Abstract
Background and Purpose:
Candida albicans (C. albicans) is an opportunistic fungus that can colonize women’s mucosal epithelial cell surfaces, causing vulvovaginitis in specific circumstances. The major genes contributing to drug resistance in C. albicans are the candida drug resistance (CDR) and multi drug resistance (MDR) genes. The purpose of this study was to evaluate the CDR-2 and MDR-1 gene expression patterns in C. albicans strains isolated from patients with recurrent vulvovaginal candidiasis.
Materials and Methods:
In this study, 40 isolates of fluconazole-resistant C. albicans were cultured on Sabouraud dextrose agar. These isolates were collected from women with vulvovaginitis who were referred to a clinic in Tehran, Iran, and transferred to a mycology laboratory. Then, RNA was extracted from the isolates using phenol-chloroform and glass beads, and the complementary DNA (cDNA) was synthetized. To detect the semi-quantitative expression of CDR-2 and MDR-1 genes, the reverse transcriptase-PCR (RT-PCR) technique was performed using specific primers.
Results:
Our findings indicated that of the 40 C. albicans isolates, 35 (87.5%) strains were positive for mRNA of the CDR-2 gene, 32 (80%) strains expressed mRNA of the MDR-1 gene, and 30 (75%) strains were confirmed to express mRNA of both the CDR-2 and MDR-1 genes simultaneously using the RT-PCR assay.
Conclusion:
According to the obtained results, the expression rates of CDR-2 and MDR-1 genes were high in fluconazole-resistant C. albicans isolates, which can cause treatments to fail and result in chronic infections. Inhibiting these important genes using novel or natural agents can help with the treatment of chronic and recurrent vaginitis.
Key Words: C. albicans, CDR-2, Gene expression, MDR-1, RT-PCR, Vulvovaginal candidiasis
Introduction
Women of reproductive age, consumers of contraceptive steroidal drugs or any of the widespread anti-bacterial agents, diabetic or pregnant women, and patients with an immunological deficiency have the predisposing factors for vulvovaginal candidiasis (VVC) [1]. The rising prevalence of fluconazole-resistant C. albicans strains is a major problem after long-term treatment of recurrent VVC (RVVC). Fluconazole resistance can occur through different mechanisms involving mutations in the drug target enzyme and sterol 14a-demethylase (14DM), alterations in sterol biosynthesis, increased expression of the ERG11 gene, as well as overexpression of genes coding membrane transport proteins of the ABC transporter (CDR-1/CDR-2) or the major facilitator (MDR1) superfamilies [2, 3].In addition, drug resistance can emerge by environmental factors, leading to fungal coloni-zation or substituting a resistant species such as Candida glabrata or Candida krusei with a sensitive one [4-6].
Previous studies illustrated that developing efflux pumps is the most frequent mechanism for azole resistance in Candida species. Efflux pumps coded by two carrier gene families include CDR-1 and CDR-2 genes belonging to the ATP-binding cassette superfamily, as well as MDR-1 genes from the major facilitator superfamily [7, 8]. It was confirmed that enhancing the expression levels of CDR-1, CDR-2, and MDR-1 in C. albicans causes fluconazole resistance [9, 10]. Activating efflux pumps coded by CDR-1 can affect all azole drugs, while efflux pumps coded by MDR are selective for fluconazole [11]. However, overexpression of several different genes contributes to fluconazole resistance in Candida species. For instance, mutations in ERG11 reduced binding of the drug target enzyme, lanosterol C14-alpha demethylase (14DM), to fluconazole and conferred higher resistance compared to the identical genes without mutation [12-14]. Moreover, fluconazole resistance protein (FLU1) is responsible for fluconazole resistance in C. albicans strain; thus, with inactivation of FLU1, fluconazole susceptibility can be increased. However, overexpression of FLU1 has not yet been approved as a cause of fluconazole resistance in clinical C. albicans isolates [15].
In recent years, by increasing the growth of azole resistant C. albicans affecting the proper treatment of VVC and regarding the major role of MDR and CDR genes as the major culprit for azole resistance in C. albicans, the present study was designed to determine the pattern of MDR-1 and CDR-2 genes in clinical samples of Candida isolated from Iranian women with VVC.
