Abstract
Background: Thyroid cancer is one of the most frequent types of endocrine cancers. In most cases, thyroid cancers are caused by deregulated miRNA expression, especially involving the miR17-92 cluster. miR17-92 transcription is altered in several different tumor types including lymphoma, leukemia, and of the breast and thyroid. As an intronic cluster, miR17-92 must be processed during splicing and therefore interaction between microprocessor and spliceosome machineries is of major importance in understanding its expression. Materials and Methods: We investigated the protein composition of spliceosomes assembled on pre-RNAs containing intronic miR18a and miR19a, components of the miR17-92 cluster, using mass spectrometry. Results: Interestingly, we observed that proteins associated with intronic miR18a and miR19a are cell-specific, and are similar for both miRNAs analyzed. The only exception is the group of heterogeneous nuclear proteins that are commonly recruited by different cells. Conclusion: miRNA processing depends on cell-specific proteins and heterogeneous nuclear proteins have a general role in miRNA processing from introns.
Keywords: Splicing, miRNA, miR17-92 cluster, thyroid cancer
Thyroid cancer is among the most frequent types of endocrine cancers. More than 56,000 new cases are expected in 2017 in the United States alone. Around 70% of thyroid cancers are papillary and mutations in V-raf murine sarcoma viral oncogene homolog B (BRAF) are the most common cause for their development, followed by the oncogene RET chromosomal rearrangements (RET/PTC) (1). It has been shown that mutated BRAFV600E can stimulate expression of the miR-17-92 cluster, especially of miR19a, disrupting transforming growth factor β (TGFβ) signaling in thyroid cancer cells (2,3). In fact, expression of miR17-92 cluster is increased in aggressive anaplastic thyroid cancer (4).
The miR17-92 cluster is found in the miR17-host gene (MIR17HG) intron on chromosome XIII. It is composed of six miRNAs (miR17a, miR18a, miR19a, miR19b, miR20a and miR92a) and reported to be one of the most potent oncogenic miRNAs (5,6). Its expression is stimulated by Myc proto-oncogene protein (c-MYC) and is altered in several different tumor types besides thyroid, including lymphoma, leukemia and solid tumors such as of the breast, lung, colon, pancreas and stomach (2,6-9). miR19a inhibitors were found to delay cell growth, indicating its activity and targets might interfere directly with tumor progression (4,10). In fact, miR19a is considered one of the most oncogenic miRNAs in this cluster (5,11).
Canonical miRNA processing depends on the activity of the microprocessor complex responsible for the initial trimming of primary miRNAs (pri-miRNAs). The microprocessor is formed by the ribonuclease III DROSHA and microprocessor complex subunit DGCR8, and promotes the first cleavages on pri-miRNAs, generating ~70 nucleotides pre-miRNAs that are able to be exported from the nucleus to be cleaved by the endoribonuclease DICER in the cytoplasm (12,13). Mature miRNAs (18-22 nucleotides) target specific mRNAs by complementary binding to their 5’ untranslated region (UTR), coding sequence or 3’ UTR sequence, strongly affecting translation and protein stability. Among the known targets for miR17-92 components are tumor suppressors and oncogenes, which helps to explain the role of this cluster in different cells. miR17-5p, for example, can inhibit cell proliferation, invasion and migration, acting as a tumor suppressor in breast cancer (14). miR19a, on the other hand, targets the tumor suppressor phosphatidylinositol 3,4,5–triphosphate 3-phosphatase and dual-specificity phosphatase PTEN, inhibiting its translation and increasing cell proliferation (5). Transcription factor E2F1 and serine/threonine protein kinase CHEK2 are among the known targets of this cluster. Interestingly, E2F1 also stimulates transcription of this cluster (10,15). It is reasonable that processing and maturation of intronic miRNAs are cooperative with the spliceosome, the macromolecular complex responsible for pre-mRNA splicing. The spliceosome is a 2-MDa complex composed of 5 small nuclear RNAs (snRNAs) and more than 100 protein components, most of which are involved in splicing regulation and pre-mRNA recognition (16). Cooperation between microprocessor and spliceosome during intronic miRNA biogenesis is just beginning to be explored (17,18).
We set out to investigate the protein composition of spliceosomes assembled on pre-RNAs containing intronic miR18a and miR19a in cells derived from papillary thyroid cancer (BCPAP and TPC-I). Our hypothesis was that miR18a and miR19a would require specific proteins for processing that could both guide splicing and promote miRNA processing. The HeLa cell line was used as an internal benchmark, since it exhibits up-regulation of expression of the miR17-92 cluster (7).
Materials and Methods
Cloning. Sequences of miR18a and miR19a were cloned into pGEM-T vector (Promega, Madison, WI, USA) and sub-cloned into the XhoI and HindIII sites within the intron derived from adenovirus major late (AdML) precursor, upstream of the branchpoint site (19). Importantly, this vector has MS2 tag-binding sites at the end of 3’ exon.
