Abstract
Integrin-mediated adhesion is a central feature of cellular adhesion, locomotion, and endothelial cell mechanobiology. Although integrins are known to be transmembrane proteins, little is known about the role of membrane biophysics and dynamics in integrin adhesion. We treated human aortic endothelial cells with exogenous amphiphiles, shown previously in model membranes, and computationally, to affect bilayer thickness and lipid phase separation, and subsequently measured single-integrin-molecule adhesion kinetics using an optical trap, and diffusion using fluorescence correlation spectroscopy. Benzyl alcohol (BA) partitions to liquid-disordered (Ld) domains, thins them, and causes the greatest increase in hydrophobic mismatch between liquid-ordered (Lo) and Ld domains among the three amphiphiles, leading to domain separation. In human aortic endothelial cells, BA increased β1-integrin-Arg-Gly-Asp-peptide affinity by 18% with a transition from single to double valency, consistent with a doubling of the molecular brightness of mCherry-tagged β1-integrins measured using fluorescence correlation spectroscopy. Accordingly, BA caused an increase in the size of focal-adhesion-kinase/paxillin-positive peripheral adhesions and reduced migration speeds as measured using wound-healing assays. Vitamin E, which thickens Lo domains and disperses them by lowering edge energy on domain boundaries, left integrin affinity unchanged but reduced binding probability, leading to smaller focal adhesions and equivalent migration speed relative to untreated cells. Vitamin E reversed the BA-induced decrease in migration speed. Triton X-100 also thickens Lo domains, but partitions to both lipid phases and left unchanged binding kinetics, focal adhesion sizes, and migration speed. These results demonstrate that only the amphiphile that thinned Ld lipid domains increased β1-integrin-Arg-Gly-Asp-peptide affinity and valency, thus implicating Ld domains in modulation of integrin adhesion, nascent adhesion formation, and cell migration.
Introduction
The adhesive interaction of a cell with its environment is mediated mainly by heterodimeric surface receptors, primarily integrins. Integrins not only work as mechanical anchors to recognize ligands but also take part in signal transduction (1). To signal, integrins are laterally clustered at the contact site and recruit intracellular signaling proteins to form an adhesion complex. These complexes are defined based on their subcellular location and size as nascent adhesions, focal complexes, focal adhesions (FAs), and fibrillar adhesions (2). The integrin-mediated adhesive strength is governed by the affinity of the integrin-ligand bonds and the number of these bonds (valency). Regulation of integrin affinity is coupled to alterations in their structural conformation (3). A general model suggests that extracellular ligand binding, outside-in signaling, rearranges the conformational state of the integrin and influences integrin affinity. Valency regulation is then governed by reorganization of multiple integrin molecules into the site of cell adhesions. Ligand-dependent reorganization occurs as a consequence of diffusivity changes of integrins and membrane reshaping (e.g., bending). Integrin binding, in turn, governs cellular locomotion (4), a principle factor in maintaining endothelium integrity (5). Due to its transmembrane nature, its dependence on lateral diffusion, and the likelihood that membrane bending is involved in adhesion, it is reasonable to hypothesize that lipids govern multiple aspects of integrin-mediated adhesion (6).
Cell membranes are complex structures composed mostly of lipids and proteins that control information transfer between the intracellular and extracellular environment. Many transmembrane transporters (such as ion channels) are known to be sensitive to the lipid environment (7). The lipid bilayer has the propensity to segregate its components laterally (8), and the domain formation is governed by bilayer thickness mismatch (9). Often termed “lipid rafts,” liquid-ordered (Lo) domains are heterogeneous small (10–200 nm) domains, which are enriched in cholesterol, sphingolipids, and a variety of signaling molecules (10) and function to compartmentalize cellular processes. The heterogeneous size of lipid rafts can be modulated by lipid composition (11). Alternatively, non-raft domains are likely liquid disordered (Ld) and are delimited by a phase boundary. The reorganization of lipid phases and integrins has been suggested to play a role in cellular adhesion (10, 12), but whether Lo or Ld phases are drivers of integrin reorganization remains a topic of debate. It has been shown that cholesterol depletion in Lo domains changes a cell’s shape and motility, and this phenomenon was specifically modulated by integrin-mediated adhesions (13). The detachment of integrin-mediated adhesions from a substrate decreased membrane order (14, 15), suggesting that adhesion causes a transition from the Ld to the Lo phase. Additional evidence that domains are involved in the early stages of focal-adhesion formation was found using ion conductance spectroscopy and precise timing of contact between a cell and the extracellular matrix (ECM) (16, 17). In that study, it was discovered that GM-1-positive domains (e.g., rafts) arrived at a fibronectin (FN)-functionalized pipette within 4 s after contact, whereas talin, an indicator of integrin activation, arrived only after 20 s. One widely used approach for the membrane domain modulation is to alter the concentration of cholesterol by depleting it with methyl-β-cyclodextrin (MβCD) or to add cholesterol using cholesterol-loaded MβCD (CHO-MβCD) (13). However, modification of cholesterol content as a strategy for modifying membrane properties is non-ideal, since protein function can depend on cholesterol directly or indirectly. In addition, because modification of cholesterol has the most impact on raft (Lo) domains (18), there remains a need for tools that target Ld domains.
Alternatively, non-lipid exogenous amphiphilic molecules are not expected to have direct effects on native proteins, and they have been shown to differentially target Lo domains, Ld domains, and domain boundaries (19). Amphiphiles regulate membrane protein function by altering the bilayer material properties (thickness, curvature, and elastic moduli). As a general mechanism, hydrophobic coupling between bilayer leaflets dominates protein function, as demonstrated with a model membrane protein, the gramicidin channel, where the hydrophobic interactions between channel and bilayer regulate the energetics and kinetics of the protein conformation (reviewed in (20, 21)). Some amphiphiles have partitioning preference to one domain. Triton X-100 (TX) selectively solubilizes the Ld domain and promotes Lo-domain formation (22). Linactants, or surfactant analogs, modify phase separation by altering interfacial free energy at the phase boundaries (23). Previous coarse-grained molecular dynamics simulations of vitamin E (VE, α-tocopherol), TX, and benzyl alcohol (BA) in the lipid bilayer (19) suggested that the partitioning preference of non-lipid amphiphiles alters lipid order and domain thickness. BA was the only amphiphile that selectively partitioned into the Ld domain and the only one that thinned that domain. Thinning of the Ld domain led to increased hydrophobic mismatch between domains and domain separation. TX had equal preference for each domain and thickened both of them, but nevertheless increased hydrophobic mismatch and fostered domain separation. VE also thickened both phases but reduced mismatch and, acting as a linactant, reduced the propensity for domain formation. Thus, these amphiphiles are useful tools for differentially tuning domain thickness and size to test hypotheses of lipid modulation of integrin function.
Changes in membrane microviscosity by amphiphiles modify endothelial cell migration (24, 25). However, research into amphiphile-induced changes in integrin functional properties has not been reported. Here, we studied the influence of three non-lipid amphiphiles, VE, TX, and BA, on integrin-mediated adhesion and human aortic endothelial cell (HAEC) migration. HAECs were selected as a model cell system because of their relevance, and the relevance of β1-integrins, to the mechanobiological origins of vascular health and disease (26, 27). We expect, however, that results here are relevant to β1-integrin function in general. Integrins exist in many cell types and lipids are found in all cell types. We focused on the analysis of single-molecule dynamics in integrin diffusion and integrin-mediated adhesion strength with specialized instruments for high spatial and temporal resolution. Fluorescence correlation spectroscopy (FCS), a powerful and sensitive analytical method of studying the translational diffusion of proteins in a small volume (∼1 μm3), was used to determine lateral diffusion and brightness of fluorescently labeled β1-integrins and Lyn kinase, a protein associated with Lo domains (e.g., rafts) (28). The brightnesses of molecules and molecular complexes were then measured to analyze protein aggregation. A force spectroscopy assay based on an optical trap with precisely timed imaging and quadrant photodiode-based detection of bead position was used to determine the timing of bead-cell interaction. This technique permitted quantification of both integrin affinity and valency changes in response to amphiphile treatment. Subsequently, alterations of integrin-mediated focal-adhesion assembly and attendant migration of HAECs were measured using total internal reflection fluorescence (TIRF) microscopy and wound-healing assays, respectively. Results suggest that although both BA and TX foster domain separation and coalescence of Lo domains, it is the action of BA on Ld domains, involving coalescence of these domains and their thinning, that lead to increases in β1-integrin affinity, valency, nascent focal-adhesion size, and attendant decreases in cell migration speed. Thus, although FAs are known to be ordered once formed (15), initial adhesion to ECM proteins and nascent focal-adhesion formation depend more strongly on the dynamics of Ld domains.
Materials and Methods
Preparation of cells and Heparan sulfate digestion on the cell surface
HAECs (C-006-5C; Gibco, Carlsbad, CA) were cultured in Medium 200 (N200500, Gibco) supplemented with Low Serum Growth Supplement (S00310; Gibco) and gentamicin/amphotericin B (50-0640; Gibco) at 37°C in a humidified incubator in an atmosphere of 5% CO2. FCS was conducted on cells in Opti-MEM (phenol-red-free reduced-serum medium, Gibco) and force spectroscopy was performed on cells in CO2-independent medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum. To maintain the medium temperature at 37°C during experiments, cells were plated into a temperature chamber (Delta T4 Culture Dish; Bioptechs, Butler, PA) and the temperature was controlled with a temperature controller (Delta T culture dish controller; Bioptechs). Before seeding cells, all surfaces were coated with 5 μg/mL FN for 1 h at 37°C.