Materials and Methods
C. albicans strains and culture conditions
Fourteen fluconazole-resistant C. albicans isolates were obtained from patients with VVC, who were admitted to gynecology centers in Tehran, Iran. The isolates were identified using the conventional method based on colony color on CHROM agar Candida, and molecular methods included polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) [16]. The isolates were stored in sterilized distilled water until the time of the experiment.
Moreover, resistance to fluconazole was shown by disk diffusion assay performed according to the Clinical Laboratory Standards Institute recommend-dations in our previous study [16]. The standard strain of C. albicans (ATCC10231) was used as a fluconazole sensitive species. This study was carried out in Medical Mycology and Parasitology laboratory in Iran University of Medical Sciences, 2016.
Total RNA extraction in Candida isolates
For this study, isolates were cultured on Sabouraud dextrose agar medium (SDA, Merck, Germany) and incubated at 37oC for 24 h. The cell wall of C. albicans was disrupted using an RNA lysis buffer and glass beads. Then, RNA was extracted using RNx-plus (Cinnagen, Tehran, Iran) and a chloroform/isoamyl alcohol solution [17]. After centrifugation, the sediment was dissolved in distilled water and stored at -20oC until use.
Elimination of genomic DNA from the total RNA
To eliminate DNA contamination from the RNA, all the samples were treated by deoxyribonuclease (DNase) enzyme (Fermentas, Paisley, England) according to the following steps: 1 µg of RNA was added to a sterilized, nuclease-free microtube; 1 µl of DNase 10X reaction buffer and 1 µl of DNase-1 were added to microtubes; 2 µl of ethylenedia-minetetraacetic acid (25 mM) was added to each microtube, and the microtubes were stored at 65oC for 10 min. The purified RNA was used for complementary DNA (cDNA) synthesis.
cDNA synthesis and reverse transcriptase-PCR (RT-PCR) assay
After DNase enzyme treatment, RNA was converted to cDNA according to the manufacturer's recommendations using a cDNA synthesis kit (Fermentas, USA). RT-PCR was performed with the reactions containing 2 μl of template cDNA, 0.6 μl of each specific primer for the MDR-1 and CDR-2 genes, 10 μl of Taq DNA polymerase, MgCl2, dNTP, and vivantis buffer), as well as 6.8 μl of diethylpyrocarbonate (DEPC) water in a final volume of 20 μl. The RT-PCR protocol was begun with an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation (94°C for 1 min), annealing (58°C for 1 min), and extension (72°C for 1 min); the protocol was terminated with a final extension step at 72°C for 3 min. The appropriate negative and positive controls were included in each test.
The primers for MDR-1 and CDR-2 were designed by Gene Runner software, and their sequence is presented in Table 1. ACT1 was used as the house-keeping gene and for confirmation of the PCR process in all the molecular tests. Ultimately, the PCR products were visualized by gel electrophoresis.
Table 1.
Nucleotide sequences of MDR-1 and CDR-2 primers
| Primer sequence | Tm (°C) | Tm (°C) | Primer name | Accession number |
|---|---|---|---|---|
| 5´-TGGCAAACAATCCAACAATAC A-3´ | 56.6 | 56.6 | CDR-2 Forward(F) | U63812 |
| 5´-AATCAAGGGAATAGATGGGTC A-3´ | 58.4 | 58.4 | CDR-2 Revers(R) | |
| 5´-TACGCGGGTTCTTTGTTGTAT G-3´ | 60.3 | 60.3 | MDR-1 Forward (F) | Y14703 |
| 5´-GATAATGTTTAGCAAGCCGAGGA-3´ | 61.1 | 61.1 | MDR-1 Revers (R) |
Results
Fluconazole susceptibility testing against C. albicans isolates
All the 40 C. albicans isolates were resistant to 25 μg of fluconazole in disk diffusion method. The inhibition zone was determined < 14 mm against fluconazole.
Patients’ age ranged between 18 and 50 years, and 57% of the patients consumed several antibiotics, 28% cases used contraceptives, and 15% of the women had diabetes.
