Abstract
Background/Aim: The post-transcriptional regulation of matrix metalloproteinases (MMPs) via microRNAs (miRNAs) has been recently described in numerous human malignancies. However, the exact mechanisms of miRNA-mediated MMPs deregulation in endometrial cancer (EC) remain unclear. Herein, we aimed to analyze the expression of MMP2, MMP16 and TIMP2 and identify miRNAs that modulate their expression. Materials and Methods: Protein expression was assessed by immunohistochemistry in formalin-fixed paraffin-embedded EC samples. Target prediction algorithms were applied to select miRNAs binding the 3’UTRs of MMP16 (miR-377, miR-382, miR-410, miR-200b) or TIMP2 (miR-200b), and their levels were measured by qPCR in laser capture-microdissected tissue fragments. Luciferase assays and western blotting were used to indicate individual miRNA- mRNA interactions. Results: Overexpression of MMP2 and MMP16 in cancerous tissues corresponded to down-regulation of miR-377, miR-382 and miR-410, while decreased expression of TIMP2 was associated with miR-200b up-regulation. In vitro experiments confirmed direct regulation of MMP16 by miR-382 and miR-410, and TIMP2 by miR-200b in EC Ishikawa cells. Conclusion: We demonstrated novel mechanisms of miRNA-mediated regulation of MMPs activity in EC.
Keywords: Endometrial cancer, microRNAs, MMP2, MMP16, TIMP2
Endometrial cancer (EC) is the most common invasive neoplasm of the female genital tract and the fourth most frequent malignancy affecting women in developed countries. Its global incidence has been increasing over the past decades, reaching approximately 60,000 new cases and 10,500 deaths in the United States in 2016 (1,2). Based on a classification system proposed by Bokhman (3), EC is divided into two main types with different histological features, clinical outcomes and prognoses. Type I tumors, accounting for 70-80% of cases, are influenced by excess or unopposed estrogen stimulation. They are typically of endometrioid morphology, often diagnosed at early stage, and associated with a favorable prognosis (3,4). On the contrary, type II cancers are usually hormone-independent and characterized by a serous or clear-cell histology or very poorly differentiated phenotypes of carcinosarcoma, or undifferentiated carcinoma. These tumors are generally associated with a more aggressive clinical course and a poorer prognosis (5). So far, numerous genetic and epigenetic aberrations involved in EC’s pathogenesis have been described (6,7). However, the knowledge about the molecular mechanisms underlying tumor invasion and metastasis, which are the primary causes of treatment failure in EC patients, remains limited.
Matrix metalloproteinases (MMPs) represent a group of structurally-related extracellular zinc endopeptidases, known for their ability to degrade components of the extracellular matrix (ECM) in both normal physiological and pathophysiological processes (8). In the past few years, MMPs have been intensively investigated as potential biomarkers and unfavorable factors in various types of human cancers (9). Of the several MMPs analyzed in endometrial tumors, matrix metalloproteinase 2 (MMP2) has been considered as the main metalloproteinase involved in the malignant behavior of EC cells (10). MMP2 is synthesized and secreted as a proenzyme (pro-MMP2) (11), and its activation depends on the proteolytic cleavage of pro-MMP2 by membrane type-1 MMP (MT1-MMP, MMP14) and membrane type-3 MMP (MT3-MMP, MMP16) (12,13). Apart from their role in soluble MMPs activation, both MMP14 and MMP16 have been demonstrated to promote cancer progression through various mechanisms including: degradation of ECM, shedding of transmembrane precursors of growth factors and growth factor-binding proteins, and activation of receptors and cell surface molecules (14). Activated MMPs are inhibited by four mammalian tissue inhibitors of metalloproteinases (TIMPs 1-4) (15). In addition to inhibiting most MMPs, TIMP2 has a crucial function in MMP2 activation, acting at low concentrations as the bridging molecule between MT-MMPs and pro-MMP2 (16).
MicroRNAs (miRNAs, miRs) are small, non-coding RNA polynucleotides, involved in post-transcriptional regulation of gene expression. By binding to regulatory sites located primarily within 3’ untranslated region (UTR) of mRNA molecules, miRNAs influence the stability and translation of their target transcripts (17,18). Over the past decade, the fundamental role of miRNAs in endometrial tumorigenesis has been established (19). Furthermore, several miRNAs have been demonstrated to directly or indirectly regulate the expression of MMPs in EC cells (20-23). In our previous study we found that a reduced level of miR-410 corresponded to an increased expression of MMP14 in EC samples (24). This observation prompted us to search for other miRNAs involved in the regulation of the MMP2 activation system components.
The aim of this study was to analyze the expression of MMP2, MMP16 and TIMP2 in type-I endometrial tumors and identify miRNAs involved in their post-transcriptional regulation. Since only limited data on MMP16 and TIMP2-targeting miRNAs is available, we applied target prediction algorithms to select miRNAs potentially binding the 3’UTR of MMP16 (miR-377, miR-382, miR-410, miR-200b) or TIMP2 mRNA (miR-200b). The expression patterns of these candidate miRNAs were assessed in laser capture-microdissected tissue fragments from cancerous, transient and adjacent normal endometrium. The predicted interactions between miRNAs and their targets were further confirmed in vitro in EC Ishikawa cells.
Materials and Methods
Patient samples. In this study we analyzed 20 formalin-fixed paraffin-embedded tissue samples obtained from 12 patients diagnosed with type-I endometrial cancer. All specimens were retrieved from the Department of Pathology, Medical University of Warsaw. The histopathological evaluation of samples was performed by a board-certified pathologist. Tumor staging and histological grading were assessed using the FIGO criteria (2009) (25). The detailed clinicopathological data of all patients has been published previously (24). A local ethics committee approved all aspects of this study in accordance with the Helsinki Declaration.
Immunohistochemistry (IHC). IHC was performed on 10 specimens. The samples were sectioned at 10 μm and treated with a standard de-paraffinization protocol with xylene and graduated alcohols. Primary antibodies: rabbit monoclonal MMP2 (ab192082), rabbit polyclonal MMP16 (ab73877), mouse monoclonal TIMP2 (ab1828) were purchased from Abcam (Cambridge, UK). Each staining was performed according to the manufacturer’s protocol using heat-mediated antigen retrieval in a citrate buffer (95˚C for 20 min). Incubations with the primary antibodies were performed overnight in a humid chamber at 4˚C. The appropriate mouse (MP-7401) or rabbit (MP-7402) secondary antibodies and the blocking solution were purchased from Vector Laboratories (Peterborough, UK). For the final color development, DAB chromogen was used (Dako, Glostrup, Denmark).
IHC scoring. After histopathological evaluation, each slide was divided into 3 areas: a cancer cell zone, a transient zone (adjacent to the neoplasm, including the infiltration areas), and a normal endometrium zone (clearly distinguishable from the cancer cells). The quantitative analysis of deconvoluted DAB pixel intensity was performed separately in each of the three zones using the IHC Profiler plugin for ImageJ (26).