In some experiments, heparan sulfate chains on HAEC surfaces were digested using heparinase III as previously described (29). Briefly, HAECs were plated at a density of 1.2 × 104 cells/cm2 on the FN-coated dish and incubated with 5 mU/mL of heparinase III (37290-86-1; Sigma Aldrich, Bellefonte, PA) in 50 mM HEPES buffer (pH 7.0) with 100 mM NaCl and 1 mM CaCl2 for 1 h at 37°C.
Plasmids and transfection
Plasmids were purified using a Qiagen Plasmid Midi kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. mCherry-Integrin-β1-N-18 (Addgene plasmid no. 55064, a gift from Michael Davidson) and LYN11-FRB-mCherry (Addgene plasmid no. 38004, a gift from Robin Irvine) were used for FCS measurement. The myristoylated and palmitoylated N-terminus of Homo sapiens Lyn kinase (NM_002350.3, residues 1–11) was used as a marker specific to lipid rafts. Enhanced green fluorescent protein (EGFP)-focal-adhesion kinase (FAK) (a gift from Song Li) and mCherry-paxillin (Addgene plasmid no. 50526, a gift from Kenneth Yamada (30)) were used for nascent focal-adhesion size analysis. For transfection, HAECs were seeded in 24-well plates or temperature chambers 1 day before the transfection and the plasmids were transfected with Lipofectamine 2000 (Invitrogen) for 3 h in Opti-MEM medium, then incubated with growth medium for 24 or 48 h.
Fabrication of ECM-coated beads
Micro beads were coated with FN (F2006; Sigma Aldrich), Arg-Gly-Asp peptide (RGD; 99895-85-2; Sigma Aldrich), or bovine serum albumin (BSA; Affymetrix, Cleveland, OH). Proteins or peptides were covalently bound to biotin (EZ-link NHS-LC-Biotin; Thermo Scientific, Skokie, IL) for 2 h at 4°C. Excess unreacted biotin molecules were removed through a Microspin column (G-25; Harvard Apparatus, Holliston, MA) for FN and BSA. Biotin-functionalized proteins or peptides were subsequently conjugated with Neutravidin polystyrene particles (2 μm polystyrene beads; Spherotech, Lake Forest, IL) for 1 h at room temperature and kept in phosphate-buffered saline supplemented with 5% BSA. Before use, the bead solution was sonicated for 30 s in an ice bath. To confirm the bead functionalization, the beads were incubated with fluorescein-isothiocyanate-conjugated anti-FN antibody (ab 72686; Abcam, Cambridge, MA) overnight at 4°C. After washing, the beads were examined using a fluorescence microscope.
Optical setup for FCS and TIRF microscopy
The optical configuration for FCS and TIRF microscopy is described in our previous work (31). An inverted Olympus IX71 microscope was equipped with a CCD camera (SensicamQE, Cooke, MI), a single-molecule fluorescence system, epifluorescence, and TIRF and was mounted on a Newport 3′ × 5′ vibration isolation workstation. For FCS, a water-cooled 532 nm, 80 MHz, 5.4 ps pulsed laser (High-Q Laser, Hohenems, Austria) was focused on a specific position in the target cell using a water-immersion objective (60×/1.2 NA; UPLAPO, Olympus, Center Valley, PA). The emission light was further filtered by an emission filter and focused onto a 0.22 NA optical fiber by a 250 mm focal length lens and the confocal probe volume was optimized with a three-axis manual micrometer stage. The light was focused onto a GaAsP photomultiplier tube (PMT) and the PMT signal was converted into an electronic pulse using time-correlated single-photon counting module (Fig.1 B). A fluorescence trace was collected into a first-in, first-out buffer in the SPC-630 board (Becker & Hickl, Berlin, Germany). For TIRF microscopy, an oil-immersion objective (PLAPO 60×/1.45 NA, TIRFM-2) was used with a continuous-wave krypton-argon-ion laser with stable TEM00 mode laser light (476, 483, 488, 496, 514, 520, 530, 568, 647, and 676 nm). The laser light was fiber-coupled to the TIRF illuminator and a micrometer on the TIRF illuminator controlled the angle of incidence of the laser beam.
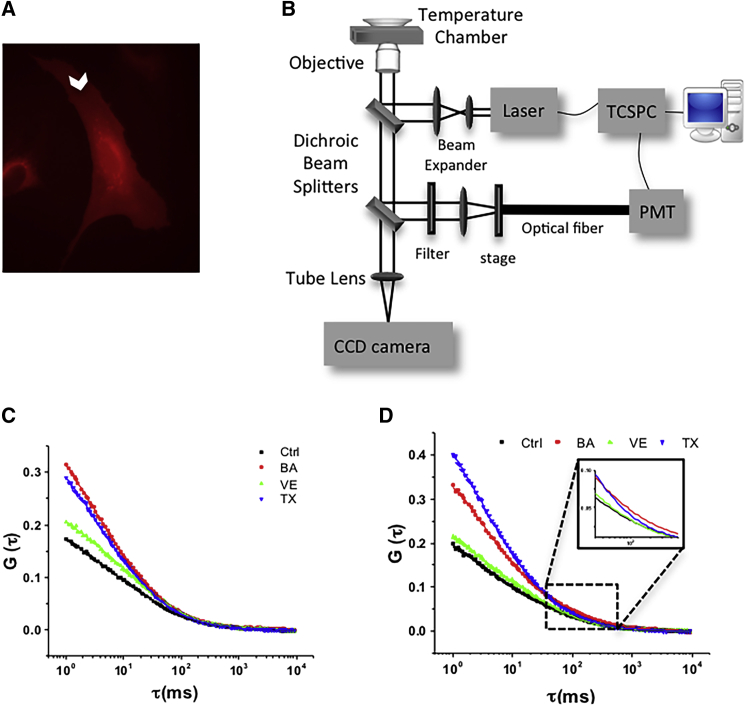
Figure 1.
FCS measurements. (A) Fluorescence image of an HAEC transfected with the mCherry-Lyn plasmid. The arrowhead indicates the point where a laser was focused for measurements. (B) Optical setup for FCS. A 532 nm pulsed laser was expanded and the expanded laser was reflected to the back aperture of the 60× objective. After excitation of the sample, the reflected emission signal was focused onto the optical fiber. The photon signal was converted into the electrical pulse by a PMT and routed to the time-correlated single-photon counting board. (C) Autocorrelation curves for mCherry-Lyn without (Ctrl) or with treatments of BA, VE, and TX. (D) Autocorrelation curves for mCherry-β1-integrin without (Ctrl) or with treatments of BA, VE, and TX. The autocorrelation curves were fit to a two-dimensional, two-component diffusion model (Eq. 3). For the display purpose, the set of FCS curves shown represents the average of measurements from ∼20 to 27 cells. For statistical purposes, every measurement was fit by Eq. 3 and the resulting curve-fit parameters were averaged and compared. The number of cells and diffusion rates are indicated in Table 1. To see this figure in color, go online.
Determination of lateral diffusion coefficients and brightness of fluorescently labeled Lyn and β1-integrin on the surface of HAECs using FCS
FCS is based on a statistical analysis of time traces of fluorescence intensity I(t). The intensity time trace can be autocorrelated to construct an autocorrelation function, G(τ):
| (1) |
where t is time, τ is lag time, and . Brackets represent averaging over all values of time t. The relationship between G(τ) and Brownian diffusion in a two-dimensional structure is theoretically given by
| (2) |
where τD is the diffusion time and N is the particle number (32). The measurement can be extended to two species of fluorescent molecules, according to the equation
| (3) |
where is the relative proportion of the noninteracting molecules. Autocorrelation curves were fit with Eq. 3 using Burst analyzer 2.0 (Becker & Hickl). The lateral diffusion coefficient (D) was calculated from D = ω2/4τD, where ω is the radial distance from the optical axis. Red fluorescent microspheres (50 nm; Fluo-max, Thermo Fisher Scientific, Waltham, MA), for which D is known (9.71 μm2/s in water), were used to compute the radius of the focal volume. All FCS measurements were performed at a location on the cell corresponding to the maximal molecular brightness to position the confocal volume on the cell surface. Measurements were performed with low-serum medium supplemented with 10 mM BA (Sigma Aldrich), 50 μM VE (V-020; Cerilliant, Round Rock, TX), or 30 μM TX (MP Biomedicals, Solon, OH). The brightnesses (ε) of fluorescently tagged Lyn and β1-integrin molecules were calculated using the relationship between the average count rate (k), the number of fluorescent molecules, and their molecular brightness, k = Nε. The same intensity-fluctuation data set for diffusion rate calculation was used to calculate molecular brightness, and the resulting N values were used as an indication of protein aggregation.
