011, 2016; Lan et al., 2013). However, 12 OsWRKY TF genes profiled in this study (OsWRKY9, 11, 22, 23, 47, 58, 60, 64, 88, 106, 113, and 114) were not described previously, possibly because only OsWRKY TF genes involved in Xa1 mediated resistance were examined in this study. Overall, these results suggest that OsWRKY TFs regulate both SA-dependent and SA-independent Xa1-mediated resistance.
In this study, SA-deficient rice plants were generated by transforming rice plants with NahG, which degrades SA to catechol. However, several studies have questioned using NahG rice to study the effect of SA on plant resistance to infection (van Wees and Glazebrook, 2003; Yang et al., 2004; Mutuku et al., 2015). van Wees and Glazebrook (2003) arrived at false conclusions concerning the role of SA in plants defense because catechol affected non-host resistance in the Arabidopsis plants expressing NahG. Another study claimed that catechol had no effect on resistance to S. hermonthica (Mutuku et al., 2015). In this study, we demonstrated that catechol had no effect on Xa1-mediated resistance, confirming that the effects observed in SA-deficient rice were not due to catechol production. The effect of catechol on plant resistance would appear to depend on the pathosystem studied.
The function of OsWRKY88, a gene whose expression was altered during SA-dependent Xa1-mediated resistance, was analyzed. OsWRKY88 ox lines showed increased resistance to Xoo and activation of the defense response. We demonstrated that OsWRKY88 directly activated the OsPR10a promoter but not the CHIT2 promoter. The higher level of the CHIT2 transcript might be due to the presence of another transcription factor induced by OsWRKY88. Some studies claim that transcription regulatory cascades of TFs are required for disease resistance (Cheng et al., 2015). OsWRKY22 and OsWRKY23 (previously known as OsWRKY31) have been previously reported to be positive regulators of resistance to M. oryzae, although the effect of OsWRKY22 and OsWRKY23 on Xoo infection was not investigated (Zhang et al., 2008; Abbruscato et al., 2012). Expression profiling of OsWRKY TF genes was performed after the infection of rice with Xoo, which revealed changes in expression of most of the genes. OsWRKY TF gene expression in Xa1-mediated resistance appeared to involve both SA-dependent and SA-independent pathways. Therefore, we propose that OsWRKY TF genes are involved in Xa1-mediated resistance.
Author contributions
NC and DH wrote the manuscript. DH designed the experiments. NC, SL, and CC performed qRT-PCR. EL analyzed the transgenic plants. SP, IA, and SB identified database OsWRKY TF gene sequences and designed primers. CH critically revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Grant PJ0108701 from the National Institute of Agricultural Sciences to DH.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01628/full#supplementary-material
References
- Abbruscato P., Nepusz T., Mizzi L., Del Corvo M., Morandini P., Fumasoni I., et al. (2012). OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol. Plant Pathol. 13, 828–841. 10.1111/j.1364-3703.2012.00795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnaresi P., Biselli C., Orrù L., Urso S., Crispino L., Abbruscato P., et al. (2012). Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS ONE 7:e51609. 10.1371/journal.pone.0051609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berri S., Abbruscato P., Faivre-Rampant O., Brasileiro A. C., Fumasoni I., Satoh K., et al. (2009). Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 9, 186–120. 10.1186/1471-2229-9-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Qiu D., Yuan T., Ding X., Li H., Duan L., et al. (2008). Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ. 31, 86–96. 10.1111/j.1365-3040.2007.01739.x [DOI] [PubMed] [Google Scholar]
- Cheng H., Liu H., Deng Y., Xiao J., Li X., Wang S. (2015). The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. 10.1104/pp.114.256016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Hwang S., Fang I., Kwon S., Park S., Ahn I., et al. (2015). Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 208, 846–859. 10.1111/nph.13516 [DOI] [PubMed] [Google Scholar]