RNA extraction
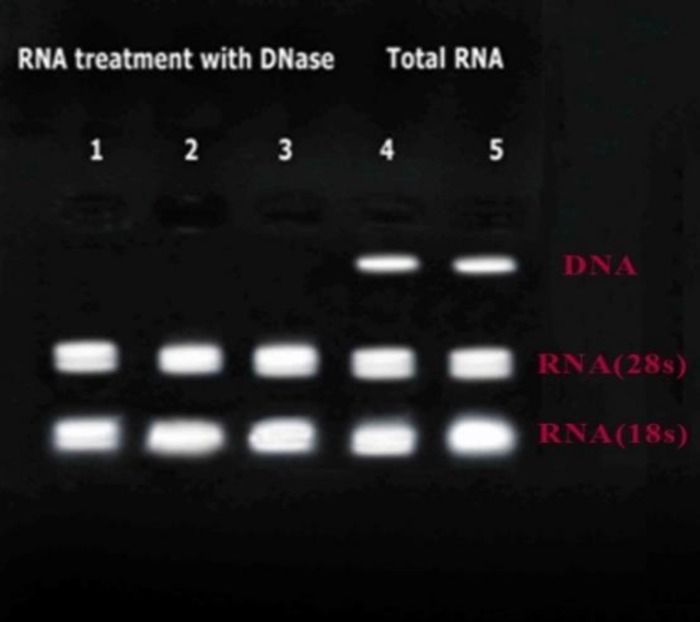
The quality of RNA was evaluated by gel electrophoresis. Figure 1 illustrates the total RNA of the isolates before and after the DNase enzyme treatment. The quantity of RNA was determined using BioPhotometer plus (Eppendorf AG, Germany), and the RNA concentration for all the samples was adjusted to 1.5 ng.
Figure 1.
Lines 1-3: Total RNA after treatment with DNase; Lines 4 and 5: RNA before treatment
MDR-1 and CDR-2 gene expression
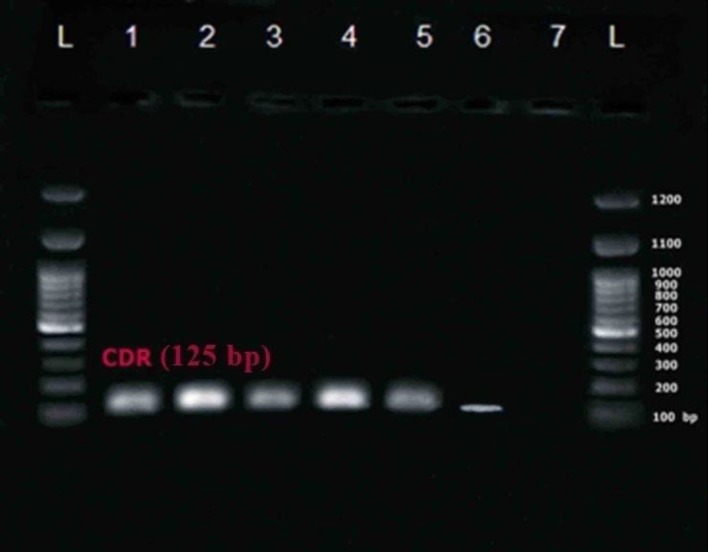
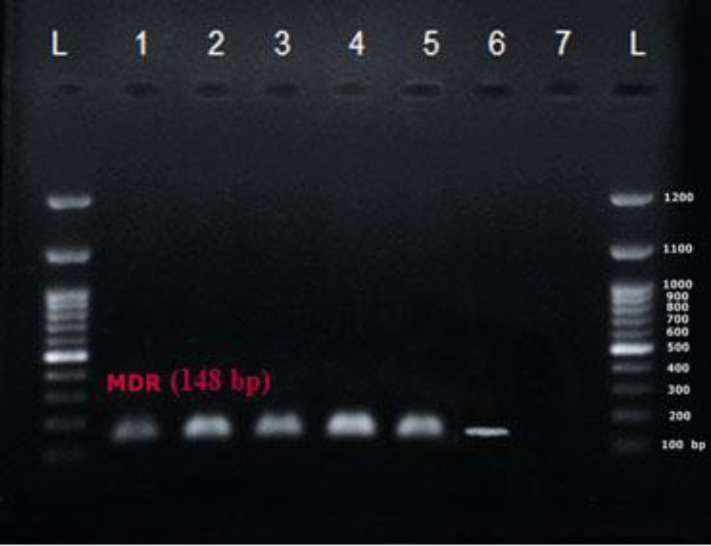
An RT-PCR reaction was carried out using special primers for MDR-1 and CDR-2 for 40 C. albicans specimens. The semi-quantitative expression of both MDR-1 and CDR-2 was assessed in C. albicans clinical isolates (figures 2, 3) using RT-PCR. The PCR product sizes were 125 bp and 148 bp for CDR-2 and MDR-1, respectively, as explained previously [18].
