Abstract
Background: The RAS association domain family protein 1a (RASSF1A) is a prominent tumor suppressor gene showing altered promoter methylation in testicular germ cell tumors (TGCT). RASSF1A promoter hypermethylation might represent an early event in TGCT tumorigenesis. We investigated whether the RASSF1A promoter methylation in peripheral blood of TGCT patients can be associated with testicular cancer risk. Materials and Methods: Following a meta-analysis, we performed a cohort study including 32 testicular cancer patients and 32 healthy controls. Promoter methylation of the RASSF1A and O6-methylguanine-DNA-methyltransferase (MGMT) genes was analyzed using bisulfite pyrosequencing of DNA from peripheral blood. Results: Meta-analysis showed an odds ratio (OR) of 7.69 for RASSF1A promoter methylation as a risk factor for TGCT. Cohort study found altered methylation of the RASSF1A promoter in blood of TGCT patients. Methylation was higher in TGCT patients before BEP chemotherapy. Conclusion: The meta-analysis indicates a role of the RASSF1A promoter hypermethylation from peripheral blood in TCGT. We confirmed that finding in our cohort study, which represents the first report of changed RASSF1A promoter methylation in peripheral blood TGCT.
Keywords: DNA methylation, RASSF1, MGMT, testicular cancer, BEP chemotherapy
Testicular germ cell tumors (TGCTs) account for most testicular cancers. Incidence of 55 new cases per 100,000 males per year world-wide, and increase in number of diagnosed man with TC in US and other Western countries, makes it an important healthcare issue (1,2). The most affected subjects are young Caucasians (between age of 25 and 29 years) and some of the most rapid trends are observed in men of European descent, especially in Scandinavian countries (3). About 23,000 new cases are predicted to occur in Europe by 2025 (4). Today, more than 90% of patients with TGCT are cured if diagnosed early, i.e. only 10-20% of patients with metastatic tumors at the time of diagnosis do not become permanently cured. The therapy depends on the type and the stage of the disease. The cure usually consists of surgery followed by chemotherapy and/or radiotherapy (5). The most commonly used chemotherapy regimens are based on the treatment protocol, which contains several cycles of antineoplastic drugs cisplatin, bleomycin and etoposide (BEP chemotherapy). Except self-inspection, there is no appropriate diagnostics that can be used in routine screening of a population under the risk for TC. The European Association of Urology recommends biopsy in high-risk patients, but non-invasive diagnostic methods, such as immunocytochemistry of semen smears are not sufficiently sensitive (6). This is why introduction of new diagnostics, that would enable recognition of TGCT at early stages, as well as estimation of therapy efficiency at the molecular level, is of special interest.
Histologically, TGCTs are characterized by substantial heterogeneity. The new updated WHO classification system divides testicular germ cell tumors into two major groups: those that are derived from germ cell neoplasia in situ (GCNIS) and those that are not (7). Germ cell neoplasia in situ cells precedes both seminoma and non-seminoma (NSGCT) in the tumors of young adults. While seminomas are morphologically homogenous (they account for around 60% of TGCT cases), non-seminomas are morphologically very heterogeneous and can be composed of one or more components (such as embryonal carcinoma, teratoma, yolk sac tumor, and choriocarcinoma and other trophoblastic tumors). Experimental evidence suggest that seminomas and non-seminomas differ in the underlying genetic lesions (8).
Besides genetic component, the familial risk of TGCT is one of the highest among cancers (9), TGCT has a strong environmental component. Genome wide association studies (GWAS) have uncovered some candidate genetic loci involved in testicular cancer susceptibility (10-13) and study of single nucleotide polymorphisms (SNPs) in the same loci showed the risk variants which predispose to both familial and bilateral TGCT, which accounted for only 11-16% of the risk for the disease (14). Various external stressors, including diagnostic procedures, contribute to TGCT pathogenesis (15). A pilot study suggests a possible familial TGCT susceptibility that may be associated with promoter methylation in previously identified TGCT risk genes in GWA studies (16).
Aberrant promoter hypermethylation of candidate tumor suppressor genes (TSG) or tumor-related genes seems not to be a frequent event in TGCT as it is the case in many other human cancers (17). Nevertheless, the RAS association domain-containing protein 1A (RASSF1A), a tumor suppressor gene located on the chromosome region 3p21, was identified as one of the main TSGs that change promoter methylation status in TGCT. This gene is functionally relevant for cell-cycle control, microtubule stabilization, cellular adhesion, motility and apoptosis, and depletion of RASSF1A is shown to be associated with higher risk of chromosome rearrangements, accelerated mitotic progression and enhanced cellular motility (18-20). Epigenetic inactivation of the RASSF1A gene by CpG methylation is recorded in many cancers (for a review see (21)), and hypermethylation of RASSF1A promoter has a prognostic value for some tumor types (22-24). The RASSF1A promoter is also found hypermethylated in both seminomas and non-seminomas, while three additional candidate genes, MGMT, APC and FHIT, were frequently found hypermethylated only in NSTGCT (25,26). Therefore, it seems that RASSF1A promoter hypermethylation might occur as the first epigenetic event in TGCT tumorigenesis and additional specific epigenetic events characterize morphologically heterogeneous non-seminoma TGCT.
Identification of sensitive and specific indicators, and possibly non-invasive biomarkers that would predict early-onset TGC tumors, are still in research focus. Peripheral whole blood is the most convenient sample for non-invasive biomarker analyses. Even though the tissue specificity of DNA methylation pattern could be a problem in such approach, there is an increasing number of publications showing that aberrant DNA methylation of candidate genes can be detected in peripheral blood of patients with cancer, even for solid tumors (27-29).