- Chu Z., Yuan M., Yao J., Ge X., Yuan B., Xu C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. 10.1101/gad.1416306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T., Kato T., Yamada K., Takai R., Akimoto-Tomiyama C., Minami E., et al. (2008). Characterization of an elicitor-induced rice WRKY gene, OsWRKY71. Biosci. Biotechnol. Biochem. 72, 240–245. 10.1271/bbb.70553 [DOI] [PubMed] [Google Scholar]
- Chujo T., Miyamoto K., Shimogawa T., Shimizu T., Otake Y., Yokotani N., et al. (2013). OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol. Biol. 82, 23–37. 10.1007/s11103-013-0032-5 [DOI] [PubMed] [Google Scholar]
- Ciolkowski I., Wanke D., Birkenbihl R. P., Somssich I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. 10.1007/s11103-008-9353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T. P., Ukness S., Vernooij B., Friedrich L., Weymann K., Negrotto D., et al. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. 10.1126/science.266.5188.1247 [DOI] [PubMed] [Google Scholar]
- Dong J., Chen C., Chen Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. 10.1023/A:1020780022549 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Gu K. Y., Yang B., Tian D. S., Wu L. F., Wang D. J., Sreekala C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. 10.1038/nature03630 [DOI] [PubMed] [Google Scholar]
- Han M., Ryu H. S., Kim C. Y., Park D. S., Ahn Y. K., Jeon J. S. (2013). OsWRKY30 is a transcription activator that enhances rice resistance to the Xanthomonas oryzae pathovar oryzae. J. Plant Biol. 56, 258–265. 10.1007/s12374-013-0160-0 [DOI] [Google Scholar]
- Huang S., Antony G., Li T., Liu B., Obasa B., Yang B., et al. (2016). The broadly effective recessive resistance gene xa5 of rice is a virulence effector-dependent quantitative trait for bacterial blight. Plant J. 86, 186–194. 10.1111/tpj.13164 [DOI] [PubMed] [Google Scholar]
- Hwang S. H., Kwon S. I., Jang J. Y., Fang I. L., Lee H., Choi C., et al. (2016). OsWRKY51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas oryzae pv. oryzae Plant Cell Rep. 35, 1975–1985. 10.1007/s00299-016-2012-0 [DOI] [PubMed] [Google Scholar]
- Hwang S. H., Lee I. A., Yie S. W., Hwang D. J. (2008). Identification of an OsPR10a promoter region responsive to salicylic acid. Planta 227, 1141–1150. 10.1007/s00425-007-0687-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. H., Yie S. W., Hwang D. J. (2011). Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 181, 316–323. 10.1016/j.plantsci.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Iyer A. S., McCouch S. R. (2004). The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 17, 1348–1354. 10.1094/MPMI.2004.17.12.1348 [DOI] [PubMed] [Google Scholar]
- Jimmy J. L., Babu S. (2015). Role of OsWRKY transcription factors in rice disease resistance. Trop. Plant Pathol. 40, 355–361. 10.1007/s40858-015-0058-0 [DOI] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Kauffman H. E., Reddy A. P. K., Hsieh S. P. Y., Merca S. D. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541. [Google Scholar]
- Kim D. Y., Kwon S. I., Choi C., Lee H., Ahn I., Park S. R., et al. (2013). Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 529, 208–214. 10.1016/j.gene.2013.08.023 [DOI] [PubMed] [Google Scholar]
- Kim Y. H., Park H. M., Choi M. S., Yun H. T., Choi I. S., Shin D. B., et al. (2009). The effects of co-cultivation medium and culture conditions on rice transformation efficiency. Korean J. Breed. Sci. 41, 252–260. 10.1186/1746-4811-7-49 [DOI] [Google Scholar]
- Koo S. C., Moon B. C., Kim J. K., Kim C. Y., Sung S. J., Kim M. C., et al. (2009). OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem. Biophys. Res. Commun. 387, 365–370. 10.1016/j.bbrc.2009.07.026 [DOI] [PubMed] [Google Scholar]
- Lan A., Huang J., Zhao W., Peng Y., Chen Z., Kang D. A. (2013). Salicylic acid-induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana. Plant Biol. 15, 452–461. 10.1111/j.1438-8677.2012.00664.x [DOI] [PubMed] [Google Scholar]
- Lee H., Ko Y. J., Cha J. Y., Park S. R., Ahn I., Hwang D. J. (2013). The C-terminal region of OsWRKY30 is sufficient to confer enhanced resistance to pathogen and activate the expression of defense-related genes. Plant Biotechnol. Rep. 18, 1–10. 10.1007/s11816-012-0252-1 [DOI] [Google Scholar]
- Li X. (2011). Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bioprotocol 1, 1–3. 10.21769/BioProtoc.95 [DOI] [Google Scholar]