Figure 2.
Lines 1-5: CDR-2 gene expression (125 bp); Line 6: CDR-2 gene expression in Candida albicans (ATCC10231) standard strain; Line 7: Negative control; L: 100 bp ladder
Figure 3.
Lines 1-5: MDR-1 gene expression (148 bp); Line 6: MDR-1 gene expression in Candida albicans (ATCC10231) standard strain; Line 7: Negative control
Table 2 indicates the semi-quantitative expression levels of CDR-2 and MDR-1.
Table 2.
Semi-quantitative expression of MDR-1 and CDR-2 genes in the isolates
| Evaluated genes | CDR-2 gene expression | MDR-1 gene expression | Expression of CDR-2 and MDR-1 | No expression of CDR-2 or MDR-1 genes |
|---|---|---|---|---|
| Number of isolates | 35 | 32 | 30 | 3 |
| Percent | 87.5 | 80 | 75 | 7.5 |
The results of the semi-quantitative expression of CDR-2 and MDR-1 genes showed that out of the 40 clinical isolates of Candida albicans, 35 (87.5%) samples expressed CDR-2, leaving only 5 (12.5%) specimens that did not express CDR-2. Further, 32 (80%) isolates expressed MDR-1, while only 8 (20%) samples did not show expression of the MDR-1 gene. Finally, 3 (7.5%) samples expressed neither the CDR-2 nor the MDR-1, whereas 30 (75%) isolates expressed both genes simultaneously.
Discussion
Recent headways in our understanding of the molecular mechanisms causing azole resistance in C. albicans revealed that increased efflux of drug, mediated mostly by the ATP-binding cassette (ABC) and the major facilitator superfamily (MFS) transporters, leads to resistance to azole anti-fungal agents [19, 20].
Our findings indicated that the expression rates of MDR and CDR genes were high in fluconazole-resistant C. albicans. In our study, the high expression rates of genes in the isolates may be due to taking high doses of fluconazole, as the patients had RVVC. The expression level of CDR-2 was higher than that of MDR-1 in the isolates, indicating that the role of CDR in forming fluconazole resistance in C. albicans is more pronounced than that of MDR-1. Regarding the assumption that CDR is specific to C. albicans, these results were favorable for our isolates.
Emerging fluconazole resistant C. albicans isolates leads to a wide range of complications in RVVC treatment, the most important of which is biofilm formation, that is, aggregate of a rigid network by Candida. The expression of MDR and CDR genes during the early phase of biofilm formation and alterations in membrane sterol composition are responsible for resistance of these biofilms against azole agents.
Although resistance is multifactorial and other molecular mechanisms participate in this phenol-menon, it is worth mentioning that the expression of drug efflux pumps during the early phase of biofilm formation and alterations in membrane sterol composition contribute to resistance of these biofilms against azoles [21, 22].
Consistent with our results, Gulat et al. assessed the expression levels of CDR-1, CDR-2, and MDR-1 in fluconazole-resistant Candida albicans isolates using real-time PCR. Our findings indicated that the expression levels of CDR-1, CDR-2, and MDR-1 genes in sensitive isolates were lower compared to resistant ones, suggesting that high expression levels of efflux genes is a major mechanism for fluconazole resistance in Candida albicans [23].
Zhang et al. evaluated the expression levels of CDR-1, CDR-2, MDR-1, and FLU-1 in 18 fluconazole-resistant isolates of Candida strains from VVC patients and reported a significant increase in CDR-1 expression, while expression levels of CDR-2, MDR-1, and FLU-1 did not significantly elevate [24].
In our study, lack of expression of MDR-1 and CDR-2 genes in 7.5% of the cases may be explained by the report presented by Lohberger et al. indicating the expression levels of drug-resistance genes (i.e., CDR-1, CDR-2, MDR-1, and ERG-11) are controlled by transcription factors such as TAC-1, which are responsible for controlling the expression of CDR-1 and CDR-2. MRR-1 and UPC-2 factors are responsible for controllingthe expression levels of MDR-1 and ERG-11, respectively. Moreover, there are some enhancing mutations (GOF) in activated alleles for increasing the expression levels of the target genes [19]. It can be concluded that the lack of MDR-1 and CDR-2 in some isolates in our study may be associated with the activation of ultra-genetic factors rather than transcription factors including ERG11 [25, 26], which can be considered in future studies.