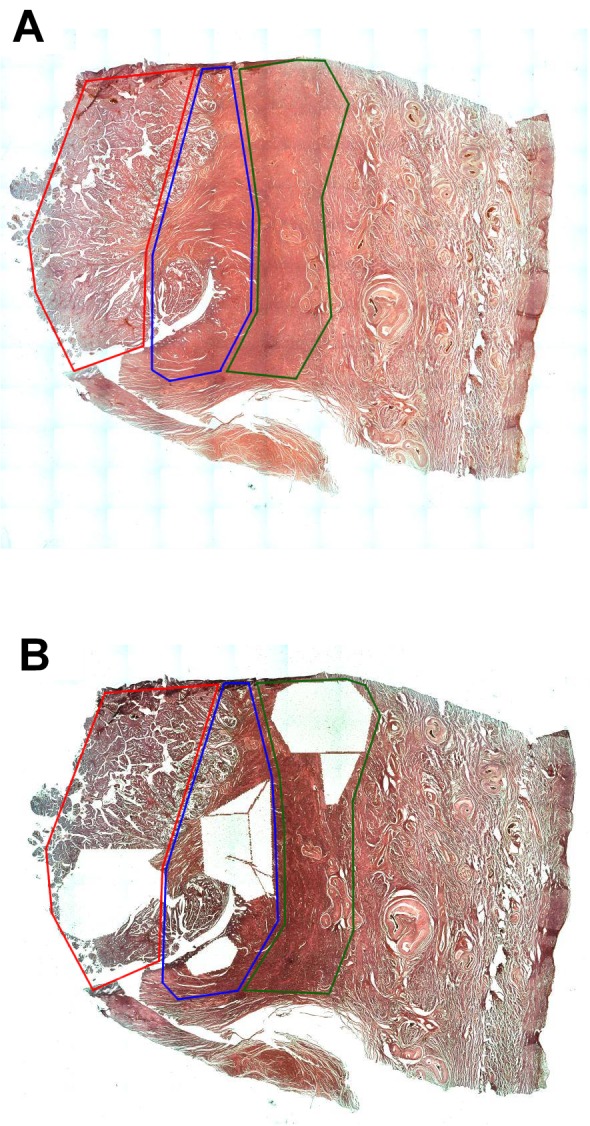
Laser capture microdissection (LCM) and RNA isolation. Laser capture microdissection and RNA isolation procedures were performed as described previously (24). Briefly, FFPE samples were cut into 10 μm sections and mounted on glass slides (SuperFrost Ultra Plus, Menzel-Glazer, Braunschweig, Germany). Subsequently, standard HE staining was performed, and approximately 10 mm2 of each predefined zone’s tissue was catapulted into separate tubes (Figure 1). Total RNA was isolated using RecoverALL Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA), according to the manufacturer’s protocol. The quantity and purity of isolated RNA were assessed by the absorbance measurements at wavelengths of 260 nm and 280 nm on NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc.). We assumed that samples with OD 260/280 ratios between 1.8 and 2.1 were appropriate for further analysis.
Figure 1. An exemplary division of the tissue section before (A) and after (B) laser capture microdissection procedure. Area of cancer, red lines; transient zone, blue lines; normal adjacent endometrium, green lines.
Reverse transcription and qPCR. Reverse transcription reactions in a total volume of 15μl were performed with miRNA-specific stem-loop RT primers using TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific Inc.), followed by qPCR reactions in a total volume of 10μl. Expression levels of miR-200b (Assay ID: 002251), miR-377 (Assay ID: 000566), miR-382 (Assay ID: 000572) and RNU43 (Assay ID: 001095) were determined using TaqMan MicroRNA Assay (Thermo Fisher Scientific Inc.). All qPCR reactions were performed in MicroAmp Fast Optical 96 Well Reaction Plates (Thermo Fisher Scientific Inc.) using Applied Biosystems 7500 Fast Real-Time PCR System with 7500 Software V2.0.6 (Thermo Fisher Scientific Inc.). Samples were assayed in duplicates, and the obtained CT values for the target genes and the housekeeping control (RNU43) were used to calculate a relative gene expression using the 2–ΔΔCT method, as described previously (27).
Cell culture, chemicals and transfection. HEK293T and human endometrial carcinoma cell line Ishikawa were maintained at 37˚C in a humidified atmosphere containing 5% CO2 in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific Inc.) supplemented with fetal bovine serum at concentrations 10% and 5%, respectively, and 100 IU/ml penicillin/streptomycin. MirVana miRNA Mimics: hsa-miR-200b-3p, hsa-miR-377-3p, hsa-miR-382-5p, hsa-miR-410-3p and mirVana miRNA Mimic Negative Control were purchased from Thermo Fisher Scientific Inc. The transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific Inc.) according to the manufacturer’s instructions. The miRNA mimics and miRNA Mimic Negative Control were used at a final concentration of 50 nM.
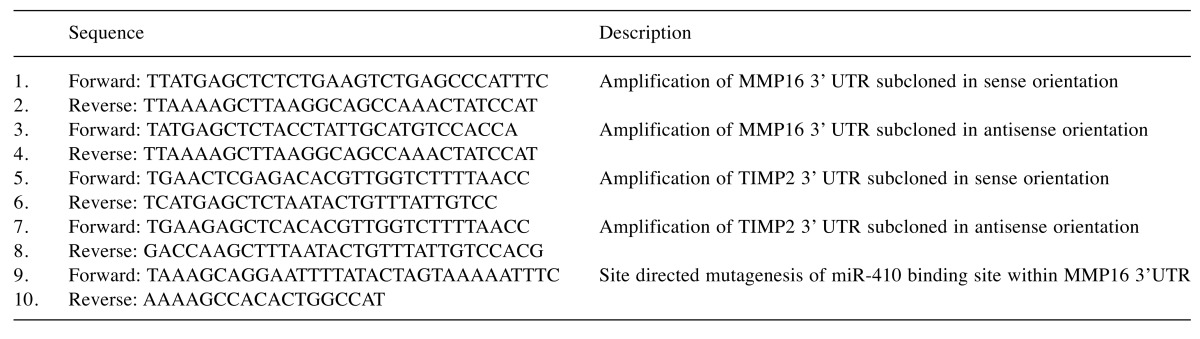
Construction of 3’-UTR reporter plasmids. RNA was isolated from human peripheral blood mononuclear cells using AxyPrep Blood Total RNA Miniprep Kit (Axygen, Scientific, Union City, CA, USA) and reverse-transcribed with GoScript Reverse Transcriptase (Promega, Madison, WI, USA). The cDNA was subjected to PCR with primer pairs amplifying the 3’UTR regions of MMP16 mRNA at positions +353 to +2370 and TIMP2 mRNA at positions +2181 to +2681. The purified PCR products were cloned utilizing SacI and HindIII sites of the pMIR-REPORT Vector, downstream of a Firefly luciferase-coding sequence, in either sense or antisense orientation. The palindromic binding site of miR-410 within MMP16 3’UTR construct in antisense orientation was further mutated using Q5 Site Directed Mutagenesis Kit (New England Biolabs, Beverly, MA, USA). The sequences of all constructs were verified by Sanger DNA sequencing. All primers used in the experiments are listed in Table I.
Table I. The list of primers used in this study.
