Abstract
Background: Podoplanin (PDPN), a mucin-type transmembrane glycoprotein, is expressed in a variety of human cancer types, and contributes to tumor progression. Our goal was to evaluate PDPN expression in hepatocellular carcinoma (HCC) using both immunohistochemistry (IHC) and RNAscope in situ hybridization. Materials and Methods: Twenty patients with HCC who underwent partial hepatectomy with curative intent were retrospectively analyzed. Results: IHC gave positive results in 11 cases, while RNAscope assay for PDPN detected amplification in 16 cases. A significant association was noted between PDPN protein expression and histological tumor grade (p=0.036). Four cases that had negative PDPN results by RNAscope were also negative by IHC, while the remaining five cases with negative results by IHC were positive by RNAscope. A positive relationship was found between PDPN mRNA protein expression (p<0.001). Conclusion: Our preliminary results suggest that PDPN contributes to the malignant potential of HCC. RNAscope proved to be a more sensitive and reliable method than IHC in PDPN detection.
Keywords: Hepatocellular carcinoma, podoplanin, RNAscope, immunohistochemistry
Hepatocellular carcinoma (HCC) is one of the most frequent cancer types worldwide, with over half a million new cases diagnosed annually (1). The global burden of HCC is estimated to increase in the next two decades, and by 2030 it is predicted to be the second most common cause of cancer-related mortality worldwide (2). The prognosis for patients with advanced disease is poor and most patients survive only a few months following diagnosis. New insights into molecular biology of this fatal disease could lead to the identification of valuable molecular biomarkers for prediction of treatment response and for development of effective anticancer therapies.
In the era of personalized medicine, research is focused on finding biomarkers based on DNA, RNA and proteins in groups of patients with similar molecular and genetic anomalies (3). RNAscope is a modern in situ hybridization (ISH) technology, compatible with the use of formalin-fixed paraffin-embedded (FFPE) tissue sections, that allows amplification of targeted-specific signals without amplifying the background (4). Compared to other RNA ISH methods, RNAscope is a highly sensitive and specific test and the ability to detect mRNA signals using standard microscopy makes it a powerful tool (4,5).
Podoplanin (PDPN), a mucin-type transmembrane protein, is expressed in a relatively large spectrum of tumours and its overexpression was found to be correlated with increased cancer cell motility and metastasis (6). However, the role of this protein in HCC tumour cells is uncertain. The purpose of the present study was to evaluate PDPN in HCC tumour cells by using both IHC and RNAscope, and to compare the sensitivity of these techniques in PDPN detection.
Materials and Methods
Patients and tissue samples. A total of 20 patients, diagnosed with HCC and who underwent partial hepatectomy at the Regional Institute of Gastroenterology and Hepatology (Cluj-Napoca, Romania) were retrospectively studied. As controls, we used liver samples from five patients with liver metastases from colonic adenocarcinoma who underwent curative hepatic resection. The control specimens were sampled from the non-tumoural areas of liver. Tumour samples, as well as clinical data, were obtained from the Institute database according to protocols approved by the Ethical Committee (number 5/5821/26.05.2016) and protocols followed the guidelines of the Helsinki Declaration.
Immunohistochemistry. Twenty FFPE tumour samples were selected for IHC. Five-micrometre-thick sections were cut for each case. The dewaxing and rehydration of the sections were followed by heat-induced epitope retrieval in citrate buffer (pH 6) for 30 min (with PT link module; Dako Cytomation, Glostrup, Denmark). The IHC technique continued with the blocking of endogenous peroxidases using 3% hydrogen peroxide. sections were then incubated with monoclonal primary antibody to PDPN (clone D2-40, ready-to-use; Dako Cytomation, Carpinteria, CA, USA) for 30 minutes. The slides were counterstained with Mayer hematoxylin. The working system was LSAB-HRP (Biotinylated Link, Code K0690; Dako Cytomation, Glostrup, Denmark) and the final product of the reaction was visualized with diaminobenzidine (Dako Cytomation, Glostrup, Denmark). The percentage of tumour cells expressing PDPN was classified into four groups as follows: 0: No staining; 1+: 1-25%; 2+: 26-50%, and 3+: 51-100%.