In this study, we conducted a meta-analysis of the association of the RASSF1A gene and TGCT and estimated its prognostic value for TGCT. We examined the odds ratio (OR) derived from eligible studies in order to establish a link between RASSF1A promoter methylation and risk for testicular cancer. Additionally, we validated the findings of the meta-analysis in our cohort of TGCT patients with seminoma and non-seminoma testicular cancer in order to ascertain if the RASSF1A promoter hypermethylation could be useful as diagnostic factor from whole blood. We also analyzed methylation status of the O6-methylguanine-DNA-methyltransferase (MGMT) gene in our cohort in order to compare diagnostic potential of the RASSF1A promoter hypermethylation (already established TSG which show promoter hypermethylation in both seminoma and non-seminoma tumor tissue), with that of the MGMT gene, which shows aberrant promoter methylation only in non-seminoma tissue (25,26).
Materials and Methods
Meta-analysis of association using odds ratio. Meta-analysis was conducted according to the PRISMA guidelines (30,31). Eligible case-control studies included investigations of the role of RASSF1A methylation in the pathogenesis and/or progression of testicular cancer. No publication date or publication status restrictions were imposed; however, only studies reported in English were included. Both seminoma and non-seminoma samples were included in the meta-analysis. Patient data, such as age, were not always available. We used relatively permissive criteria for study eligibility due to the small overall number of available reports. Eligibility assessment was performed independently by two reviewers (IS and AV) and the final decision was reached by consensus.
The PubMed database was searched for studies published up to March 1st, 2017. The following search strategy was used: (testicular OR testis) AND (cancer OR carcinoma OR tumor) AND (RASSF1A methylation). The following information was extracted from the studies: first author’s name, year of publication, material for methylation analysis, control type, the measuring method of methylation, the number of individuals in the case and control groups.
To assess heterogeneity of the included studies, we calculated Cochran’s Q with the corresponding p-value and the associated I2 value (measure of inconsistency) for the meta analysis. As a visual aid for assessing bias across studies, we constructed a “funnel plot” of the effect (natural logarithm of odds ratio) by its standard error in order to intuitively demonstrate symmetry; to formally assess the degree of asymmetry, we used Egger’s test.
Meta-analysis of prognostic value of the RASSF1A gene expression. Association between the RASSF1A gene expression levels and prognosis in different types of cancer was analyzed using the PrognoScan database, which employs the minimum p-value approach for grouping patients for survival analysis, thus enabling systematic analysis of multiple datasets (32). Briefly, patients were ordered by expression levels of a gene of interest and then divided into high- and low-expression groups at all possible cut-off points. The risk differences of all groups were then estimated by the log-rank test. Finally, the grouping that gave the most significant p-value was selected.
Cohort study. To verify results from the meta analysis, we performed our own cohort study. We collected all available Croatian TC samples and matching controls.
Patients and sample collection. Our cohort included 32 testicular cancer patients and the same number of healthy controls from the general population matched according to age (±4 years) and smoking status, since it is well known that aging and smoking alter DNA methylation status in general, and particularly of the RASSF1A gene promoter (33,34). Control subjects were not occupationally exposed to ionizing radiation or known chemical mutagens/carcinogens and were not diagnosed or treated for any neoplastic disease. According to the histological type of tumor 11 patients had seminoma and 21 had non-seminoma tumor (7 out of 21 had combined seminoma and non-seminoma tumors which is considered by pathologist as non-seminoma).
All patients were, following the surgical procedure, subjected to two rounds of chemotherapy. Both rounds included etoposide and cisplatin, while the first round also included antibiotic treatment with bleomycin (BEP). The time interval between the two rounds of chemotherapy was three weeks. The first blood sample was obtained from patients after the surgical procedure (i.e. orchiectomy), but before the use of chemotherapy. The second blood sample was taken after two rounds of chemotherapy. Sampling took place in the years 2012 and 2013. The research has been approved by the appropriate ethics committee.
DNA methylation analysis. DNA was extracted from 200 μL of blood using the QIAamp DNA Blood Mini Kit (Qiagen) in accordance with the manufacturer’s protocol. For DNA methylation analysis, 500 ng of DNA was subjected to bisulfite conversion, which was performed according to manufacturer’s instructions (EZ DNA Methylation-Gold Kit, Zymo Research).
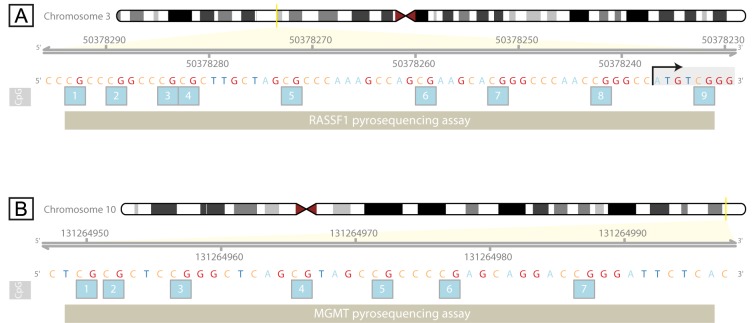
Polymerase chain reaction (PCR) was performed using PyroMark PCR Kit (Qiagen) according to the manufacturer’s instructions. Specific regions rich in CpG dinucleotides of the genes RASSF1A and MGMT were amplified using the primers specific for bisulfite modified DNA. The RASSF1A fragment encompassed by the pyrosequencing assay was mostly within the UTR, located 57 nucleotides upstream to 6 nucleotides downstream from the translation initiation site (TIS), within a CpG island spanning the promoter, the first exon and a part of the first intron (Figure 1A). We chose this fragment because first CpG site (cg04743654) from the assay was found differentially methylated on Illumina 450K for different cell lines including human testicular embryonic carcinoma cell line NT2/D1 in which this CpG site was moderately methylated. The MGMT fragment encompassed by pyrosequencing assay was located 531-484 nucleotides upstream from the TIS, within a CpG island (Figure 1B). Previously described primers were used for RASSF1A pyrosequencing (35). Primers used for MGMT were: 5’-GGT GAT TGT AGT TTT TGG A-3’ and 5’-TCC TAT CAC AAA AAT AAT CC-3’ (biotinylated at the 5’ end). PCR was initiated by pre-denaturation for 15 min at 95˚C followed by 50 cycles of 94˚C (MGMT, RASSF1A) for 30 s, 51˚C (MGMT) or 65˚C (RASSF1A) for 30 s and 72˚C for 30 s, with final extension at 72˚C for 10 min. PCR products were separated by agarose gel electrophoresis to ascertain correct amplification and sufficient product quality.