Optical trap design and calibration
The optical trap was designed to be operated on an inverted Olympus IX71 microscope equipped with epifluorescence, a CCD camera (Sensicam), and a Nanodrive piezoelectric computer controlled stage (Mad City Labs, Dane County, WI) on a Newport 3′ × 5′ vibration isolation workstation. A continuous TEM00 laser (1064 nm; Arroyo Instruments, San Luis Obispo, CA) was focused by an Olympus 60×, 1.45 NA oil immersion objective. The inverted microscope was also set up for differential interference contrast (DIC) microscopy. The location of the bead was precisely controlled with a custom LabVIEW software program that monitored both the DIC image and quadrant photodiode voltage (QPD; NoahCorp, Melbourne, FL) located at the back focal plane of the condenser. The optical trap system was calibrated using the drag-force method, which involved calculating the Stokes drag force by sinusoidal motion of fluid applied to a trapped bead. Applied force of a sphere distance (h) from the surface can be calculated using the Stokes drag on a sphere (F = 6πμrv) or by using a correction for diffusion near a surface, described by Faxen's Law (33):
| (4) |
where r is the radius of the bead, ν is the velocity of the bead, and μ is the viscosity of the liquid. In this equation, the denominator is a correction for the Stokes drag on a sphere. In the experiments described herein, the location of the bead (h) was 10 μm above the coverslip. Considering a 2 μm bead, the value of F was <6% different than what would be calculated from the Stokes drag. This error was below the standard error of the measurement. Therefore, the correction was negligible in these experiments. As the stage oscillated according to a sine wave, the CCD camera imaged the location of the bead and particle-tracking software was used to track the motion of the bead (34). For the calibration of trap stiffness, the position of the stage was moved by Asin(2πft), where A is amplitude and f is frequency. The velocity, v, of the stage (and fluid past the bead) was v = A2πfcos(2πft). The force applied to the bead was calculated by Stokes’s law (F = 6πμrv), and the trap stiffness, k, was calculated by Hooke’s law, F = kx. A customized LabVIEW program, which included a frequency analyzer, was used to measure the position of the trapped bead, x (as described below), calculate the amplitude of the cosine position waveforms, and automate all steps. Because we both imaged the bead and collected voltage from the QPD, this approach provided simultaneously the spring constant and the value of bead position as a function of QPD voltage. The spring constant was calculated before each set of experiments. On average, the spring constant was k = 0.109 ± 0.013 pN/nm. With a known optical-trap spring constant, experimental forces could subsequently be calculated using only QPD voltage.
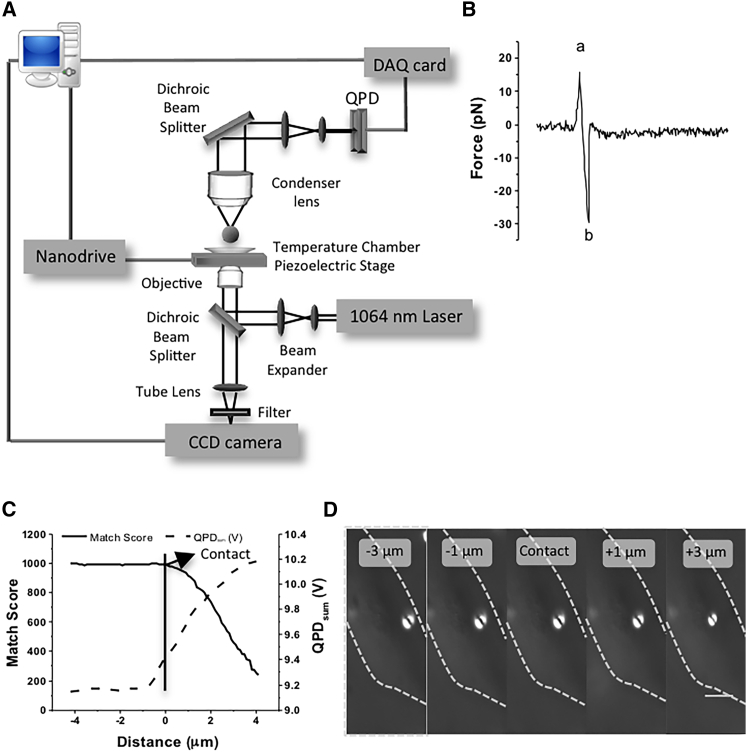
To define the precise timing (t = 0) of bead-cell contact, a quantitative method with match score and QPD sum was developed (Fig. 2, C and D). In detail, an optically trapped bead was brought to an individual cell by moving the stage position in increments of 0.1 μm using LabVIEW-controlled output voltages via a computer D/A interface. DIC images and QPD sum voltages were recorded for each step. As soon as the cell surface began to push a bead out of the optical trap focal point, the bead changed appearance. The changing appearance of the bead was quantified using a centroid-based particle-tracking LabVIEW program that used cross-correlation-based particle tracking methods and quantified the match score relative to the first image taken (34). A perfect match score was 1000. Typically, the QPD voltage increased before the decrease of the match score. Since the decrease of the match score was indicative of the bead touching the cell, the increase of the QPD voltage could be used to predict this contact. The initial contact point occurred when the normalized QPD value was on average 1.03 times the reference value, as shown Fig. 2 C. Thus, in subsequent experiments, when the QPD reached 3% above its initial value, the custom LabVIEW program could automatically stop the approach and begin the experimental process and data acquisition.
Figure 2.
Setup for optical traps. (A) Optical scheme of the optical trap. The optical trap was generated with a 1064 nm laser. Displacements of the trapped bead were measured with a QPD placed at a plane optically conjugate to the back focal plane of the condenser. (B) A portion of digitized data showing the voltage exerted by the trap on the bead during the approach phase and after detaching from the cell surface. The laser trap exerted a positive compressive force on the RGD-coated 2 μm bead (peak a), then the trap motion was reversed. The force on the bead was increased in the negative direction until the cell-bead bond ruptured (peak b). (C) A quantitative method of defining contact point based on the values of QPD sum voltages and match scores. A bead was trapped next to the cell. The piezo-electric stage was moved to allow the cell to approach from the side in increments of 0.1 μm until the bead became out of focus. QPD sum voltages and images were recorded for each step and a match score was calculated from the images using image correlation between the original image and the image of the bead distorted by its interaction with the cell (out of focus). (D) DIC images at the given position. The dashed line represents the cell boundary. The scale bar represents 10 μm. To see this figure in color, go online.
Force measurement procedure
The optical-trap force measurements were performed on an inverted microscope with a 1064 nm laser set to 1000 mW. This setting resulted in 205 mW of power at the back aperture of the objective. All experiments were conducted at 37°C in a temperature-controlled chamber. HAECs were transferred onto chambers one day before the experiments. Before a measurement, the medium was exchanged for CO2-independent medium containing one of three amphiphiles (10 mM BA, 50 μM VE, or 30 μM TX). After the dish was placed on a microscope stage, ECM-functionalized beads were introduced into the dish and trapped by the laser beam. Using a manual stage, the trapped bead was brought to a position 10 μm above the glass and the calibration was conducted as described above. A bead was brought into proximity of an individual cell and the stage was programmed to stop moving when the bead touched the cell, which was indicated by a 3% increase in the QPD sum voltage. When the piezo-stage position stopped, the duration of contact was set to 500 or 1500 ms and then the bead was pulled away from the cell by moving the piezo stage. An increase in the QPD X voltage was converted into piconewton force with the calibrated optical-trap stiffness and voltage/nm value. The HAEC was withdrawn from beads at a rate of 5 μm/s unless otherwise indicated. For the integrin affinity test, various loading rates were acquired by adjusting the pulling speed to 5, 10, 30, and 50 μm/s.
Analysis of nascent FA sizes
Before experiments, HAECs were co-transfected with mCherry-paxillin and EGFP-FAK plasmids. After 2 days of recovery, HAECs were detached with trypsin, resuspended in growth medium with or without amphiphiles, and incubated for 1 h at 37°C in a humidified incubator (5% CO2), separately. Then the cells were seeded on a FN-coated glass dish for 2 h in the same condition of suspension. Fluorescently tagged paxillin and FAK in HAECs was observed via TIRF microscope and FA size was analyzed using the open-source software “Focal Adhesion Analysis Server” (35).
Cell migration assay
For scratch-induced migration assays, one side of a confluent cell monolayer was scratched with a sterile razor blade. After washing out floating cells, growth medium was added to each well without or with one of the three amphiphiles. The cells migrated for 24 h at 37°C in a humidified atmosphere with 5% CO2. Migrated cells were counted with the ImageJ cell counting plugin. To test migration recovery, cells were incubated in growth medium supplemented by BA for 24 h and the medium was changed to growth medium with or without VE for 24 h.
Statistical analysis
Peaks in rupture-force histograms were fitted with a Gaussian function via a peak analysis tool in OriginPro. All fits were converged until a χ2 tolerance value of 10−6 was reached. Box plots were used to show the mean, median, and 25th–75th percentiles for each group. The results are expressed as the mean ± SE or mean ± SD (36). Mean ± SE values were calculated for the diffusion rates of proteins, the number of fluorescent molecules, and molecular brightness. Mean ± SD values were computed for cell migration experiments. The normality of the distribution was tested with the Shapiro-Wilk method. Analysis of variance was performed for the normally distributed data. When analysis of variance indicated a significant difference in the mean values of main effects, we performed the post hoc test using Tukey’s honestly significant difference. Non-normally distributed data were analyzed with the Kruskal-Wallis test, which was followed by Dunn’s multiple comparison tests.