In 2013, Guo et al. assessed the correlation between alcohol dehydrogenase (ADH-1) gene expression and CDR-1, CDR-2, and FLU-1 in Candida albicans collected from patients with VVC. Expression of CDR-1, CDR-2, MDR-1, and ERG-11 showed a positive correlation between the expression levels of ADH-1 mRNA and CDR-1, CDR-2, and FLU-1 [27].
Ariana et al. evaluated the expression of CDR-1, CDR-2, and MDR-1 in resistant Candida albicans isolates compared to fluconazole susceptible isolates. Their outcomes indicated moderate expression of CDR-1, CDR-2, and MDR-1 genes, while resistant isolates showed slight or no expression [28].
Our findings were in line with those of Salari et al. who evaluated the CDR-1, CDR-2, MDR-1, and ERG11 genes expression in C. albicans clinically isolated from HIV-infected patients in Iran by real-time PCR. Their results indicated that the CDR-1 gene expression in fluconazole-resistant C. albicans increased significantly compared to other known genes [29].
This finding was not in congruence with the results of the current study. This discrepancy could be related to the source of infection, the number of isolates, and genetic diversity of isolates in different geographic areas.
Conclusion
The high expression levels of MDR-1 and CDR-2 genes in C. albicans isolates in RVVC highlights the important role of these genes in developing fluconazole resistance, causing treatment attempts to fail and leading to chronic infections. Therefore, inhibition of the key genes involved in the disease as well as combination therapy using novel synthetic or natural drugs could help patients with chronic and recurrent VVC.
Acknowledgments
The authors wish to thank the Research Admini-stration of the international campus of Iran University of Medical Sciences, Tehran, Iran.
Author’s contribution
K. K. performed the tests. M. F. and M. R. designed and managed the research project. S. F. helped with molecular testing and S. N. analyzed the data.
Conflict of interest
None declared.
Financial disclosure
This study was financially supported by Iran University of Medical Sciences (Grant No: 25647).
References
- 1.Zhao X, Oh SH, Cheng G, Green CB, Nuessen JA, Yeater K, et al. ALS3 and ALS8 represent a single locus that encodes a Candidaalbicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150(Pt 7):2415–28. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- 2.Marichal P, Vanden Bossche H. Mechanisms of resistance to azole antifungals. Acta Biochim Pol. 1995;42(4):509–16. [PubMed] [Google Scholar]
- 3.Morschhäuser J. The genetic basis of fluconazole resistance development in Candida albicans. Biochim Biophys Acta. 2002;1587(2-3):240–8. doi: 10.1016/s0925-4439(02)00087-x. [DOI] [PubMed] [Google Scholar]
- 4.Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. Multicenter comparison of the VITEK 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing amphotericin B, flucytosine, and voriconazole against Candida spp. J Clin Microbiol. 2007;45(11):3522–8. doi: 10.1128/JCM.00403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wayne PA. Zone diameter interpretive standards, corresponding minimal inhibitory concentration (MIC) interpretive breakpoints, and quality control limits for antifungal disk diffusion susceptibility testing of yeasts; Third International Supplement CLSI document-M444-S3. New York, US: 2009. [Google Scholar]
- 6.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, et al. Clinical practice guidelines for the management candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(5):503–35. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143(Pt 2):405–16. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 8.Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39(11):2378–86. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White TC. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41(7):1482–7. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanglard D. Current understanding of the modes of action of and resistance mechanisms to conventional and emerging antifungal agents for treatment of Candida infections. Candida and Candidiasis. Washington, DC: ASM Press; 2002. pp. 349–83. [Google Scholar]