Figure 1. Location of the pyrosequencing assay within the RASSF1A (A) and MGMT (B) genes used for analysis of CpG methylation status. Position of the analyzed CpG sites is shown with absolute genomic coordinates according to the human genome GRCh37/hg19 assembly. Positions of the pyrosequencing assays on the chromosome 3 (A) and chromosome 10 (B) are indicated by the yellow bar. Translation start is indicated with the arrow above the ATG start codon (A).
For quantitative measurement of DNA methylation levels in the genomic fragments of interest, bisulfite converted and PCR amplified DNA was sequenced using pyrosequencing technology (PyroMark Q24 Advanced, Qiagen) according to manufacturer’s instructions. Sequencing primers for MGMT and RASSF1A were: 5’-GGT ATT AGG AGG GGA GAG ATT-3’ and 5’-GTT TTG TGG TTT-3’, respectively. The pyrosequencing assay comprised 7 CpG sites for MGMT and 9 CpG sites for RASSF1A.
Statistical analysis. Differences in methylation level of the RASSF1A and MGMT genes were assessed separately for each tested CpG site. Differences between the patients before and after therapy, as well as corresponding controls, were tested using the Mann-Whitney U-test. For matched samples before and after therapy, the Wilcoxon signed-rank test was used. Data processing and visualization was done using the R statistical environment (36). Meta analysis was done using the “metafor” package (37).
Results
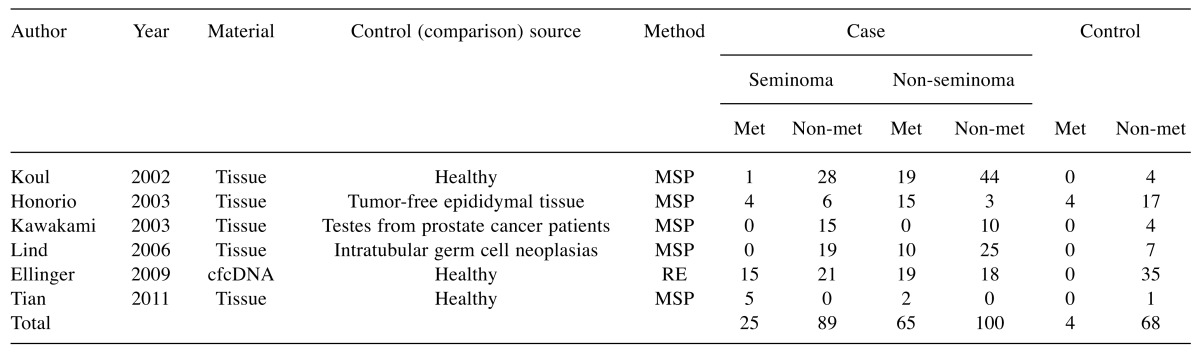
Meta-analysis. Study selection and characteristics. To identify the association between the RASSF1A promoter methylation and TGCT, we conducted a meta-analysis of 6 eligible studies (25,38-42) involving a total of 279 TGCT cases and 72 controls to calculate the odds ratio (OR) and the corresponding 95% confidence intervals where the RASSF1A promoter methylation was modelled as a risk factor for TGCT.
Details of the eligible studies are summarized in Table I. Five studies analyzed testicular cancer tissue and one used cell-free circulating DNA. For the analysis, all studies used methylation specific PCR (MSP), except for one that used restriction endonuclease (RE) based assay (Table I). As a control source, three studies used healthy testicular tissue of different types: tumor-free epididymal tissue, testes from prostate cancer patients and intra-tubular germ cell neoplasia which are considered a precursor lesion for many types of TGCT.
Table I. The studies selected for meta analysis of association between the RASSF1A promoter methylation and testicular cancer risk.
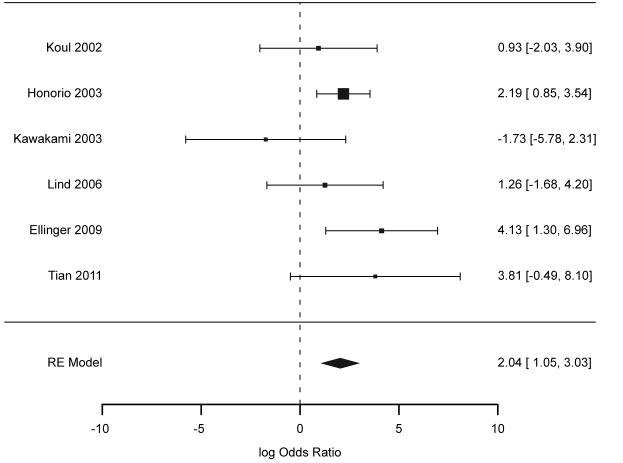
Results of individual studies and their synthesis. Odds ratio (OR) for TGCT – considering the RASSF1A promoter methylation – was taken as the outcome, which is shown in the forest plot (Figure 2). Only the study by Kawakami (39) showed negative log OR, contradicting the results of other studies; yet, this value was accompanied by wide confidence limits. Other studies supported increased odds for TGCT with increased RASSF1A promoter methylation, as seen in Figure 2. Summarized OR under random effects model was 7.69 (95% CI=2.86-20.70). Heterogeneity based on Cochran’s Q for the six studies was 5.34 (p=0.225), giving an I2 value of 0.00%.
Figure 2. Forest plot of the logarithm of odds ratio (OR) for the association between the RASSF1A promoter methylation and testicular cancer. Selected publications are listed on the Y-axis with the summary (random effects model) at the bottom. Error bars represent 95% confidence intervals.