Results
Amphiphiles alter membrane viscosity and govern domain aggregation
We previously demonstrated by coarse-grained molecular dynamics simulations and in giant unilamellar vesicles that three non-lipid amphiphiles, BA, VE, and TX, induce domain formation or dispersion (19). To examine whether amphiphiles can modulate membrane domains of HAECs in ways that mirror those found in model membranes, we first analyzed the lateral mobility and brightness of Lyn-kinase-positive domains (here called lipid rafts) upon treatment with BA, VE, and TX. Lipid rafts are ordered nanoscale domains of assemblies of sphingolipids, cholesterol, and proteins that form platforms to function in signal transduction and cell migration. The dynamics of lipid rafts and their modulation by the amphiphiles were tracked by measuring the diffusion and molecular brightness of Lyn kinase (Lyn), which localizes to rafts via myristoylation and palmitoylation. HAECs were transfected with mCherry-tagged Lyn plasmid (Fig. 1 A). Lyn diffused in the plane of the membrane in and out of the confocal detection volume where it was excited by a 532 nm laser. Emission of mCherry fluorophores was detected by a photon-counting PMT connected to a time-correlated single-photon counting module (Fig. 1 B). The photons of mCherry were collected, and autocorrelation curves were computed after cellular treatment with each of the three amphiphiles (Fig. 1 C). Data from measurements were fit to a two-dimensional, two-component diffusion model (Eq. 3), which gave two groups of diffusion coefficients ranging from 37.5 to 62.9 μm2/s and 1.6 to 2.3 μm2/s (Table. 1). The small diffusion coefficient was assumed to reflect diffusion of protein in the plane of the membrane (Dmem), whereas the large diffusion coefficient was assumed to reflect diffusion of protein in the cytoplasm (Dcyt). The rationale for these assumptions is as follows. First, diffusion coefficients of fluorescently labeled proteins in mammalian cell cytoplasm were normally distributed between 20 and 100 μm2/s, which was in good agreement with the fast component of a two-diffusing-components model (37). Second, in FCS measurements, the diffusivity of the fast component was ∼25-fold faster than that of the slow component, consistent with the 25-fold lower viscosity one obtains using the Stokes Einstein equation for diffusion of a sphere in a medium with a viscosity similar to cytoplasm. Third, each treatment, which favors lipid membranes, affected the slow component only. For example, TX, which is known to make pores in the membrane, only significantly increased diffusion in the slow component, suggesting that the slow component reflects membrane diffusion (38). As mentioned above, the values of Dcyt were not statistically different among amphiphile treatments. However, the values of Dmem were significantly increased after TX and VE treatment.
Table 1.
Diffusion Coefficient and Molecular Brightness for mCherry-Tagged Lyn and β1-Integrin on the Membrane of HAECs
| N | Dcyt (μm2/s) | Dmem (μm2/s) | Brightness | Cells | |
|---|---|---|---|---|---|
| mCherry-Lyn | |||||
| Ctrl | 5.2 (0.5) | 37.5 (6.4) | 1.6 (0.1) | 128.0 (9.8) | 20 |
| BA | 2.9 (0.3)a | 38.2 (9.4) | 2.0 (0.2) | 279.1 (23.1)c | 20 |
| VE | 4.1 (0.3) | 59.2 (11.5) | 1.9 (0.2) | 168.6 (13.3) | 22 |
| TX | 3.0 (0.3)a | 62.9 (19.2) | 2.3 (0.2)b | 242.9 (19.9)c | 21 |
| mCherry-β1-Integrin | |||||
| Ctrl | 4.8 (0.4) | 32.6 (5.7) | 1.0 (0.1) | 113.0 (6.7) | 24 |
| BA | 3.2 (0.4)a | 18.2 (1.5) | 0.7 (0.1) | 224.6 (18.5)a | 27 |
| VE | 4.1 (0.3) | 40.7 (5.5) | 1.5 (0.2)a | 129.8 (10.1) | 25 |
| TX | 2.1 (0.2)a | 32.5 (7.9) | 1.9 (0.2)a | 287.3 (17.7)a | 24 |
Molecular brightness values were calculated from the FCS data and are expressed as counts per second per molecule. Diffusion rates were fit using Burst Analyzer 2.0 (Becker & Hickl) with a two-dimensional, two-component diffusion model (Eq. 3). Dcyt represents the cytoplasmic proteins and Dmem represents the diffusing proteins on the cell membrane. Data are represented as the mean, with the standard error in parentheses. Statistical analyses for brightness were done by one-way ANOVA followed by Tukey’s test, and others were completed by the Kruskal-Wallis test followed by Dunne’s multiple comparison test. All significance values are relative to control.
p < 0.0001.
p < 0.05.
p < 0.01.
For fluorescently tagged proteins, the molecular brightness of a particle and an average number of particles within the observation volume are useful indicators of protein clustering. If two fluorescently tagged proteins aggregate and diffuse together through the observation volume, the complex will transport two fluorescent molecules and twice as many photons will be detected, but FCS will interpret this to be one diffusing species. The average brightness of single mCherry molecules in solution (115 counts per second per molecule (CPSM)) was comparable to the values from single mCherry-Lyn molecules in control HAECs, suggesting that the cellular environment, itself, did not artificially change brightness. Furthermore, the brightness of the single mCherry fluorescent proteins in solution was unchanged after adding amphiphiles (Table S1), demonstrating that the amphiphiles did not affect fluorescence directly. In cells, the brightness increased 2.2-fold and 1.9-fold with BA and TX treatments, respectively, and the average number of fluorescent molecules (N) in the observation volume accordingly decreased to 0.6 of control values. Thus, we conclude that diffusing Lyn-positive domains in control cells contain one Lyn molecule each, and BA and TX caused aggregation of two lipid rafts to one larger one, resulting in two Lyn molecules per raft. Additionally, BA left Lyn diffusion unchanged, suggesting that it does not target Lo domains, consistent with previous results in model membranes (20).
The manipulation of lipid domains with amphiphiles leads to changes in integrin clustering
We next applied FCS to mCherry-β1-integrin-transfected HAECs to elucidate the effect of amphiphiles on the diffusion of β1-integrins, which is the most dominantly expressed integrin subunit (39) and regulates adhesion strength (40). The brightness of mCherry-β1-integrin was increased 2.0-fold and 2.5-fold and the N values decreased to 0.7 and 0.4 of control values with BA and TX treatment, respectively, following a pattern similar to that of mCherry-Lyn-transfected HAECs. In contrast to the effects of BA on Lyn-domains, the mean value of Dmem of β1-integrins (1.0 μm2/s) increased to 1.5 and 1.9 μm2/s with VE and TX treatments, respectively (Table 1). Accordingly, autocorrelation curves were shifted (Fig. 1 D). Thus, both BA and TX treatments caused clustering of integrin-positive domains with attendant doubling of integrin molecules, consistent with domain-forming actions of the amphiphiles in model membranes.
Optical-trap force spectroscopy determines single-integrin adhesion strength
To analyze whether amphiphiles have an effect on the initial binding response of integrins, an optically trapped bead was chosen as a force transducer, and the force was measured relative to a precisely defined t = 0 contact point using a newly developed quantitative measurement of the surface-bead contact deflection (Fig. 2, A and B). Rupture forces ranging from 5 to 55 pN were generated between the FN-coated bead and HAECs for a 0.5 s contact, but rupture forces >30 pN significantly decreased after removing the heparan sulfate proteoglycan on the cell surface (Fig. S1). Along with an integrin-recognition sequence, FN has heparin-binding sites that interact with heparan sulfate proteoglycans (41) on the cell surface. To eliminate unexpected bonds from the heparin-binding domain of FN, all the rupture forces were measured with RGD-coated beads and the rupture forces were obtained under the condition of single bond formation between integrins and RGD peptides, which was supported by a single well-defined peak on the force spectrum and low adhesion frequency (10–20%) (42). Rupture forces <10 pN were considered non-specific binding, as quantified by adhesion with BSA-coated beads (Fig. S1) (43).