Dual luciferase assays. HEK293T cells were cultured in 24-well plates and co-transfected with miRNA mimics or miRNA mimic negative control, together with 100 ng of an indicated reporter plasmid and 50 ng of a pRL-TK Renilla luciferase plasmid per well. The cells were lysed 24 h after transfection, and subsequently Firefly and Renilla luciferase activities were measured with the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. The Firefly luciferase activity was normalized using the Renilla luciferase readings. The assays were performed in triplicates in two independent experiments.
Western blot. Ishikawa cells were harvested 48 h after transfection with miRNA mimics or miRNA mimic negative control. The proteins were extracted using ice-cold RIPA lysis buffer, resolved on 12% polyacrylamide gels, and transferred to polyvinyl difluoride membranes (Millipore, Temecula, CA, USA). Membranes were blocked in 5% fat-free milk in TBST buffer and incubated overnight at 4˚C with indicated primary antibodies against MMP16 and TIMP2, and subsequently incubated with appropriate horseradish peroxidase–labeled secondary antibodies for 1 h at room temperature. The primary antibodies used in these experiments were also used for IHC (see Immunohistochemistry section in Methods). The blots were visualized with Western Lighting Ultra ECL (PerkinElmer, Surrey, UK) and ChemiDoc Imaging System (Biorad, Hercules, CA, USA). Densitometric analyses were performed using ImageJ. The band intensities were expressed relative to the loading control β-actin. All experiments were performed in triplicates.
Statistical analyses. A two-tailed Wilcoxon signed-rank test was used for comparing matched groups of samples. Pearson correlation coefficient was calculated to measure the statistical dependence between the expression of examined IHC markers and miRNAs. Student’s t-test was used to analyze the results of Dual Luciferase Assays. All statistical tests were performed using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA). The values are presented as mean±SEM unless indicated otherwise. A p-value of <0.05 was considered statistically significant.
Results
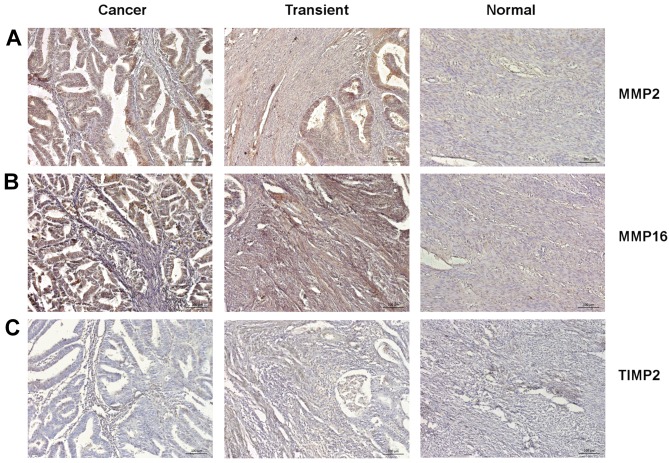
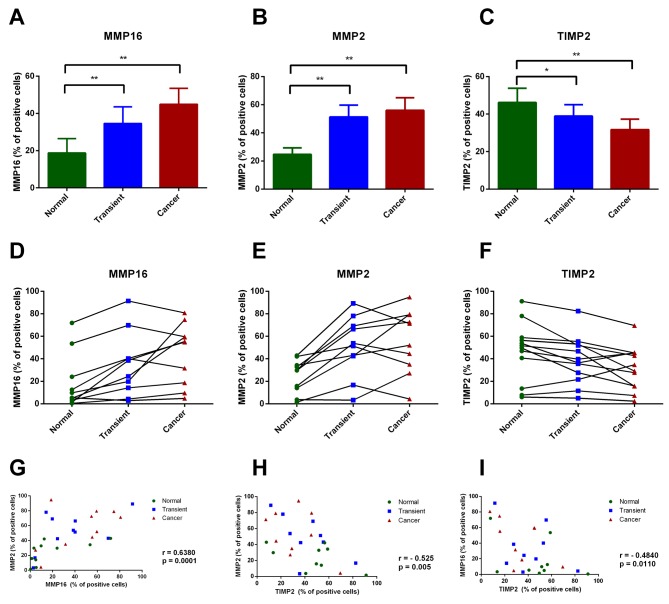
Endometrial cancer cells exhibit high MMP2 and MMP16, along with low TIMP2 expression. We assessed the percentage of MMP2, MMP16 and TIMP2-immunopositive cells in cancerous tissue, transient zone (including the infiltration area), and normal adjacent endometrium (Figure 2A-C). Both MMP2 and MMP16 were more abundantly expressed in histologically distinguishable cancer area and transient zone, comparing to normal endometrium (p<0.01) (Figure 3A-B). Conversely, TIMP2 showed a higher expression in normal endometrium than in transient and cancer zones (p<0.05 and p<0.01, respectively) (Figure 3C). Although the percentage of cells positively stained for MMP2, MMP16 and TIMP2 varied between different tumors, a general trend toward increased MMP2 and MMP16, together with decreased TIMP2 expression in cancer area was observed across all examined samples (Figure 3D-F). Notably, MMP2 and MMP16 showed a strong positive reciprocal correlation in analyzed tumors (r=0.6380; p=0.0001), and the expression of both MMPs negatively correlated with TIMP2 levels. (r=–0.525; p=0.005 and r=–0.484; p=0.0110 respectively) (Figure 3G-I).
Figure 2. Representative photographs of IHC staining for MMP2 (A), MMP16 (B) and TIMP2 (C) within cancer tissue, transient zone and normal adjacent endometrium.
Figure 3. Immunohistochemical analysis of MMP16, MMP2 and TIMP2 expression in type-I endometrial cancer samples. (A-C) The percentage of MMP16, MMP2 and TIMP2 immunopositive cells in three pre-defined areas of histological sections: cancer tissue, transient zone and normal adjacent endometrium. The p-values were calculated using Wilcoxon signed-rank test: *p<0.05, **p<0.01, ***p<0.001. (D-F) The solid lines connect points representing IHC score of each zone from individual histological sections. (G-H) Scatter plots showing the correlation between analyzed proteins in examined samples. The correlation was calculated using Pearson’s coefficient.
MMP16 is a putative target of miR-377, miR-382, miR-410, and miR-200b. We used target prediction algorithms (miRWalk, Microt4, Targetscan) to identify miRNAs functionally implicated in the modulation of MMP16 and TIMP2 expression. The performed analysis revealed two conserved binding sites for miR-377 and one conserved binding site for miR-382 and miR-410 within the 3’ UTR sequence of MMP16 mRNA. Furthermore, we found that miR-200b, previously identified as a regulator of TIMP2 expression, was also predicted to have a binding site within the 3’UTR of MMP16 mRNA. Based on these findings, we selected miR-377, miR-382 and miR-200b for further analysis in EC samples. Since the expression pattern of miR-410 was examined in the same set of samples previously (24), we included this data in the following correlation analyses.