- Liu Q., Yuan M., Zhou Y., Li X., Xiao J., Wang S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969. 10.1111/j.1365-3040.2011.02391.x [DOI] [PubMed] [Google Scholar]
- Liu X. Q., Bai X. Q., Qian Q., Wang X. J., Chen M. S., Chu C. C. (2005). OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15, 593–603. 10.1038/sj.cr.7290329 [DOI] [PubMed] [Google Scholar]
- Liu X., Bai X., Wang X., Chu C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. 10.1016/j.jplph.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Mutuku J. M., Yoshida S., Shimizu T., Ichihashi Y., Wakatake T., Takahashi A., et al. (2015). The WRKY45-dependent signaling pathway is required for resistance against Striga hermonthica parasitism. Plant Physiol. 168, 1152–1163. 10.1104/pp.114.256404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S. P., Somssich I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. 10.1104/pp.109.138990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D., Xiao J., Ding X., Xiong M., Cai M., Cao Y., et al. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20, 492–499. 10.1094/MPMI-20-5-0492 [DOI] [PubMed] [Google Scholar]
- Qiu Y., Yu D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47. 10.1016/j.envexpbot.2008.07.002 [DOI] [Google Scholar]
- Rice WRKY Working Group. (2012). Nomenclature report on rice WRKY's – conflict regarding gene names and its solution. Rice 5, 3–5. 10.1186/1939-8433-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H. S., Han M., Lee S. K., Cho J. I., Ryoo N., Heu S., et al. (2006). A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 25, 836–847. 10.1007/s00299-006-0138-1 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Shimono M., Sugano S., Nakayama A., Jiang C. J., Ono K., Toki S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. 10.1105/tpc.106.046250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W. Y., Wang G. L., Chen L., Kim K. S., Holstein T., Wang B., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. 10.1126/science.270.5243.1804 [DOI] [PubMed] [Google Scholar]
- Sun C., Palmqvist S., Olsson H., Borén M., Ahlandsberg S., Jansson C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15, 2076–2092. 10.1105/tpc.014597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Cao Y., Yang Z., Xu C., Li X., Wang S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527. 10.1046/j.1365-313X.2003.01976.x [DOI] [PubMed] [Google Scholar]
- Tao Z., Liu H., Qiu D., Zhou Y., Li X., Xu C., et al. (2009). A Pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 151, 936–948. 10.1104/pp.109.145623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wang J., Zeng X., Gu K., Qiu C., Yang X., et al. (2014). The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26, 497–515. 10.1105/tpc.113.119255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees S. C., Glazebrook J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. 33, 733–742. 10.1046/j.1365-313X.2003.01665.x [DOI] [PubMed] [Google Scholar]
- Wang C., Zhang X., Fan Y., Gao Y., Zhu Q., Zheng C., et al. (2015). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8, 290–302. 10.1016/j.molp.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Wang H., Meng J., Peng X., Tang X., Zhou P., Xiang J., et al. (2015). Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 89, 157–171. 10.1007/s11103-015-0360-8 [DOI] [PubMed] [Google Scholar]
- Wei T., Ou B., Li J., Zhao Y., Guo D., Zhu Y., et al. (2013). Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PLoS ONE 8:e59720. 10.1371/journal.pone.0059720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Qi M., Mei C. (2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40, 909–919. 10.1111/j.1365-313X.2004.02267.x [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Yamanouchi U., Katayose Y., Toki S., Wang Z. X., Kono I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. U.S.A. 95, 1663–1668. 10.1073/pnas.95.4.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Peng Y., Guo Z. (2008). Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 18, 508–521. 10.1038/cr.2007.104 [DOI] [PubMed] [Google Scholar]
- Zhang Y. J., Wang L. J. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. 10.1186/1471-2148-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.