Cell culture and transfection. Three cell lines were used HeLa, BCPAP and TPC-I. HeLa and TPC-I were kindly provided by Dr. James A. Fagin (Memorial Sloan-Kettering Cancer Center, MSKCC, USA) (1), and BCPAP was kindly provided by Dr. Massimo Santoro (University “Federico II” of Naples, Italy). All three lineages were cultivated in 100 mm plates in 4-5 ml of Dulbecco’s modified Eagle’s/ Ham’s nutrient mixture F12 medium with 10% fetal bovine serum; 1.2 g/l sodium bicarbonate (NaHCO3), in a humidified incubator with a controlled atmosphere (5% CO2) at 37˚C. Transfection of plasmids was performed in triplicate, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions.
Immunoprecipitation and mass spectrometry. In order to identify the spliceosome proteins associated with intronic miRNAs, transfected cells were submitted to precipitation using MS2-MBP protein. The cellular extract was incubated with MS2-MBP for 2 hours before incubation with dextrose-sepharose resin (GE Healthcare, Little Chalfont, UK). Samples were washed in sizing column buffer 2 (SCB2) [20 mM HEPES (pH 7.9), 150 mM KCl and 0.5 mM EDTA] and eluted with SCB2 supplemented with 10 mM maltose. These samples were digested with trypsin sequencing grade (Promega), purified and submitted to mass spectrometric analysis on a quadrupole time-of-flight mass spectrometer Premier instrument (LNBio, CNPEM, Campinas, Brazil).
RNA extraction. Total RNA from the transfected cells was extracted using Trizol reagent (Invitrogen). cDNA was synthesized with the Superscript System (Invitrogen) and random primers according to the manufacturer’s instructions. cDNAs (100 μg) were analyzed by quantitative real-time reverse transcription–polymerase chain reaction (qRT-PCR), using SYBR Green® reagent (Thermo Fischer, Waltham, MA, USA) to verify splicing efficiency. Two pairs of primers were used: for the exon junction and exon 2 (‘mature’ RNA) and a pair annealing at both ends of the precursor (‘total’ RNA). The mature RNA amplicon was detected only when the splicing occurred; the efficiency of this reaction was measured as a ratio between mature and total RNA cycle thresholds (CTs).
MS2-MBP expression and purification. In order to express and purify MS2-MBP, recombinant E. coli BL21 containing pMS2:MBP was grown in Luria-Bertani medium supplemented with ampicillin. Addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside and growing cells for 4 h at 37˚C induced expression of MS2:MBP. Cell extracts were incubated for 2 hours with dextrose-sepharose resin (GE Healthcare) and eluted in 0.5 M maltose buffer. In order to remove nucleic acid contamination, the sample was further purified in a heparin FF column (GE Healthcare) and eluted with a 0.1-1 M NaCl gradient.
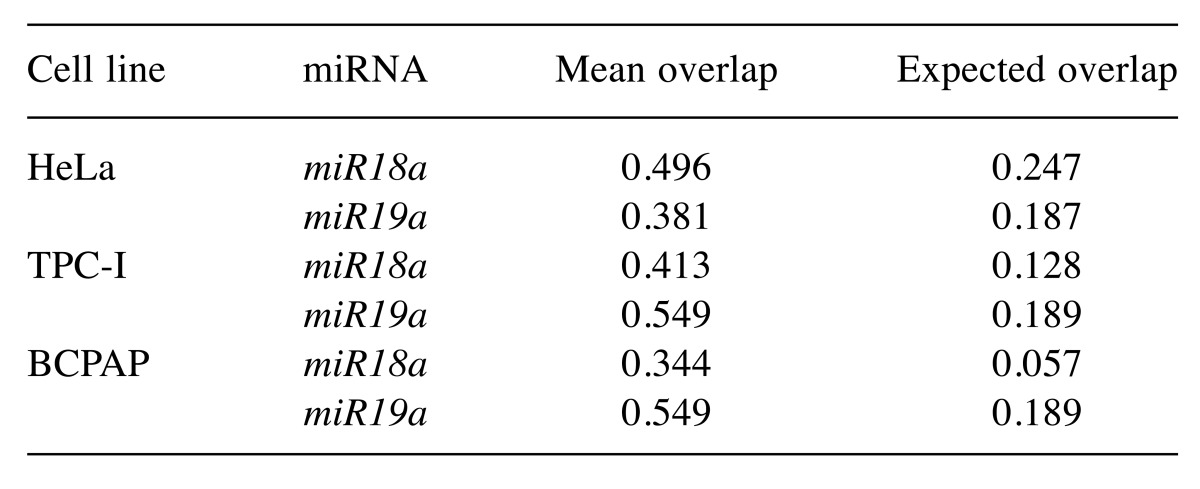
Statistical analysis. We addressed the variation of proteins associated with miRNAs across the different cell lines. Our analysis combines the statistical description of protein overlapping between miRNAs (miR18a and miR19a) and cells (HeLa, TPC-I and BCPAP) with randomization tests (20). In all analyses, we measured the overlap between the composition of proteins in each pair of samples using the Jaccard index (21). The Jaccard index ranges from 0, with no protein occurring in both samples (miR18a and miR19a), to 1, in which all proteins found for miR18a were also present for miR19a. We then compared the empirical value recorded for the Jaccard index with the expected value using a randomization test that assumes proteins can be recorded in any sample by chance. Firstly, we tested if biological triplicates were more similar to each other than to samples from different cells and miRNAs. We compared the mean overlap for pairs of the same triplicates with the expected value generated using a randomization procedure. We randomized the protein recorded in a given pair of triplicates across samples from any combination of cell type and miRNA (n=1,000 randomizations). Alternative randomization procedures led to similar results. Because similarity between pairs of triplicates was higher than expected by the randomization test (Table I), we pooled the data of triplicates to generate a single protein list with proteins recorded for each combination of miRNA and cell type.