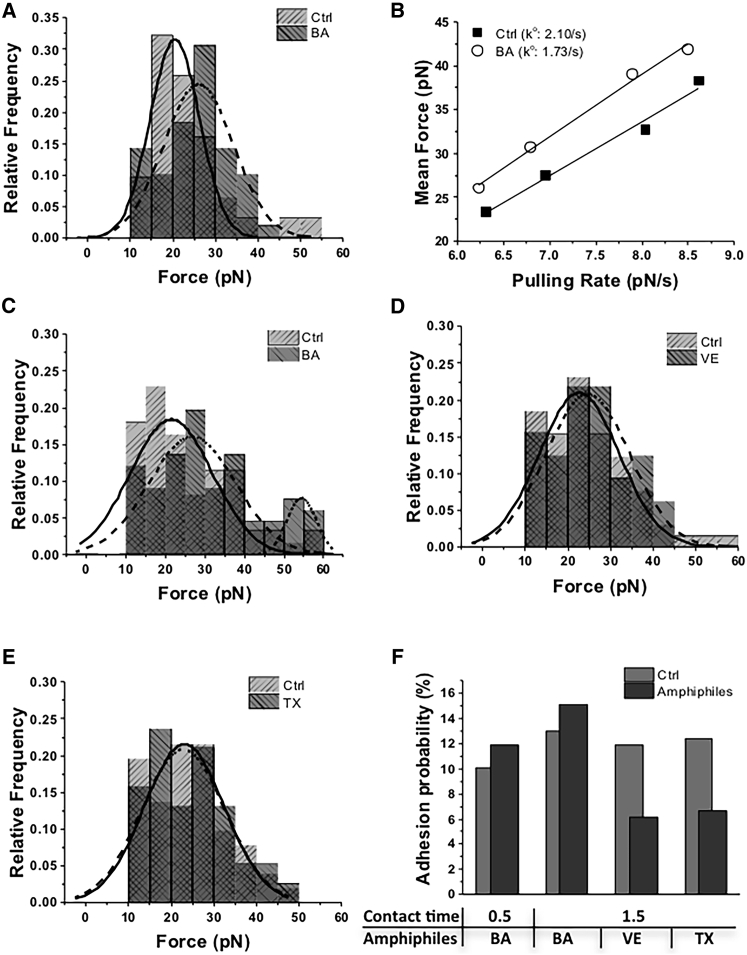
BA causes enhanced integrin affinity and valency
Rupture forces were measured with the optically trapped RGD-coated bead after a 0.5 s contact on HAECs. We observed a well-defined single peak in the force histogram (Fig. 3 A) that implied that forces were generated from a single bond between an integrin and an RGD peptide. After treating HAECs with BA, the force histogram showed a single peak, but the force distribution shifted to higher forces (maximum peak from 20.5 to 26.1 pN) relative to control. These data suggested that the rupture forces were still generated by a single bond, but the treatment of BA increased the affinity of integrin to the RGD-coated bead. To gain additional insight into the effect of BA on integrin affinity, the dissociation rate constant of the integrins was calculated. A generally accepted model for understanding the relationship between force and an adhesion complex is that the applied force distorts the energy landscape of the integrin/ECM bond, which increases the dissociation rate constant. The dissociation rate constant, ko, can be obtained from the relationship between rupture-force distribution, fm, and loading rate, rf (42, 44),
| (5) |
where γ is the position of the transition state, T is temperature, and kB is the Boltzmann constant. The force spectra of mean force versus log rf are presented in Fig. 3 B. This analysis indicated that decreased by 18% (from 2.10/s to 1.73/s) after treatment with BA. Together, these results demonstrate that BA treatment of HAECs increases the affinity of β1-integrins for RGD peptides.
Figure 3.
Force measurements. (A) The rupture-force histograms from integrins and RGD-coated bead interactions with (dashed line) or without (solid line) BA treatment for 0.5 s bead-HAEC contact time. The numbers of ruptures included in the analysis were 31 for control cells and 49 for BA-treated cells. (B) Dynamic force spectra for integrin-RGD adhesion without (solid squares) and with (open circles) BA treatment as a function of pulling rates of 551.1 (n = 48; 8.5% of the total contact number), 1045.6 (n = 44; 15.3%), 3086.5 (n = 35; 8.6%), and 5493.8 (n = 30; 10.3%) pN/s for BA-treated cells and 507.3 (n = 54; 14.4%), 887.7 (n = 42; 17.6%), 2679.4 (n = 29; 11.6%), and 4870.2 (n = 34; 10.6%) pN/s for control cells. The ko was obtained using Eq. 5. The other unknown from Eq. 5, γ, changed from 0.69 to 0.60 nm after BA treatment. (C–E) Rupture-force histograms of interactions between integrins and RGD-coated beads (C) with (dashed line) and without BA (solid line), (D) with (dashed line) and without VE (solid line), and (E) with (dashed line) and without TX (solid line) for 1.5 s contact time of the beads with the HAECs. The numbers of ruptures included in the graph in (B) were 61 for control cells and 66 for BA-treated cells, and those in the graph in (C) were 65 for control cells and 32 for VE-treated cells. The numbers of ruptures included in (D) were 51 for control cells and 38 for VE-treated cells. (F) Adhesion probability depended on the contact time between the optically trapped RGD-coated beads and integrins, and the amphiphiles on the HAECs. All histogram data were fitted by Origin Pro “Peak Analyzer” using Gaussian curves.
To provide additional time for the lateral redistribution of integrins to the contact site with ECM, the force measurements were carried out after increasing surface contact time from 0.5 to 1.5 s. Under this setting, the force distribution of the control group peaked at 21.53 pN. The BA-treated cells generated two peaks at 26.81 and 54.39 pN, with the value of the second peak approximately double that of the first peak (Fig. 3 C). This force doubling indicates that two integrins were engaged and suggests that BA caused clustering of integrin domains containing single integrin molecules to larger domains containing two integrin molecules.
VE and TX do not change integrin affinity or valency, but they lower adhesion probability
We examined whether VE and TX influence the binding response of integrins. The addition of VE and TX to HAECs did not change the rupture-force distribution between integrins and RGD-coated beads in the 1.5 s contact experiments (Fig. 3, D and E). However, the adhesion probability for both conditions significantly decreased from 11.9 to 6.1% with VE addition and from 12.4 to 6.7% with TX addition, whereas BA treatment increased the probability (Fig. 3 F). The decreased adhesion probability with a single peak indicates that the VE and TX treatment reduced the chance of interaction between integrins and RGD peptides, but a single bond was generated during each event.
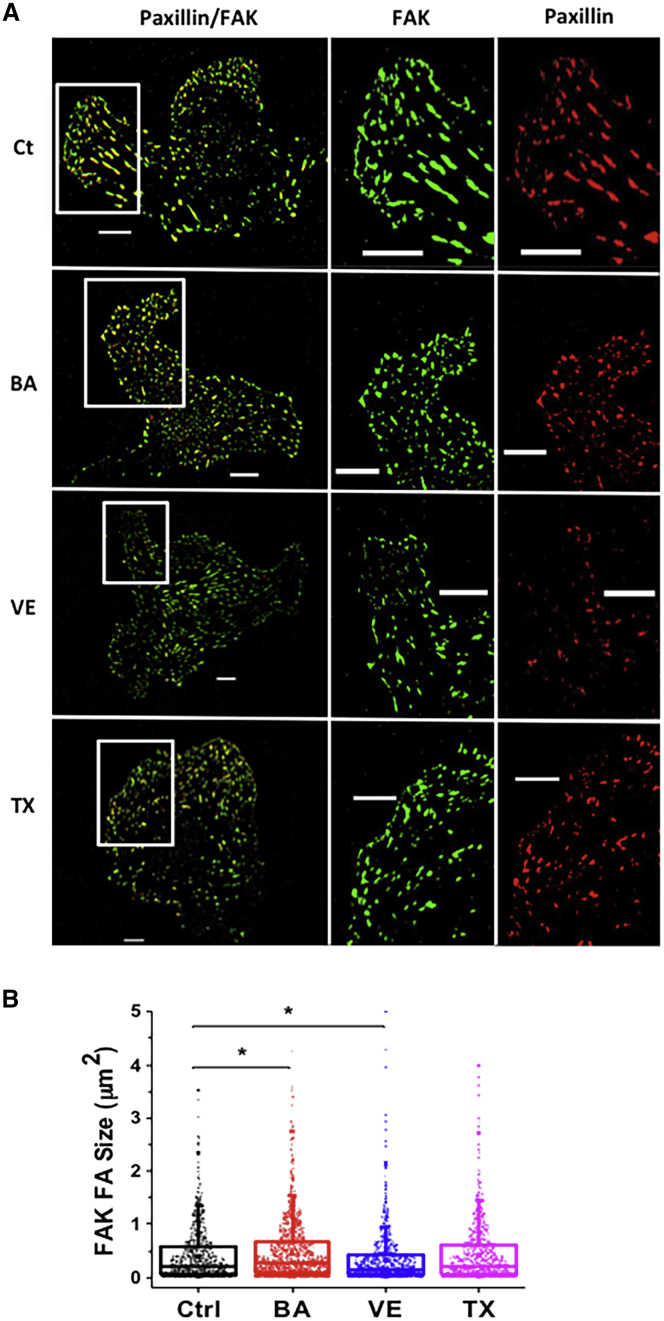
Amphiphiles modulated nascent FA size and cell mobility
FAs are integrin-containing multiprotein complexes carrying out important functions such as force transduction and signaling. FA kinase (FAK) and paxillin are important adhesion proteins recruited at the cell’s leading edge during cell migration, and normalized FAK/paxillin fluorescence intensity ratio is >1 at the cell front and <1 at the cell rear (45). TIRF microscopy was used to monitor dual-color images of EGFP-FAK and mCherry-paxillin after treatment with three amphiphiles. The images of cells with FAK and paxillin are shown in Fig. 4 A and the boxed areas are magnified. To compare the effect of amphiphiles on the newly generated FA size, we defined FAs as nascent when FAK size was larger than paxillin size on the first layer (relative to the cell edge) of FAs at the HAEC edge and subsequently compared FAK sizes. BA-treated HAECs had significantly larger nascent FAs than the control HAECs. In contrast, VE treatment significantly reduced the nascent FA size (Fig. 4 B).