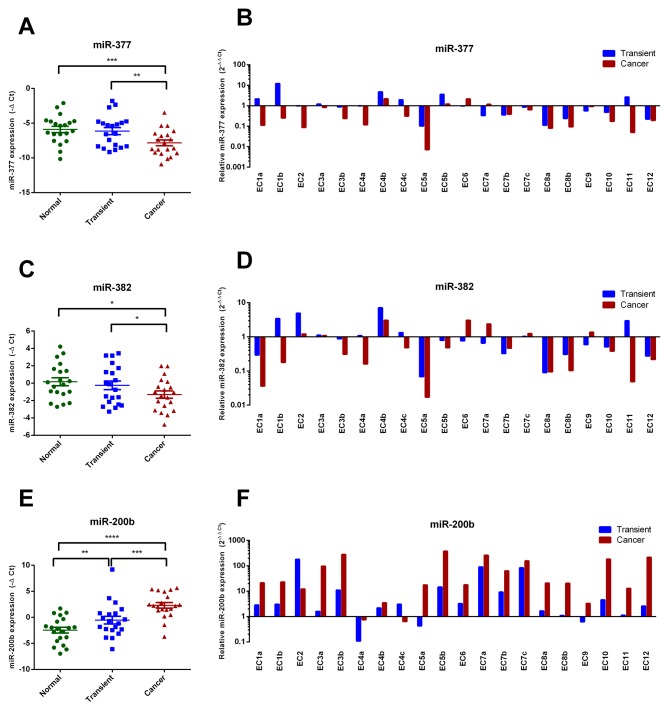
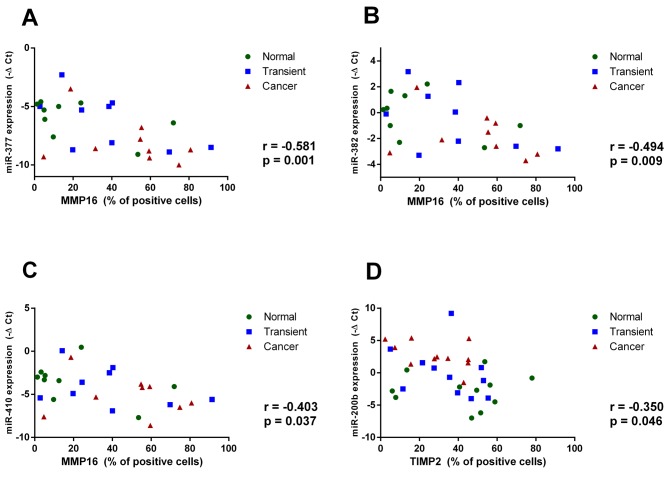
Down-regulation of miR-377, miR-382, and miR-410 corresponds to increased MMP16 levels, and up-regulation of miR-200b is associated with reduced TIMP2 expression in cancer tissue. The miRNA expression analysis performed separately for each predefined zone showed that both miR-377 and miR-382 were down-regulated in cancerous tissues comparing to matched transient (p<0.01 and p<0.05), and normal endometrium areas (p<0.001 and p<0.05, respectively) (Figure 4A-D). Oppositely, a significantly higher expression of miR-200b was observed in cancer cells comparing to adjacent normal tissues and transient zones (p<0.0001 and p<0.001, respectively) (Figure 4E-F). To further explore the association between miRNAs and their predicted targets we utilized Pearson correlation analysis, incorporating IHC data calculated separately for each histological zone. We observed a significant negative relationship between the percentage of MMP16-positively stained cells and the expression of miR-377 (r=–0.581; p=0.001), miR-382 (r=–0.494; p=0.009) and miR-410 (r=–0.403; p=0.037) in analyzed tumors (Figure 5A-C). On the contrary, miR-200b expression level did not show any significant correlation with MMP2 or MMP16 immunostaining (data not shown). However, an inverse correlation between miR-200b and TIMP2 expression was observed. (r=–0.35; p=0.046) (Figure 5D).
Figure 4. miR-377 and miR-382 are down-regulated in tumor tissues, while miR-200b is up-regulated in tumor tissues and transient zone. (A, C, E) The mean expression of miR-377, miR-382 and miR-200b in three analyzed zones of the histological sections. The p-values were calculated using Wilcoxon signed-rank test: *p<0.05, **p<0.01, ***p<0.001. (B, D, F) The miRNAs expression fold change over normal endometrium in all studied specimens.
Figure 5. The expression of miR-377, miR-382 and miR-410 is inversely correlated with MMP16, while the expression of miR-200b negatively correlates with TIMP2. Scatter plots showing the relationship between the expression of miR-377 (A), miR-382 (B), miR-410 (C) and the quantified expression of MMP16. (D) The correlation between miR-200b and the quantified TIMP2 expression. The correlation was calculated using Pearson’s coefficient.
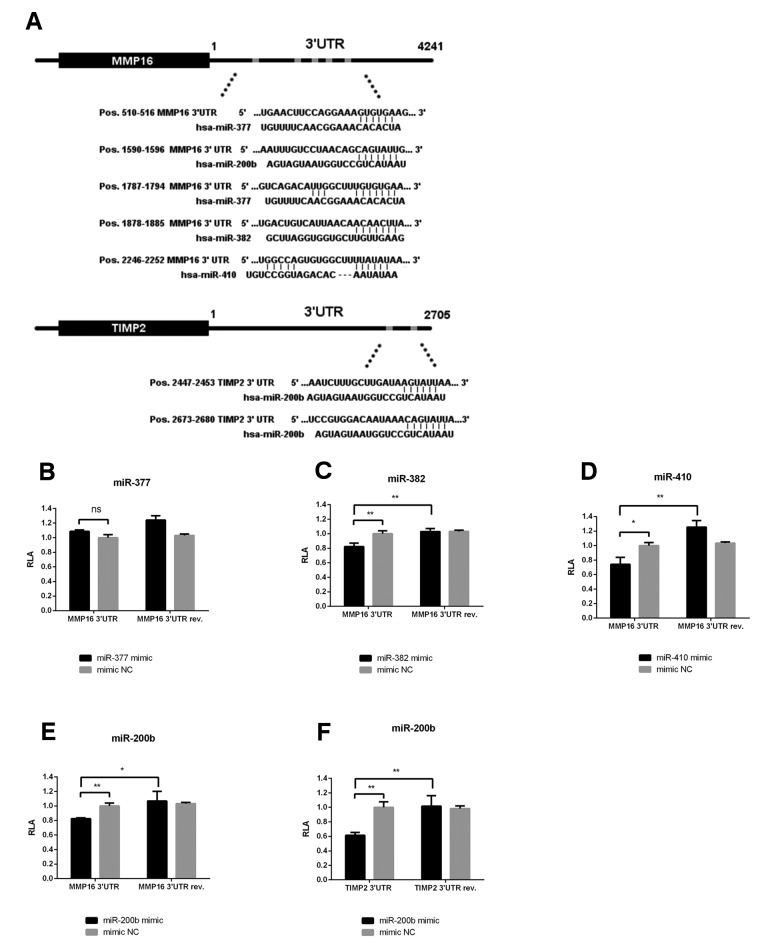
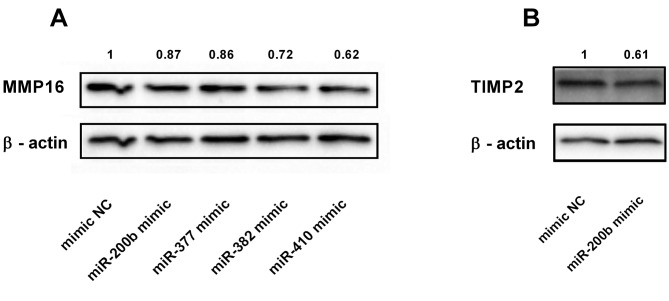
miR-382 and miR-410 directly target MMP16, while miR-200b inhibits TIMP2 in endometrial cancer cells. To investigate the interaction between indicated miRNAs and their presumed binding sites within the 3’UTR of MMP16 or TIMP2 mRNA, we performed a series of luciferase reporter assays (Figure 6). A significant decrease in luciferase activity was observed in cells co-transfected with vectors containing matching sequences from 3’UTR of MMP16 and miR-382, miR-410 or miR-200b, but not miR-377 (Figure 6B-E). Luciferase activities from reporter vectors carrying 3’UTRs in reverse orientation were not diminished, indicating the specificity of examined interactions. Furthermore, luciferase activity was reduced by approximately 40% in cells co-transfected with miR-200b and TIMP2 3’UTR construct compared to the control groups (Figure 6F). As demonstrated by western blotting experiments, the overexpression of miR-382 and miR-410 in Ishikawa cells resulted in a significant reduction of MMP16 protein level, while only a minor effect was observed upon transfection with miR-200b (Figure 7A). On the other hand, miR-200b overexpression markedly reduced TIMP2 level (Figure 7B). These results suggest that, although miR-200b has a functional binding site within both TIMP2 and MMP16 3’UTR, its overexpression does not translate into significantly reduced MMP16 levels, but results in TIMP2 down-regulation in EC cells.