Table I. The observed mean overlap, measured by means of the Jaccard index, between pairs of triplicates for each combination of miRNA and cell line. Expected mean overlap between triplicates was computed randomizing the number of proteins recorded for a given pair of triplets across the entire protein list (n=1,000 randomizations).
We then described the patterns of overlap using Venn diagrams comparing the protein lists in each cell line and the protein list recorded for the miRNAs within cell lines. Venn diagrams were computed using the package VennDiagram in R (22). We then performed a randomization test (20) to investigate if the protein lists associated with each miRNA was more similar or distinct than expected by chance. We computed the connectance of our dataset, c, in which c=E/[6P], in which 6 is the number of combinations of miRNAs (miR18a and miR19a) and cell types (HeLa, BCPAP, and TPC-I) used in the experiment, P is the number of proteins recorded in the experiment, 6P is the maximal number of records of proteins for all combinations of miRNAs and cell types, i.e. if all proteins were recorded in all samples, and E is the empirical number of records of proteins for each combination of mRNA and cell type. We used c as a probability in the randomization test. In each randomization (n=1000 randomizations) and for each combination of proteins recorded, miRNA, and cell type, we sampled a number from a uniform distribution [0,1] and if the number sampled was smaller than c, we recorded the protein in that combination of cell type and miRNA in the randomized dataset. We then computed the mean overlap and compared it with the empirical estimates of the mean overlap among pairs of miRNAs and cell types. The computation of Jaccard indices, and the randomization analysis were performed using a MATLAB script (available upon request).
Results
Cells containing plasmids with miR18a and miR19a in its introns were subjected to MS2 immunoprecipitation to isolate the spliceosomes. This experiment was performed in triplicate and samples were analyzed by the quadrupole time-of-flight mass spectrometer (LNBio-CNPEM, Campinas, São Paulo, Brazil). We first investigated whether the similarity of proteins recorded in the biological triplicates for each combination of miRNA and cell line was higher than expected by chance. Since the mean overlap in protein composition between pairs of triplicates was higher than expected by chance (Table I), we pooled the data of triplicates into a single protein list for each combination of miRNA and cell line (Table II).