RNAscope assay. The same 20 FFPE tumour samples were analysed with RNAscope assay (Advanced Cell Diagnostics, Hayward, CA, USA) using target probes for PDPN. The technique was performed manually following the manufacturer’s instructions. In brief, 5 µm sections were cut for each case, deparaffinised and treated with pretreatment solution s: #1 for 10 minutes at room temperature, #2 for 15 min at 98˚C and #3 for 15 min at 40˚C. The FFPE sections with targeted probes were then incubated in a HybEZ oven (Advanced Cell Diagnostics) for 2 h at 40˚C. Slides were then washed and incubated with the signal amplification solutions: #1 for 30 minutes at 40˚C, #2 for 15 min at 15˚C, #3 for 30 minutes at 40˚C, #4 for 15 min at 15˚C, #5 for 30 min at room temperature, and #6 for 15 min at room temperature. Hybridization signals were visualized with RNAscope 2.5 HD-Brown detection kit (Advanced Cell Diagnostics, Hayward, CA, USA) and the slides were counterstained with hematoxylin (2 minutes). RNAscope results were examined under a standard bright-field microscope at ×20-40 magnification. We used the scoring system provided by the vendor: 0: negative, 0-1 dot/10 tumour cells, 1+: 1-3 dots/cell visible at ×20-40 magnification, 2+: 4-10 dots/cell visible at ×20-40 magnification, 3+: >10 dots/cell and <10% positive cells with dot clusters visible at ×20 magnification, and 4+: >10 dots/cell and >10% positive cells with dot clusters visible at ×20 magnification). The PDPN miRNA signals were evaluated in one representative slide from each case, which was previously selected for IHC. For each slide, three areas containing the highest number of positive cells or dot clusters were selected. All tumor cells within each field were counted and then the percentage of positive PDPN mRNA signals was used as the final score. Image acquisition and analysis were performed using Nikon Eclipse E 600 microscope (Microscopes/Instruments Division, Vienna, Austria) and Lucia G software (Laboratory Imaging, Prague, Czech Republic) for microscopic image analysis.
Statistical analysis. SPSS 22.0 software for Windows (IBM Corp., Armonk, NY, USA) was used for statistical analysis. The relation between PDPN expression at mRNA and protein levels was analysed by Pearson’s test, and association between PDPN expression and clinicopathological parameters was analysed by Spearman’s test. p-Values of less than 0.05 were considered statistically significant.
Results
Clinical data. The present study included samples from 20 patients, comprising of 12 males (60%) and 8 females (40%), with age between 40 and 91 years (mean=64.7 years). All patients selected for this study had three or few tumoural nodules with a maximal diameter of 6 cm.
According to the Edmondson and Steiner system (7), the tumors were graded as follows: grade 1: 2 cases, grade 2: 8 cases, grade 3: 10 cases, and grade 4: 0 cases. Tumor stages were classified according to TNM classification of tumors of the liver (8) as follows: I: 5 cases, II: 8 cases, III: 7 cases, and IV: 0 cases. Among the 20 HCCs, cirrhosis was present in five cases (25%) and vascular invasion was noted in eight cases (40%).
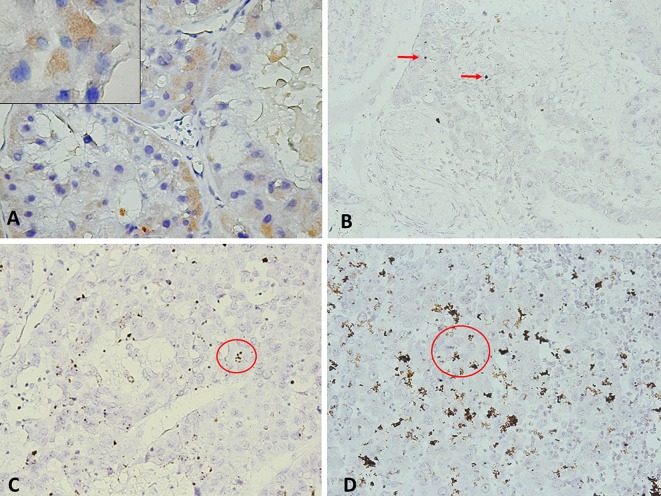
PDPN protein expression. In the control samples, PDPN expression was identified in lymphatic vessels (Figure 1A), while hepatocytes were negative for this marker. Cytoplasmatic expression of PDPN protein in tumour cells was identified in 11 HCCs (55%), including score 1+ in seven cases (64%), score 2+ in three cases (27%), and score 3+ in one case (9%). PDPN-expressing cells were significantly correlated with the histological grade (p=0.036). Looking at the whole group of patients, we noted that high PDPN expression was more often found in high-grade tumours. Compared to grade 1 tumours, where all cases were PDPN-negative, most grade 3 tumours (80%) expressed PDPN. No significant correlation was seen between PDPN protein expression and the other clinicopathological parameters examined.
Figure 1. A: Podoplanin (PDPN) immunohistochemistry in showing cytoplasmic staining of tumour cells at low magnification (×40) and at high magnification (inset; ×100), score 2+. B-D: PDPN mRNA signal was identified in tumour cells as multiple small brown dots as indicated by red arrow: score 1, with 0-1 dots/cell, (B); score 2, 4-10 dots/cell (C); score 4, >10 dots/cell and more than 10% positive cells with dot clusters (D).