Figure 6. miR-382 and miR-410 directly target MMP16 3’UTR, while miR-200b targets both MMP16 and TIMP2 3’UTR. A schematic representation of the predicted seed regions in the MMP16 and TIMP2 3’UTRs (A). Dual luciferase assays of Hek293T cells co-transfected with miR-377, miR-382, miR-410 and miR-200b mimics and luciferase reporters containing matching sequence of MMP16 3’UTR subcloned in sense or anti-sense orientation (B-E), and dual luciferase assay of cells co-transfected with miR-200b mimic and vectors containing matching sense or anti-sense 3’UTR of TIMP2 (F). A normalized firefly luciferase activity (firefly luciferase activity/Renilla luciferase activity) was calculated. The values were normalized to a relative luciferase activity from cells transfected with non-targeting control mimic. Error bars represent SD of three independent replicates in a representative experiment. *p<0.05, **p<0.01, ***p<0.001 by Student’s t-test.
Figure 7. MMP16 expression is reduced mostly by miR-382 and miR-410, while overexpression of miR-200b inhibits TIMP2 expression. Western blot analysis of MMP16 (A) and TIMP2 (B) in EC Ishikawa cells after transfection with miRNAs predicted to regulate their expression.
Discussion
In this study we demonstrated the expression pattern of MMP16, along with MMP2 and TIMP2 in type-I endometrial cancer, and presented a potential post-transcriptional regulation mechanism of MMP16 and TIMP2 by miRNA. Our results indicated a significant up-regulation of MMP16 and MMP2 together with a down-regulation of TIMP2 in cancerous tissues in comparison to normal endometrium. We showed that an increased MMP16 expression corresponds to a down-regulation of miR-382 and miR-410, and validated both molecules as novel MMP16-targeting miRNAs in EC cells. Furthermore, we confirmed that TIMP2 expression in tumor tissue is restricted by miR-200b. To our knowledge, this is the first report describing the expression pattern of MMP16 in EC and its regulation via miR-382 and miR-410 in cancer cells.
The results presented in our study are in concordance with previously published reports. MMP2 expression in endometrial tumors has been analyzed in different patient populations, and several studies described its increased expression in EC cells, as well as a link between MMP2 immunostaining and tumor histological grade, depth of myometrial invasion, vascular or lymphatic invasion, and shorter overall survival (28-32). Despite the ambiguous role of TIMP2 in the regulation of MMP2 activity, its down-regulation in EC cells has been associated with MMP2 hyperactivation and malignant tumor behavior. These observations suggested a suppressive role of TIMP2 in endometrial tumorigenesis (33,34). Similarly, a strong MMP2 and weak TIMP2 immunostaining has been reported as significant prognostic marker of poor survival in EC patients (10,34). MMP2 has been demonstrated to play a crucial role in the remodeling of the collagenous ECM by degrading a broad spectrum of ECM molecules, including: collagen types I, IV, V, VII, X, and IX, elastin, fibronectin, aggrecan, vitronectin, and laminin (23). Furthermore, it has been shown to modulate tumor growth factor-beta (TGF-β) activity by cleaving latent form of TGF-β and releasing inactive TGF-β precursor that is sequestered by ECM (35). Therefore, MMP2 could promote tumor invasion and metastasis by diverse mechanisms, including TGF-β-dependent induction of epithelial to mesenchymal transition and stimulation of angiogenesis.
Notably, our analysis revealed a significant correlation between the expression of MMP2 and MMP16 in cancerous tissues. These results might suggest a substantial involvement of MMP16 in MMP2 activation in EC cells. MMP16 not only activates pro-MMP2 by proteolytic cleavage, but is also known to degrade ECM proteins, especially basement membrane components, which facilitates cancer cell migration and tumor spreading (36). Although MMP16 protein has not been previously analyzed in endometrial tumors, its overexpression has been associated with malignant behavior and poor prognosis in numerous human malignancies, including colorectal cancer, gastric cancer, hepatocellular carcinoma, melanoma and renal cell carcinoma (36-41). For instance, MMP16 has been reported as a β-catenin target gene in human gastric cancer, which induction is relevant for the Wnt-mediated invasion and metastasis in gastric cancer cells (38). Similarly, increased MMP16 expression has been demonstrated to promote the invasion and metastasis of melanoma cells by decreasing cell adhesion, inhibiting collagen alignment and inducing lymphatic invasion (36). Interestingly, silencing of MMP16 by miRNAs significantly diminished invasion and migration of glioma, as well as pancreatic cancer, and osteosarcoma cells (42-46). These observations, together with the data presented herein, suggest a possible proto-oncogenic role of MMP16 in endometrial tumorigenesis and indicate that its overexpression might depend on a down-regulation of tumor suppressive miRNAs.
Because of the established role of MMPs in cancer invasion and metastasis, these proteins have been considered as attractive therapeutic targets. However, pharmacological MMPs inhibitors have failed in multiple clinical trials (23,47). Although there are many factors possibly contributing to the failure of MMP inhibitors in the clinic, among the most frequently discussed reasons are the high structural homology at the active site shared by proteases of this family and their overlapping specificity (48). Moreover, MMPs are involved in a variety of biological processes other than tumor development, and therefore the design of MMP inhibitors that are highly selective and have low side effect profiles seems to be particularly challenging. In comparison to small inhibitor molecules, miRNAs can target MMPs more selectively without the interference of the structural similarities of MMP catalytic domains (23). Furthermore, a single miRNA may simultaneously target multiple genes and signaling pathways critical for a malignant tumor behavior (49). Accordingly, the comprehensive characterization of miRNAs involved in a regulation of MMPs and their suppressors in cancer cells might greatly inform new drug trials and constitutes a first step toward the development of new anticancer therapeutic approaches (50).