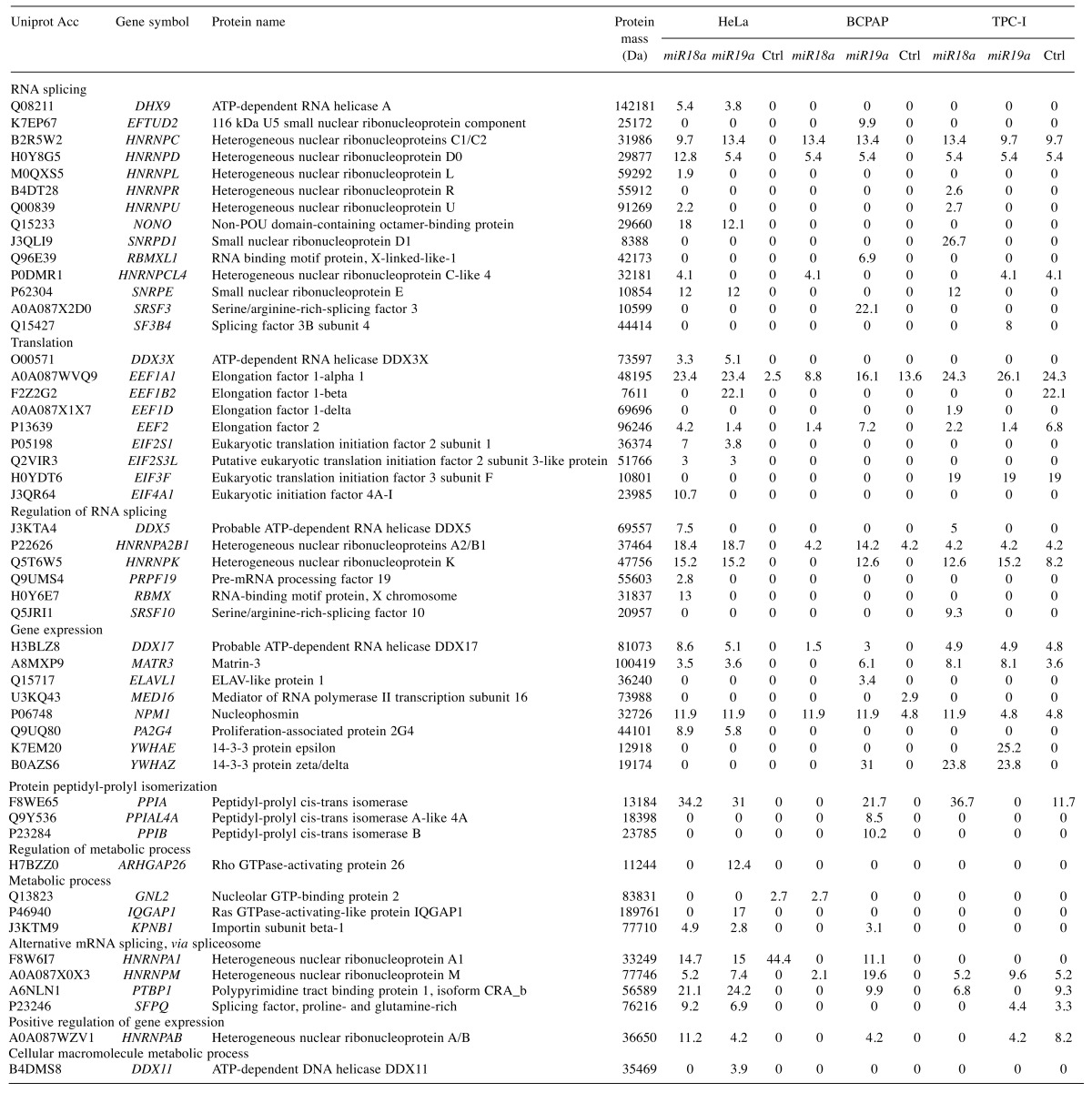
Table II. Proteins identified by mass spectrometry in HeLa, BCPAP and TPC-I cells transfected with preRNA containing intronic miR18a or miR19a. Control (Ctrl): PreRNA without miRNA. Columns show the molecular mass of proteins (Da), and percentage of protein coverage as recorded by the peptides identified in mass spectrometry.
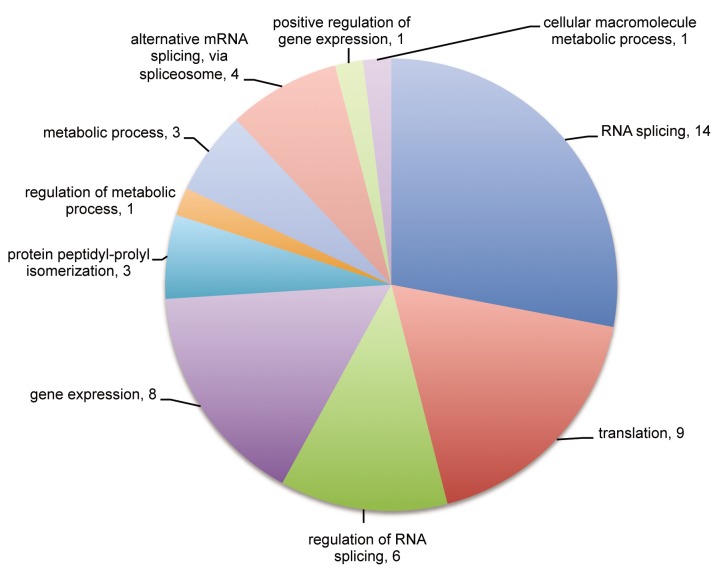
The great majority of the identified proteins were found to be related to RNA binding, including proteins involved in transcription, translation and splicing. Twenty-four out of the 50 identified proteins are directly involved with RNA splicing, regulation and spliceosome assembly, according to the Gene Ontology classification (Figure 1). Among these are several heterogeneous nuclear proteins (hnRNP), A2/B1, A1, C, D, L, M, K, R, U, and general splicing proteins such as small nuclear ribonucleoproteins E and D (SNRPE1 and SNRPD), serine/arginine-rich splicing factor 3 (SRSF3), splicing factor 3B subunit 4 (SF3B4), pre-mRNA processing factor 19 (PRPF19) and non-POU domain-containing octamer-binding protein (NONO). The presence of these proteins strongly indicates we successfully isolated spliceosomes. We also detected proteins involved with translation and metabolic processes, such as the peptidyl-prolyl-isomerases (PPIs). Interestingly, the PPIs (cyclophilins) are also related to spliceosome assembly, as previously described (23).