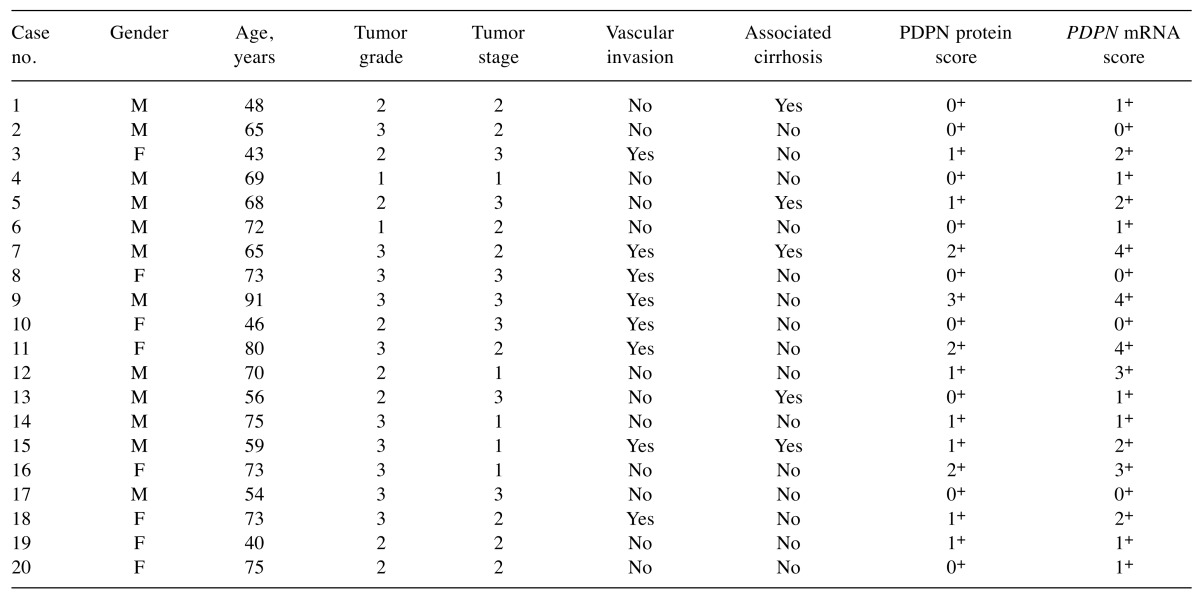
PDPN mRNA expression. PDPN mRNA signals were detected in HCC tumour cells as brown dotted or clustered patterns (Figure 1B-D). Of the 20 HCC samples, 16 (80%) were PDPN-positive, including score 1+ in seven (44%), score 2+ in four (25%), score 3+ in two (12%), and score 4+ in three (19%). The four cases that had negative PDPN results by RNAscope were also negative by IHC. The remaining five PDPN cases with negative results by IHC were positive by RNAscope, all of them with a low but present amplification, scored as 1+ (Table I). A positive relationship was found between PDPN mRNA and protein expression (p<0.001).
Table I. RNAscope and protein scores for podoplanin and clinicopathological characteristics of the study patients.
F: Female; M: male.
Discussion
The prognosis of patients with advanced HCC remains poor due to the lack of specific symptoms in the early stages of the disease. For these patients, treatment options are limited to chemoembolization or systemic chemotherapy which, unfortunately, have a low efficacy (9). By contrast, for patients diagnosed at an early stage, the 5-year survival rate with surgery as therapy is >93% (10). Therefore, it is crucial to identify new molecular markers for diagnosis at an early stage, that may improve treatment outcome.
PDPN, a small glycoprotein with 'many faces', is receiving increasing interest due to its various functions in development, immunology and cancer (11). In addition to playing a significant role in lymphangiogenesis, PDPN expression has been reported in tumour cells themselves in diverse cancer types (6). We previously showed that PDPN evaluated by IHC is expressed in tumour cells of HCC (12). In the present study, we demonstrated PDPN expression in HCC tumour cells using both IHC (protein) and RNAscope assay (mRNA). As in our previous research, in this study, PDPN was highly expressed in HCCs with a poor grade of differentiation. Given the fact that PDPN is involved in epithelial–mesenchymal transition and contributes to invasion and metastasis (6,13,14), we speculate that this protein could be a prognostic marker of malignancy in HCC. However, functions of PDPN differ between distinct types of cancer. For instance, overexpression of PDPN in glioblastoma, head and neck, oesophageal, and skin squamous cell carcinomas (SCCs) contributed to a poor prognosis (11), while a low expression in cervical carcinoma and lung squamous cell carcinoma was associated with a favourable prognosis (15,16). Therefore, further studies are necessary to clarify the exact mechanism by which PDPN expression is regulated in HCCs.