- 11.Löffler J, Kelly SL, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151(2):263–8. doi: 10.1111/j.1574-6968.1997.tb12580.x. [DOI] [PubMed] [Google Scholar]
- 12.Orozco AS, Higginbotham LM, Hitchcock CA, Parkinson T, Falconer D, Ibrahim AS, et al. Mechanism of fluconazole resistance in Candida krusei. Antimicrob Agents Chemother. 1998;42(10):2645–9. doi: 10.1128/aac.42.10.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42(2):241–53. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Ribot JL, McAtee RK, Lee LN, Kirkpatrick WR, White TC, Sanglard D, et al. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemothera. 1998;42(11):2932–7. doi: 10.1128/aac.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calabrese D, Bille J, Sanglard D. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology. 2000;146(Pt 11):2743–54. doi: 10.1099/00221287-146-11-2743. [DOI] [PubMed] [Google Scholar]
- 16.Roudbary M, Roudbarmohammadi S, Bakhshi B, Farhadi Z. Relation of ALS1 and ALS3 genes and fluconazole resistance in candida albicans isolated from vaginal candidiasis. Inter J Mol Clin Microbiol. 2012;2(2):170–4. [Google Scholar]
- 17.Roudbarmohammadi S, Roudbary M, Bakhshi B, Katiraee F, Mohammadi R, Falahati M. ALS1 and ALS3 gene expression and biofilm formationin Candida albicans isolated from vulvovaginal candidiasis. Adv Biomed Res. 2016;5:105. doi: 10.4103/2277-9175.183666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen LM, Xu YH, Zhou CL, Zhao J, Li CY, Wang R. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Med Res. 2010;38(2):536–45. doi: 10.1177/147323001003800216. [DOI] [PubMed] [Google Scholar]
- 19.Lohberger A, Coste AT, Sanglard D. Distinct roles of Candida albicans drug resistance transcription factors TAC1, MRR1, and UPC2 in virulence. Eukaryot Cell. 2014;13(1):127–42. doi: 10.1128/EC.00245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider S, Morschhäuser J. Induction of Candida albicans drug resistance genes by hybrid zinc cluster transcription factors. Antimicrob Agents Chemother. 2015;59(1):558–69. doi: 10.1128/AAC.04448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramage G, Bachmann S, Patterson TF, Wickes BL, López-Ribot JL. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother. 2002;49(6):973–80. doi: 10.1093/jac/dkf049. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee PK, Chandra J. Candida biofilm resistance. Drug Resist Updat. 2004;7(4-5):301–9. doi: 10.1016/j.drup.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Gulat S, Doluca Dereli M. Investigation of the expression levels of efflux pumps in fluconazole-resistant Candida albicans isolates. Microbiol Bul. 2014;48(2):325–34. doi: 10.5578/mb.7213. [DOI] [PubMed] [Google Scholar]
- 24.Zhang JY, Liu JH, Liu FD, Xia YH, Wang J, Liu X, et al. Vulvovaginal candidiasis: species distribution, fluconazole resistance and drug efflux pump gene overexpression. Mycoses. 2014;57(10):584–91. doi: 10.1111/myc.12204. [DOI] [PubMed] [Google Scholar]
- 25.Oliveria Carvalho V, Okay TS, Melhem MS, Walderez Szeszs M, del Negro GM. The new mutation L321F in Candida albicans ERG11 gene may be associated with fluconazole resistance. Rev Iberoam Micol. 2013;30(3):209–12. doi: 10.1016/j.riam.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Xu Y, Li C. Association of T916C (Y257H) mutation in Candida albicans ERG11 with fluconazole resistance. Mycoses. 2013;56(3):315–20. doi: 10.1111/myc.12027. [DOI] [PubMed] [Google Scholar]
- 27.Guo H, Zhang XL, Gao LQ, Li SX, Song YJ, Zhang H. Alcohol dehydrogenase I expression correlates with CDR1, CDR2 and FLU1 expression in Candida albicans from patients with vulvovaginal candidiasis. Chin Med J. 2013;126(11):2098–102. [PubMed] [Google Scholar]
- 28.Ariana N, Nazemi A, Nasrollahi Omran A. Using PCR to compare the expression of CDR1, CDR2, and MDR1 in Candida albicans isolates resistant and susceptible to fluconazole. Med Lab J. 2015;9(4):33–7. [Google Scholar]
- 29.Salari S, Khosravi AR, Mousavi SA, Nikbakht-Brogeni GH. Mechanisms of resistance to fluconazole in Candida albicans clinical isolates from Iranian HIV-infected patients with oropharyngeal candidiasis. J Mycol Med. 2016;26(1):35–41. doi: 10.1016/j.mycmed.2015.10.007. [DOI] [PubMed] [Google Scholar]