Figure 1. Gene Ontology classification for the 50 identified proteins in this study. Twenty-four proteins were found to be related to RNA splicing, regulation and spliceosome assembly.
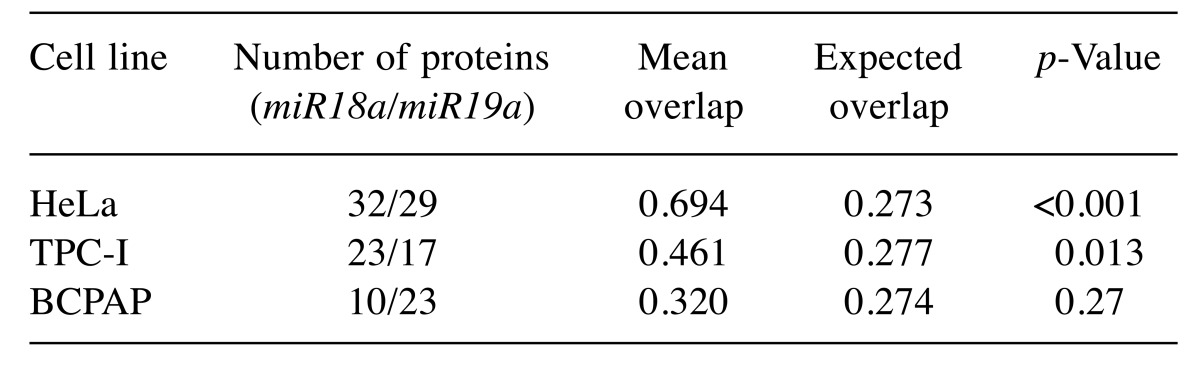
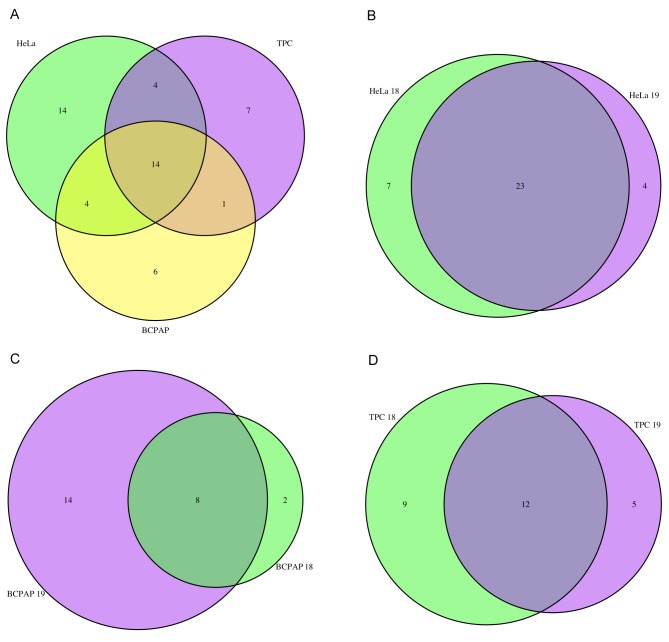
Our analysis of the level of overlap between miRNAs using Venn diagrams with randomization tests indicated that protein lists were more similar between miRNAs in the same cell than expected by chance (Table III). The degree of overlap, however, varied with cell type. The protein lists of miRNAs from HeLa were more than twofold more similar than expected by chance, and the list of proteins of miRNAs in TPC-I was 1.7-fold more similar than expected by chance (Table III). In contrast, the list of proteins found associated with miRNAs in BCPAP cell line was as similar as expected by chance. This latter result was a consequence of the fact that there were fewer proteins associated with miR18a in BCPAP than with miR19a (Table III). In fact, the proteins found associated with miR18a in BCPAP were almost a perfect subset of the group of proteins found to be associated with miR19a in the same cell line, as indicated in Figure 2C.
Table III. The number of proteins associated with each miRNA in a given cell line and the observed mean overlap, measured by means of the Jaccard index. p-Values were estimated as the number of randomizations in which the randomized mean overlap of proteins was higher than the empirical mean overlap in proteins (see Materials and Methods for details). p-Values less than 0.05 indicate that the observed overlap in protein lists was higher than expected by chance.