Figure 4.
FA analysis. (A) Representative binary TIRF images of FAs, which were labeled with FAK-EGFP (green) and mCherry-paxillin (red). Superimposition of FAK and mCherry appears as yellow and the regions of interest are magnified and separated by color. The scale bar represents 10 μm. (B) Box plots of FAK FA size; only FAs near the cell boundary (first row of FAs) are included for data analysis (n = 733 for control cells and 1135, 869, and 733 for VA-, VE-, and TX-treated cells, respectively). The medians are represented by the horizontal lines in the rectangular boxes; the lower and upper sides of the boxes represent the 25th and 75th percentiles of sizes, respectively, and the ends of the horizontal lines represent the 5th and 95th percentiles. The sizes of single adhesions were analyzed by the open-source software Focal Adhesion Analysis Server (35). Statistical analyses were done by the Kruskal-Wallis test; ∗p < 0.0001. To see this figure in color, go online.
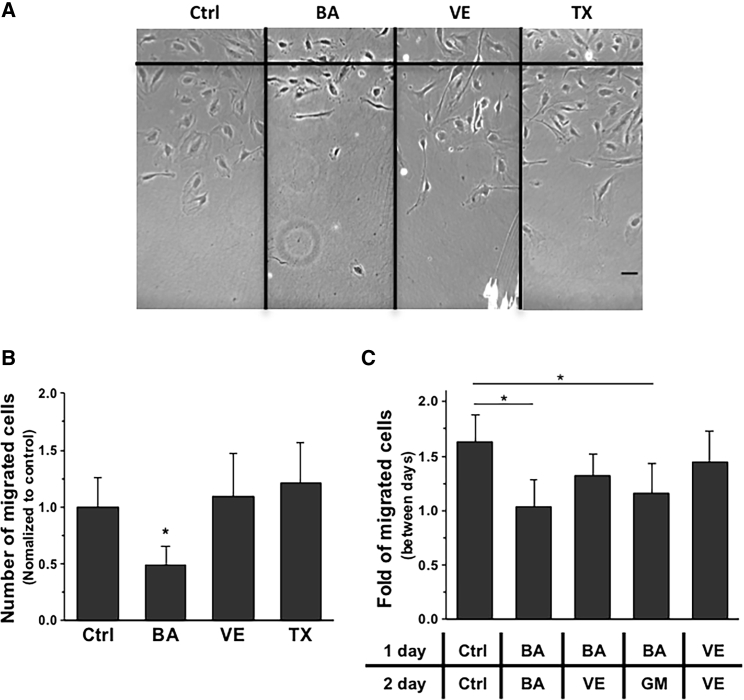
Studies have suggested that amphiphilic compounds influence not only FA formation but also cell migration (24, 25). Cell migration was analyzed using the scratch-induced-migration assay after treating cells with BA, VE, and TX for 24 h, separately. BA treatment significantly decreased cell motility, but VE and TX treatment did not result in a significant change in cell migration when compared with the control cells (Fig. 5, A and B). However, reduced mobility with BA treatment could be reversed by changing to medium supplemented with VE (Fig. 5 C).
Figure 5.
Cell migration assay. (A) Phase-contrast micrographs after wounding the monolayer of HAECs, which was incubated in the media containing BA, VE, and TX. (B) Numbers of cells that migrated 24 h after wounding with a sterile razor blade. Around 3000 μm of images were stitched together and quantified by ImageJ. The cell numbers were normalized with the stitched image length and the final results were normalized to the control and expressed as the mean ± SD. (C) Fold of the migrated cells between days. At the first day, HAECs migrated with medium supplemented with BA in three plates and the medium was changed to growth medium (GM) or VE on the second day. The final results were expressed as a fold increase between days and expressed as the mean ± SD. Statistical analyses were done by one-way analysis of variance followed by Tukey’s test; ∗p < 0.0001. The scale bars represent 10 μm.
Discussion
By controlling lipid headgroups, acyl chain lengths, and saturation states, cells can generate thousands of different lipid types that spontaneously form cell membranes to segregate the cell’s internal components from the environment. Lipids allow membrane proteins to aggregate or disperse and alter the conformational states of membrane proteins (7). The regulation of membrane protein function through membrane lipids may take place because of specific chemical interactions between proteins and lipid molecules or non-specific interactions between proteins and the physical properties of the bilayer (46, 47), such as bilayer thickness. Since proteins do not have binding sites for non-native amphiphiles, these compounds likely regulate membrane proteins in a non-specific manner (48). Although manipulation of cholesterol concentration is a widely used approach for membrane modulation, exogenous amphiphiles have some advantages over cholesterol as a means to manipulate membrane structure and lipid dynamics. First, amphiphiles have been shown to differentially target Lo domains, Ld domains, and domain boundaries, whereas modification of cholesterol has the most impact on raft (Lo) domains because Lo domains are known to be cholesterol rich. Second, amphiphiles leave intact the existing, and highly regulated, native membrane composition. Third, previous coarse-grained molecular dynamics simulations establish the mechanisms of amphiphile-induced membrane phase separation. The specific partitioning preferences of amphiphilic molecules to lipid domains lead to changes in domain thickness and domain edge energy, leading to phase separation (19). Therefore, it is likely that the amphiphiles used in this study cause changes in integrin function via changes in bilayer thickness, phase separation, and attendant changes in dimerization and diffusion.
Molecular brightness studies with mCherry-Lyn and mCherry-β1-integrins demonstrated that BA treatment promoted aggregation of lipid rafts and β1-integrin domains, where the brightness was twofold and N was decreased to half, as predicted when two molecules diffuse as one unit together in the observation volume. The average N values calculated by fitting autocorrelation curves with Eq. 3 and molecular brightness were calculated by using the relationship between the average count rate, N, and brightness (Table 1). The membrane aggregation is consistent with previous studies in model membranes where BA induced coalescence of domains into large patches of Lo domains separate from Ld domains (19). This clustering of integrins by BA treatment likely is responsible for the two force peaks on the force spectroscopy distribution of integrin-mediated adhesion. Our study used optical trap-based force spectroscopy to demonstrate that single-integrin force production was modified by BA, which targets Ld domains in lipid bilayers. An alternative way to measure the effects of amphiphiles on force production overall is traction force microscopy (reviewed in (49)), and single-integrin forces on substrates have been detected (50). In our study using an optical trap, we could determine directly how many integrin molecules were bound, along with their force kinetics. Thus, the optical trap was the most appropriate tool for identifying the relationship between the membrane, integrin-RGD bond formation, and force production (51). In traction force microscopy, there are other aspects of force generation, such as actin-myosin contraction, that might be indirectly affected by amphiphiles, necessitating additional control experiments. Nevertheless, traction force microscopy could be an excellent next step to measure the effects of amphiphiles on overall integrin-mediated force generation.
Although FN specifically binds to many different types of integrins, cell adhesion studies have focused on FN binding to β1 and β3 integrins. The bond between FN and α5β1-integrins has been shown to exhibit catch-bond behavior (52), whereas β3 integrins exhibit slip bonds (53). In our experiments, bonds between integrins and RGD-coated beads exhibited catch-bond behavior (Fig. S2) in which bond lifetime was prolonged with increasing force. Affinity is the binding strength between a single molecule and its ligand. This is reported by the equilibrium dissociation constant, a ratio of dissociation rate and association rate. Interestingly, the rate of dissociation of β1-integrin from RGD peptide decreased by 18% with BA treatment, which was interpreted as an increase of affinity between β1-integrin and RGD peptides (Fig. 3 B). Advanced understanding of integrin structure has shown that integrin affinity is highly related to integrin structure. The ligand-binding headpiece is connected to the plasma membrane by two stalks. In the resting state, the stalks are severely bent and the ligand-binding domains are unfavorable for binding. The integrin converts from an intermediate state, which is a switchblade-like expansion exhibiting cross stalks and a closed headpiece, to a fully activated state with extended conformation, with straight stalks and an open headpiece. Molecular dynamics models suggest that interaction between integrin αIIbβ3 and the RGD sequence dissociates the outer membrane clasp between the head domain and proximal domain to the plasma membrane on the β-subunit of the integrin, thus permitting the straight active form (54). The geometry and composition of transmembrane β-subunit of integrin αIIbβ3 potentially regulate the integrin clustering process (55), and the β-subunit maintains a 25° angle with the membrane surface normal. The stalk angle of β1-integrin may be related to the packing of lipids around the transmembrane stalk (56) and thus may be influenced by bilayer thickness. Whereas the β subunits of integrins exhibit sequence homology in transmembrane and cytoplasmic domains (57), we suggest that β1-integrins observed in our study behave in a similar manner to β3-integrins, referenced above, with respect to the transmembrane domain. In model membranes, β1-integrins preferentially partition into the Ld domain before activation (58), and BA treatment decreased the thickness of the Ld domain (19). Thus, we hypothesize that β1-integrins diffuse in Ld domains and the decreased membrane thickness around the helix caused by BA treatment may shift integrin conformational equilibrium toward an increase in the proportion of the straight-stalk (open-state) configuration by modulating the stalk angle, resulting in enhanced affinity of integrin to RGD in the initial binding response. After activation and binding, it is possible that the resulting organization could induce a transition of the lipids from Ld to Lo.