A major class of cancer biomarkers are represented by RNA biomarkers or gene-expression signatures (17). One of the most widely used methods of gene-expression analysis, considered currently the gold-standard technique, is real-time polymerase chain reaction (RT-PCR) (18). While this technique has many benefits over other methods, its clinical application has not yet provided satisfactory results. RNAscope, one of the newest ISH methods, has the advantages of in situ analysis of RNA biomarkers (4). In our study, the results of RNAscope assay for PDPN showed detectable mRNA signals in 80% of the HCC cases. By contrast, detection of PDPN by IHC was demonstrated in 55% cases. In addition, five cases with negative PDPN results by IHC were positive by RNAscope. Our data suggest that RNA scope is a more sensitive and reliable method than IHC, and can be very useful for cases in which IHC fails to identify detectable signals. However, in the present study we did find a positive relationship between PDPN mRNA and protein (p<0.001). Our results are in line with a recent study that demonstrated a significant correlation between these two techniques in breast cancer (19).
In conclusion, our preliminary results suggest that PDPN expression may contribute to the malignant potential of HCC. In addition, we demonstrated that RNAscope is a highly reproducible and sensitive method for the detection of PDPN expression. Since the present research is unique and restricted to a small patient cohort, further studies are necessary to elucidate the molecular functions of PDPN, which might not only lead to validation of PDPN as a prognostic marker in HCC, but also for development of new therapeutic strategies based on anti-PDPN models.
Acknowledgements
The present work was partly supported by the Victor Babes University of Medicine and Pharmacy Timisoara, Romania
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 3.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 4.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itzkovitz S, van Oudenaarden A. Validating transcripts with probes and imaging technology. Nat Methods. 2011;8:S12–19. doi: 10.1038/nmeth.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Astarita JL, Acton SE, Turley SJ. Podoplanin: emerging functions in development, the immune system, and cancer. Front Immunol. 2012;3:283. doi: 10.3389/fimmu.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmonson HA, Steiner PE. Primary carcinoma of the liver. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 8.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. Fourth Edition. IARC Press, Lyon. 2010:322–326. [Google Scholar]
- 9.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takayama T, Makuuchi M, Kojiro M, Lauwers GY, Adams RB, Wilson SR, Jang HJ, Charnsangavej C, Taouli B. Early hepatocellular carcinoma: pathology, imaging, and therapy. Ann Surg Oncol. 2008;15:972–978. doi: 10.1245/s10434-007-9685-0. [DOI] [PubMed] [Google Scholar]
- 11.Ugorski M, Dziegiel P, Suchanski J. Podoplanin – a small glycoprotein with many faces. Am J Cancer Res. 2016;6:370–386. [PMC free article] [PubMed] [Google Scholar]
- 12.Cioca A, Ceausu RA, Marin I, Raica M, Cimpean AM. The multifaced role of podoplanin expression in hepatocellular carcinoma. Eur J Histochem. 2017;61:2707. doi: 10.4081/ejh.2017.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín-Villar E, Megías D, Castel S, Yurrita MM, Vilaró S, Quintanilla M. Podoplanin binds ERM proteins to activate RhoA and promote epithelial-mesenchymal transition. J Cell Sci. 2006;119:4541–4553. doi: 10.1242/jcs.03218. [DOI] [PubMed] [Google Scholar]
- 14.Renart J, Carrasco-Ramirez P, Fernandez-Munoz B, Martin-Villar E, Montero L, Yurrita MM, Quintanilla M. New insights into the role of podoplanin in epithelial-mesenchymal transition. Int Rev Cell Mol Biol. 2015;317:185–239. doi: 10.1016/bs.ircmb.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Dumoff KL, Chu CS, Harris EE, Holtz D, Xu X, Zhang PJ, Acs G. Low podoplanin expression in pretreatment biopsy material predicts poor prognosis in advanced-stage squamous cell carcinoma of the uterine cervix treated by primary radiation. Mod Pathol. 2006;19:708–716. doi: 10.1038/modpathol.3800580. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Ishii G, Nagai K, Nagano T, Kojika M, Murata Y, Atsumi N, Nishiwaki Y, Miyazaki E, Kumamoto T, Ochiai A. Low podoplanin expression of tumor cells predicts poor prognosis in pathological stage IB squamous cell carcinoma of the lung, tissue microarray analysis of 136 patients using 24 antibodies. Lung Cancer. 2009;63:418–424. doi: 10.1016/j.lungcan.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: When will molecular signatures become relevant to patient care. Nat Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 18.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Guo S, Song W, Xianq C, Yang C, Tao K, Zhou L, Cao Y, Liu S. Estrogen receptor α (ERα) status evaluation using RNAscope in situ hybridization: a reliable and complementary method for IHC in breast cancer tissues. Hum Pathol. 2017;61:121–129. doi: 10.1016/j.humpath.2016.12.005. [DOI] [PubMed] [Google Scholar]