Previous reports have shown that miR-377, miR-382 and miR-410 could exert tumor-suppressive effects in different types of cancer cells (51-53). However, little is known regarding their functional implication in endometrial tumorigenesis. The involvement of miR-382 and miR-410 in the regulation of MMP16 expression has not been studied in cancer yet, but a single report indicated that low levels of miR-382, along with MMP16 up-regulation, contributed to human intervertebral disc degeneration (54). In accordance with our findings, global miRNA expression profiling by Lee et al., performed on four endometrioid adenocarcinoma samples and four normal endometrial tissues, revealed that miR-377, miR-382 and miR-410 were significantly down-regulated in tumor samples (55). Similarly, Torres et al. found a down-regulation of miR-410 in EC compared to normal endometrium, and reported that microRNA signatures consisting of miR-410/miR-92 or miR-410/miR-200b/miR-92 accurately differentiated between cancerous and healthy tissues (56). Interestingly, miR-377, miR-382 and miR-410 belong to one of the largest miRNA clusters located within the imprinted Dlk1-Dio3 locus on chromosome 14q32 (57). The aberrant expression of this cluster has been implicated in numerous human malignancies and several studies revealed a tumor suppressive function for a subset of miRNAs located within 14q32 region (58-61). Moreover, Guo et al. demonstrated that the tumor suppressor lncRNA MEG3 (maternally expressed 3), derived from the same genomic locus, is underexpressed in EC samples compared to normal endometrium (62). The simultaneous down-regulation of three miRNAs from Dlk1-Dio3 imprinted locus in EC cells observed in our study suggested that decreased expression of the genes located within this cluster might promote endometrial tumors formation. Therefore, further studies on the role of miRNAs located within Dlk1-Dio3 locus in endometrial tumors are warranted.
The miRNA family of miR-200 consists of five members (miRNAs: 200b, 200a, 429, 200c and 141) organized as two genomic clusters located on chromosome 1 and 12 (63). Although the expression pattern and function of these miRNAs have been widely studied in different types of human cancers, their influence on migration and invasion of tumor cells remains controversial. All miR-200 family members have been demonstrated to act as tumor suppressors inhibiting epithelial to mesenchymal transition (EMT) in both normal and malignant cells through double-negative feedback regulation with the ZEB transcription factors and regulation of cadherin and vimentin expression (64). Moreover, miR-200b has been reported as a tumor suppressor negatively regulating MMP16 expression in bladder cancer cells (65). Nevertheless, several reports demonstrated that miR-200 family members were up-regulated in endometrial tumors, and their overexpression was associated with malignant cells growth and invasion (66). In a study presented by Dai et al. (22), miR-200b enhanced MMP2 activity in EC cells via TIMP2 down-regulation. In agreement with the these findings, our analysis revealed a prominent up-regulation of miR-200b in tumor tissues compared to normal adjacent endometrium and a significant inverse correlation between miR-200b expression and TIMP2 protein levels in analyzed samples. Although our luciferase reporter assays indicated that miR-200b could interact with both MMP16 and TIMP2 3’UTRs, its overexpression in Ishikawa cells resulted in a considerable reduction of only TIMP2 protein. These results may suggest that miR-200b promotes EC cells invasion through TIMP2 down-regulation. However, its possible role in fine-tuning MMP16 levels could not be excluded. Interestingly, in recent studies miR-200b together with miR-200a and miR-429 have been identified as oncogenic miRNAs negatively regulating tumor suppressor - phosphatase and tensin homolog (PTEN) in EC cells (67,68). Functional loss of PTEN expression leads to the constitutive activation of PI3K/Akt/mTOR pathway in EC cells, which is one of the main mechanism driving endometrial tumorigenesis (5), therefore overexpression of miRNAs belonging to miR-200 family can be associated with numerous aspects of endometrial cancer pathogenesis, including proliferation, invasion, metastasis and drug resistance.
In conclusion, our study showed an increased expression of MMP16 in endometrial tumors and identified novel miRNAs, aberrantly expressed in neoplastic cells, and involved in the regulation of MMPs activity. However, due to the limited number of patients included in this study and its retrospective character, further investigations on a larger cohort of patients are required to validate our results and verify the diagnostic and prognostic potential of MMP16 in EC. Moreover, additional cell line-based in vitro studies are necessary to explore the mechanisms of miRNA-mediated regulation of MMPs expression in tumor cells and characterize the possible suppressive role of miR-382 and miR-410 in endometrial tumorigenesis. Identification of the novel epigenetic mechanisms underlying tumor invasion and metastases could provide further insights into the diagnosis and treatment of EC (69).
Conflicts of Interest
The Authors declare no conflict of interest.
Acknowledgements
This work was supported by a grant from Medical University of Warsaw: 1M15/PM2/16/16.
References
- 1.Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 4.Rutgers JKL. Update on pathology, staging and molecular pathology of endometrial (uterine corpus) adenocarcinoma. Future Oncol. 2015;11:3207–3218. doi: 10.2217/fon.15.262. [DOI] [PubMed] [Google Scholar]
- 5.Merritt MA, Cramer DW. Molecular pathogenesis of endometrial and ovarian cancer. Cancer Biomark. 2010;9:287–305. doi: 10.3233/CBM-2011-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banno K, Kisu I, Yanokura M, Masuda K, Ueki A, Kobayashi Y, Susumu N, Aoki D. Epigenetics and genetics in endometrial cancer: new carcinogenic mechanisms and relationship with clinical practice. Epigenomics. 2012;4:147–162. doi: 10.2217/epi.12.13. [DOI] [PubMed] [Google Scholar]
- 7.Network CGAR, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 9.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honkavuori-Toivola M, Santala M, Soini Y, Turpeenniemi-Hujanen T, Talvensaari-Mattila A. Combination of strong MMP-2 and weak TIMP-2 immunostainings is a significant prognostic factor in endometrial carcinoma. Dis Markers. 2013;35:261–266. doi: 10.1155/2013/416870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagase H. Cell surface activation of progelatinase A (proMMP-2) and cell migration. Cell Res. 1998;8:179–186. doi: 10.1038/cr.1998.18. [DOI] [PubMed] [Google Scholar]