With respect to the gylcocalyx, the increases in integrin affinity and valency with BA treatment were not observed in heparinase-III-treated cells, and rupture-force distribution and adhesion probability were similar to those in non-treated cells, as shown in Fig. S3. Thus, we conclude that removal of the glycocalyx prevents integrin clustering without influencing integrin binding. This result is consistent with previous studies demonstrating that the glycocalyx has an important role in integrin clustering (59). Integrin clustering could be responsible for observations that BA treatment of HAECs causes an increase in the size of nascent FAs. Such increases in FA size arise from integrin-mediated assembly of signaling adaptors, a hierarchical process (60) responsible for signaling leading to migration. Specifically, nascent FAs are generated at the cell periphery by ECM interactions and FAK is recruited before paxillin (45). BA further caused clustering of integrins and an attendant decrease in the diffusion rate (Fig. 3 C; Table 1). Consistent with the role of BA in FA assembly, when we calculated FAK assembly and disassembly rates in FAs using Focal Adhesion Analysis Server software (35), we found that both rates increased after treatment with BA (Fig. S4). Thus, we assume that the adhesion turnover rate may not change with amphiphile treatment. However, the number of assembling FAs increased from 9 to 44, whereas the number of disassembling FAs decreased from 69 to 31 after 1 min in 20 mM BA. Thus, BA treatment may not affect the assembly or disassembly rate of FAs but may affect the number of FAs. Quantified newly generated nascent FAs revealed that BA-treated HAECs exhibited significantly larger nascent FAs than did untreated HAECs (Fig. 4, A and B). Larger FAs are associated with lower migration rates in cells (61), consistent with the inhibitory effect of BA on cell migration observed in our study.
VE and TX treatments of HAECs affect integrin-mediated adhesion differently. Both VE and TX treatments resulted in single bond generation between β1-integrins and RGD peptides (Fig. 3, D and E) while decreasing the probability of adhesion relative to that observed for control cells (Fig. 3 F). The decreased adhesion probabilities for both VE and TX treatment correspond to enhanced diffusion rates of β1-integrins (Table 1). Thus, the initial responses between β1-integrin and RGD may be mainly controlled by the diffusion rate of β1-integrins on HAEC membranes. The important factors underlying diffusion rates in the cellular membrane are the size of a protein and the viscosity of the membrane. The decrease in the adhesion probability upon addition of VE may be caused by reduced domain sizes, whereas the reduction in adhesion frequency with the addition of TX may come from higher diffusion rates due to the lowered viscosity of the membrane with surfactant-mediated pores. In the integrin-mediated FA formation, VE treatment decreased nascent FA size and TX did not result in a significant change (Fig. 4 B).
Taken together, these studies provide insights into the role of integrin functional changes caused by amphiphiles in endothelial cell migration on FN-coated substrates. By conducting studies on FN, we avoided complications of non-specific binding. We propose that the number of bonds between integrins and the ECM may play an important role in cell mobility. BA treatment on cells, which yields higher-affinity double bonds and results in larger FAs, delayed their mobility. TX- and VE-treated cells, which, similar to control cells, generated single bonds and did not increase FA size, exhibited mobility that was similar to that observed for control cells (Fig. 5, A and B). Interestingly, the effects of amphiphiles on cell migration can be reversed. Reduced mobility of BA-treated cells can be recovered by changing the medium to VE-supplemented medium, which is similar to the reversibility of TX-induced domain formation by VE in model membranes (19). Our study was focused on integrins in endothelial cells, in particular, the relationship between β1-integrins and FN. Because β1-integrin partners (α5 and α8) recognize the RGD tripeptide in FN (62), this study is likely relevant to other cell types that express α5β1 or α8β1 integrins, such as fibroblasts (63), human umbilical vein endothelial cells (64), epithelial cells (65), and tumor cells (66).
Conclusions
This study shows that alterations of lipid bilayer thickness and diffusion can exert functional changes in β1-integrin-mediated adhesion, nascent FA formation, and cell migration. Although others have investigated the role of the membrane in integrin-mediated adhesion in an indirect manner, we identified membrane-mediated changes in integrin function directly. Even with respect to clustering, we have identified a mechanism of clustering and established an approximate time constant for clustering that might be useful in the development of models for integrin-mediated adhesion in the future. Modulation of the HAEC membrane phase by amphiphiles clusters or disperses integrins. Changes in the diffusion rate of integrins in the plane of the membrane altered the probability that a bond between β1-integrins and RGD peptide would form. Decreasing Ld domain thickness with BA increased integrin affinity for RGD. In turn, transitions of integrin affinity and valency affected integrin-related nascent adhesion, formation, and attendant migration. Such results demonstrate that the lipid bilayer, beyond being a passive bystander to protein function and organization, plays an important role in modulating integrin function and consequent adhesion-related cellular behavior.
Author Contributions
S.S. designed and conducted all of the experiments, analyzed the data, and wrote the manuscript. G.J.M. developed the optical-trap technique, wrote software for it, and tested and provided preliminary data. P.J.B. outlined the goals of the study, designed the experiments, interpreted the data, and co-wrote the manuscript.
Acknowledgments
The authors acknowledge support from National Science Foundation grant CMMI-1334847.
Editor: Philip LeDuc.
Footnotes
Four figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30807-X.
Supporting Material
References
- 1.Ross T.D., Coon B.G., Schwartz M.A. Integrins in mechanotransduction. Curr. Opin. Cell Biol. 2013;25:613–618. doi: 10.1016/j.ceb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardel M.L., Schneider I.C., Waterman C.M. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takagi J., Springer T.A. Integrin activation and structural rearrangement. Immunol. Rev. 2002;186:141–163. doi: 10.1034/j.1600-065x.2002.18613.x. [DOI] [PubMed] [Google Scholar]
- 4.Slater J.H., Boyce P.J., Frey W. Modulation of endothelial cell migration via manipulation of adhesion site growth using nanopatterned surfaces. ACS Appl. Mater. Interfaces. 2015;7:4390–4400. doi: 10.1021/am508906f. [DOI] [PubMed] [Google Scholar]
- 5.Tardy Y., Resnick N., Dewey C.F., Jr. Shear stress gradients remodel endothelial monolayers in vitro via a cell proliferation-migration-loss cycle. Arterioscler. Thromb. Vasc. Biol. 1997;17:3102–3106. doi: 10.1161/01.atv.17.11.3102. [DOI] [PubMed] [Google Scholar]
- 6.Pierres A., Monnet-Corti V., Bongrand P. Do membrane undulations help cells probe the world? Trends Cell Biol. 2009;19:428–433. doi: 10.1016/j.tcb.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 8.Simons K., Sampaio J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heberle F.A., Petruzielo R.S., Katsaras J. Bilayer thickness mismatch controls domain size in model membranes. J. Am. Chem. Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 10.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 11.Heberle F.A., Feigenson G.W. Phase separation in lipid membranes. Cold Spring Harb. Perspect. Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leitinger B., Hogg N. The involvement of lipid rafts in the regulation of integrin function. J. Cell Sci. 2002;115:963–972. doi: 10.1242/jcs.115.5.963. [DOI] [PubMed] [Google Scholar]
- 13.Ramprasad O.G., Srinivas G., Pande G. Changes in cholesterol levels in the plasma membrane modulate cell signaling and regulate cell adhesion and migration on fibronectin. Cell Motil. Cytoskeleton. 2007;64:199–216. doi: 10.1002/cm.20176. [DOI] [PubMed] [Google Scholar]
- 14.Norambuena A., Schwartz M.A. Effects of integrin-mediated cell adhesion on plasma membrane lipid raft components and signaling. Mol. Biol. Cell. 2011;22:3456–3464. doi: 10.1091/mbc.E11-04-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaus K., Le Lay S., Schwartz M.A. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 2006;174:725–734. doi: 10.1083/jcb.200603034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuentes D.E., Bae C., Butler P.J. Focal adhesion induction at the tip of a functionalized nanoelectrode. Cell. Mol. Bioeng. 2011;4:616–626. doi: 10.1007/s12195-011-0214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuentes D.E., Butler P.J. Coordinated mechanosensitivity of membrane rafts and focal adhesions. Cell. Mol. Bioeng. 2012;5:143–154. doi: 10.1007/s12195-012-0225-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabouillot T., Muddana H.S., Butler P.J. Endothelial cell membrane sensitivity to shear stress is lipid domain dependent. Cell. Mol. Bioeng. 2011;4:169–181. doi: 10.1007/s12195-010-0136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muddana H.S., Chiang H.H., Butler P.J. Tuning membrane phase separation using nonlipid amphiphiles. Biophys. J. 2012;102:489–497. doi: 10.1016/j.bpj.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundbæk J.A. Regulation of membrane protein function by lipid bilayer elasticity—a single molecule technology to measure the bilayer properties experienced by an embedded protein. J. Phys. Condens. Matter. 2006;18:S1305–S1344. doi: 10.1088/0953-8984/18/28/S13. [DOI] [PubMed] [Google Scholar]