When patients were stratified according to histology type (seminoma vs. non-seminoma), we found a difference in the association between DNA methylation and TGCT, with OR 2.89 (95% CI=0.54-15.33; I2=41.05%) for seminoma and 12.55 (95% CI 4.22-37.71; I2=0.00%) for non-seminoma. Large heterogeneity and wide confidence intervals for the seminoma group are due to the reduced sample size of that group after stratification (Table I).
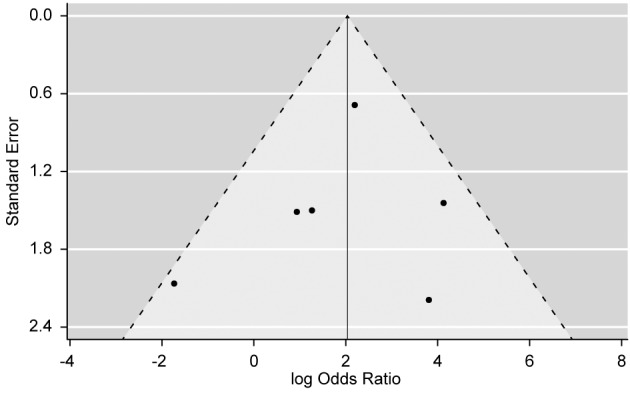
Risk of bias across studies. We found no bias in the included studies, which can be seen from the symmetry of the funnel plot in Figure 3. Egger’s test gave a p-value of 0.679, which formally confirms the absence of significant bias across included publications. When cases were stratified according to histology type into seminoma and non-seminoma group, evidence against bias was even stronger in the seminoma group (p=0.795), although not as strong for non-seminoma group (p=0.306).
Figure 3. Funnel plot of the logarithm of the odds ratio with its corresponding standard error (Y-axis), where each black dot represents one of the six studies included in the meta-analysis, shows no apparent asymmetry would have indicated bias across studies. The absence of bias was formally confirmed using Egger’s test (p=0.679).
Prognostic value of RASSF1A promoter methylation. In the PrognoScan database, we found 25 statistically significant (Cox p-value <0.05) probes indicating a possible prognostic value of the RASSF1 gene in various types of cancers, 20 of which (80%) were associated with better survival in cases of high expression. Therefore, high expression of RASSF1 gene in cancer seems to indicate better prognosis.
Cohort study. In our cohort study, we wanted to verify if RASSF1A hypermethylation could be diagnosed from blood of TC patients. The MGMT gene was chosen as a “control gene” for methylation difference between patients with histologically-different testicular tumors and healthy controls since RASSF1A hypermethylation was signature for both seminomas and NSTGCT tissue and the MGMT gene was found hypermethylated only in tissue of NSTGCT (25,26). Additionally, we were interested on the effects of BEP chemotherapy on RASSF1A promoter methylation.
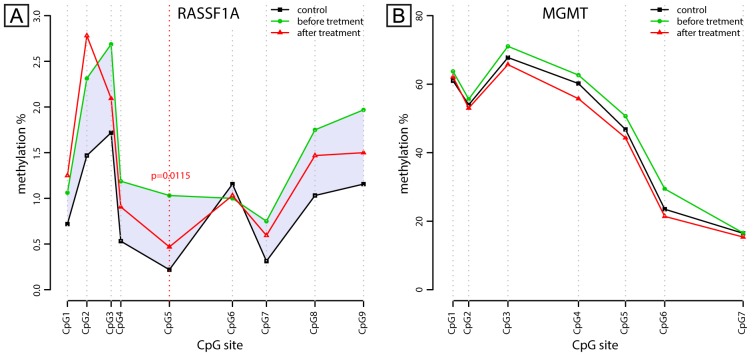
RASSF1A and MGMT promoter methylation was analyzed from DNA of peripheral blood nucleated cells (PBNC) using the pyrosequencing method after bisulfite conversion of DNA. The pyrosequencing assay for RASSF1A comprised 9 CpG sites and the methylation levels were generally low (less than 3%) at this fragment. However, the methylation levels at 5 out of 9 CpG sites in the RASSF1A fragment were significantly higher in the TC patients before BEP chemotherapy compared to the healthy controls (CpG sites number 2, 3, 4, 5 and 7; Figure 4A). In addition, CpG site number 5 (CpG5) showed statistically significant difference (p=0.0115) in the methylation level between patients before and after the BEP chemotherapy. The general trend was the that in CpG methylation in patients, which had a tendency to return to the methylation level found in healthy controls after the BEP therapy, as illustrated in Figure 4A. No significant difference in CpG methylation was found after stratification of patients into the seminoma and non-seminoma, which is probably due to too small number of individuals in each group.
Figure 4. Methylation levels of the individual CpG sites within the RASSF1A (A) and MGMT (B) gene promoters. Methylation level is determined by pyrosequencing and is expressed here as a percentage. The distance between the CpG sites corresponds to their relative position within the analyzed DNA fragment. Points (triangle, circle and square) represent the mean methylation level for each group. Lines are drawn as a visual aid for illustrating the spatial organization of methylation levels along the analyzed region. A) The CpG sites 2, 3, 4, 5 and 7 in the analyzed fragment within the RASSF1A promoter show statistically significant difference between patients (before chemotherapy) and healthy controls. Methylation at the site CpG5 shows statistically significant difference (p=0.0115) between the patients before and after BEP chemotherapy (indicated by vertical dashed line). Shaded area emphasizes CpG methylation difference between healthy controls and testicular cancer patients. B) No significant difference was found in the MGMT promoter methylation.
The pyrosequencing assay for the MGMT gene comprised of 7 CpG sites (Figure 4B). The highest methylation level was recorded at the CpG site number 3 (CpG3; around 70%), while other CpG sites had a lower methylation level in all studied groups. We could not find any statistically significant difference in CpG methylation status in the blood of the TGCT patients and healthy controls.