Figure 2. Venn diagrams comparing overlap of identified proteins. A: Comparison between proteins identified in HeLa, BCPAP and TPC-I cell lines. B-D: Comparison between proteins associated with miR18a and miR19a for HeLa (B), BCPAP (C) and TPC-I (D).
A comparison of the proteins identified in the three cell lines confirms a high degree of similarity among proteins associated with miRNAs within each cell line (Figure 2A). This result might have at least one important functional implication: each cell line has its own set of proteins involved in maturation of these miRNAs. HeLa-specific proteins included ATP-dependent DNA helicase DDX11, RNA-binding motif protein X chromosome (RBMX) and ATP-dependent RNA helicase A (DHX9), involved in RNA splicing and regulation, and proteins involved in translation (Table II). Six proteins were specifically found in association with the miRNAs analyzed for BCPAP. The great majority of these are involved with splicing and gene expression, for example ELAV-like protein 1 (ELAVL1), RNA-binding motif protein X chromosome-like 1 (RBMXL1), SRSF3, PPIB and PPIAL4. Seven proteins were TPC-I-specific, including some translation factors and some components of the splicing machinery, such as SNRPD1, SF3B4 and SRSF10. The only protein common to papillary thyroid lineage cell lines BCPAP and TPC-I was found to be YWHAZ, or protein 14-3-3, involved with gene expression and RNA binding (Table II).
However, 14 out of the 50 identified proteins were common to all three cell lines, including some components of the splicing machinery such as polypyrimidine tract-binding protein 1 (PTBP1) and matrin-3 (MATR3), but mainly hnRNP proteins (AB, M, C, D, CL4 and A2/B1) (Figure 2A). Interestingly, hnRNP proteins were consistently present, independently of the miRNA or cell line, which is also consistent with the hypothesis that these proteins participate in miRNA splicing and maturation (24,25).
In HeLa cells, 34 proteins were found to be associated with miRNAs, of which 23 were common to both miR18a and miR19a (Figure 2B). In this group we found proteins such as PTBP1, MATR3, SNRPE and NONO, which are core components of the spliceosome and probably participate in splicing for the majority of transcripts. We also found hnRNPA1 and hnRNPA2/B1. Our results corroborate previous reports that indicated hnRNPA1 was associated with miR18a processing (26). This suggests that splicing and processing of intronic miR18a and miR19a in HeLa cells is performed by a similar set of proteins.
Twenty-four proteins in BCPAP were identified in association with miRNAs, eight of which were common to both miR18a and miR19a (Figure 2C). Interestingly, the great majority of proteins found to be associated with miRNAs in BCPAP were associated with miR19a, which may indicate a preferential splicing of genes bearing this miRNA in this cell line. In fact, miR19a is considered the most oncogenic miRNA of the cluster, probably associated with tumor development and cell proliferation in several cell types, especially in thyroid cancer cells (4,5). The prevalence of splicing proteins associated with introns containing miR19a might be due to the frequency or higher need for maturation of this miRNA in BCPAP cells, but further studies are necessary to determine this.
A total of 26 proteins were detected in association with miRNAs in TPC-I, and 12 of these were common to miR18a and miR19a (Figure 2D). These included proteins involved in splicing and in translation and gene expression in general. Some hnRNPs were also present in association with both miRNAs, including A2/B1, M, C, D and K, again reinforcing the idea that these might participate in miRNA splicing and maturation, as observed in the other cells.
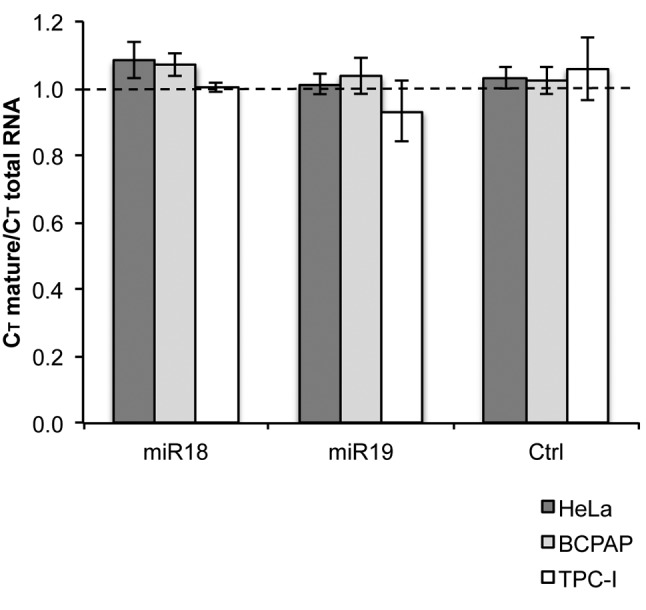
We also asked whether the presence of miRNA in the intron would interfere with the splicing reaction. In order to analyze how efficiently these pre-RNAs were spliced, we used PCR analysis. We used primers complementary to both pre-RNA endings (total pre-RNA) and to exon junctions. If amplification was observed with exon junction primers, we considered mature RNA to have been produced. Total numbers of threshold cycles observed in qRT-PCR were used to calculate a ratio between mature and total RNA (Figure 3). A ratio of 1 indicates the sample had the same amounts of mature and total RNA. A ratio higher than 1 indicates the sample had a lower amount of mature RNA. Importantly, our results indicate that the presence of either miR18a or miR19a in the intron did not affect the efficiency of the reaction, providing ratios close to 1 in the experiments. This means that, as expected, reporter precursors with the miRNAs recruited spliceosome and performed splicing just as efficiently as precursors without intronic miRNAs. This indicates that the isolated complexes do not have any bias toward increased or reduced splicing efficiency.