- 12.Sato H, Takino T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances peri-cellular proteolysis and invasion. Cancer Sci. 2010;101:843–847. doi: 10.1111/j.1349-7006.2010.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison CJ, Overall CM. TIMP independence of matrix metalloproteinase (MMP)-2 activation by membrane type 2 (MT2)-MMP is determined by contributions of both the MT2-MMP catalytic and hemopexin C domains. J Biol Chem. 2006;281:26528–26539. doi: 10.1074/jbc.M603331200. [DOI] [PubMed] [Google Scholar]
- 14.Sounni NE, Noel A. Membrane type-matrix metalloproteinases and tumor progression. Biochimie. 2005;87:329–342. doi: 10.1016/j.biochi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Jackson HW, Defamie V, Waterhouse P, Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer. 2017;17:38–53. doi: 10.1038/nrc.2016.115. [DOI] [PubMed] [Google Scholar]
- 16.Brew K, Nagase H. The tissue inhibitors of metallo-proteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanokura M, Banno K, Iida M, Irie H, Umene K, Masuda K, Kobayashi Y, Tominaga E, Aoki D. MicroRNAS in endometrial cancer: recent advances and potential clinical applications. EXCLI J. 2015;14:190–198. doi: 10.17179/excli2014-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Che Q, Qiu H, Bao W, Chen X, Lu W, Li B, Wan X. Elevated MiR-222-3p promotes proliferation and invasion of endometrial carcinoma via targeting ERα. PLoS One. 2014;9:e87563. doi: 10.1371/journal.pone.0087563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D, Zhou H, Xun Q, Xu X, Ling J, Hu Y. MicroRNA-103 regulates the growth and invasion of endometrial cancer cells through the down-regulation of tissue inhibitor of metalloproteinase 3. Oncol Lett. 2012;3:1221–1226. doi: 10.3892/ol.2012.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai Y, Xia W, Song T, Su X, Li J, Li S, Chen Y, Wang W, Ding H, Liu X, Li H, Zhao Q, Shao N. MicroRNA-200b is overexpressed in endometrial adenocarcinomas and enhances MMP2 activity by down-regulating TIMP2 in human endometrial cancer cell line HEC-1A cells. Nucleic Acid Ther. 2013;23:29–34. doi: 10.1089/nat.2012.0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Li H. Role of microRNA-mediated MMP regulation in the treatment and diagnosis of malignant tumors. Cancer Biol Ther. 2013;14:796–805. doi: 10.4161/cbt.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rak B, Garbicz F, Paskal W, Pełka K, Marczewska JM, Wołosz D, Włodarski P. The expression of MMP-14 and microRNA-410 in FFPE tissues of human endometrial adenocarcinoma. Histol Histopathol. 2016;31:911–920. doi: 10.14670/HH-11-728. [DOI] [PubMed] [Google Scholar]
- 25.Creasman W. Revised FIGO staging for carcinoma of the endometrium. Int J Gynaecol Obstet. 2009;105:109. doi: 10.1016/j.ijgo.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Varghese F, Bukhari AB, Malhotra R, De A. IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One. 2014;9:e96801. doi: 10.1371/journal.pone.0096801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Yilmaz E, Koyuncuoglu M, Görken IB, Okyay E, Saatli B, Ulukus EC, Saygili U. Expression of matrix metalloproteinase-2 and survivin in endometrioid and nonendometrioid endometrial cancers and clinicopathologic significance. J Gynecol Oncol. 2011;22:89–96. doi: 10.3802/jgo.2011.22.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart CJR, Crook ML. CD147 (EMMPRIN) and matrix metalloproteinase-2 expression in uterine endometrioid adenocarcinoma. Pathol Res Pract. 2011;207:30–36. doi: 10.1016/j.prp.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Aglund K, Rauvala M, Puistola U, Angström T, Turpeenniemi-Hujanen T, Zackrisson B, Stendahl U. Gelatinases A and B (MMP-2 and MMP-9) in endometrial cancer-MMP-9 correlates to the grade and the stage. Gynecol Oncol. 2004;94:699–704. doi: 10.1016/j.ygyno.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 31.Honkavuori M, Talvensaari-Mattila A, Soini Y, Turpeenniemi-Hujanen T, Santala M. MMP-2 expression associates with CA 125 and clinical course in endometrial carcinoma. Gynecol Oncol. 2007;104:217–221. doi: 10.1016/j.ygyno.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Talvensaari-Mattila A, Santala M, Soini Y, Turpeenniemi-Hujanen T. Prognostic value of matrix metalloproteinase-2 (MMP-2) expression in endometrial endometrioid adenocarcinoma. Anticancer Res. 2005;25:4101–4105. [PubMed] [Google Scholar]
- 33.Honkavuori-Toivola M, Talvensaari-Mattila A, Soini Y, Turpeenniemi-Hujanen T, Santala M. Immunoreactivity for TIMP-2 is associated with a favorable prognosis in endometrial carcinoma. Tumour Biol. 2012;33:935–941. doi: 10.1007/s13277-012-0321-7. [DOI] [PubMed] [Google Scholar]
- 34.Graesslin O, Cortez A, Uzan C, Birembaut P, Quereux C, Daraï E. Endometrial tumor invasiveness is related to metalloproteinase 2 and tissue inhibitor of metalloproteinase 2 expressions. Int J Gynecol Cancer. 2006;16:1911–1917. doi: 10.1111/j.1525-1438.2006.00717.x. [DOI] [PubMed] [Google Scholar]
- 35.Dallas SL, Rosser JL, Mundy GR, Bonewald LF. Proteolysis of latent transforming growth factor-beta (TGF-beta)-binding protein-1 by osteoclasts. A cellular mechanism for relase of TGF-beta from bone matrix. J Biol Chem. 2002;277:21352–21360. doi: 10.1074/jbc.M111663200. [DOI] [PubMed] [Google Scholar]
- 36.Tatti O, Gucciardo E, Pekkonen P, Holopainen T, Louhimo R, Repo P, Maliniemi P, Lohi J, Rantanen V, Hautaniemi S, Alitalo K, Ranki A, Ojala PM, Keski-Oja J, Lehti K. MMP16 Mediates a Proteolytic Switch to Promote Cell-Cell Adhesion, Collagen Alignment, and Lymphatic Invasion in Melanoma. Cancer Res. 2015;75:2083–2094. doi: 10.1158/0008-5472.CAN-14-1923. [DOI] [PubMed] [Google Scholar]
- 37.Wu S, Ma C, Shan S, Zhou L, Li W. High expression of matrix metalloproteinases 16 is associated with the aggressive malignant behavior and poor survival outcome in colorectal carcinoma. Sci Rep. 2017;7:46531. doi: 10.1038/srep46531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowy AM, Clements WM, Bishop J, Kong L, Bonney T, Sisco K, Aronow B, Fenoglio-Preiser C, Groden J. beta-Catenin/Wnt signaling regulates expression of the membrane type 3 matrix metalloproteinase in gastric cancer. Cancer Res. 2006;66:4734–4741. doi: 10.1158/0008-5472.CAN-05-4268. [DOI] [PubMed] [Google Scholar]
- 39.Cao L, Chen C, Zhu H, Gu X, Deng D, Tian X, Liu J, Xiao Q. MMP16 is a marker of poor prognosis in gastric cancer promoting proliferation and invasion. Oncotarget. 2016;7:51865–51874. doi: 10.18632/oncotarget.10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanjmyatav J, Steiner T, Wunderlich H, Diegmann J, Gajda M, Junker K. A specific gene expression signature characterizes metastatic potential in clear cell renal cell carcinoma. J Urol. 2011;186:289–294. doi: 10.1016/j.juro.2011.03.033. [DOI] [PubMed] [Google Scholar]