Discussion
Aberrant gene-specific promoter methylation occurs in virtually every type of tumor. Potential for measuring such gene-specific methylation in different tumors and its diagnostic and predictive value have already been demonstrated for many cancers. Previous studies have identified associations between methylation levels of several genes with TGCT, which could be a molecular mechanism for aberrant activation/inactivation of these genes in this cancer (16,25). Even though seminomas and non-seminomas demonstrate (cyto)genetic similarities, it appears that the two types of tumors differ significantly at the epigenetic level (16,25,43,44). The RASSF1A gene has been identified as the main tumor-suppressor gene silenced in testicular cancer tissue by aberrant hypermethylation and the first epigenetic event in the TGCT tumorigenesis (25). Moreover, the methylation status of the RASSF1A promoter appears to play a role in cisplatin chemotherapy resistance and sensitivity (45) and correlates with clinical parameters (46) suggesting that this gene might be profoundly implicated in molecular mechanisms of TGCT.
Several meta-analysis studies showed an association between RASSF1A promoter hypermethylation and different types of tumors as well as biomarker value of the RASSF1A methylation status (47-49). This study represents the first meta-analysis of published studies to evaluate the association of RASSF1A promoter hypermethylation with testicular cancer. The association and the odds ratio (OR) with TGCT were indeed different when patients were stratified according to histology type, with stronger association appearing for NSTGCT. This is in accordance with the finding of Honorio (25) who detected RASSF1A hypermethylation in 83% of non-seminoma, and only in 40% of seminoma components. In addition, seminomas are more highly hypomethylated than non-seminoma throughout the genome, with almost unmethylated CpG islands in seminomas in contrast to hypermethylated CpG islands in non-seminomas (43).
This study has some limitations such as the relatively small total number of patients with TC included in the meta-analysis. However, there was no apparent risk of bias in the included studies. The only exception was the non-seminoma group after stratification, where the additional reduction in sample size introduced by stratification decreased the statistical power in analysis of the non-seminoma group, thus making conclusions about this type of tumor less reliable.
Heterogeneity among studies was expected at least to a certain degree, due to typically small sample size of each individual study (which is the result of the rarity of TCGC) and a relatively small overall number of studies that matched our inclusion criteria. However, two complementary measures of inconsistency across studies, Cochran’s Q and I2, both indicated that the included studies were in agreement with each other and as such could provide material for a valid meta-analysis. In order to accommodate for variation between study outcomes, we opted for the less statistically powerful random effects model and still obtained a significant value for the combined odds ratio.
Even though TCGC is the most common type of testicular cancer, it is still a relatively rare disease. Therefore, the total number of included studies and the total sample size, both in the meta-analysis and the cohort study, was limited by this fact. It should also be noted that the methods for methylation analysis were different – MSP and RE based assays were used in studies included in the meta-analysis, while bisulfite pyrosequencing was applied in our cohort analysis. Even though sequencing techniques are generally superior to PCR based methodologies or those based on restriction with methylation sensitive enzymes, pyrosequencing method has a limitation in the length of the analyzed sequence (approx. 120 bp) and in “fishing” the functionally relevant CpG sites. The CpG islands within gene promoters are large and contain many CpG dinucleotides potentially involved in transcriptional regulation; however, they are unidentified in most of the gene promoters.
The results of methylation level difference in promoter of the MGMT gene between TGCT patients and healthy controls in our cohort study showed no statistically significant differences, and this finding could be interpreted in two ways. Either the difference in the methylation level for this gene, found in testicular tissue of the NSTGCT patients (25,26) is not strong enough to be detectable in PBNC (in our cohort study), or different methods used in our study and other studies led to different results. Namely, in two studies that report differences in the methylation level in the MGMT promoter of TC tissue and in whole blood of patients with non-seminomas (but not seminoma), the method of methylation-specific PCR was used (25,26,44). Our pyrosequencing assay covered seven CpG sites located within CpG island of the MGMT gene, and it is possible that these sites do not change methylation level in TGCT. It is documented that either solitary CpGs can change methylation level in cancer or many CpGs within large fragments change methylation level and this would depend on specific TSG and the specific cancer (50-52). Therefore, some other, unidentified regions within the CpG island of the MGMT gene could be involved in transcriptional regulation of this gene and might contain differentially methylated CpG sites.
Diverse sample types employed in the studies used for meta-analysis, which includes testicular cancer tissue, whole blood and cell free circulating DNA, highlight the requirement for careful interpretation. In our cohort study, we chose to analyze methylation from whole blood since this type of analysis could be useful in non-invasive early diagnostics. It is known that any tumor has its tissue specific methylomes and even if they can give insights on the molecular pathology of a particular cancer, they cannot serve as prognostic and/or diagnostic biomarker due to unavailability of target tissue, and invasiveness of methods that would provide such tissue. Recent studies suggest that DNA methylation markers from peripheral blood might serve as diagnostic, early detection/risk or prognostic and recurrence cancer markers and a number of publications show such good correlation in different cancers (53-56). In our cohort study, we confirmed that RASSF1A promoter hypermethylation could be diagnosed from blood of TGCC patients. The level of RASSF1A methylation at several CpG sites, located within the CpG island, was significantly increased in patients before BEP chemotherapy compared to healthy controls, suggesting that hypermethylation of RASSF1A play role in the disease. Even though absolute differences in CpG methylation levels between cases and controls were small, they were statistically significant. Since we analyzed methylation from whole blood, one should in principle account for the lack of discrimination between different cell types within PBMCs. However, our necessary assumption was that every tissue reflects a degree of global RASSF1A methylation, including methylation of the target (testicular) cells that are candidates for malignant transformation in the context of our study.