Figure 3. Splicing efficiency analysis using quantitative real-time reverse transcription–polymerase chain reaction. cDNA samples were derived from cells transfected with miR18a, miR19a or without miRNA (x-axis). Ratios higher than 1 (dashed line) indicate a lower amount of mature RNA, and therefore lower splicing efficiency.
Discussion
Intronic miRNAs must be processed in the midst of splicing reactions. Although 82% of human miRNAs are found in introns (18,27), the interaction between the miRNA processing complex, the microprocessor, and the spliceosome, responsible for intron removal, is poorly understood (17). Previous reports implicated DROSHA partners in the interplay between microprocessor and spliceosome. Interestingly, the presence of DROSHA was found to be essential for miRNA processing from introns (13,17). We attempted to identify proteins associated with intronic miR18a and miR19a during splicing in thyroid cancer cells. Our results indicate three important implications for processing and maturation of these miRNAs.
Firstly, pre-RNAs with and without miRNA in the intron exhibit no difference in splicing efficiency. Interestingly, this result is coherent with a previous report which showed miRNAs are processed along with splicing (18). We used a rather conservative approach to perform our study, since we opted for an optimal intron, derived from AdML precursor, to verify splicing efficiency. This was important to control for minor cellular differences that could interfere with splicing catalysis, leading to different splicing efficiencies. Since spliceosomes assembled upon pre-RNAs containing miRNAs have similar catalytic activation rates, we assumed regulation of splicing in miRNA-containing introns might be related to protein partners of microprocessor and spliceosome complexes, as previously reported (17).
We then investigated the set of proteins recruited for miR18a and miR19a processing. Interestingly, we found that protein composition of these spliceosomes is similar within a cell but differs between different cell lines. This implies miRNA maturation is dependent on different pathways in different cells. Previous reports have indicated that miRNA expression is specifically regulated in different types of cell (25). Our result has an important implication: cancer pathogenesis derived from altered expression of miR18a and miR19a is highly dependent on specific proteins and factors typical of the cell machinery. In fact, thyroid cancers are mostly caused by genetic alterations, some of which lead to increased expression of the miR17-92 cluster (2,4). The altered expression of this cluster is probably caused by the activity of different proteins in BCPAP and TPC-I cells. Among the specific proteins we identified in BCPAP was ELAVL1. ELAVL1 (HuR) targets several mRNAs, modulating their translation, and it can also cooperate with miRNAs in regulation of gene expression (28,29). Interestingly, it can recruit LET-7 to c-MYC 3’UTR, repressing its translation. Binding of ELAVL1 probably alters RNA conformation enabling LET-7 binding and translational inhibition of c-MYC, which would also interfere with miR17-92 expression (30). The increase in ELAVL1/HuR levels has been previously observed in thyroid cancers, and also correlated with poor prognosis (31). Therefore, ELAVL1 might be an important regulatory factor of cancer pathogenesis, ultimately regulating miR17-92 expression.
Thirdly, our analysis provided a set of 14 proteins that appear to always be recruited by miRNA-containing introns, in all three cell lines tested and with both miRNAs. Among these are ATP-dependent RNA helicase DDX17 and hnRNP proteins. Previous literature data indicated that hnRNP proteins establishes communication between the microprocessor and the spliceosome (13). It is noteworthy that nuclear proteins such as TAR DNA-binding protein 43 (TDP43) and RNA-binding protein FUS, homologous to hnRNPs, were previously found to complex with DROSHA (25). In fact, it is possible that spliceosome and microprocessor interactions depend on DROSHA activity and its partners. Interestingly, among the common set of proteins we found DDX17 (P72) also predicted as a microprocessor-associated factor by DROSHA immunoprecipitation experiments (13). Besides their possible activity in miRNA processing, hnRNP proteins associate with pre-RNAs early in spliceosome assembly, and have been related to alternative splicing control (32,33). Binding of these proteins can also block splice site recognition by spliceosome components, therefore inhibiting or delaying splicing in specific pre-RNAs. Further experiments should confirm the direct association of these proteins with intronic miRNA processing and maturation.