- 41.Arai I, Nagano H, Kondo M, Yamamoto H, Hiraoka N, Sugita Y, Ota H, Yoshioka S, Nakamura M, Wada H, Damdinsuren B, Kato H, Marubashi S, Miyamoto A, Takeda Y, Dono K, Umeshita K, Nakamori S, Wakasa K, Sakon M, Monden M. Overexpression of MT3-MMP in hepatocellular carcinoma correlates with capsular invasion. Hepatogastroenterology. 2007;54:167–171. [PubMed] [Google Scholar]
- 42.Wang H, Li X-T, Wu C, Wu Z-W, Li Y-Y, Yang T-Q, Chen G-L, Xie X-S, Huang Y-L, Du Z-W, Zhou Y-X. miR-132 can inhibit glioma cells invasion and migration by target MMP16 in vitro. Onco Targets Ther. 2015;8:3211–3218. doi: 10.2147/OTT.S79282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, Ge R, Jiang S, Li G, Chen Y, He M-L, Kung H-f, Lai L, Lin MC. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Wang Y, Yu L, Sun C, Cheng D, Yu S, Wang Q, Yan Y, Kang C, Jin S, An T, Shi C, Xu J, Wei C, Liu J, Sun J, Wen Y, Zhao S, Kong Y. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer Lett. 2013;339:260–269. doi: 10.1016/j.canlet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 45.Lin F, Wang X, Jie Z, Hong X, Li X, Wang M, Yu Y. Inhibitory effects of miR-146b-5p on cell migration and invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ Sci Technolog Med Sci. 2011;31:509–514. doi: 10.1007/s11596-011-0481-5. [DOI] [PubMed] [Google Scholar]
- 46.Chen B, Huang Z, Zhang Y, Chen Y, Li Z. MicroRNA-145 Suppresses Osteosarcoma Metastasis via Targeting MMP16. Cell Physiol Biochem. 2015;37:2183–2193. doi: 10.1159/000438575. [DOI] [PubMed] [Google Scholar]
- 47.Fingleton B. MMPs as therapeutic targets--still a viable option. Semin Cell Dev Biol. 2008;19:61–68. doi: 10.1016/j.semcdb.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldberg GI. Renaissance of MMPs as therapeutic targets? Maybe. Structure. 2015;23:6–7. doi: 10.1016/j.str.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 49.D’Angelo B, Benedetti E, Cimini A, Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36:5571–5575. doi: 10.21873/anticanres.11142. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Tseng S-H. The potential of RECK inducers as antitumor agents for glioma. Anticancer Res. 2012;32:2991–2998. [PubMed] [Google Scholar]
- 51.Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee NPY, Law S, Xu LY, Li EM, Chan KW, Qin YR, Guan XY, He QY, Cheung ALM. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986–4000. doi: 10.1038/onc.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu M, Jin H, Xu C-X, Sun B, Mao Z, Bi W-Z, Wang Y. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5:9472–9483. doi: 10.18632/oncotarget.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen J, Niu W, Zhou M, Zhang H, Ma J, Wang L, Zhang H. MicroRNA-410 suppresses migration and invasion by targeting MDM2 in gastric cancer. PLoS One. 2014;9:e104510. doi: 10.1371/journal.pone.0104510. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Wang H-Q, Zhang Y-Z, Cheng Y-F, Zhang W-L, Sun Z, Samartzis D, Luo Z-J. Down-regulated MIR-382 contributes to human intervertebral disc degeneration by targeting MMP-16: Gp53. Spine Journal Meeting. 2014;Abstracts:136–137. [Google Scholar]
- 55.Lee TS, Jeon HW, Kim YB, Kim YA, Kim M, Kang SB. Aberrant microRNA expression in endometrial carcinoma using formalin-fixed paraffin-embedded (FFPE) tissues. PLoS One. 2013;8:e81421. doi: 10.1371/journal.pone.0081421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, Zamboni G, Maciejewski R. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer. 2013;132:1633–1645. doi: 10.1002/ijc.27840. [DOI] [PubMed] [Google Scholar]
- 57.Dimmeler S, Ylä-Herttuala S. 14q32 miRNA cluster takes center stage in neovascularization. Circ Res. 2014;115:680–682. doi: 10.1161/CIRCRESAHA.114.304973. [DOI] [PubMed] [Google Scholar]
- 58.Lehner B, Kunz P, Saehr H, Fellenberg J. Epigenetic silencing of genes and microRNAs within the imprinted Dlk1-Dio3 region at human chromosome 14.32 in giant cell tumor of bone. BMC Cancer. 2014;14:495. doi: 10.1186/1471-2407-14-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gattolliat C-H, Thomas L, Ciafrè SA, Meurice G, Le Teuff G, Job B, Richon C, Combaret V, Dessen P, Valteau-Couanet D, May E, Busson P, Douc-Rasy S, Bénard J. Expression of miR-487b and miR-410 encoded by 14q32.31 locus is a prognostic marker in neuroblastoma. Br J Cancer. 2011;105:1352–1361. doi: 10.1038/bjc.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Devor EJ, DE Mik JN, Ramachandran S, Goodheart MJ, Leslie KK. Global dysregulation of the chromosome 14q32 imprinted region in uterine carcinosarcoma. Exp Ther Med. 2012;3:677–682. doi: 10.3892/etm.2012.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thayanithy V, Sarver AL, Kartha RV, Li L, Angstadt AY, Breen M, Steer CJ, Modiano JF, Subramanian S. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo Q, Qian Z, Yan D, Li L, Huang L. LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by repressing Notch signaling. Biomed Pharmacother. 2016;82:589–594. doi: 10.1016/j.biopha.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 63.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abba ML, Patil N, Leupold JH, Allgayer H. MicroRNA regulation of epithelial to mesenchymal transition. J Clin Med. 2016;5:8. doi: 10.3390/jcm5010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen MF, Zeng F, Qi L, Zu XB, Wang J, Liu LF, Li Y. Transforming growth factor-β1 induces epithelial-mesenchymal transition and increased expression of matrix metalloproteinase-16 via miR-200b down-regulation in bladder cancer cells. Mol Med Rep. 2014;10:1549–1554. doi: 10.3892/mmr.2014.2366. [DOI] [PubMed] [Google Scholar]
- 66.Lee J-W, Park Y-A, Choi J-J, Lee YY, Kim C-J, Choi C, Kim T-J, Lee NW, Kim B-G, Bae D-S. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol Oncol. 2011;120:56–62. doi: 10.1016/j.ygyno.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 67.Wu Q, Lu R-L, Li J-X, Rong L-J. MiR-200a and miR-200b target PTEN to regulate the endometrial cancer cell growth in vitro. Asian Pac J Trop Med. 2017;10:498–502. doi: 10.1016/j.apjtm.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 68.Yoneyama K, Ishibashi O, Kawase R, Kurose K, Takeshita T. miR-200a, miR-200b and miR-429 are onco-miRs that target the PTEN gene in endometrioid endometrial carcinoma. Anticancer Res. 2015;35:1401–1410. [PubMed] [Google Scholar]
- 69.Caplakova V, Babusikova E, Blahovcova E, Balharek T, Zelieskova M, Hatok J. DNA methylation machinery in the endometrium and endometrial cancer. Anticancer Res. 2016;36:4407–4420. doi: 10.21873/anticanres.10984. [DOI] [PubMed] [Google Scholar]