Other authors (45) found that, among other genes, promoter hypermethylation of the RASSF1A gene plays a role in resistance of TGCT to chemotherapy. In our study, we analyzed DNA methylation status in peripheral blood of drug naïve and cisplatin treated TC patients. We investigated whether cisplatin treatment also affects the methylation of this gene in the non-target tissue (i.e. blood) and whether this effect can be a marker of successful treatment. The methylation level that we recorded returned to background levels, further suggesting a role of RASSF1A methylation in TGCT pathogenesis (57,58). The MGMT promoter methylation did not show this effect. Even if there might be a potential of RASSF1A promoter methylation as prognostic factor after chemotherapy, the study needs to be extended to include a larger sample in order to obtain conclusive information within a narrow confidence limit.
To our knowledge, this is the first report of changed RASSF1A promoter methylation in peripheral blood in any TGCT cohort. Due to the scarcity of adequate samples and the relatively low incidence of the disease in Croatia, our cohort was too small for statistically powerful results when we performed stratification of the patients into subgroups according to tumor type (seminomas and non-seminomas). However, the pooling of the similar results and conducting a meta-analysis of different TC datasets (279 TGCT patients from eligible studies) helped us to strengthen the case for potential role (or at least a significant association) of RASSF1A gene promoter hypermethylation found in peripheral blood of both seminoma and non-seminoma TC patients. This is further strengthened by the results of our “control gene”, the MGMT, which did not show any methylation differences between the TGCT patients and healthy controls. The MGMT promoter hypermethylation was found only in non-seminomas, therefore, it is considered to constitute additional epigenetic alteration subsequent to hypermethylation of the RASSF1A gene, which was suggested to represent an early epigenetic event in the pathway of NSTGCT development (25,26). Since we could not obtain any conclusive results when we performed stratification of our TGCT patients into the subgroups, it was not possible to replicate the results reported by others who found differences in the methylation level in the MGMT promoter in whole blood of NSTGCT patients (44). Larger studies, both observational cohorts and clinical trials are required to test whether RASSF1A promoter hypermethylation can be useful diagnostic and prognostic factors for testicular cancer.
Acknowledgements
The research presented here was supported by European Commission (FP7) project RegPot Integra-Life (Grant Agreement № 315997).
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int J Cancer. 2015;136(5):E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Vasdev N, Moon A, Thorpe AC. Classification, epidemiology and therapies for testicular germ cell tumours. Int J Dev Biol. 2013;57(2-4):133–139. doi: 10.1387/ijdb.130031nv. [DOI] [PubMed] [Google Scholar]
- 3.Stevenson SM, Lowrance WT. Epidemiology and diagnosis of testis cancer. Urol Clin North Am. 2015;42(3):269–275. doi: 10.1016/j.ucl.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Le Cornet C, Lortet-Tieulent J, Forman D, Beranger R, Flechon A, Fervers B, Schuz J, Bray F. Testicular cancer incidence to rise by 25% by 2025 in europe? Model-based predictions in 40 countries using population-based registry data. Eur J Cancer. 2014;50(4):831–839. doi: 10.1016/j.ejca.2013.11.035. [DOI] [PubMed] [Google Scholar]
- 5.Illuminati G, Calio FG, Angelici AM, Pizzardi G, Pasqua R, Masci F, Vietri F. Outcome of resection and chemotherapy versus chemotherapy alone for retroperitoneal recurrence of testicular cancer involving the inferior vena cava: A retrospective cohort study of 22 consecutive patients. Anticancer Res. 2016;36(7):3483–3488. [PubMed] [Google Scholar]
- 6.Rajpert-De Meyts E, McGlynn KA, Okamoto K, Jewett MA, Bokemeyer C. Testicular germ cell tumours. Lancet. 2016;387(10029):1762–1774. doi: 10.1016/S0140-6736(15)00991-5. [DOI] [PubMed] [Google Scholar]
- 7.Williamson SR, Delahunt B, Magi-Galluzzi C, Algaba F, Egevad L, Ulbright TM, Tickoo SK, Srigley JR, Epstein JI, Berney DM, Members of the ITTP. The world health organization 2016 classification of testicular germ cell tumours: A review and update from the international society of urological pathology testis consultation panel. Histopathology. 2017;70(3):335–346. doi: 10.1111/his.13102. [DOI] [PubMed] [Google Scholar]
- 8.Vladusic T, Hrascan R, Kruslin B, Pecina-Slaus N, Perica K, Bicanic A, Vrhovac I, Gamulin M, Franekic J. Histological groups of human postpubertal testicular germ cell tumours harbour different genetic alterations. Anticancer Res. 2014;34(8):4005–4012. [PubMed] [Google Scholar]
- 9.Kharazmi E, Hemminki K, Pukkala E, Sundquist K, Tryggvadottir L, Tretli S, Olsen JH, Fallah M. Cancer risk in relatives of testicular cancer patients by histology type and age at diagnosis: A joint study from five nordic countries. Eur Urol. 2015;68(2):283–289. doi: 10.1016/j.eururo.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J, Albano A, Chen C, Starr JR, Rader DJ, Godwin AK, Reilly MP, Hakonarson H, Schwartz SM, Nathanson KL. Common variation in kitlg and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41(7):811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litchfield K, Levy M, Orlando G, Loveday C, Law PJ, Migliorini G, Holroyd A, Broderick P, Karlsson R, Haugen TB, Kristiansen W, Nsengimana J, Fenwick K, Assiotis I, Kote-Jarai Z, Dunning AM, Muir K, Peto J, Eeles R, Easton DF, Dudakia D, Orr N, Pashayan N, Collaboration UKTC, Consortium P, Bishop DT, Reid A, Huddart RA, Shipley J, Grotmol T, Wiklund F, Houlston RS, Turnbull C. Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor. Nat Genet. 