Finally, our results revealed that independently of the miRNA used, the proteins involved in miRNA maturation were cell-specific. This indicates that development of thyroid cancer derived from miR18a and miR19a altered expression depends primarily on cell-specific proteins. But, interestingly, our data suggest a set of 14 proteins commonly used for miRNA maturation, independent of the miRNA identity or cell line, including mostly hnRNP proteins.
Acknowledgements
The Authors thank CAPES, CNPq (474672/2013-1) and FAPESP (2013/02738-7; 2017/06994-9) for financial support. We are grateful to LNBio (CNPEM, Campinas, SP, Brazil) for mass spectrometry analysis (MAS-17241 and MAS-18357). We thank Dr. James A. Fagin (Memorial Sloan-Kettering Cancer Center, MSKCC, USA) and Dr. Massimo Santoro (University “Federico II” of Naples, Italy) for TPC-I, HeLa and BCPAP cell lines. We are indebted to Paulo R. Guimarães Jr., Ana Paula Aprigio Assis, Cesar Fuziwara, Kelly Saito, Carla C. Oliveira and Gisela T. Ramos for excellent support.
References
- 1.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- 2.Fuziwara CS, Kimura ET. High Iodine Blocks a NOTCH/miR-19 Loop Activated by the BRAFV600E Oncoprotein and Restores the Response to TGFβ in Thyroid Follicular Cells. Thyroid. 2014;24:453–462. doi: 10.1089/thy.2013.0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dews M, Fox J, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, Cleary MA, Thomas-Tikhonenko A. The myc-miR-17-92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent antiangiogenic factors. Cancer Res. 2010;70:8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takakura S, Mitsutake N, Nakashima M, Namba H, Saenko VA, Rogounovitch TI, Nakazawa Y, Hayashi T, Ohtsuru A, Yamashita S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–1154. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Vecchiarelli-Federico LM, Li YJ, Egan SE, Spaner D, Hough MR, Ben-David Y. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood. 2012;119:4486–4498. doi: 10.1182/blood-2011-09-378687. [DOI] [PubMed] [Google Scholar]
- 7.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihailovich M, Bremang M, Spadotto V, Musiani D, Vitale E, Varano G, Zambelli F, Mancuso FM, Cairns DA, Pavesi G, Casola S, Bonaldi T. miR-17-92 fine-tunes MYC expression and function to ensure optimal B cell lymphoma growth. Nat Commun. 2015;6:8725. doi: 10.1038/ncomms9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17-92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS. Functional Anatomy of the Human Microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 14.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 16.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka N, Fujita M, Ohno M. Functional association of the microprocessor complex with the spliceosome. Mol Cell Biol. 2009;29:3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YK, Kim VN. Processing of intronic microRNAs. Embo J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manly BF. Randomization, bootstrap and Monte Carlo methods in biology. CRC Press. 2006 [Google Scholar]
- 21.Jaccard P. The distribution of the flora in the alpine zone. New phytologist. 1912;11:37–50. [Google Scholar]
- 22.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams BM, Coates MN, Jackson SR, Jurica MS, Davis TL. Nuclear cyclophilins affect spliceosome assembly and function in vitro. Biochem J. 2015;469:223–233. doi: 10.1042/BJ20150396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci USA. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 27.Franca GS, Vibranovski MD, Galante PA. Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat Commun. 2016;7:11438. doi: 10.1038/ncomms11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14:10015–10041. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uren PJ, Burns SC, Ruan J, Singh KK, Smith AD, Penalva LO. Genomic analyses of the RNA-binding protein Hu antigen R (HuR) identify a complex network of target genes and novel characteristics of its binding sites. J Biol Chem. 2011;286:37063–37066. doi: 10.1074/jbc.C111.266882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits LET-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldan F, Mio C, Allegri L, Conzatti K, Toffoletto B, Puppin C, Radovic S, Vascotto C, Russo D, Di Loreto C, Damante G. Identification of tumorigenesis-related mRNAs associated with RNA-binding protein HuR in thyroid cancer cells. Oncotarget. 2016;7:63388–63407. doi: 10.18632/oncotarget.11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, Siziopikou KP, Peng J, Xiao X, Cheng C. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191–1203. doi: 10.1101/gad.241968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akerman M, Fregoso OI, Das S, Ruse C, Jensen MA, Pappin DJ, Zhang MQ, Krainer AR. Differential connectivity of splicing activators and repressors to the human spliceosome. Genome Biol. 2015;16:119. doi: 10.1186/s13059-015-0682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]