- 21.Lundbaek J.A., Collingwood S.A., Andersen O.S. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. J. R. Soc. Interface. 2010;7:373–395. doi: 10.1098/rsif.2009.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heerklotz H. Interactions of surfactants with lipid membranes. Q. Rev. Biophys. 2008;41:205–264. doi: 10.1017/S0033583508004721. [DOI] [PubMed] [Google Scholar]
- 23.Trabelsi S., Zhang S., Schwartz D.K. Linactants: surfactant analogues in two dimensions. Phys. Rev. Lett. 2008;100:037802. doi: 10.1103/PhysRevLett.100.037802. [DOI] [PubMed] [Google Scholar]
- 24.Jensen L.D.E., Hansen A.J., Lundbaek J.A. Regulation of endothelial cell migration by amphiphiles—are changes in cell membrane physical properties involved? Angiogenesis. 2007;10:13–22. doi: 10.1007/s10456-006-9060-y. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh P.K., Vasanji A., Fox P.L. Membrane microviscosity regulates endothelial cell motility. Nat. Cell Biol. 2002;4:894–900. doi: 10.1038/ncb873. [DOI] [PubMed] [Google Scholar]
- 26.Chiu J.J., Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol. Rev. 2011;91:327–387. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler P.J. Mechanotransduction of Shear Stress by the Endothelium. In: Tanishita K., Yamamoto K., editors. Vascular Engineering: New Prospects of Vascular Medicine and Biology with a Multidiscipline Approach. Springer; 2016. pp. 159–197. [Google Scholar]
- 28.Lu S., Ouyang M., Wang Y. The spatiotemporal pattern of Src activation at lipid rafts revealed by diffusion-corrected FRET imaging. PLOS Comput. Biol. 2008;4:e1000127. doi: 10.1371/journal.pcbi.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonaka R., Iesaki T., Arikawa-Hirasawa E. Perlecan deficiency causes endothelial dysfunction by reducing the expression of endothelial nitric oxide synthase. Physiol. Rep. 2015;3:e12272. doi: 10.14814/phy2.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammond G.R., Fischer M.J., Irvine R.F. PI4P And PI(4,5)P2 are essential but independent lipid determinants Of membrane identity. Science. 2013;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gullapalli R.R., Tabouillot T., Butler P.J. Integrated multimodal microscopy, time-resolved fluorescence, and optical-trap rheometry: toward single molecule mechanobiology. J. Biomed. Opt. 2007;12:014012. doi: 10.1117/1.2673245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benda A., Fagul’ová V., Hof M. Fluorescence lifetime correlation spectroscopy combined with lifetime tuning: new perspectives in supported phospholipid bilayer research. Langmuir. 2006;22:9580–9585. doi: 10.1021/la061573d. [DOI] [PubMed] [Google Scholar]
- 33.Svoboda K., Block S.M. Biological applications of optical forces. Annu. Rev. Biophys. Biomol. Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 34.Dangaria J.H., Butler P.J. Macrorheology and adaptive microrheology of endothelial cells subjected to fluid shear stress. Am. J. Physiol. Cell Physiol. 2007;293:C1568–C1575. doi: 10.1152/ajpcell.00193.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berginski M.E., Vitriol E.A., Gomez S.M. High-resolution quantification of focal adhesion spatiotemporal dynamics in living cells. PLoS One. 2011;6:e22025. doi: 10.1371/journal.pone.0022025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altman D.G., Bland J.M. Standard deviations and standard errors. BMJ. 2005;331:903. doi: 10.1136/bmj.331.7521.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kühn T., Ihalainen T.O., Timonen J. Protein diffusion in mammalian cell cytoplasm. PLoS One. 2011;6:e22962. doi: 10.1371/journal.pone.0022962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Digman M.A., Brown C.M., Gratton E. Paxillin dynamics measured during adhesion assembly and disassembly by correlation spectroscopy. Biophys. J. 2008;94:2819–2831. doi: 10.1529/biophysj.107.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 40.Roca-Cusachs P., Gauthier N.C., Sheetz M.P. Clustering of α5β1 integrins determines adhesion strength whereas αvβ3 and talin enable mechanotransduction. Proc. Natl. Acad. Sci. USA. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mostafavi-Pour Z., Askari J.A., Humphries M.J. Identification of a novel heparin-binding site in the alternatively spliced IIICS region of fibronectin: roles of integrins and proteoglycans in cell adhesion to fibronectin splice variants. Matrix Biol. 2001;20:63–73. doi: 10.1016/s0945-053x(00)00131-1. [DOI] [PubMed] [Google Scholar]
- 42.Li F., Redick S.D., Moy V.T. Force measurements of the α5β1 integrin-fibronectin interaction. Biophys. J. 2003;84:1252–1262. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Litvinov R.I., Bennett J.S., Shuman H. Multi-step fibrinogen binding to the integrin αIIbβ3 detected using force spectroscopy. Biophys. J. 2005;89:2824–2834. doi: 10.1529/biophysj.105.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tees D.F., Waugh R.E., Hammer D.A. A microcantilever device to assess the effect of force on the lifetime of selectin-carbohydrate bonds. Biophys. J. 2001;80:668–682. doi: 10.1016/S0006-3495(01)76047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y.L., Lu S., Chien S. FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci. Rep. 2014;4:6024. doi: 10.1038/srep06024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Lee A.G. How lipids and proteins interact in a membrane: a molecular approach. Mol. Biosyst. 2005;1:203–212. doi: 10.1039/b504527d. [DOI] [PubMed] [Google Scholar]
- 48.Lundbaek J.A. Lipid bilayer-mediated regulation of ion channel function by amphiphilic drugs. J. Gen. Physiol. 2008;131:421–429. doi: 10.1085/jgp.200709948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polacheck W.J., Chen C.S. Measuring cell-generated forces: a guide to the available tools. Nat. Methods. 2016;13:415–423. doi: 10.1038/nmeth.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rabinovitz I., Gipson I.K., Mercurio A.M. Traction forces mediated by α6β4 integrin: implications for basement membrane organization and tumor invasion. Mol. Biol. Cell. 2001;12:4030–4043. doi: 10.1091/mbc.12.12.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neuman K.C., Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong F., García A.J., Zhu C. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Litvinov R.I., Barsegov V., Shuman H. Dissociation of bimolecular αIIbβ3-fibrinogen complex under a constant tensile force. Biophys. J. 2011;100:165–173. doi: 10.1016/j.bpj.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehrbod M., Trisno S., Mofrad M.R.K. On the activation of integrin αIIbβ3: outside-in and inside-out pathways. Biophys. J. 2013;105:1304–1315. doi: 10.1016/j.bpj.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehrbod M., Mofrad M.R.K. Localized lipid packing of transmembrane domains impedes integrin clustering. PLOS Comput. Biol. 2013;9:e1002948. doi: 10.1371/journal.pcbi.1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lau T.L., Partridge A.W., Ulmer T.S. Structure of the integrin β3 transmembrane segment in phospholipid bicelles and detergent micelles. Biochemistry. 2008;47:4008–4016. doi: 10.1021/bi800107a. [DOI] [PubMed] [Google Scholar]
- 57.Srichai M., Zent R. Integrin Structure and Function. In: Zent R., Pozzi A., editors. Cell-Extracellular Matrix Interactions in Cancer. Springer; 2010. pp. 19–41. [Google Scholar]
- 58.Siegel A.P., Kimble-Hill A., Naumann C.A. Native ligands change integrin sequestering but not oligomerization in raft-mimicking lipid mixtures. Biophys. J. 2011;101:1642–1650. doi: 10.1016/j.bpj.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paszek M.J., DuFort C.C., Weaver V.M. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaidel-Bar R., Cohen M., Geiger B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem. Soc. Trans. 2004;32:416–420. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- 61.Kim D.H., Wirtz D. Focal adhesion size uniquely predicts cell migration. FASEB J. 2013;27:1351–1361. doi: 10.1096/fj.12-220160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Humphries J.D., Byron A., Humphries M.J. Integrin ligands at a glance. J. Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dalton S.L., Marcantonio E.E., Assoian R.K. Cell attachment controls fibronectin and α5β1 integrin levels in fibroblasts. Implications for anchorage-dependent and -independent growth. J. Biol. Chem. 1992;267:8186–8191. [PubMed] [Google Scholar]
- 64.Cascone I., Napione L., Bussolino F. Stable interaction between α5β1 integrin and Tie2 tyrosine kinase receptor regulates endothelial cell response to Ang-1. J. Cell Biol. 2005;170:993–1004. doi: 10.1083/jcb.200507082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J.W., Juliano R.L. α5β1 integrin protects intestinal epithelial cells from apoptosis through a phosphatidylinositol 3-kinase and protein kinase B-dependent pathway. Mol. Biol. Cell. 2000;11:1973–1987. doi: 10.1091/mbc.11.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roman J., Ritzenthaler J.D., Han S. α5β1-integrin expression is essential for tumor progression in experimental lung cancer. Am. J. Respir. Cell Mol. Biol. 2010;43:684–691. doi: 10.1165/rcmb.2009-0375OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.