2017;49(7):1133–1140. doi: 10.1038/ng.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, Renwick A, Hughes D, Hines S, Seal S, Morrison J, Nsengimana J, Deloukas P, Collaboration UKTC, Rahman N, Bishop DT, Easton DF, Stratton MR. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41(7):807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, Huddart RA, Bishop DT, Easton DF, Stratton MR, Rahman N, Collaboration UKTC. Variants near dmrt1, tert and atf7ip are associated with testicular germ cell cancer. Nat Genet. 2010;42(7):604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kratz CP, Han SS, Rosenberg PS, Berndt SI, Burdett L, Yeager M, Korde LA, Mai PL, Pfeiffer R, Greene MH. Variants in or near kitlg, bak1, dmrt1, and tert-clptm1l predispose to familial testicular germ cell tumour. J Med Genet. 2011;48(7):473–476. doi: 10.1136/jmedgenet-2011-100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salminen E, Niiniviita H, Jarvinen H, Heinavaara S. Cancer death risk related to radiation exposure from computed tomography scanning among testicular cancer patients. Anticancer Res. 2017;37(2):831–834. doi: 10.21873/anticanres.11385. [DOI] [PubMed] [Google Scholar]
- 16.Mirabello L, Kratz CP, Savage SA, Greene MH. Promoter methylation of candidate genes associated with familial testicular cancer. Int J Mol Epidemiol Genet. 2012;3(3):213–227. [PMC free article] [PubMed] [Google Scholar]
- 17.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant cpg-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24(2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 18.Agathanggelou A, Cooper WN, Latif F. Role of the ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65(9):3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 19.Dallol A, Cooper WN, Al-Mulla F, Agathanggelou A, Maher ER, Latif F. Depletion of the ras association domain family 1, isoform a-associated novel microtubule-associated protein, c19orf5/map1s, causes mitotic abnormalities. Cancer Res. 2007;67(2):492–500. doi: 10.1158/0008-5472.CAN-06-3604. [DOI] [PubMed] [Google Scholar]
- 20.Song MS, Song SJ, Ayad NG, Chang JS, Lee JH, Hong HK, Lee H, Choi N, Kim J, Kim H, Kim JW, Choi EJ, Kirschner MW, Lim DS. The tumour suppressor rassf1a regulates mitosis by inhibiting the apc-cdc20 complex. Nat Cell Biol. 2004;6(2):129–137. doi: 10.1038/ncb1091. [DOI] [PubMed] [Google Scholar]
- 21.Hesson LB, Cooper WN, Latif F. The role of rassf1a methylation in cancer. Dis Markers. 2007;23(1-2):73–87. doi: 10.1155/2007/291538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai Y, Sakano S, Suehiro Y, Okada T, Korenaga Y, Hara T, Naito K, Matsuyama H, Hinoda Y. Methylation level of the rassf1a promoter is an independent prognostic factor for clear-cell renal cell carcinoma. Ann Oncol. 2010;21(8):1612–1617. doi: 10.1093/annonc/mdp577. [DOI] [PubMed] [Google Scholar]
- 23.Martins AT, Monteiro P, Ramalho-Carvalho J, Costa VL, Dinis-Ribeiro M, Leal C, Henrique R, Jeronimo C. High rassf1a promoter methylation levels are predictive of poor prognosis in fine-needle aspirate washings of breast cancer lesions. Breast Cancer Res Treat. 2011;129(1):1–9. doi: 10.1007/s10549-010-1160-0. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Wang B, Chen X, Bi J. The prognostic value of rassf1a promoter hypermethylation in non-small cell lung carcinoma: A systematic review and meta-analysis. Carcinogenesis. 2011;32(3):411–416. doi: 10.1093/carcin/bgq266. [DOI] [PubMed] [Google Scholar]
- 25.Honorio S, Agathanggelou A, Wernert N, Rothe M, Maher ER, Latif F. Frequent epigenetic inactivation of the rassf1a tumour suppressor gene in testicular tumours and distinct methylation profiles of seminoma and nonseminoma testicular germ cell tumours. Oncogene. 2003;22(3):461–466. doi: 10.1038/sj.onc.1206119. [DOI] [PubMed] [Google Scholar]
- 26.Martinelli CM, Lengert AV, Carcano FM, Silva EC, Brait M, Lopes LF, Vidal DO. Mgmt and calca promoter methylation are associated with poor prognosis in testicular germ cell tumor patients. Oncotarget. 2016 doi: 10.18632/oncotarget.11167. doi: 10.18632/oncotarget.11167. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Moundhri MS, Al-Nabhani M, Tarantini L, Baccarelli A, Rusiecki JA. The prognostic significance of whole blood global and specific DNA methylation levels in gastric adenocarcinoma. PLoS One. 2010;5(12):e15585. doi: 10.1371/journal.pone.0015585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59(1):67–70. [PubMed] [Google Scholar]
- 29.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16(1):108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 30.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The prisma statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. PLoS Med. 2009;6(7):e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: The prisma statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno H, Kitada K, Nakai K, Sarai A. Prognoscan: A new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters I, Vaske B, Albrecht K, Kuczyk MA, Jonas U, Serth J. Adiposity and age are statistically related to enhanced rassf1a tumor suppressor gene promoter methylation in normal autopsy kidney tissue. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2526–2532. doi: 10.1158/1055-9965.EPI-07-0203. [DOI] [PubMed] [Google Scholar]
- 34.Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the european prospective investigation into cancer and nutrition (epic-turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22(5):843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 35.Pasquali L, Bedeir A, Ringquist S, Styche A, Bhargava R, Trucco G. Quantification of cpg island methylation in progressive breast lesions from normal to invasive carcinoma. Cancer Lett. 2007;257(1):136–144. doi: 10.1016/j.canlet.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 36.R Core Team RA language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria, 2015 [Google Scholar]
- 37.Viechtbauer W. Conducting meta-analyses in r with the metafor package. J Stat Softw. 2010;36(3):48. [Google Scholar]
- 38.Ellinger J, Albers P, Perabo FG, Muller SC, von Ruecker A, Bastian PJ. Cpg island hypermethylation of cell-free circulating serum DNA in patients with testicular cancer. J Urol. 2009;182(1):324–329. doi: 10.1016/j.juro.2009.02.106. [DOI] [PubMed] [Google Scholar]
- 39.Kawakami T, Okamoto K, Kataoka A, Koizumi S, Iwaki H, Sugihara H, Reeve AE, Ogawa O, Okada Y. Multipoint methylation analysis indicates a distinctive epigenetic phenotype among testicular germ cell tumors and testicular malignant lymphomas. Genes Chromosomes Cancer. 2003;38(1):97–101. doi: 10.1002/gcc.10234. [DOI] [PubMed] [Google Scholar]
- 40.Koul S, Houldsworth J, Mansukhani MM, Donadio A, McKiernan JM, Reuter VE, Bosl GJ, Chaganti RS, Murty VV. Characteristic promoter hypermethylation signatures in male germ cell tumors. Mol Cancer. 2002;1:8. doi: 10.1186/1476-4598-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lind GE, Skotheim RI, Fraga MF, Abeler VM, Esteller M, Lothe RA. Novel epigenetically deregulated genes in testicular cancer include homeobox genes and scgb3a1 (hin-1) J Pathol. 2006;210(4):441–449. doi: 10.1002/path.2064. [DOI] [PubMed] [Google Scholar]
- 42.Tian Y, Hou Y, Zhou X, Cheng H, Zhou R. Tumor suppressor rassf1a promoter: P53 binding and methylation. PLoS One. 2011;6(2):e17017. doi: 10.1371/journal.pone.0017017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smiraglia DJ, Szymanska J, Kraggerud SM, Lothe RA, Peltomaki P, Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21(24):3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- 44.Smith-Sorensen B, Lind GE, Skotheim RI, Fossa SD, Fodstad O, Stenwig AE, Jakobsen KS, Lothe RA. Frequent promoter hypermethylation of the o6-methylguanine-DNA methyltransfe-rase (mgmt) gene in testicular cancer. Oncogene. 2002;21(57):8878–8884. doi: 10.1038/sj.onc.1205978. [DOI] [PubMed] [Google Scholar]
- 45.Koul S, McKiernan JM, Narayan G, Houldsworth J, Bacik J, Dobrzynski DL, Assaad AM, Mansukhani M, Reuter VE, Bosl GJ, Chaganti RS, Murty VV. Role of promoter hypermethylation in cisplatin treatment response of male germ cell tumors. Mol Cancer. 2004;3:16. doi: 10.1186/1476-4598-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuberi M, Mir R, Dholariya S, Najar I, Yadav P, Javid J, Guru S, Mirza M, Gandhi G, Khurana N, Ray PC, Saxena A. Rassf1 and pten promoter hypermethylation influences the outcome in epithelial ovarian cancer. Clinical Ovarian and Other Gynecologic Cancer. 2014;7(1-2):33–39. [Google Scholar]
- 47.Jiang Y, Cui L, Chen WD, Shen SH, Ding LD. The prognostic role of rassf1a promoter methylation in breast cancer: A meta-analysis of published data. PLoS One. 2012;7(5):e36780. doi: 10.1371/journal.pone.0036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng RW, Li YC, Chen X, Huang YX, Shi H, Du DD, Niu X, Lu C, Lu MX. Aberrant methylation of rassf1a closely associated with hnscc, a meta-analysis. Sci Rep. 2016;6:20756. doi: 10.1038/srep20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao C, Dai W, Li H, Tang W, Jia S, Wu X, Luo Y. The relationship between rassf1a gene promoter methylation and the susceptibility and prognosis of melanoma: A meta-analysis and bioinformatics. PLoS One. 2017;12(2):e0171676. doi: 10.1371/journal.pone.0171676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cameron EE, Baylin SB, Herman JG. P15(ink4b) cpg island methylation in primary acute leukemia is heterogeneous and suggests density as a critical factor for transcriptional silencing. Blood. 1999;94(7):2445–2451. [PubMed] [Google Scholar]
- 51.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38(5):540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 52.Mamrut S, Harony H, Sood R, Shahar-Gold H, Gainer H, Shi YJ, Barki-Harrington L, Wagner S. DNA methylation of specific cpg sites in the promoter region regulates the transcription of the mouse oxytocin receptor. PLoS One. 2013;8(2):e56869. doi: 10.1371/journal.pone.0056869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brennan K, Garcia-Closas M, Orr N, Fletcher O, Jones M, Ashworth A, Swerdlow A, Thorne H, Investigators KC, Riboli E, Vineis P, Dorronsoro M, Clavel-Chapelon F, Panico S, Onland-Moret NC, Trichopoulos D, Kaaks R, Khaw KT, Brown R, Flanagan JM. Intragenic atm methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Res. 2012;72(9):2304–2313. doi: 10.1158/0008-5472.CAN-11-3157. [DOI] [PubMed] [Google Scholar]
- 54.Iwamoto T, Yamamoto N, Taguchi T, Tamaki Y, Noguchi S. Brca1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with brca1 promoter methylation. Breast Cancer Res Treat. 2011;129(1):69–77. doi: 10.1007/s10549-010-1188-1. [DOI] [PubMed] [Google Scholar]
- 55.Koestler DC, Marsit CJ, Christensen BC, Accomando W, Langevin SM, Houseman EA, Nelson HH, Karagas MR, Wiencke JK, Kelsey KT. Peripheral blood immune cell methylation profiles are associated with nonhematopoietic cancers. Cancer Epidemiol Biomarkers Prev. 2012;21(8):1293–1302. doi: 10.1158/1055-9965.EPI-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langevin SM, Koestler DC, Christensen BC, Butler RA, Wiencke JK, Nelson HH, Houseman EA, Marsit CJ, Kelsey KT. Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: An epigenome-wide association study. Epigenetics. 2012;7(3):291–299. doi: 10.4161/epi.7.3.19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan D, Delbes G, Landry M, Robaire B, Trasler JM. Epigenetic alterations in sperm DNA associated with testicular cancer treatment. Toxicol Sci. 2012;125(2):532–543. doi: 10.1093/toxsci/kfr307. [DOI] [PubMed] [Google Scholar]
- 58.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31(15):1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]